Abstract
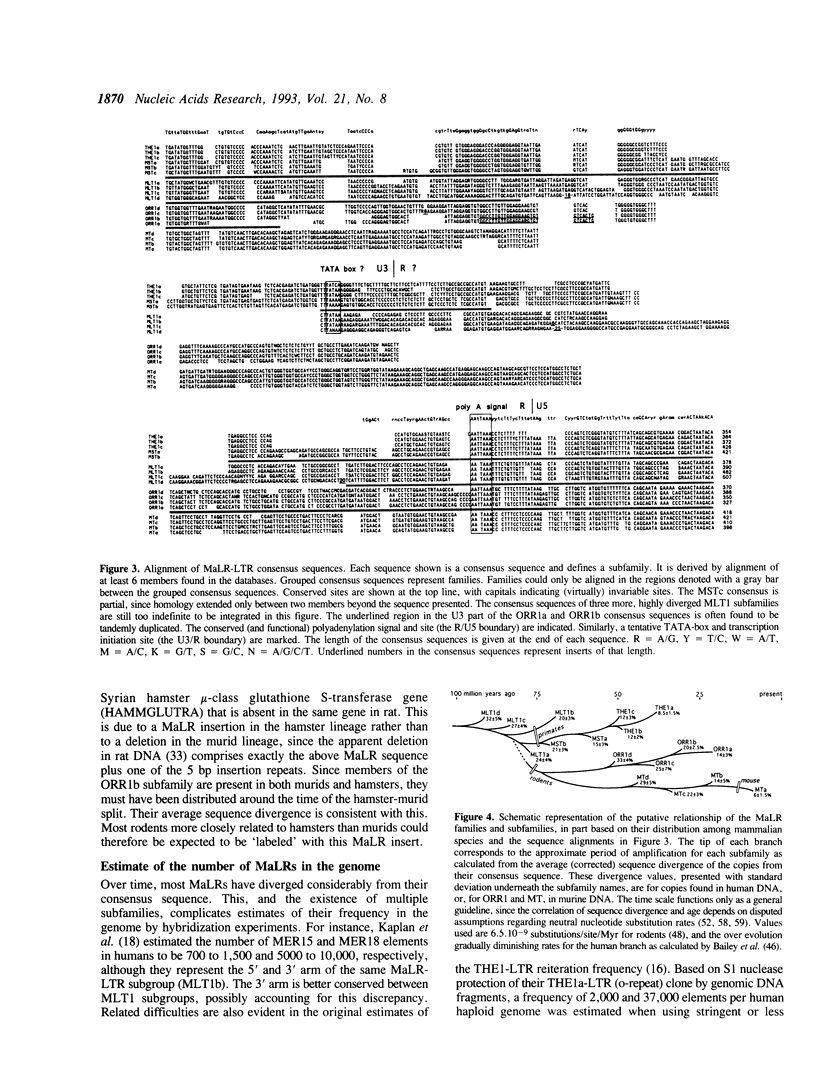
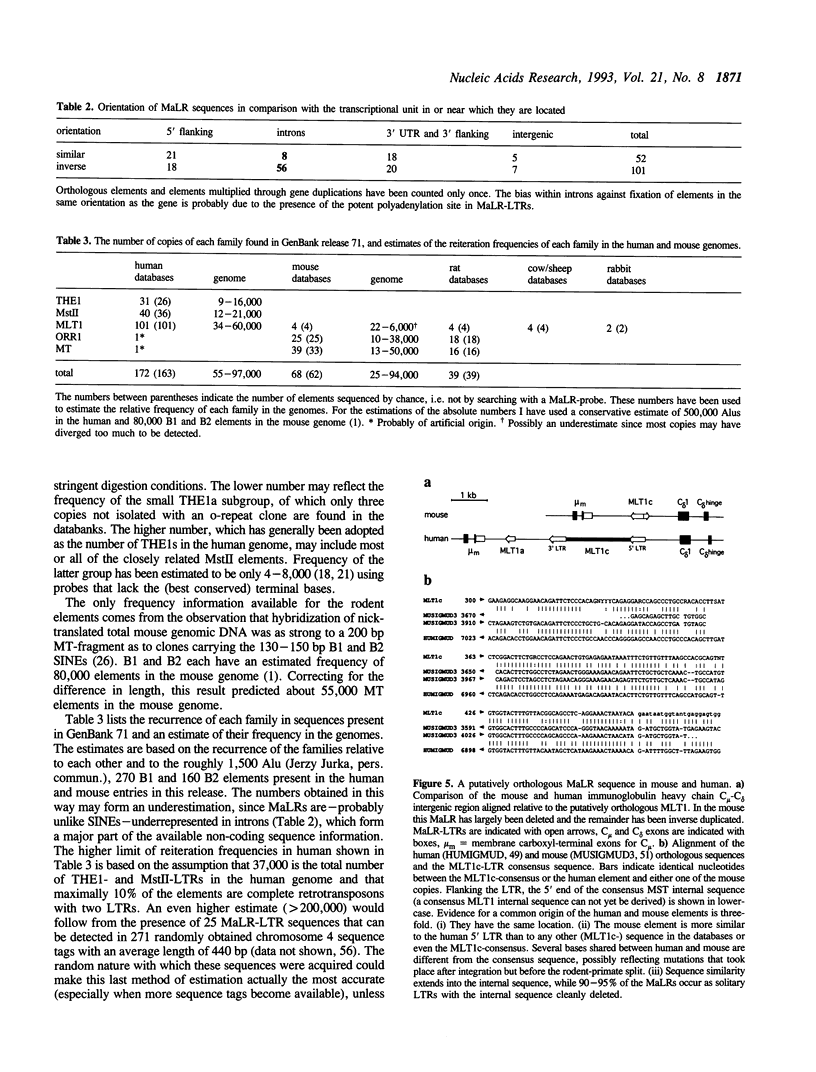
A new superfamily of mammalian transposable genetic elements is described with an estimated 40,000 to 100,000 members in both primate and rodent genomes. Sequences known before as MT, ORR-1, MstII, MER15 and MER18 are shown to represent (part of) the long terminal repeats of retrotransposon-like elements related to THE1 in humans. These transposons have structural similarities to retroviruses. However, the putative product of a 1350 base pair open reading frame detected in the consensus internal sequence of THE1 does not resemble retroviral proteins. The elements are named 'Mammalian apparent LTR-retrotransposons' (MaLRs). The internal sequence is usually found to be excised. Their presence in rodents, artiodactyls, lagomorphs, and primates, the divergence of the individual elements from their consensus, and the existence of a probably orthologous element in mouse and man suggest that the first MaLRs were distributed before the radiation of eutherian mammals 80-100 million years ago. MaLRs may prove to be very helpful in determining the evolutionary branching pattern of mammalian orders and suborders.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams M. D., Dubnick M., Kerlavage A. R., Moreno R., Kelley J. M., Utterback T. R., Nagle J. W., Fields C., Venter J. C. Sequence identification of 2,375 human brain genes. Nature. 1992 Feb 13;355(6361):632–634. doi: 10.1038/355632a0. [DOI] [PubMed] [Google Scholar]
- Akahori Y., Handa H., Imai K., Abe M., Kameyama K., Hibiya M., Yasui H., Okamura K., Naito M., Matsuoka H. Sigma region located between C mu and C delta genes of human immunoglobulin heavy chain: possible involvement of tRNA-like structure in RNA splicing. Nucleic Acids Res. 1988 Oct 25;16(20):9497–9511. doi: 10.1093/nar/16.20.9497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey W. J., Fitch D. H., Tagle D. A., Czelusniak J., Slightom J. L., Goodman M. Molecular evolution of the psi eta-globin gene locus: gibbon phylogeny and the hominoid slowdown. Mol Biol Evol. 1991 Mar;8(2):155–184. doi: 10.1093/oxfordjournals.molbev.a040641. [DOI] [PubMed] [Google Scholar]
- Bastien L., Bourgaux P. The MT family of mouse DNA is made of short interspersed repeated elements. Gene. 1987;57(1):81–88. doi: 10.1016/0378-1119(87)90179-x. [DOI] [PubMed] [Google Scholar]
- Birnstiel M. L., Busslinger M., Strub K. Transcription termination and 3' processing: the end is in site! Cell. 1985 Jun;41(2):349–359. doi: 10.1016/s0092-8674(85)80007-6. [DOI] [PubMed] [Google Scholar]
- Bodrug S. E., Ray P. N., Gonzalez I. L., Schmickel R. D., Sylvester J. E., Worton R. G. Molecular analysis of a constitutional X-autosome translocation in a female with muscular dystrophy. Science. 1987 Sep 25;237(4822):1620–1624. doi: 10.1126/science.3629260. [DOI] [PubMed] [Google Scholar]
