Abstract
Mitochondrial dysfunction can lead to diverse cellular and organismal responses. We used DNA microarrays to characterize the transcriptional responses to different mitochondrial perturbations in Saccharomyces cerevisiae. We examined respiratory-deficient petite cells and respiratory-competent wild-type cells treated with the inhibitors of oxidative phosphorylation antimycin, carbonyl cyanide m-chlorophenylhydrazone, or oligomycin. We show that respiratory deficiency, but not inhibition of mitochondrial ATP synthesis per se, induces a suite of genes associated with both peroxisomal activities and metabolite-restoration (anaplerotic) pathways that would mitigate the loss of a complete tricarboxylic acid cycle. The array data suggested, and direct microscopic observation of cells expressing a derivative of green fluorescent protein with a peroxisomal matrix-targeting signal confirmed, that respiratory deficiency dramatically induces peroxisome biogenesis. Transcript profiling of cells harboring null alleles of RTG1, RTG2, or RTG3, genes known to control signaling from mitochondria to the nucleus, suggests that there are multiple pathways of cross-talk between these organelles in yeast.
INTRODUCTION
Mitochondria are essential organelles whose primary function is the synthesis of ATP by oxidative phosphorylation. Mitochondria are also the site of many important metabolic and biosynthetic reactions, such as the tricarboxylic acid (TCA) cycle, amino acid, and heme biosynthesis. The biogenesis of mitochondria requires products from both the nuclear and mitochondrial genomes. The latter contributes a minimal genetic system dedicated to the expression of about a dozen polypeptides, most of which are components of the oxidative phosphorylation apparatus. Alterations in mitochondrial function and mitochondrial damage have been linked to a variety of cellular and organismal responses including apoptosis, neuromuscular disease, tumor pathogenesis, and aging (Green and Reed, 1998; Cortopassi and Wong, 1999; Wallace, 1999; Baysal et al., 2000).
In the budding yeast, Saccharomyces cerevisiae, mitochondrial DNA (mtDNA) is dispensable for growth as long as cells are supplied with a fermentable carbon source. This provides a convenient experimental system for analyzing how cells respond to changes in the functional state of mitochondria. One such response is retrograde regulation, a pathway of interorganelle communication whereby the expression of some nuclear genes is altered in cells with dysfunctional mitochondria (Parikh et al., 1987). In cells lacking mtDNA (ρo petites), for example, expression of the CIT2 gene encoding a peroxisomal isoform of citrate synthase is dramatically up-regulated (Liao et al., 1991). CIT2 expression is dependent on a set of nonessential genes, RTG1, RTG2, and RTG3, which are central players in the retrograde response pathway (Liao and Butow, 1993; Jia et al., 1997). The expression of four other genes, CIT1, ACO1, IDH1, and IDH2, encoding TCA cycle enzymes that lead to the synthesis of α-ketoglutarate (α-KG), is also dependent on the RTG genes, but only in cells with compromised or dysfunctional mitochondria (Liu and Butow, 1999); in cells with robust mitochondrial function, expression of those genes is dependent on the Hap 2,3,4,5 transcription complex. Glutamate, a precursor to nucleotides and other amino acids, is synthesized directly from the TCA cycle intermediate α-KG and is a potent inhibitor of RTG-dependent gene expression (Liu and Butow, 1999). Thus, glutamate levels may be a key signaling component in the retrograde response pathway.
RTG1 and RTG3 encode bHLH/Zip transcription factors that activate target gene transcription by binding as a complex to a novel upstream activation sequence called an R box (GTCAC) (Jia et al., 1997). RTG2 encodes a protein with an N-terminal ATP binding domain similar to the hsp70/actin/sugar kinase superfamily (Bork et al., 1992). Although the biochemical activity of Rtg2p is unknown, it is required for the translocation of Rtg1p and Rtg3p from the cytoplasm to the nucleus when the retrograde response is activated (Sekito et al., 2000). The retrograde response pathway and RTG2 in particular have been implicated in yeast aging: ρo cells with a robust retrograde response have a significantly longer life span than their ρ+ counterparts, and that life span extension requires RTG2 (Kirchman et al., 1999). These findings suggest that the retrograde response may affect a broad range of cellular activities.
