Abstract
Liver fibrosis is an active process that involves changes in cell-cell and cell-extracellular matrix (ECM) interaction. Secreted protein, acidic and rich in cysteine (SPARC) is an ECM protein with many biological functions that is overexpressed in cirrhotic livers and upregulated in activated hepatic stellate cells (aHSCs). We have recently shown that SPARC downregulation ameliorates liver fibrosis in vivo. To uncover the cellular mechanisms involved, we have specifically knocked down SPARC in two aHSC lines [the CFSC-2G (rat) and the LX-2 (human)] and in primary cultured rat aHSCs. Transient downregulation of SPARC in hepatic stellate cells (HSCs) did not affect their proliferation and had only minor effects on apoptosis. However, SPARC knockdown increased HSC adhesion to fibronectin and significantly decreased their migration toward PDFG-BB and TGF-β1. Interestingly, TGF-β1 secretion by HSCs was reduced following SPARC small interfering RNA (siRNA) treatment, and preincubation with TGF-β1 restored the migratory capacity of SPARC siRNA-treated cells through mechanisms partially independent from TGF-β1-mediated induction of SPARC expression; thus SPARC knockdown seems to exert its effects on HSCs partially through modulation of TGF-β1 expression levels. Importantly, collagen-I mRNA expression was reduced in SPARC siRNA-transfected HSCs. Consistent with previous results, SPARC knockdown in aHSCs was associated with altered F-actin expression patterns and deregulation of key ECM and cell adhesion molecules, i.e., downregulation of N-cadherin and upregulation of E-cadherin. Our data together suggest that the upregulation of SPARC previously reported for aHSCs partially mediates profibrogenic activities of TGF-β1 and PDGF-BB and identify SPARC as a potential therapeutic target for liver fibrosis.
Keywords: fibrogenesis, migration, adhesion, cadherin, small interference RNA
liver cirrhosis is the consequence of an excessive accumulation of extracellular matrix (ECM), which ultimately leads to distortion of parenchymal architecture and hepatic failure (11). Liver fibrogenesis is an active process that involves the interaction of different cell types; among these, activated hepatic stellate cells (aHSCs) play a central role (3). In the injured liver, aHSCs have the ability to migrate to and to accumulate at the site of damage wherein they start to produce matrix fibrillar components (3) as well as other ECM components, such as proteoglycans and glycoproteins (16). In addition, aHSCs actively synthesize inflammatory cytokines and receptors, which recruit inflammatory cells (16). The presence of inflammation further increases the recruitment of aHSCs, creating a positive feedback that results in an enhanced and perpetuated liver damage (12). The profibrogenic activity of aHSCs is stimulated by soluble factors that include TGF-β1 and PDGF-BB (20). Moreover, aHSCs are the main source of TGF-β1 in the fibrotic liver (4, 17). TGF-β1 and PDGF-BB were found to modulate a network of cytokines and are regarded as master molecules controlling proliferation, migration, and activation of HSCs during liver fibrogenesis (20, 26). Accordingly, aHSCs are considered as a prominent target for antifibrotic therapies.
SPARC (secreted protein, acidic and rich in cysteine), also called osteonectin, is a secreted multifunctional matrix-associated protein that has the ability to promote cell detachment from the ECM as well as to enhance cell proliferation and motility (2, 30, 32). SPARC is upregulated in aHSCs (5) and in cirrhotic livers from rats and humans (14, 23, 24). We have recently demonstrated that SPARC expression levels correlate with the severity of liver fibrosis and shown that inhibition of SPARC by an adenovirus encoding an antisense mRNA (AdasSPARC) ameliorates hepatic fibrosis in rats (6). These data strongly support a profibrogenic role of SPARC in chronic liver injury. However, the underlying biological mechanisms have not been sufficiently addressed.
We herein show that SPARC knockdown in HSCs results in a partial reduction in their activation phenotype and in their capacity to migrate, involving increased expression of cell adhesion molecules, disruption of the actin filament cytoskeleton pattern, and a shift in the cadherin expression profile. In addition, the secretion of TGF-β1 by HSCs is significantly reduced in SPARC small interfering RNA (siRNA)-treated cells; furthermore, preincubation of these cells with TGF-β1 rescues their migratory phenotype independent from TGF-β1-mediated SPARC induction. Altogether, our data suggest that SPARC downregulation in HSCs exerts its antifibrotic effects by reducing their migratory ability and attenuating their activated phenotype.
MATERIALS AND METHODS
Cell culture.
The CFSC-2G hepatic stellate cell (HSC) line, obtained from cirrhotic rat liver, was kindly provided by Dr. Marcos Rojkind [Albert Einstein College of Medicine, New York, NY (15)]. CFSC-2G cells were cultured in MEM (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS) (Invitrogen) and nonessential amino acids. LX-2 cells, an immortalized human HSC line, were cultured in DMEM (Invitrogen) plus 5% FBS and 1% penicillin-streptomycin (37). Primary cultured HSCs were isolated and incubated as previously described with minor modifications (13). Briefly, HSCs were isolated from adult male Sprague-Dawley rats (400 g) by in situ perfusion. Subsequently, liver cells were dispersed in pronase and collagenase, and HSCs were then obtained by Nycodenz gradient centrifugation and cultured in DMEM 10% FBS. Seven days after plating, aHSCs were transfected with 10 nmol/l of SPARC or control siRNAs and experimental conditions were similar to those used for CFSC-2G cells (see below). Transfection efficiency was assessed 2 days after transfection using a FITC-labeled control siRNA (siRNA-FITC) (Dharmacon; Chicago, IL; catalog no D-001630-01-05). Cells were tested for adhesion and collagen mRNA expression at 3 days after transfection.
Preparation and transfection of siRNA constructs.
Rat SPARC siRNAs (siSPARC; 4 constructs used in combination) and a control siRNA (siCtr; D-001210-05-05) were purchased from Dharmacon (Chicago, IL). The sequences of the siSPARC constructs are shown in Fig. 1, A and B. A FITC-labeled control siRNA (siRNA-FITC) (Dharmacon; Chicago, IL) was used for transfection efficiency analyses. CFSC-2G cells were cultured in antibiotic-free medium for 16 h prior to transfection. Cells were transfected using Lipofectamine 2000 (Invitrogen) (375 pmol/2 × 105 cells) in serum-free Opti-MEM I (Invitrogen). After 24 h, culture medium was replaced by MEM-2% FBS. For transfection efficiency calculation, control siRNA-FITC-transfected cells were trypsinized after 24, 48, and 72 h of incubation and directly examined by fluorescence microscopy.
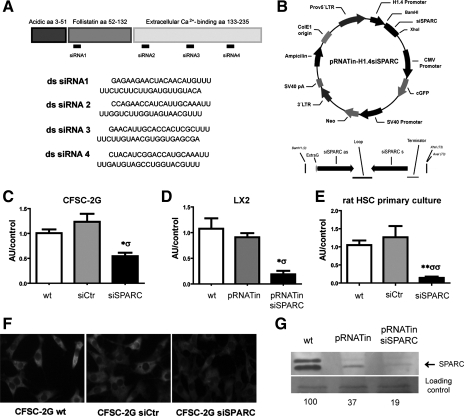
Fig. 1.