- Brown P. O. Integration of retroviral DNA. Curr Top Microbiol Immunol. 1990;157:19–48. doi: 10.1007/978-3-642-75218-6_2. [DOI] [PubMed] [Google Scholar]
- Bulmer M., Wolfe K. H., Sharp P. M. Synonymous nucleotide substitution rates in mammalian genes: implications for the molecular clock and the relationship of mammalian orders. Proc Natl Acad Sci U S A. 1991 Jul 15;88(14):5974–5978. doi: 10.1073/pnas.88.14.5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caddle M. S., Lussier R. H., Heintz N. H. Intramolecular DNA triplexes, bent DNA and DNA unwinding elements in the initiation region of an amplified dihydrofolate reductase replicon. J Mol Biol. 1990 Jan 5;211(1):19–33. doi: 10.1016/0022-2836(90)90008-A. [DOI] [PubMed] [Google Scholar]
- Deininger P. L., Batzer M. A., Hutchison C. A., 3rd, Edgell M. H. Master genes in mammalian repetitive DNA amplification. Trends Genet. 1992 Sep;8(9):307–311. doi: 10.1016/0168-9525(92)90262-3. [DOI] [PubMed] [Google Scholar]
- Deka N., Willard C. R., Wong E., Schmid C. W. Human transposon-like elements insert at a preferred target site: evidence for a retrovirally mediated process. Nucleic Acids Res. 1988 Feb 11;16(3):1143–1151. doi: 10.1093/nar/16.3.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle R. F., Feng D. F., Johnson M. S., McClure M. A. Origins and evolutionary relationships of retroviruses. Q Rev Biol. 1989 Mar;64(1):1–30. doi: 10.1086/416128. [DOI] [PubMed] [Google Scholar]
- Easteal S. A mammalian molecular clock? Bioessays. 1992 Jun;14(6):415–419. doi: 10.1002/bies.950140613. [DOI] [PubMed] [Google Scholar]
- Fan W., Trifiletti R., Cooper T., Norris J. S. Cloning of a mu-class glutathione S-transferase gene and identification of the glucocorticoid regulatory domains in its 5' flanking sequence. Proc Natl Acad Sci U S A. 1992 Jul 1;89(13):6104–6108. doi: 10.1073/pnas.89.13.6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields C. A., Grady D. L., Moyzis R. K. The human THE-LTR(O) and MstII interspersed repeats are subfamilies of a single widely distributed highly variable repeat family. Genomics. 1992 Jun;13(2):431–436. doi: 10.1016/0888-7543(92)90264-s. [DOI] [PubMed] [Google Scholar]
- Fisher E. M., Alitalo T., Luoh S. W., de la Chapelle A., Page D. C. Human sex-chromosome-specific repeats within a region of pseudoautosomal/Yq homology. Genomics. 1990 Aug;7(4):625–628. doi: 10.1016/0888-7543(90)90209-d. [DOI] [PubMed] [Google Scholar]
- Gonzalez F. J., Kasper C. B. Cloning and characterization of the rat NADPH-cytochrome P-450 oxidoreductase gene. J Biol Chem. 1983 Jan 25;258(2):1363–1368. [PubMed] [Google Scholar]
- Heinlein U. A., Lange-Sablitzky R., Schaal H., Wille W. Molecular characterization of the MT-family of dispersed middle-repetitive DNA in rodent genomes. Nucleic Acids Res. 1986 Aug 26;14(16):6403–6416. doi: 10.1093/nar/14.16.6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollis G. F., Hieter P. A., McBride O. W., Swan D., Leder P. Processed genes: a dispersed human immunoglobulin gene bearing evidence of RNA-type processing. Nature. 1982 Mar 25;296(5855):321–325. doi: 10.1038/296321a0. [DOI] [PubMed] [Google Scholar]