To obtain a more comprehensive view of cellular responses to mitochondrial dysfunction and to identify potential downstream targets of RTG1, RTG2, and RTG3, we used cDNA-based microarrays to examine genome-wide changes in gene expression induced by a variety of mitochondrial perturbations and by inactivation of RTG1, RTG2, and RTG3. Our results suggest that, to overcome the absence of a complete TCA cycle in respiratory-deficient cells, metabolism has been reconfigured by activation of peroxisomal activities and by reactions that serve to maintain supplies of biosynthetic intermediates (i.e. anaplerotic pathways). The activation of only some of these pathways is dependent on the RTG genes. The increase in peroxisomal activity inferred from transcript profiling was confirmed by the direct observation that respiratory deficiency is an inducer of peroxisome biogenesis.
MATERIALS AND METHODS
Yeast Strains and Growth Conditions
Except as noted, strain PSY142 (MATα leu2 lys2 ura3ρ+) and its isogenic derivatives were grown at 30°C in YP (1% yeast extract, 2% bacto peptone, Difco, Detroit, MI) 2% raffinose medium. The ρo derivatives were obtained by several passages of ρ+ cells in YP dextrose medium containing 20 μg/ml ethidium bromide. For plating assays (Figure 3) and microarray experiments evaluating the effect of added propionate, YNB plus cas medium (0.67% yeast nitrogen base supplemented with 1% casamino acids) was used. Acid-treated YNB media were adjusted to pH 5.5 with 5 N KOH before autoclaving. Null alleles of rtg1, rtg2, and rtg3 were LEU2 disruptants described previously (Rothermel et al., 1995; Liu and Butow, 1999). PDH1 was deleted in strain CEY1131 (a/α his3Δ1/his3Δ1 ura3Δ0/ura3Δ0 LEU2/leu2Δ0 LYS2/lys2Δ0 TRP1/trp1Δ63) derived from “designer deletion” strains isogenic with the S288C background (Brachmann et al., 1998) by transplacement with URA3 using PDH1-URA3 hybrid primers for polymerase chain reaction (PCR) amplification of URA3. The deletion was confirmed by Southern blotting of haploid Ura+ segregants. The plating assay (Figure 3, B and C) was performed on a haploid segregant, YCE1131-11-4C (MATα pdh1Δ::URA3 leu2Δ0 his3Δ1 ura3Δ0 lys2Δ0) and a Ura+ PDH1 control with identical auxotrophies (CEY1118-3B). Strain MMYO11-GFP-AKL (MATα ura3-1 leu2-3, 112 his3-1 trp1-1 can1-100 ade2::GFP-AKL) contains two tandem copies of the coding region of green fluorescent protein (GFP) with a C-terminal AKL extension (Marshall et al., 1996) under control of the constitutive PGK1 promoter integrated into the ADE2 locus.
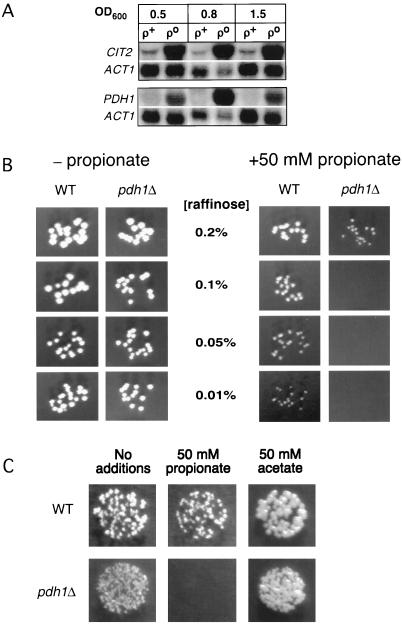
Figure 3.
PDH1 is a retrograde regulated gene and may function in propionate metabolism. (A) Northern blots showing increased abundance of PDH1 and CIT2 transcripts in ρo compared with ρ+ cells at three different cell densities. (B) Growth of wild-type (WT) and pdh1Δ cells on YNB 1% casein medium with or without propionate and containing limiting amounts of raffinose as indicated. (C) Growth of wild-type (WT) and pdh1Δ cells on YNB, 1% casein, 0.05% raffinose medium with or without propionate or acetate as indicated.
RNA Isolation and Northern Blot Analysis
RNA isolation for microarrays and Northern blots was performed as described by Kohrer and Domdey (1991). Northern blots were done essentially as described by Liu and Butow (1999).