Specific SPARC (secreted protein, acidic and rich in cysteine) small interfering RNAs (siRNAs) and a SPARC siRNA-modified plasmid strongly reduce SPARC expression levels in cultured hepatic stellate cells. A: schematic showing the target regions for siRNA constructs in the SPARC mRNA molecule and the sequences of the 4 SPARC siRNA constructs that were used in combination in our studies. B: schematic showing the modified pRNATinH1.4 construct used in our studies to knock down SPARC in LX-2 cells; note the site wherein the SPARC siRNA sequence was inserted. C–E: quantitative RT-PCR (qPCR) analysis of SPARC mRNA expression levels in CFSC-2G (C), LX-2 (D), and primary cultured rat hepatic stellate cells (HSCs) (E) was performed at day 3 after transfections. *,σP < 0.05; **P < 0.01; *vs. nontransfected (wt); σvs. control siRNA (siCtr) or control pRNATinH1.4-transfected (pRNATin) cells. F: representative images showing changes in SPARC expression levels among experimental conditions, in CFSC-2G cells. Original magnification: ×400. G: representative image of the Western blot analysis for SPARC in LX-2 cells, corresponding to 1 of 3 independent experiments; values are shown as percentage of wt band intensity. siSPARC, SPARC siRNA-transfected cells; pRNATin siSPARC, SPARC siRNA-modified pRNATinH1.4 transfected cells. aa, Amino acids; ds, double-stranded.
For SPARC knockdown in LX-2 cells, a modified pRNATin-H1.4 reporter plasmid (Genscript) was used (Fig. 1B). A pRNATin.H1.4-siSPARC vector was constructed following the Genscript cloning protocol. Briefly, three different oligos against SPARC with cohesive BamHI and XhoI sites were designed and purchased (IDT). Complementary siRNAs were annealed in SSC at 95°C and gel purified. pRNATinH1.4 plasmid (Genscript cat. no. SD1260) was digested with BamHI and XhoI. Ligation was performed with T4Ligase (Invitrogen, cat. no. 15224-041) in a molar ratio of insert to vector 3:1. One Shot Stbl3 competent cells (Invitrogen, cat. no. C7373-03) were transformed with the ligation mixture. At least 20 clones were analyzed and the positive ones were sequenced to confirm the presence of the insert. The siRNA51 prepared against the region 52 to 72 bp of SPARC mRNA (NID:NM_003118) showed the best results in terms of downregulation of SPARC mRNA levels. LX-2 cells were grown on six-well plates (3 × 105 cells/well) to 90% confluence in medium without antibiotics. Transfection was performed with use of Lipofectamine 2000, following manufacturer instructions (4 μl Lipofectamine/250 μl Opti-MEM; 3 μg plasmid/250 μl of Opti-MEM).
Real-time polymerase chain reaction.
Quantitative real-time reverse transcriptase-polymerase chain reaction (qPCR) analyses of SPARC, TGF-β1, or collagen mRNA expression in experimental conditions were carried out at 72 h after transfection. Total RNA was extracted from nontransfected (wt; CFSC-2G, LX-2, or primary HSCs), siCtr- or siSPARC-transfected (CFSC-2G or primary HSCs), or pRNATinH1.4- or pRNATinH1.4 siSPARC-transfected (LX-2) cells by using TRIzol reagent (Invitrogen). Two micrograms of RNA were reverse-transcribed with 200 U of SuperScript II Reverse Transcriptase (Invitrogen) using 500 ng of oligo(dT) primers. cDNAs were subjected to qPCR (Stratagene Mx3005p, Stratagene, La Jolla, CA) using SYBR green (Invitrogen) with the following primers: rat SPARC, 5′-CCACTCGCTTCTTTGAGACC-3′ (forward), 5′-TAGTGGAAG TGGGTGGGGAC-3′ (reverse); human SPARC, 5′-AAACCGAAGAGGAGGTGGTG-3 (forward), 5′-GCAAAGAAGTGGCAGGAAGA-3′ (reverse); rat TGF-β1, 5′-ACCAACTACTGCTTCAGCTC-3′ (forward), 5′-TGTTGGTTGTAGAGGGCAAG-3′ (reverse); rat collagen, 5′-CCTACATGGACCAACAGACTG-3′ (forward), 5′-GGAGGTCTTGGTGGTTTTGTA-3′ (reverse); human collagen, 5′-CCTACATGGACCAGCAGACTG-3′ (forward), 5′-GGAGGTCTTGGT GGTTTTGTA-3′ (reverse); and glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
To confirm the deregulation found in the expression of some genes in the ECM and adhesion molecules array, qPCR was also performed (see ECM and adhesion molecules PCR-array). The primers used were CdhE (Cdh1), 5′-AGCCAATCCTGATGAAATCG-3′ (forward), 5′-CCATACATATCGGCCAGCTT-3′ (reverse); CdhN (Cdh2), 5′-ACTGAGG AGCCGATGAAGGAACCAC-3′ (forward), 5′-GTTGATGATGAAGATGCCCGTTGGA-3′ (reverse); Mmp13, 5′-ACCTGGACA AGCAGCTCCAA-3′ (forward), 5′-GAGTGGTC CAGACCGAGGG-3′ (reverse); Adamts8, 5′-TCCTGACTGTGTCTGGTGAGGT-3′ (forward), 5′-GATGTTGGTGCT TGCTCTTTCTT-3′ (reverse). After a cycle of 95°C for 10 min, the reactions were cycled 40 times under the following parameters: 95°C for 30 s, corresponding melting temperature for 30 s, 72°C for 1 min. At the end of the PCR reaction, the temperature was increased from 60 to 95°C at a rate of 2°C/min, and the fluorescence was measured every 15 s to construct the melting curve. Values were normalized to levels of GAPDH transcript. The relative amount of the PCR product amplified from untreated cells was set as 1. A nontemplate control was run in every assay, and all determinations were performed in triplicate in two or three separated experiments.
Western blot analysis.
Supernatants from HSCs were collected, centrifuged, and concentrated with an Amicon Ultra-2 centrifugal filter device (Millipore). Total protein was measured by Bradford assay by using standard techniques. SDS-PAGE with 10% resolving gel was used to separate proteins and SPARC was detected by incubating the gel first with a goat polyclonal anti-SPARC antibody (R&D Systems, Minneapolis, MN) (1:400; Tris-buffered saline, Tween, and 1% BSA) and later with horseradish peroxidase-conjugated horse anti-mouse IgG (Vector, Burlingame, CA). Protein loading and transfer was monitored by staining the membranes with Ponceau Red before staining with antibodies. Bands intensities were measured by densitometry analysis using the Scion Image (Scion, Frederick, MD).
ECM and adhesion molecules PCR-array.
A rat ECM and adhesion molecules RT2 profiler PCR array (SuperArray Bioscience, Frederick, MD) was used to search for differential expression among experimental conditions. This array includes 84 relevant genes important for cell-cell and cell-matrix interactions. Total RNA was isolated from siCtr- or siSPARC-transfected CFSC-2G cells using a RNeasy RNA isolation kit (Qiagen). Single-stranded cDNA was synthesized with SuperScript II reverse transcriptase (Invitrogen). Genomic DNA contamination was eliminated by DNase I treatment prior to reverse transcription. RT2 Profiler PCR array (PARN-013) and RT2 Real-Timer SYBR green/ROX PCR mix were purchased from SuperArray. PCR was performed on Mx3005p (Stratagene). For data analysis the ΔΔCt method was used; for each gene fold changes were calculated as differences in gene expression. A positive value is indicative of gene upregulation whereas a negative value indicates gene downregulation.
Immunofluorescence.