- Hwu H. R., Roberts J. W., Davidson E. H., Britten R. J. Insertion and/or deletion of many repeated DNA sequences in human and higher ape evolution. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3875–3879. doi: 10.1073/pnas.83.11.3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal A. K., Gonzalez F. J., Nebert D. W. Human P1-450 gene sequence and correlation of mRNA with genetic differences in benzo[a]pyrene metabolism. Nucleic Acids Res. 1985 Jun 25;13(12):4503–4520. doi: 10.1093/nar/13.12.4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J. Novel families of interspersed repetitive elements from the human genome. Nucleic Acids Res. 1990 Jan 11;18(1):137–141. doi: 10.1093/nar/18.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan D. J., Jurka J., Solus J. F., Duncan C. H. Medium reiteration frequency repetitive sequences in the human genome. Nucleic Acids Res. 1991 Sep 11;19(17):4731–4738. doi: 10.1093/nar/19.17.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshet E., Schiff R., Itin A. Mouse retrotransposons: a cellular reservoir of long terminal repeat (LTR) elements with diverse transcriptional specificities. Adv Cancer Res. 1991;56:215–251. doi: 10.1016/s0065-230x(08)60482-0. [DOI] [PubMed] [Google Scholar]
- Kimura S., Hanioka N., Matsunaga E., Gonzalez F. J. The rat clofibrate-inducible CYP4A gene subfamily. I. Complete intron and exon sequence of the CYP4A1 and CYP4A2 genes, unique exon organization, and identification of a conserved 19-bp upstream element. DNA. 1989 Sep;8(7):503–516. doi: 10.1089/dna.1.1989.8.503. [DOI] [PubMed] [Google Scholar]
- Law M. L., Gao J. Z., Puck T. T. A nuclear protein associated with human cancer cells binds preferentially to a human repetitive DNA sequence. Proc Natl Acad Sci U S A. 1989 Nov;86(21):8472–8476. doi: 10.1073/pnas.86.21.8472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law M. L., Xu Y. S., Berger R., Tung L. Molecular analysis of human repetitive sequence family and its use as genetic marker. Somat Cell Mol Genet. 1987 Jul;13(4):381–389. doi: 10.1007/BF01534936. [DOI] [PubMed] [Google Scholar]
- Lawrance S. K., Das H. K., Pan J., Weissman S. M. The genomic organisation and nucleotide sequence of the HLA-SB(DP) alpha gene. Nucleic Acids Res. 1985 Oct 25;13(20):7515–7528. doi: 10.1093/nar/13.20.7515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leib-Mösch C., Brack-Werner R., Werner T., Bachmann M., Faff O., Erfle V., Hehlmann R. Endogenous retroviral elements in human DNA. Cancer Res. 1990 Sep 1;50(17 Suppl):5636S–5642S. [PubMed] [Google Scholar]