Microarray Analysis
Microarrays consisting of 6219 yeast genes were prepared essentially as described by DeRisi et al. (1997) and were based on PCR amplification of S288C yeast genomic DNA using gene-specific oligo pairs supplied by Research Genetics (Birmingham, AL). A custom-built spotting robot was used (http://pompous.swmed.edu/exptbio/microarrays/index.htm). PCR was performed with 10 cycles of melting for 15 s at 94°C, annealing for 30 s at 54°C, and extension for 4 min at 68°C, followed by 25 cycles in which extension time was increased by 20 s per cycle. The PCR reaction mixture contained 10 mM Tris-Cl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.2 μM each oligo, 0.15 ng/μl genomic DNA template, 0.2 mM each deoxyribonucleotide triphosphate, 0.025 U/μl TAQ (Life Technologies, Grand Island, NY), and 0.0001 U/μl Pfu polymerase (Stratagene, La Jolla, CA). For the 192 longest genes in the genome (those exceeding 4073 base pairs [bp] in length) we prepared and arrayed additional PCR products using custom oligos designed to amplify 342–859 bp (average length = 458 bp) near the 3′-end of the open reading frame. Before arraying, we analyzed all of the DNAs by agarose gel electrophoresis, to confirm PCR success and product lengths. Overall, from the 6219 Research Genetics oligo pairs, we found 3% PCR failures and an equivalent rate of trace yields (<13 ng/μl spotted on the array).
Numerical Analyses
The web companion to this article (containing all numerical data and graphical images of raw data) may be found at http://hamon.swmed.edu/butow_array/petite.html. Methods for background subtraction, low value rejection, and normalization are described in detail elsewhere (Epstein et al., 2000) and are available at the above web site.
Each perturbation described in this paper is represented by a pair of replicate hybridizations (two microarrays). For each gene, we computed the signed geometric mean (SGM) response, defined as the antilog of the root mean square of the two log expression ratios; however, we assign a negative value to the geometric mean log ratio before taking the antilog when both log ratios are negative (consistent down-regulation) and assign a value of 1 when the two observations are of opposite sign (discrepant response). To restrict more stringently the genes we consider to be part of a response set, we require a twofold change in SGM response from both early and late logarithmic phase growth (ρ+ versus ρo; Table 1A) or from both 1- and 2-h time points (antimycin and oligomycin; Table 2A). In the case of carbonyl cyanide m-chlorophenylhydrazone (CCCP) treatment (Table 2A), we consider a gene to be part of the response set based on a single SGM of replicate hybridizations, but we use a slightly higher threshold (2.19-fold change) to render the size of the CCCP response set approximately the same as the oligomycin and antimycin response sets.
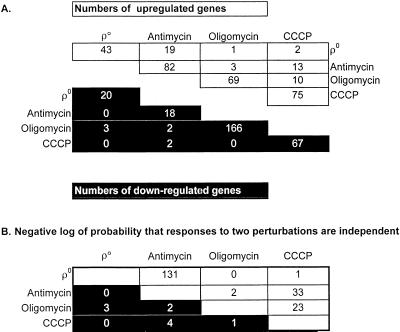
Table 2.
Genes showing consistent response to each perturbation
Overlapping effects of four perturbations of mitochondrial respiration and ATP synthesis. (A) Numbers of genes induced or repressed by four perturbations of mitochondrial ATP synthesis and by all pairwise combinations of perturbations. The numbers on the upper and lower main diagonals count the genes induced or repressed, respectively, in each perturbation. The numbers off the main diagonals count the genes induced or repressed by each pair of perturbations. Resipiratory competent ρ+ cells were treated with either 1 μm/ml antimycin, 3 μg/ml oligomycin, or 4.1 μg/ml CCCP. (B) χ2 analysis of the overlap between the sets of genes induced or repressed by each of the individual perturbations.
The Cluster and TreeView programs (Eisen et al., 1998) were used to make Figures 1 and 5, using uncentered correlation as the similarity metric for average linkage clustering. For cluster analysis, we analyzed the unaveraged, normalized expression ratios and included all genes showing at least a threefold change in at least two hybridizations.