SPARC immunostaining was performed in nontransfected and siCtr- and siSPARC-transfected CFSC-2G cells, previously grown in MEM-2% FBS for 72 h. Cells were washed with PBS and fixed with 4% paraformaldehyde for 20 min. They were subsequently incubated with a mouse anti-human SPARC antibody (1:250) (kindly provided by Dr. Podhajcer) for 2 h at room temperature, followed by extensive washings in PBS and incubation (2 h, 37°C) with FITC-labeled anti-mouse antibody (1:100; Jackson Immunoresearch). Images were captured via a Nikon Eclipse E800 microscope (Nikon, Melville, NY) coupled to a Nikon DN100 CCD camera. Control experiments without primary antibody showed only a faint background staining (not shown).
Proliferation and apoptosis assays.
Untreated CSFC-2G cells or siCtr- or siSPARC-transfected CFSC-2G cells were grown for 72 h on 96 plates. CFSC-2G cells were then serum-starved for 18 h and subsequently pulsed with l μCi/ml [methyl-3H] thymidine (specific activity 20 Ci/mmol; Perkin Elmer) for the last 6 h. Cells were then lysed and the incorporation of radioactivity was measured in a liquid scintillation β-counter (Beckman LS 6500).
Morphological features associated with apoptosis were analyzed by acridine orange and ethidium bromide staining. Briefly, 72 h after transfection pellets were resuspended in the dye mixture of 100 μg/ml acridine orange and 100 μg/ml ethidium bromide (in phosphate-buffered saline) and visualized by fluorescence microscopy. In two independent experiments, a minimum number of 100 cells were counted and number of fragmented nuclei, enlarged cytoplasm, and condensed chromatin were determined. Early apoptosis, a bright green nucleus showing condensation of chromatin as dense green areas (conformational relaxation of DNA); late apoptosis, orange nucleus showing condensation of chromatin (degradation of DNA). Total percentage of apoptotic cells was calculated as subtraction of spontaneous apoptosis from induced apoptosis (% apoptosis of treated cells − % apoptosis of untreated cells) × 100.
Migration assay.
The migratory responses of nontransfected (wt; CFSC-2G or LX-2), siCtr- (CFSC-2G), siSPARC- (CFSC-2G), pRNATinH1.4- (LX-2) and pRNATinH1.4 siSPARC- (LX-2) transfected cells toward different chemotactic stimuli were assayed by using a 48-Transwell microchemotaxis Boyden Chamber system (Neuro Probes, Gaithersburg, MD). Eighteen-hour-starved cells (25,000 cells for CFSC-2G; 50,000 cells for LX-2) were placed into the upper chamber of the Transwell unit, separated from the lower chamber by 5-μm porosity polycarbonate filters (Neuro Probes, Gaithersburg, MD). The bottom chambers of the system were filled with either FBS or MEM supplemented with PDGF-BB (10 ng/ml) or TGF-β1 (5 ng/ml). In other experiments, siSPARC-transfected CFSC-2G cells were or not preincubated with 0.1 ng/ml of TGF-β1 during 24 h. In these assays, cells were allowed to migrate through the filter pores into the lower chambers, for 4 or 12 h depending on the cell line (CFSC-2G or LX-2, respectively). Then the membrane was carefully removed and cells on the upper side of the membrane were scraped out. Cells attached to the lower side of the membrane were fixed in 2% formaldehyde and stained in May-Grunwald/Giemsa. Migrated CFSC-2G cells were counted in a phase-contrast microscope, 10 fields per well were recorded, and mean number of cells per field was calculated. All experiments were repeated at least three times.
ELISA.
Nontransfected or siCtr- or siSPARC-transfected CFSC-2G cells were incubated for 72 h and serum starved for 24 h prior to the assay. Conditioned media were collected and centrifuged at 5,000 g for 10 min at 4°C. Levels of TGF-β1 in the conditioned media were analyzed in the supernatants by ELISA (R&D Systems), following manufacturer protocol. Samples were pretreated with 1 M HCl for 15 min at room temperature prior to neutralization with 1 M NaOH, a procedure that converts all latent TGF-β1 into the active form. The optical density of each well was determined at 540 or 570 nm within 30 min.
Adhesion experiments.
Ninety-six-well MaxiSorp plates (NUNC, Rochester, NY) were coated with fibronectin (10 μg/ml) for 18 h at 37°C. As a negative control, wells were coated with 10% BSA. After extensive washing with PBS, plates were blocked with 1% BSA in PBS for 1 h at room temperature and washed with PBS. Nontransfected, siCtr- or siSPARC-transfected CFSC-2G cells (50,000 cell/well), and pRNATinH1.4- or pRNATinH1.4 siSPARC-transfected LX-2 cells or primary HSCs (10,000 cell/well) were incubated in serum-free medium for 2 h at 37°C, in a humidified incubator containing 5% CO2. After removal of nonadherent cells, cells were fixed with 100% methanol, stained with crystal violet, solubilized with 1% SDS, and quantified with an ELISA plate reader, at 600 nm.
Phalloidin staining.
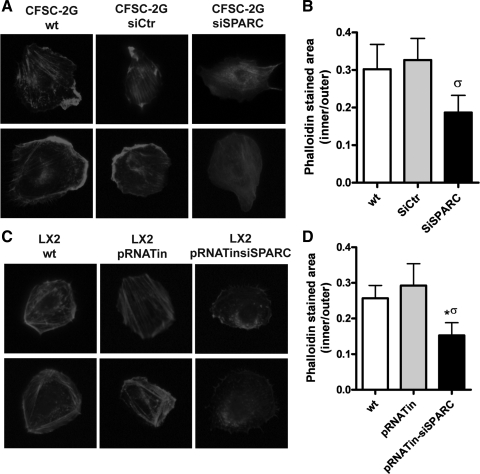
Nontransfected, siCtr- or siSPARC-transfected CFSC-2G cells, as well as pRNATinH1.4- or pRNATinH1.4 siSPARC-transfected LX-2 cells were trypsinized, plated at low density (8,000 cells/well) on PLl/fibronectin-coated coverslips, and further incubated for 4 h. Cultures were then fixed in 4% paraformaldehyde during 15 min and cells were permeabilized with 0.1% Triton X-100/PBS for 10 min. After three washes in PBS, cells were incubated with Alexa Fluor 647-conjugated phalloidin (Molecular Probes) for 2 h at room temperature. Coverslips were then mounted by use of a glycerol-gelatin mounting medium (Sigma). For phalloidin staining area quantification, 30 cells per condition were analyzed. Values were obtained from two regions (the inner half and the outer half) in each of spread cells using the ImageJ software (National Institutes of Health). A cell was considered to have spread when at least one process was larger than its nucleus diameter. The quotient among inner and outer staining area values was calculated per cell and used for statistical comparisons, in both cell lines. A cell was considered to be polarized when the presence of an anchorage site and a migratory front was recognized at opposite poles. Pictures were taken with a Nikon DN100 CCD camera mounted onto a Nikon Eclipse E800 microscope.
Statistical analyses.
Data are expressed as means ± SE, unless stated. Statistical analyses were performed by Student's t, Mann-Whitney, or Kruskal-Wallis tests, when appropriate. Differences were considered to be significant when P < 0.05. Data analyses were performed by using the Prism GraphPad (GraphPad Software, San Diego, CA).
RESULTS
SPARC efficient downregulation in HSCs by specific siRNA constructs.