- Levin H. L., Boeke J. D. Demonstration of retrotransposition of the Tf1 element in fission yeast. EMBO J. 1992 Mar;11(3):1145–1153. doi: 10.1002/j.1460-2075.1992.tb05155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. H., Gouy M., Sharp P. M., O'hUigin C., Yang Y. W. Molecular phylogeny of Rodentia, Lagomorpha, Primates, Artiodactyla, and Carnivora and molecular clocks. Proc Natl Acad Sci U S A. 1990 Sep;87(17):6703–6707. doi: 10.1073/pnas.87.17.6703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. H., Tanimura M., Sharp P. M. An evaluation of the molecular clock hypothesis using mammalian DNA sequences. J Mol Evol. 1987;25(4):330–342. doi: 10.1007/BF02603118. [DOI] [PubMed] [Google Scholar]
- Lloyd J. A., Lamb A. N., Potter S. S. Phylogenetic screening of the human genome: identification of differentially hybridizing repetitive sequence families. Mol Biol Evol. 1987 Mar;4(2):85–98. doi: 10.1093/oxfordjournals.molbev.a040429. [DOI] [PubMed] [Google Scholar]
- Marić S. C., Crozat A., Jänne O. A. Structure and organization of the human S-adenosylmethionine decarboxylase gene. J Biol Chem. 1992 Sep 15;267(26):18915–18923. [PubMed] [Google Scholar]
- Martin S. L. LINEs. Curr Opin Genet Dev. 1991 Dec;1(4):505–508. doi: 10.1016/s0959-437x(05)80199-6. [DOI] [PubMed] [Google Scholar]
- McClure M. A. Evolution of retroposons by acquisition or deletion of retrovirus-like genes. Mol Biol Evol. 1991 Nov;8(6):835–856. doi: 10.1093/oxfordjournals.molbev.a040686. [DOI] [PubMed] [Google Scholar]
- Mermer B., Colb M., Krontiris T. G. A family of short, interspersed repeats is associated with tandemly repetitive DNA in the human genome. Proc Natl Acad Sci U S A. 1987 May;84(10):3320–3324. doi: 10.1073/pnas.84.10.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milstein C. P., Deverson E. V., Rabbitts T. H. The sequence of the human immunoglobulin mu-delta intron reveals possible vestigial switch segments. Nucleic Acids Res. 1984 Aug 24;12(16):6523–6535. doi: 10.1093/nar/12.16.6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'hUigin C., Li W. H. The molecular clock ticks regularly in muroid rodents and hamsters. J Mol Evol. 1992 Nov;35(5):377–384. doi: 10.1007/BF00171816. [DOI] [PubMed] [Google Scholar]
- Okada N. SINEs. Curr Opin Genet Dev. 1991 Dec;1(4):498–504. doi: 10.1016/s0959-437x(05)80198-4. [DOI] [PubMed] [Google Scholar]
- Pascale E., Valle E., Furano A. V. Amplification of an ancestral mammalian L1 family of long interspersed repeated DNA occurred just before the murine radiation. Proc Natl Acad Sci U S A. 1990 Dec;87(23):9481–9485. doi: 10.1073/pnas.87.23.9481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson K. E., Deka N., Schmid C. W., Misra R., Schindler C. W., Rush M. G., Kadyk L., Leinwand L. A transposon-like element in human DNA. Nature. 1985 Jul 25;316(6026):359–361. doi: 10.1038/316359a0. [DOI] [PubMed] [Google Scholar]