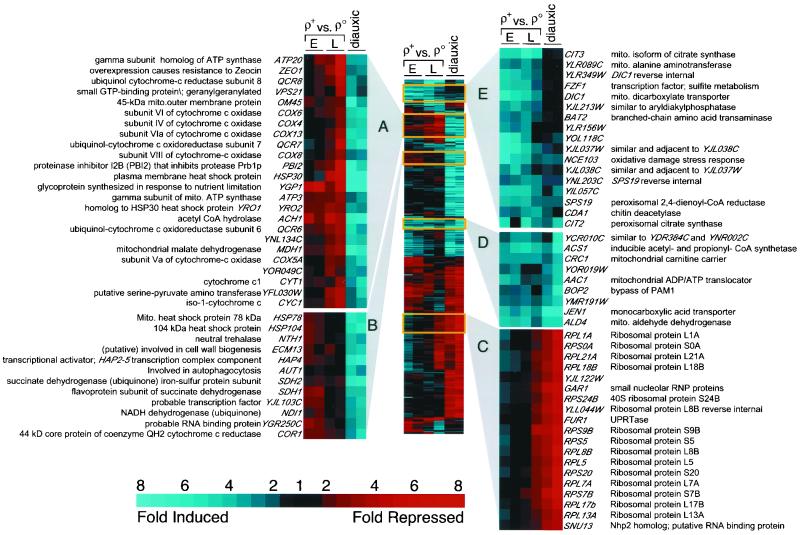
Figure 1.
2-D cluster analysis of gene expression in ρo versus ρ+ cells. The first four columns from left to right are replicate hybridizations of cDNAs prepared from early (E; OD600 = 0.5) and late (L; OD600 = 1.5) log phase cultures of ρ+ versus ρo cells differentially labeled with Cy3 and Cy5. Data from DeRisi et al. (1997) for the last two time points in the diauxic shift of glucose-grown ρ+ cells (OD600 = 6.9 and 7.3) are included in the last two columns on the right, indicated as diauxic. All 402 genes showing at least a threefold change in at least two hybridizations are shown. Blue denotes genes induced in ρo relative to ρ+ or induced during the diauxic shift, and red refers to repressed genes. Mito, mitochondrial; RNP, ribonucleoprotein; UPRT, uracil phosphoribosyl transferase.
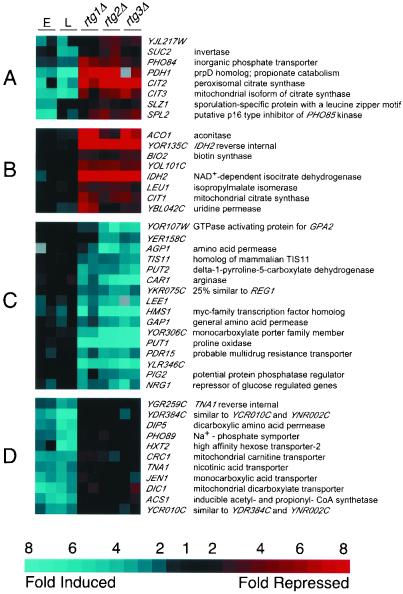
Figure 5.
Effects of RTG mutations in ρo cells. The four columns on the left are replicate comparisons of early (E) and late (L) log phase ρ+ and ρo cultures, as detailed in the legend to Figure 1. The six columns on the right are replicate comparisons of ρo cells and ρo cells containing deletions of rtg1, 2, or 3, respectively, and are based on cultures harvested at an OD600 = 0.8. (A–C) Genes whose expression is affected by RTG1, 2, and 3 in ρo cells. (D) A group of genes induced by respiratory deficiency in an RTG-independent manner. Blue refers to genes induced in ρo relative to ρ+ or induced in ρo rtg relative to ρo RTG. Red refers to repressed genes. All 273 genes that changed by at least threefold in at least two experiments are shown.
To evaluate the probability that the genes jointly affected by two perturbations would arise by chance alone (Table 2B), we computed the expectation of the size of the overlapping set as the product of the fractional representation of the genome in each of the component sets, multiplied by the size of the genome (∼6200). The expectation was compared with the actual size of the overlapping set based on χ2.
Inhibitors of Oxidative Phosphorylation
Antimycin A (Sigma, St. Louis, MO; A-8674), oligomycin (Sigma O-4876), or CCCP (Sigma C-2759) were added to log phase cultures of ρ+ cells at final concentrations of 1, 3 and 4.1 μg/ml, respectively. Cells from a portion of the culture were harvested for RNA preparation at an OD600 of 0.68 (antimycin) or 0.5 (oligomycin and CCCP), and the remaining cells were treated with the inhibitor. Additional samples were collected after ∼1 and 2 h of further culture for oligomycin and antimycin or after 1.5 h for CCCP. Labeled cDNAs prepared from each inhibitor-treated culture were mixed with labeled cDNA from the zero-time controls for microarray analysis.