To evaluate the mechanisms involved in SPARC antifibrotic effects, specific siRNA constructs were designed to silence SPARC in rat (Fig. 1A) and human (Fig. 1B) HSCs. The efficiency of siRNA transfection was of 50 ± 10% (FITC-labeled control siRNA) and 68 ± 11% (reporter pRNATinH1.4 plasmid), respectively (Supplementary Fig. S1).
SPARC mRNA was expressed in rat HSCs based on qPCR (CSFC-2G and primary cultured cells; Fig. 1, C and E) and immunofluorescence (CSFC-2G; Fig. 1F). SPARC siRNA treatment resulted in a significant reduction in SPARC mRNA levels in these cells (CSFC-2G: 0.54 ± 0.06 vs. 1.24 ± 0.16, siSPARC vs. siCtr, respectively; primary cultured HSCs: 0.14 ± 0.04 vs. 1.26 ± 0.3, siSPARC vs. siCtr, respectively) at 3 days after transfection (Figs. 1, C and E). Similarly, SPARC was also expressed by human HSCs (LX-2) (Fig. 1D) and its expression levels were efficiently knocked down at 3 days after pRNATinH1.4 siSPARC transfection (0.19 ± 0.07 vs. 0.91 ± 0.08, siSPARC vs. siCtr, respectively). Western blot analysis of supernatants from LX-2 cultures confirmed that SPARC expression was significantly downregulated in cells treated with pRNATinH1.4 siSPARC at 3 days after transfection (Fig. 1G). Therefore, both siRNA strategies were effective in reducing SPARC expression levels in HSC lines from different origins, as well as in rat primary HSC cultures.
SPARC knockdown reduces HSC migration ability.
Chemokines and cytokines drive key biological processes in HSCs such as their activation, proliferation, and migration that result in the accumulation of these cells at the sites of injury (12). Because proliferation of aHSCs is considered a key event in the hepatic wound healing process, we first investigated the effects of SPARC knockdown on CFSC-2G cells mitotic activity. Proliferation rate was measured by incorporation of [3H]thymidine. SPARC siRNA treatment did not significantly affect HSC proliferation (dpm: 15,087 ± 669.8 vs. 13,590 ± 677.5 vs. 15,177 ± 1,460; wt vs. siCtr vs. siSPARC, respectively; n = 5; P = 0.96, siSPARC vs. wt; P = 0.38, siSPARC vs. siCtr; statistical comparisons were done by applying Student's t-test). Consistently, only a mild increase in late apoptosis was observed (% apoptotic cells: 5.5 ± 2.5 vs. 7.5 ± 0.5 vs. 11 ± 1; wt vs. siCtrl vs. siSPARC, respectively; two independent experiments) with no apparent changes in early apoptosis (% apoptotic cells: 6 ± 0 vs. 7 ± 1 vs. 8.5 ± 1.5; wt vs. siCtrl vs. siSPARC, respectively; two independent experiments).
Since proliferation and apoptosis were largely unaffected by siSPARC treatment in HSCs, we next asked whether HSCs migration ability might be involved in the antifibrotic effect of SPARC downregulation.
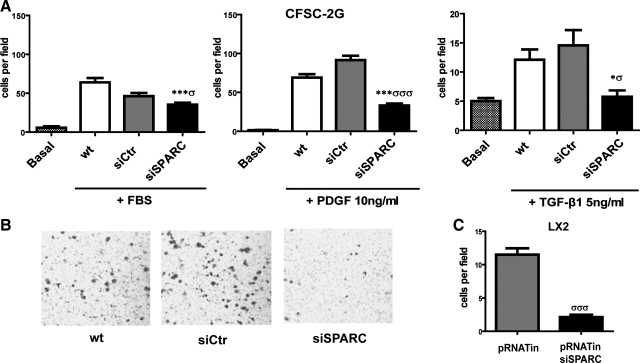
A number of proinflammatory mediators produced in liver damaged sites can act as chemoattractants, with PDGF-BB being the most potent one for HSCs (38). Therefore, we used this cytokine to assess the effects of SPARC silencing on HSC migratory activity in a Boyden chamber two-compartment system. The optimal concentration of PDGF for migration of both CFSC-2G and LX-2 cells was 10 ng/ml (determined by serial 10× dilutions; 1–100 ng/ml; not shown). SPARC downregulation significantly reduced HSC migration (by 2- to 3-fold) compared with nontransfected or control siRNA-transfected cells, toward either FBS (Fig. 2A) or PDGF-BB (Fig. 2, A and B). Similar results were found in LX-2 cells, using PDGF-BB as chemoattractant (Fig. 2C). TGF-β1 is a multifunctional cytokine with known profibrogenic activity (9): SPARC silencing in rat HSCs additionally reduced their migration capacity toward this cytokine (Fig. 2A). These findings suggest that SPARC downregulation in HSCs causes a migratory deficient phenotype, which is likely to be cell-type independent.
Fig. 2.
Migratory defect of HSCs in response to PDGF-BB and TGF-β1 following SPARC siRNA knockdown. A: quantitative data showing the migratory capacity of CFSC-2G cells toward FBS, PDGF-BB, or TGF-β1, in Boyden chamber assays. *P < 0.05, ***P < 0.001 vs. wt; σP < 0.05, σσσP < 0.001 vs. siCtr. B: representative pictures showing a reduced number of cells at the bottom side of filters in the siSPARC group. C: quantification of the migratory capacity of LX-2 cells toward PDGF-BB. σσσP < 0.001. Values correspond to 3 independent experiments and were therefore normalized to the mean value of their respective wt controls. Error bars represent means ± SD.
SPARC knockdown in rat HSCs attenuates migration by modulating TGF-β1 expression and secretion.
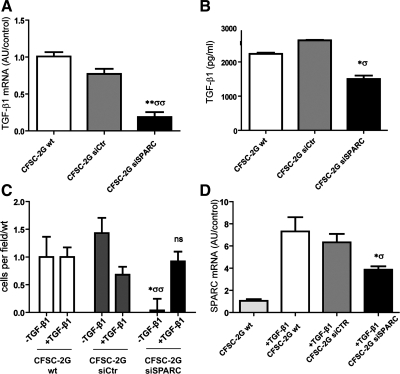
We then investigated whether SPARC knockdown might modulate TGF-β1 mRNA and protein expression in rat HSCs (CFSC-2G). A significant reduction in TGF-β1 mRNA levels was observed in SPARC siRNA-treated cells (0.18 ± 0.06 vs. 0.77 ± 0.07, siSPARC vs. siCtr, respectively) 3 days after transfection (Fig. 3A; compared with nontransfected and siCtr-transfected cells). Similarly, TGF-β1 secretion was strongly reduced in supernatants obtained from cultures treated with siSPARC compared with baseline or control siRNA treatments (Fig. 3B) as measured by ELISA. The reduction in TGF-β1 expression and secretion levels by SPARC-deficient HSCs might contribute to their in vivo antifibrotic effects. We next asked whether the reduced migratory capacity of siSPARC-treated HSCs might result from the reduced TGF-β1 levels. To this end, HSCs were preincubated with TGF-β1 (5 ng/ml) for 24 h prior to migratory assay, which rescued the migratory ability of SPARC-deficient cells in response to PDGF-BB (Fig. 3C). Interestingly, TGF-β1 pretreatment was only able to partially (∼60%) restore SPARC mRNA levels (Fig. 3D); therefore, these results suggest that rescue of the migratory capacity of SPARC siRNA-treated cells by TGF-β1 preincubation is only partially dependent on restoration of SPARC expression levels. From this we hypothesize that, during activation of HSCs, SPARC likely acts upstream of TGF-β1 to allow or enhance their ability to respond to chemotactic stimuli such as PDGF-BB.