- Paulson K. E., Matera A. G., Deka N., Schmid C. W. Transcription of a human transposon-like sequence is usually directed by other promoters. Nucleic Acids Res. 1987 Jul 10;15(13):5199–5215. doi: 10.1093/nar/15.13.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards J. E., Gilliam A. C., Shen A., Tucker P. W., Blattner F. R. Unusual sequences in the murine immunoglobulin mu-delta heavy-chain region. Nature. 1983 Dec 1;306(5942):483–487. doi: 10.1038/306483a0. [DOI] [PubMed] [Google Scholar]
- Ryan S. C., Dugaiczyk A. Newly arisen DNA repeats in primate phylogeny. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9360–9364. doi: 10.1073/pnas.86.23.9360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid C. W., Wong E. F., Deka N. Single copy sequences in galago DNA resemble a repetitive human retrotransposon-like family. J Mol Evol. 1990 Aug;31(2):92–100. doi: 10.1007/BF02109478. [DOI] [PubMed] [Google Scholar]
- Sekiguchi T., Nohiro Y., Nakamura Y., Hisamoto N., Nishimoto T. The human CCG1 gene, essential for progression of the G1 phase, encodes a 210-kilodalton nuclear DNA-binding protein. Mol Cell Biol. 1991 Jun;11(6):3317–3325. doi: 10.1128/mcb.11.6.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobel E., Martinez H. M. A multiple sequence alignment program. Nucleic Acids Res. 1986 Jan 10;14(1):363–374. doi: 10.1093/nar/14.1.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Paulson K. E., Schmid C. W., Kadyk L., Leinwand L. Non-Alu family interspersed repeats in human DNA and their transcriptional activity. Nucleic Acids Res. 1984 Mar 26;12(6):2669–2690. doi: 10.1093/nar/12.6.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagle D. A., Stanhope M. J., Siemieniak D. R., Benson P., Goodman M., Slightom J. L. The beta globin gene cluster of the prosimian primate Galago crassicaudatus: nucleotide sequence determination of the 41-kb cluster and comparative sequence analyses. Genomics. 1992 Jul;13(3):741–760. doi: 10.1016/0888-7543(92)90150-q. [DOI] [PubMed] [Google Scholar]
- Temin H. M. Reverse transcriptases. Retrons in bacteria. Nature. 1989 May 25;339(6222):254–255. doi: 10.1038/339254a0. [DOI] [PubMed] [Google Scholar]
- Wang G. H., Seeger C. The reverse transcriptase of hepatitis B virus acts as a protein primer for viral DNA synthesis. Cell. 1992 Nov 13;71(4):663–670. doi: 10.1016/0092-8674(92)90599-8. [DOI] [PubMed] [Google Scholar]
- Wilbur W. J., Lipman D. J. Rapid similarity searches of nucleic acid and protein data banks. Proc Natl Acad Sci U S A. 1983 Feb;80(3):726–730. doi: 10.1073/pnas.80.3.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Word C. J., White M. B., Kuziel W. A., Shen A. L., Blattner F. R., Tucker P. W. The human immunoglobulin C mu-C delta locus: complete nucleotide sequence and structural analysis. Int Immunol. 1989;1(3):296–309. doi: 10.1093/intimm/1.3.296. [DOI] [PubMed] [Google Scholar]
- Xiong Y., Eickbush T. H. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990 Oct;9(10):3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