Microscopy
Early log phase cells of strain PGK1-GFP-AKL grown at 30°C in YP 2% raffinose medium were fixed by addition of methanol-free formaldehyde to 4% (vol/vol). After 10 min, the cells were collected by centrifugation at 700 × g and resuspended in phosphate-buffered saline (PBS) with 4% formaldehyde for 1 h. Fixed cells were washed four times in PBS and stained for 15 min with 2 μg/ml Calcofluor White (Sigma, Saint Louis, MO) in PBS followed by three washes in PBS. Three microliters of cells were mounted on 2% agarose pads in water (Waddle et al., 1996). Three-dimensional images (28- × 0.108-nm steps) were collected, contrast scaled to 0–255 intensity levels using a range from 0 to 1500, and projected into a single focal plane using the maximum intensity at a given pixel. Overlays between the GFP and Calcofluor images were done using custom software (EditView4D, http://hamon.swmed.edu/∼jwaddle/).
For time-lapse microscopy, 3 μl of log phase cells grown at 25°C on YP 2% raffinose medium were mounted on 3% agarose pads containing YP 2% raffinose (Waddle et al., 1996). Where indicated, metabolic inhibitors were added to the culture and to the molten agarose solution (∼65°C) just before mounting. Microscopy was performed on an Olympus BMAX-60F with Nomarski differential interference contrast (DIC) and fluorescence optics, a 60× 1.4 NA UPlanApo lens with additional 1.6× magnification, a Princeton Instruments charge-coupled device (EEV-37-BFT), and shutters, filter wheels, and focus control from Ludl Electronic Products (Hawthorne, NY). Automated microscopy was performed with custom software (Jimage4D; complete description of hardware and software can be accessed at http://hamon.swmed.edu/∼jwaddle/jimage4d.html). For each experiment, cells in a 35- × 35- × 9-μm3 volume (135 nm/pixel) were imaged every 15 min for 8–12 h. Neutral density filters blocked 95 and 98.5% of the incident light from the xenon and halogen lamps, respectively. A 250-ms exposure was used for each optical channel. Primary image data were stored as sequentially numbered files, each a stack of images at a given time point, at the full dynamic range of the camera (4096 intensity levels). The images were corrected for uneven illumination and camera bias and then scaled to 8 bits/pixel using a fixed, linear gray scale from 50 to 255. Values >255 were set to white, those <50 to black. Using these image acquisition and scaling parameters, GFP fluorescence from a ρ+ strain is barely visible above background, while the ρo strain produces bright, fluorescence patches. To produce movies, fluorescence from a given volume was projected into a single 2-D image and then positioned to the right of an equatorial slice from the DIC image stack. The DIC-fluorescence image pairs were then placed into a new stack to view the movie in time-lapse mode, or further montaged with the DIC-fluorescence pair stacks from other movies. QuickTime movies of each can be viewed at http://hamon.swmed.edu/∼jwaddle/scratch/.
RESULTS AND DISCUSSIONS
Differential Gene Expression between ρ+ and ρo Petite Cells
To identify genes whose expression changes as a result of mitochondrial dysfunction, we first carried out microarray analysis of the differential, genome-wide expression profile of a respiratory-competent ρ+ strain versus an isochromosomal ρo petite derivative. Two independent pairs of ρ+ and ρo cultures grown in rich medium containing 2% raffinose, a nonrepressing carbon source, were analyzed, with one pair harvested in early (OD600 = 0.5) and the other pair harvested in late (OD600 = 1.5) logarithmic growth phase. Because we are comparing partially oxidative and obligatorily fermentative strains both grown under derepressing conditions, we elected to compare our results with a transcript profile based on a comparison of robustly oxidative versus fully fermentative, glucose-repressed ρ+ yeast cultures. Complete data of this sort are available from the experiments of DeRisi et al. (1997), who documented the genome-wide changes in gene expression of yeast cells undergoing the transition from glucose fermentative to oxidative metabolism—the diauxic shift. Many proteins, including enzymes of oxidative metabolism, become derepressed during the diauxic shift, reaching a maximum level of expression when the glucose is exhausted from the medium and the cells are utilizing ethanol for growth.