Fig. 3.
SPARC knockdown reduces TGF-β1 gene expression and secretion by HSCs. Rescue of siSPARC cells migration ability by TGF-β1 pretreatment. A: qPCR analysis of TGF-β1 mRNA expression levels in CFSC-2G cells. B: quantitative data showing levels of TGF-β1 in HSC culture medium under different experimental conditions measured by ELISA. Note that levels of this cytokine are decreased in SPARC-deficient cells. C: quantification showing a rescue in the migratory capacity in response to PDGF-BB (10 ng/ml) of siSPARC HSCs by pretreatment with TGF-β1 (0.1 ng/ml; added 24 h before chemotaxis assay). Values correspond to 3 independent experiments and were therefore normalized to the mean value of their respective wt controls (−TGF-β1 or +TGF-β1). D: quantification of SPARC mRNA by qPCR when HSCs were pretreated with TGF-β1 (0.1 ng/ml). Error bars represent SD values. *P < 0.05 vs. CFSC-2G wt; σP < 0.05 vs. CFSC-2G siCtr; **P < 0.01 vs. wt; σσP < 0.01 vs. CFSC-2G siCtr; Mann-Whitney test.
Reduced SPARC expression increases HSC adhesiveness to fibronectin and alters their actin filament cytoskeleton pattern.
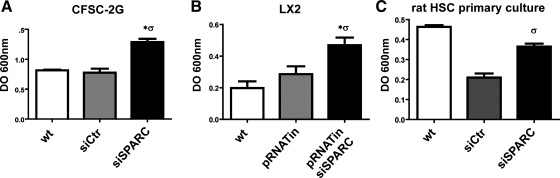
Cellular migration relies on an adequate balance between forces to govern adhesion and detachment from the substratum, which requires ECM-dependent intracellular signaling pathways and dynamic changes in the actin filament cytoskeleton. As mentioned above, migration is crucially involved in the development of a number of pathological processes including liver fibrosis (27, 38). To assess whether the defective migration of SPARC siRNA-treated HSCs might result from changes in cell adhesiveness, we assayed their ability to adhere to fibronectin, an ECM component that promotes the formation of adhesive complexes through engagement of integrins on the cell surface. After a 2-h incubation, siSPARC-treated CFSC-2G cells showed a significantly higher adherence to fibronectin compared with control conditions (Fig. 4A). Similar results were obtained in SPARC-deficient LX-2 cells (Fig. 4B) and in primary rat HSCs (Fig. 4C). From these results we conclude that SPARC downregulation in HSCs significantly enhances their adhesiveness thereby impairing the ability of HSCs to migrate in response to chemoattractant signals.
Fig. 4.
SPARC knockdown increases HSC adhesion to fibronectin. Quantitative data showing a significant increase in the adhesiveness of SPARC-deficient CFSC-2G (A), LX-2 (B), and primary cultured rat HSCs (C). OD, optic density. Error bars represent SD values. *P < 0.05 vs. CFSC-2G wt; σP < 0.05 vs. CFSC-2G siCtr; σσP < 0.01 vs. CFSC-2G siCtr; Mann-Whitney test.
Changes in cell adhesive properties and in migratory ability are usually accompanied by alterations in actin filament cytoskeleton dynamics. To address this issue, HSCs were incubated in PLL/fibronectin-coated surfaces for 4 h and then stained with phalloidin. A significant reduction in both the proportion of cells with a thick fibrillar pattern, known as stress fibers, as well as in the phalloidin staining distribution were found in SPARC siRNA-treated cells compared with naive or control siRNA-treated ones (Fig. 5, A–D). SPARC inhibition also decreased cellular migration compared with the control groups (percentage of polarized, migratory phenotype: 70 vs. 60 vs. 40%, wt vs. siCtrl vs. siSPARC-treated, CFSC-2G cells; 60 vs. 64 vs. 40%, wt vs. pRNATin vs. pRNAtin-siSPARC, P < 0.05 Fisher test, LX-2 cells) (Fig. 5A, bottom, for CFSC-2G cells; Fig. 5B, bottom, for LX-2 cells). From these results we can conclude that SPARC knockdown in HSCs alters the actin filament cytoskeleton, which is likely to be linked to deficiencies in their migratory ability.
Fig. 5.
SPARC knockdown alters actin filament cytoskeleton pattern in HSCs. Representative images showing differential actin filament expression pattern in CFSC-2G and LX-2 cells (phalloidin staining). Representative microphotographs showing changes in the appearance of stress fiber patterns on CFSC-2G (A, top) and LX-2 (C, top) cells. Changes in a polarized/migratory phenotype are shown in A and C, bottom, for CFSC-2G and LX-2 cells, respectively. B and D: quantitative data showing differences in phalloidin staining distribution in CFSC-2G cells (B) and LX-2 cells (D) (values are expressed as the quotient among inner and outer stained area). *P < 0.05 vs. wt; σP < 0.05 vs. control siRNA; Mann-Whitney test.
SPARC transient downregulation in HSCs reduces collagen type I mRNA expression.
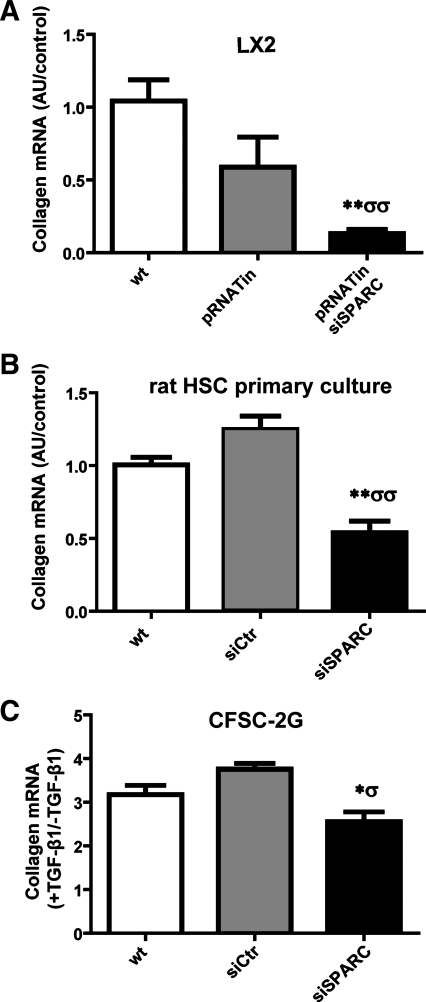
Because HSCs are the major source of collagen during hepatic fibrogenesis, we investigated the effect of SPARC inhibition on collagen mRNA expression in HSCs. LX-2 cells were cultured and transfected with pRNATin or pRNATinsiSPARC for up to 72 h. SPARC knockdown significantly reduced collagen type I mRNA expression levels (Fig. 6A). A similar result was observed in rat primary cultured SPARC siRNA-treated HSCs compared with mock-transfected cells (Fig. 6B). Consistently, in CFSC-2G cells TGF-β1 was found to upregulate collagen type I expression, but this was inhibited when cells were transfected with SPARC siRNA (Fig. 6C).
Fig. 6.