To present our results, we used a 2-D clustering algorithm (Eisen et al., 1998) that groups similarly responsive genes and displays them as a two-color graphic with color intensities proportional to the fold change in gene expression (Figure 1). Selected regions of the cluster representation have been magnified to display groups of genes in the ρ+-ρo comparison and the late diauxic shift. Figure 1, clusters A and B, represent a group of genes that are down-regulated in ρo cells and up-regulated in cells that have switched from fermentative to oxidative metabolism. A large proportion of the genes in this group encode mitochondrial proteins of the oxidative phosphorylation apparatus, such as subunits of the cytochrome c oxidase and ATP synthase complexes. These data imply that petites make no (futile) attempt to compensate for their respiratory-deficient state by up-regulating the expression of oxidative phosphorylation genes. This contrasts with the observation that some genes of the oxidative phosphorylation apparatus are up-regulated in human cells harboring deleterious mtDNA mutations (Heddi et al., 1999). In those cases, mitochondrial gene expression was also elevated, suggesting activation of a general pathway of increased mitochondrial biogenesis to compensate for mitochondrial dysfunction.
Figure 1, cluster C, contains mainly cytoplasmic ribosomal protein genes, which are substantially down-regulated in the diauxic shift (DeRisi et al., 1997), and which show an interesting difference in their regulation between early and late logarithmic phase ρo cultures. Coordinate down-regulation of genes involved in ribosome biogenesis is generally observed in cells as growth rate slows, as in the diauxic shift, e.g., when there is a switch from an optimal carbon source (glucose) to a poor one (ethanol) (DeRisi et al., 1997). In medium containing 2% raffinose, the doubling time of ρo petite cells is ∼40–50% of their isochromosomal ρ+ counterparts. Nevertheless, these same ribosomal protein genes are apparently down-regulated in ρo cells only when those cells traverse late logarithmic phase.
Figure 1, clusters D and E, represent a group of genes whose expression is up-regulated in ρo petites and, for the most part, in cells late in the diauxic shift. Most of the genes within this group, which includes CIT2, are involved in intermediary metabolism and small molecule transport pathways. This group is potentially the most interesting class of genes whose expression is affected in ρo petite cells, because differences in their expression could reflect metabolic changes that would compensate for the loss of respiration.
As a further refinement of the analysis of genes up-regulated in ρo petites, we focus on those genes whose geometric mean expression level (defined in MATERIALS AND METHODS) increased at least twofold in both the low and high OD replicate comparisons of ρ+ and ρo cultures; these are the genes whose expression is consistently affected by the ρo mitochondrial genotype in a cell density-independent manner. By these criteria, we identified 43 genes that are up-regulated in the respiratory-incompetent ρo petite cells (Table 1A).
Transcript Profiling Suggests Metabolic Remodeling in Respiratory-deficient Cells
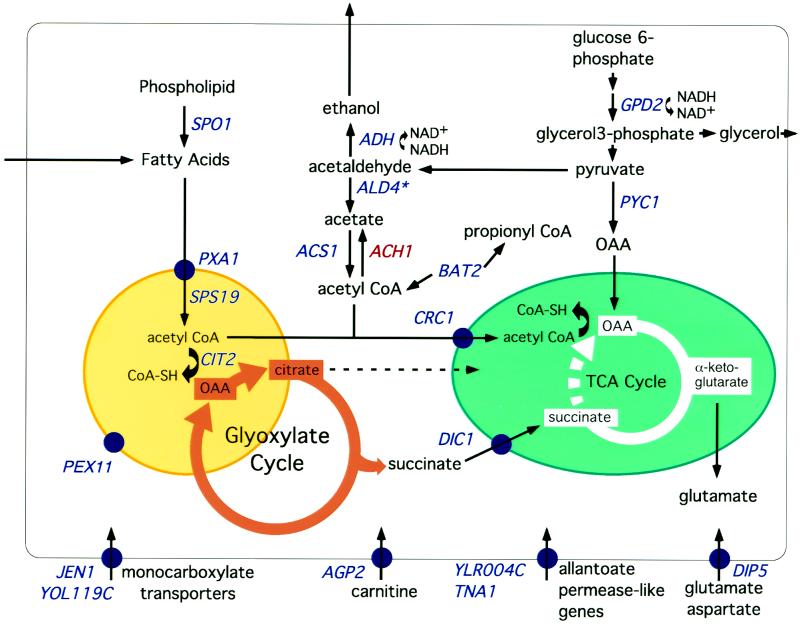
Intermediates of the TCA cycle are required for the biosynthesis of amino acids and nucleotides. Because part of the TCA cycle (succinate oxidation) cannot proceed in respiratory-deficient cells, oxaloacetate (OAA) is not regenerated and must therefore be supplied stoichiometrically. The genome-wide transcript profile suggests that metabolism in ρo cells has been reconfigured to provide increased supplies of OAA and its condensation partner, acetyl-CoA for these biosynthetic reactions (Figure 2 and Table 1A). For example, the expression of PYC1, encoding pyruvate carboxylase, which catalyzes the synthesis of OAA from pyruvate and CO2, and ACS1, encoding an acetyl-CoA synthase, are up-regulated in ρo petites. ACH1, however, which encodes an acetyl-CoA hydrolase, is strongly down-regulated in these cells. Additionally, many of the genes up-regulated in ρo cells encode proteins that function in the conversion and flux of metabolites generated from β-oxidation of fatty acids (a peroxisomal activity in yeast) to intermediates of the TCA and glyoxylate cycles (Figure 2). The central role of the peroxisomes in these metabolic interconversions is also reflected by the finding that 13 of the up-regulated genes indicated in Table 1A are localized to the peroxisome, induced by oleate (a potent peroxisomal proliferator), or support peroxisomal activities (Veenhuis et al., 1987; Kunau et al., 1988; Skoneczny et al., 1988).
Figure 2.
Metabolic pathways affected in respiratory-deficient cells as inferred from transcript profiling. Nineteen of the 34 genes induced in ρo petites (Table 1A) and having some functional description are indicated in blue. Also included in the display in red is ACH1, whose expression is strongly down-regulated in ρo cells. A peroxisome is indicated by the yellow circle and a mitochondrion by the green ellipse. TCA cycle flux from succinate to OAA is blocked (broken line) in respiratory-deficient cells. Gene products associated with a particular membrane are indicated by blue circles. On our array, the spots for ADH1 and ADH2 both showed induction in ρo cells. However, because of 157 nucleotides of contiguous identity between these two genes, transcripts of ADH1 and ADH2 cannot be distinguished by microarray probes targeting entire reading frames.
Crucial to the replenishment of acetyl-CoA and OAA, and metabolites derived from OAA, is their transport from extramitochondrial sources to mitochondria. This transport requires carboxylic acid carriers and acylcarnitine transferases. It is striking therefore that in ρo cells there is an up-regulation of expression not only of DIC1, encoding a mitochondrial dicarboxylic acid transporter, and CRC1, encoding an inner mitochondrial membrane acylcarnitine transporter, but of AGP2, which encodes a plasma membrane carnitine transporter. It has been suggested that yeast cells are unable to synthesize carnitine de novo and must therefore rely on extracellular sources of carnitine to provide the substrates for the acylcarnitine carriers (van Roermund et al., 1999). The up-regulation of CIT2 could provide additional citrate directly from the glyoxylate cycle and the OAA replenished by OAA derived from the pyruvate carboxylase reaction.
Because petite cells lack mitochondrial electron transport and are therefore dependent on glycolysis for biomass and energy, they require alternative mechanisms for reoxidation of NADH. The major pathways for these steps in nonoxidative cells would be α-glycerol-3-phosphate and alcohol dehydrogenase activities. Genes encoding enzymes carrying out these reactions, GPD2 and ADH, are both up-regulated in those cells. In addition, the array data suggest that the ρo petite cells have further optimized nutrient availability by up-regulation of transporters for amino acids (DIP5), poor nitrogen sources (TNA1 and YLR004C), sugars (HXT10), and monocarboxylic acids (JEN1 and YOL119C). Finally, ρo petites up-regulated two amino acid transaminases, BAT2 (branched chain amino acids, see below) and YLR089C (a potential mitochondrial alanine amino transferase) that would lead to increased supplies of pyruvate, acetyl-CoA, and propionyl-CoA. In summary, transcript profiling suggests that, to overcome blocks in the TCA cyle, ρo petites reconfigure metabolism by recruiting peroxisomal activities, small molecule transport systems and lipid, sugar, and amino acid turnover to increase the availability of OAA, acetyl-CoA, and propionyl-CoA for biosynthetic reactions.
Genes That May Function in Propionate Metabolism
One of the genes whose expression in respiratory-deficient cells most closely correlates with CIT2 is PDH1 (YPR002W). Coregulation of expression of these genes in cells with certain mitochondrial perturbations has also been noted recently by Hughes et al. (2000). PDH1 is 62% identical to the prpD genes of Escherichia coli and Salmonella typhimurium, which play an unknown but essential role in propionate catabolism (Horswill and Escalante-Semerena, 1997; Textor et al., 1997). Given that ρo cells also up-regulate BAT2, which leads to the production of propionyl-CoA as a product of valine and isoleucine catabolism (Figure 2) and ACS1, the acetyl-CoA synthase isoform that also functions as a propionyl-CoA synthase (van den Berg et al., 1996), we investigated a possible role for PDH1 in propionate metabolism. We confirmed by Northern blot analysis that PDH1 is strongly induced in early, middle, and late log phase cultures of ρo petites, in a manner similar to CIT2 (Figure 3A).