Inhibition of SPARC in HSCs attenuates collagen mRNA expression. A: quantitative data showing collagen type I mRNA expression levels in nontransfected or pRNATinH1.4 or pRNATin-siSPARC transfected LX-2 cells, as measured by qPCR. B and C: quantitative data showing collagen type I mRNA expression levels in primary cultured and CFSC-2G rat HSCs. In CFSC-2G cells collagen was induced by TGF-β1 pretreatment and a stimulation index in each condition is shown. The relative amount of the PCR product amplified from wt cells samples was set at 1 for A and B. *P < 0.05 vs. CFSC-2G wt; **P < 0.01 vs. wt; σP < 0.05 vs. CFSC-2G siCtr; σσP < 0.01 vs. pRNATin (A) or siCtr (B); Mann-Whitney test.
Expression profile analyses: SPARC deficiency in HSCs results in a loss in mesenchymal features and in the upregulation of several cell adhesion proteins.
To further uncover the mechanisms involved in the phenotype of siSPARC-treated HSCs, the expression profile of siCtr- and of siSPARC-transfected cells were compared by an ECM and cell adhesion PCR-array approach (Fig. 7).
Fig. 7.
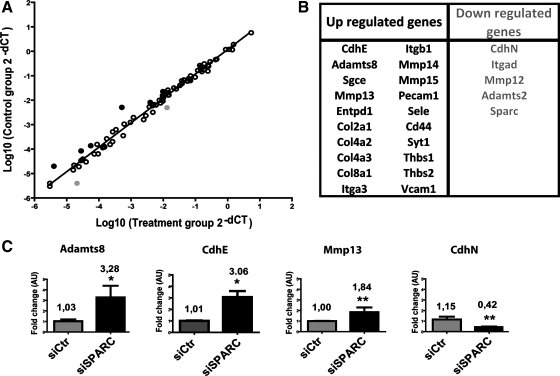
SPARC downregulation in HSCs increased the expression of CdhE, cell adhesion, and metalloproteinase genes and decreased expression of CdhN. A rat extracellular matrix and adhesion molecules PCR array was used to investigate differences in expression of 84 genes involved in cell-cell and cell-matrix interactions. A: graph showing the correlation between fold changes of siCtr- and siSPARC-transfected CFSC-2G cells. Black and gray dots represents upregulated or downregulated genes with a fold change >2, respectively. B: list of upregulated (black letters) and downregulated (gray letters) in the PCR array, fold change >1.5. C: quantification of mRNA levels of those genes found most upregulated or downregulated in PCR array, obtained by qPCR analyses. Values were normalized to levels of GAPDH transcript and analyzed by the ΔΔCt method. Fold change mean is represented. *P < 0.05, **P < 0.001 vs. siCtr; Mann-Whitney test.
From the 84 genes involved in cell-cell and cell-matrix interactions included in the array, many more genes were found upregulated in siSPARC-treated HSCs (≥1.5 fold change; n = 20) than downregulated (n = 5). Consistent with previous functional results showing an increased adhesive propensity of SPARC-deficient HSCs, 50% of upregulated genes are involved in cell-ECM adhesion (Sgce, Col2a1, Col4a2, Col4a3, Col8a1, Itga3, Itgb1, Cd44, Thbs1, and Thbs2). In contrast, only two genes related to cell adhesion to the ECM were downregulated (≥1.5 fold change; CdhN and Itgad). In addition, several metalloproteinases were also found upregulated (n = 4; Adamst8, MMP13, MMP14, MMP15) whereas only two were downregulated (MMP12, Adamst2). Changes in the expression of those genes found most highly deregulated in the array analysis were further confirmed by qPCR (Fig. 1C for SPARC; Fig. 7C for Adamts8, CdhE, MMP13, and CdhN).
It is worth noting that the protein most highly upregulated in SPARC-deficient HSCs was E-cadherin (CdhE) (fold change: 9.7, array; 3.03, qPCR; Fig. 7 and Supplementary Table S1; the online version of this article contains supplemental data) while the most highly downregulated was N-cadherin (CdhN)(fold change: 0.37, array and qPCR; Fig. 7 and Supplementary Table S1). In addition, in SPARC-deficient HSCs four other genes involved in cell-cell adhesion were also upregulated in the array analysis (Pecam1, Sele, Syt1, and Vcam1). From these results and the observed shift from CdhN to CdhE expression, we conclude that SPARC downregulation in HSCs likely induces a loss in mesenchymal properties, which is thought to be intimately linked to cell migration decreased capability.
DISCUSSION
During liver fibrogenesis, SPARC upregulation has previously been demonstrated in aHSCs. Our group also reported that specific SPARC downregulation ameliorates liver fibrosis in an in vivo rat model. Herein we have further analyzed the mechanisms involved by using in vitro hepatic fibrosis models, frequently employed to assess HSCs biology and function (19). To reduce SPARC expression in HSCs, specific siRNA constructs were used (8).
Neither HSCs proliferation nor their apoptosis was largely affected by siSPARC treatment. In contrast, our SPARC knockdown in rat and human HSCs lines as well as in primary cultured rat HSCs significantly increases their adhesiveness to fibronectin and reduces their migratory capability toward known chemoattractant signals, in agreement with previous studies supporting a role for SPARC in fibroblast and cancer cell migration (28, 36).
As a potent profibrogenic cytokine, TGF-β1 also stimulates collagen mRNA in HSCs (18). As previously published, siSPARC treatment significantly reduced TGF-β1 mRNA and protein levels in HSCs (10). As expected, SPARC knockdown also led to downregulation of collagen type I expression in both LX-2 and rat primary cultured HSCs (34, 39). Since only a 50% reduction in SPARC levels was observed in SPARC siRNA-treated CFSC-2G cells, downregulation of collagen type I was not significant in this cell line; however, the expression of this ECM protein was decreased when SPARC-deficient CFSC-2G cells were treated with TGF-β1. Therefore, it seems that SPARC knockdown confers a relative resistance to profibrogenic activity of TGF-β1. Interestingly, the deficient migratory phenotype in these cells was rescued by preincubating them with TGF-β1, suggesting that this cytokine is required for HSC migration. Our data suggest that the rescue in the migratory phenotype of SPARC-deficient HSCs by TGF-β1 preincubation involves mechanisms likely partially independent from SPARC silencing. Moreover, siSPARC treatment resulted in the loss of mesenchymal features in HSCs, as reflected in the observed shift from a CdhN to a CdhE expression profile. Thus our results are consistent with a known role of TGF-β1 as inductor of a mesenchymal phenotype in HSCs, which is required for their migratory phenotype (18).
In our gene array analysis, several genes involved in cell-ECM adhesion processes were found upregulated. It is worth noting that although the expression levels of some collagen genes were increased, they were not among those comprising the fibrotic matrix of injured liver (21) but rather those commonly found in the epithelial basal membrane, such as collagen type IV. Consistent with the upregulation of several molecules related to cell adhesion to the ECM, SPARC siRNA treatment significantly increased the adhesiveness of HSCs to the substrate. This imbalance between cell adhesion and detachment likely contributes to the HSCs migratory deficiency observed upon SPARC downregulation. In addition, the F-actin expression pattern was altered in siSPARC-treated HSCs, which this likely adds to the observed migratory defect.
Expression of several metalloproteinases was found upregulated in SPARC-deficient HSCs. Metalloproteinases are enzymes that can disrupt ECM; therefore their increased expression in these cells might contribute to the liver antifibrotic role resulting from SPARC downregulation (6). Only two enzymes with a metalloproteinase function, MMP12 and Adamst2, were downregulated in siRNA-treated cells; however, these two enzymes were previously shown to be induced in HSCs during activation in vitro and/or in vivo (7).
Among all genes upregulated in SPARC siRNA-treated cells, the marked and significant upregulation of Adamst8 was noteworthy. The preferred name for this gene is “A disintegrin and metalloproteinase with thrombospondin motifs 8” and, as suggested by the name, the codified protein has a metalloproteinase domain, a disintegrin-like domain, and a thrombospondin type 1 (TS) motif. It has multiple binding functions and an endopeptidase activity (33). Considering the importance of TGF-β1 for HSC activation and its requirement for intracellular and then extracellular proteolytic activation (22), the observed increase in Adamst8 and other metalloproteinases, as well as in thrombospondin 1 (Thbs1), might be an adaptive mechanism in SPARC-deficient HSCs to increase the availability of sufficient levels of TGF-β1 active forms to avoid cell death. Nevertheless, the biological significance of increased Adamst8 expression in the context of liver fibrosis requires further investigation.
As noted above, CdhE mRNA, encoding the E-cadherin protein, was among those most highly upregulated in SPARC-deficient HSCs. Consistently, CdhN mRNA, which encodes N-cadherin, was the most downregulated gene in these cells. These data are consistent with previous reports showing decreased expression of N-cadherin with SPARC silencing (31) and of E-cadherin with SPARC overexpression (29). In other cell types, especially epithelial cells, this shift in cadherin expression is considered a hallmark of a mesenchymal-to-epithelial transition (MET) process (35). Since HSCs are not epithelial cells, this transition can be appropriately referred to as simply a loss of mesenchymal features. In addition, because N-cadherin upregulation is a marker for HSC activation (25), we can additionally conclude that reduced levels of SPARC in HSCs likely results in the inhibition of the HSCs activation process.
Changes in SPARC expression levels have been previously reported in several malignant tumors including hepatocellular carcinoma. Nevertheless, the role of SPARC in cancer is controversial and seems to depend on the type of tumor. For instance, we have previously shown that SPARC overexpression in liver cancer cells results in E-cadherin upregulation and N-cadherin downregulation (1); these results are in contrast with earlier observations in melanoma cells, in which high levels of SPARC were associated with N-cadherin upregulation and E-cadherin downregulation (29, 31). Thus SPARC overexpression in different types of cancer cells seems to be differently involved in modulating EMT or MET, likely affecting migratory properties of cells.
In summary, SPARC downregulation in activated HSCs exerts its antifibrotic effects likely through a reduction in their capability to migrate toward regions of injury. Since TGF-β1 expression was reduced in SPARC siRNA-treated HSCs and the migratory phenotype was rescued by preincubating cells with this cytokine, SPARC knockdown seems to reduce migratory ability of HSCs through the modulation of TGF-β1 gene activity in these cells. A loss of mesenchymal properties in SPARC-deficient HSCs, characterized by the shift in cadherin expression profile and the downregulation of collagen type I expression, likely due to TGF-β1 downregulation, appears to be an important determinant of the migratory phenotype. In addition, both an enhancement in cellular adhesion and alterations in actin cytoskeleton were present in SPARC-deficient HSCs. The upregulation of many genes involved in cell adhesiveness suggests that SPARC's contribution to the observed migratory phenotype might be partially independent of the loss of mesenchymal features in HSC. Whether or not this is also a consequence of TGF-β1 downregulation in SPARC-deficient HSCs remains to be addressed. Altogether our data further suggest SPARC as a target molecule in HSCs with potential clinical interest in the treatment of liver cirrhosis.
GRANTS
This work was supported by grants from Universidad Austral and Agencia Nacional de Promoción Científica y Tecnológica (PICTO-CRUP-2005-31179; PICT-2005-34788; PICT-2006-1882; PICT-2008-00123), CTE-06, and AECI 2008 and from the National Institutes of Health (DK56621, AA017067, and AA018408-01 to S. L. Friedman).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. M. Rojkind (Marion Bessin Liver Research Center, Albert Einstein College of Medicine, Bronx, New York) for kindly providing us with cell line CFSC-2G used in this work. We thank Soledad Arregui and Guillermo Gastón for expert technical assistance.
REFERENCES
- 1. Atorrasagasti C, Malvicini M, Aquino JB, Alaniz L, Garcia M, Bolontrade M, Rizzo M, Podhajcer OL, Mazzolini G. Overexpression of SPARC obliterates the in vivo tumorigenicity of human hepatocellular carcinoma cells. Int J Cancer 126: 2726–2740, 2010 [DOI] [PubMed] [Google Scholar]
- 2. Barker TH, Baneyx G, Cardo-Vila M, Workman GA, Weaver M, Menon PM, Dedhar S, Rempel SA, Arap W, Pasqualini R, Vogel V, Sage EH. SPARC regulates extracellular matrix organization through its modulation of integrin-linked kinase activity. J Biol Chem 280: 36483–36493, 2005 [DOI] [PubMed] [Google Scholar]
- 3. Bataller R, Brenner DA. Liver fibrosis. J Clin Invest 115: 209–218, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bissell DM, Wang SS, Jarnagin WR, Roll FJ. Cell-specific expression of transforming growth factor-beta in rat liver. Evidence for autocrine regulation of hepatocyte proliferation. J Clin Invest 96: 447–455, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blazejewski S, Le Bail B, Boussarie L, Blanc JF, Malaval L, Okubo K, Saric J, Bioulac-Sage P, Rosenbaum J. Osteonectin (SPARC) expression in human liver and in cultured human liver myofibroblasts. Am J Pathol 151: 651–657, 1997 [PMC free article] [PubMed] [Google Scholar]
- 6. Camino AM, Atorrasagasti C, Maccio D, Prada F, Salvatierra E, Rizzo M, Alaniz L, Aquino JB, Podhajcer OL, Silva M, Mazzolini G. Adenovirus-mediated inhibition of SPARC attenuates liver fibrosis in rats. J Gene Med 10: 993–1004, 2008 [DOI] [PubMed] [Google Scholar]
- 7. De Minicis S, Seki E, Uchinami H, Kluwe J, Zhang Y, Brenner DA, Schwabe RF. Gene expression profiles during hepatic stellate cell activation in culture and in vivo. Gastroenterology 132: 1937–1946, 2007 [DOI] [PubMed] [Google Scholar]
- 8. Dykxhoorn DM, Palliser D, Lieberman J. The silent treatment: siRNAs as small molecule drugs. Gene Ther 13: 541–552, 2006 [DOI] [PubMed] [Google Scholar]
- 9. Framson PE, Sage EH. SPARC and tumor growth: where the seed meets the soil? J Cell Biochem 92: 679–690, 2004 [DOI] [PubMed] [Google Scholar]
- 10. Francki A, Bradshaw AD, Bassuk JA, Howe CC, Couser WG, Sage EH. SPARC regulates the expression of collagen type I and transforming growth factor-beta1 in mesangial cells. J Biol Chem 274: 32145–32152, 1999 [DOI] [PubMed] [Google Scholar]
- 11. Friedman SL. Liver fibrosis — from bench to bedside. J Hepatol 38, Suppl 1: S38–S53, 2003 [DOI] [PubMed] [Google Scholar]
- 12. Friedman SL. Molecular regulation of hepatic fibrosis, an integrated cellular response to tissue injury. J Biol Chem 275: 2247–2250, 2000 [DOI] [PubMed] [Google Scholar]
- 13. Friedman SL, Roll FJ. Isolation and culture of hepatic lipocytes, Kupffer cells, and sinusoidal endothelial cells by density gradient centrifugation with Stractan. Anal Biochem 161: 207–218, 1987 [DOI] [PubMed] [Google Scholar]
- 14. Frizell E, Liu SL, Abraham A, Ozaki I, Eghbali M, Sage EH, Zern MA. Expression of SPARC in normal and fibrotic livers. Hepatology 21: 847–854, 1995 [PubMed] [Google Scholar]
- 15. Greenwel P, Schwartz M, Rosas M, Peyrol S, Grimaud JA, Rojkind M. Characterization of fat-storing cell lines derived from normal and CCl4-cirrhotic livers. Differences in the production of interleukin-6. Lab Invest 65: 644–653, 1991 [PubMed] [Google Scholar]
- 16. Gressner AM. The cell biology of liver fibrogenesis - an imbalance of proliferation, growth arrest and apoptosis of myofibroblasts. Cell Tissue Res 292: 447–452, 1998 [DOI] [PubMed] [Google Scholar]
- 17. Gressner AM. Cytokines and cellular crosstalk involved in the activation of fat-storing cells. J Hepatol 22: 28–36, 1995 [PubMed] [Google Scholar]
- 18. Gressner AM, Weiskirchen R, Breitkopf K, Dooley S. Roles of TGF-beta in hepatic fibrosis. Front Biosci 7: d793–d807, 2002 [DOI] [PubMed] [Google Scholar]
- 19. Herrmann J, Gressner AM, Weiskirchen R. Immortal hepatic stellate cell lines: useful tools to study hepatic stellate cell biology and function? J Cell Mol Med 11: 704–722, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ikeda K, Wakahara T, Wang YQ, Kadoya H, Kawada N, Kaneda K. In vitro migratory potential of rat quiescent hepatic stellate cells and its augmentation by cell activation. Hepatology 29: 1760–1767, 1999 [DOI] [PubMed] [Google Scholar]
- 21. Inagaki Y, Okazaki I. Emerging insights into transforming growth factor beta Smad signal in hepatic fibrogenesis. Gut 56: 284–292, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kisseleva T, Brenner DA. Mechanisms of fibrogenesis. Exp Biol Med (Maywood) 233: 109–122, 2008 [DOI] [PubMed] [Google Scholar]
- 23. Kristensen DB, Kawada N, Imamura K, Miyamoto Y, Tateno C, Seki S, Kuroki T, Yoshizato K. Proteome analysis of rat hepatic stellate cells. Hepatology 32: 268–277, 2000 [DOI] [PubMed] [Google Scholar]
- 24. Lamireau T, Le Bail B, Boussarie L, Fabre M, Vergnes P, Bernard O, Gautier F, Bioulac-Sage P, Rosenbaum J. Expression of collagens type I and IV, osteonectin and transforming growth factor beta-1 (TGFbeta1) in biliary atresia and paucity of intrahepatic bile ducts during infancy. J Hepatol 31: 248–255, 1999 [DOI] [PubMed] [Google Scholar]
- 25. Lim YS, Lee HC, Lee HS. Switch of cadherin expression from E- to N-type during the activation of rat hepatic stellate cells. Histochem Cell Biol 127: 149–160, 2007 [DOI] [PubMed] [Google Scholar]
- 26. Marra F, Gentilini A, Pinzani M, Choudhury GG, Parola M, Herbst H, Dianzani MU, Laffi G, Abboud HE, Gentilini P. Phosphatidylinositol 3-kinase is required for platelet-derived growth factor's actions on hepatic stellate cells. Gastroenterology 112: 1297–1306, 1997 [DOI] [PubMed] [Google Scholar]
- 27. Melton AC, Soon RK, Jr, Park JG, Martinez L, Dehart GW, Yee HF., Jr Focal adhesion disassembly is an essential early event in hepatic stellate cell chemotaxis. Am J Physiol Gastrointest Liver Physiol 293: G1272–G1280, 2007 [DOI] [PubMed] [Google Scholar]
- 28. Podhajcer OL, Benedetti LG, Girotti MR, Prada F, Salvatierra E, Llera AS. The role of the matricellular protein SPARC in the dynamic interaction between the tumor and the host. Cancer Metastasis Rev 27: 691–705, 2008 [DOI] [PubMed] [Google Scholar]
- 29. Robert G, Gaggioli C, Bailet O, Chavey C, Abbe P, Aberdam E, Sabatie E, Cano A, Garcia de Herreros A, Ballotti R, Tartare-Deckert S. SPARC represses E-cadherin and induces mesenchymal transition during melanoma development. Cancer Res 66: 7516–7523, 2006 [DOI] [PubMed] [Google Scholar]
- 30. Sage EH, Bornstein P. Extracellular proteins that modulate cell-matrix interactions. SPARC, tenascin, and thrombospondin. J Biol Chem 266: 14831–14834, 1991 [PubMed] [Google Scholar]
- 31. Sosa MS, Girotti MR, Salvatierra E, Prada F, de Olmo JA, Gallango SJ, Albar JP, Podhajcer OL, Llera AS. Proteomic analysis identified N-cadherin, clusterin, and HSP27 as mediators of SPARC (secreted protein, acidic and rich in cysteines) activity in melanoma cells. Proteomics 7: 4123–4134, 2007 [DOI] [PubMed] [Google Scholar]
- 32. Termine JD, Kleinman HK, Whitson SW, Conn KM, McGarvey ML, Martin GR. Osteonectin, a bone-specific protein linking mineral to collagen. Cell 26: 99–105, 1981 [DOI] [PubMed] [Google Scholar]
- 33. Vazquez F, Hastings G, Ortega MA, Lane TF, Oikemus S, Lombardo M, Iruela-Arispe ML. METH-1, a human ortholog of ADAMTS-1, and METH-2 are members of a new family of proteins with angio-inhibitory activity. J Biol Chem 274: 23349–23357, 1999 [DOI] [PubMed] [Google Scholar]
- 34. Wang JC, Lai S, Guo X, Zhang X, de Crombrugghe B, Sonnylal S, Arnett FC, Zhou X. Attenuation of fibrosis in vitro and in vivo with SPARC siRNA. Arthritis Res Ther 12: R60, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wells A, Yates C, Shepard CR. E-cadherin as an indicator of mesenchymal to epithelial reverting transitions during the metastatic seeding of disseminated carcinomas. Clin Exp Metastasis 25: 621–628, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wu RX, Laser M, Han H, Varadarajulu J, Schuh K, Hallhuber M, Hu K, Ertl G, Hauck CR, Ritter O. Fibroblast migration after myocardial infarction is regulated by transient SPARC expression. J Mol Med 84: 241–252, 2006 [DOI] [PubMed] [Google Scholar]
- 37. Xu LHA, Albanis E, Arthur MJ, O'Byrne SM, Blaner WS. Human hepatic stellate cell lines, LX-1 and LX-2: new tools for analysis of hepatic fibrosis. Gut 54: 142–151, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yang C, Zeisberg M, Mosterman B, Sudhakar A, Yerramalla U, Holthaus K, Xu L, Eng F, Afdhal N, Kalluri R. Liver fibrosis: insights into migration of hepatic stellate cells in response to extracellular matrix and growth factors. Gastroenterology 124: 147–159, 2003 [DOI] [PubMed] [Google Scholar]
- 39. Zhou X, Tan FK, Guo X, Wallis D, Milewicz DM, Xue S, Arnett FC. Small interfering RNA inhibition of SPARC attenuates the profibrotic effect of transforming growth factor beta1 in cultured normal human fibroblasts. Arthritis Rheum 52: 257–261, 2005 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.