Exogenous propionate reportedly augments the growth of yeast in glucose-limited, aerobic chemostat cultures (Pronk et al., 1994) but is toxic under anaerobic conditions (Verduyn et al., 1990), presumably because the energetic stress of anaerobiosis exceeds the capacity of the plasma membrane ATP-dependent proton pump to excrete H+ efficiently when propionic acid is taken up by cells. We deleted PDH1 and determined the effects of propionate on the growth of the wild type and pdh1Δ strains cultured in medium containing low concentrations of raffinose to limit the energy supply. Below 0.2% raffinose, 50 mM propionate is far more toxic to pdh1Δ than to wild-type cells (Figure 3B). In contrast, 50 mM acetate augments the growth of both wild type and pdh1Δ strains grown in limiting raffinose (Figure 3C), demonstrating that pdh1Δ cells are specifically sensitive to propionate treatment. These data suggest that PDH1 functions in propionate utilization in yeast.
Yeast and E. coli appear to catabolize propionate via the methyl citrate pathway, which results in a net partial oxidation of propionate to pyruvate (Pronk et al., 1994); (Textor et al., 1997). The methyl citrate pathway proceeds by reactions resembling the synthase, isomerase, and lyase steps of the glyoxylate cycle, followed by the regeneration of OAA from succinate via the TCA cycle (Tabuchii et al., 1974). However, the genes encoding the enzymes that metabolize propionate in S. cerevisiae have not been determined. We used microarrays to compare transcripts between wild-type yeast grown in 2% raffinose medium with or without supplemental 50 mM propionate. We found a strong induction of PDH1, CIT3 (but not CIT2 or CIT1), ACO1 (but not YJL200C), and ICL2 (but not ICL1) in response to propionate (data available at http://hamon.swmed.edu/butow_array/petite.html). The organic acid-transporting ATPase, PDR12, was also dramatically induced, consistent with the model for propionate toxicity under conditions of energy limitation mentioned earlier (Verduyn et al., 1990). Possibly, CIT3, ACO1, and ICL2 perform the anticipated synthase, isomerase, and lyase steps of the methyl-citrate pathway, and PDH1 plays an as yet undefined role in propionate metabolism. Interestingly, PDH1 orthologs in prokaryotic operons are adjacent to a citrate synthase-like gene (prpC) whose production was shown to have methyl citrate synthase activity (Textor et al., 1997), and in yeast, PDH1 is adjacent to CIT3 on chromosome VI. The induction of ACS1, BAT2, CIT3, and PDH1 in ρo petites may represent a further example of the metabolic reorganization of respiratory-deficient cells, with branched chain amino acid turnover leading to propionyl-CoA and ultimately to pyruvate via the methyl citrate pathway.
Different Mitochondrial Inhibitors Elicit Different Genome-wide Responses in Gene Expression
The absence of oxidative phosphorylation in ρo cells is the result of their failure to synthesize components of both the mitochondrial electron transport chain and the ATP synthase complex. To gain insight into the effects of specific, acute perturbations of mitochondrial function on global patterns of gene expression, we exposed ρ+ cells to three different inhibitors of oxidative phosphorylation—antimycin, CCCP, and oligomycin. The effects of these inhibitors on mitochondrial function are well documented, and they are known to inhibit oxidative phosphorylation in fundamentally different ways. Antimycin is a specific inhibitor of mitochondrial electron transport, blocking the reoxidation of reduced cytochrome b. CCCP uncouples electron transport from ATP synthesis by enabling free movement of protons across the inner mitochondrial membrane, resulting in a collapse of the H+ gradient necessary to drive ATP synthesis via the ATP synthase complex. In contrast to antimycin treatment, cells treated with CCCP have maximal (uncoupled) rates of mitochondrial electron transport activity. Finally, oligomycin is a specific inhibitor of the F0 component of the F1-F0 ATP synthase complex and inhibits both ATP synthesis and ATP synthesis-associated (state 4) respiration.
Log phase cultures of ρ+ cells grown in YP 2% raffinose medium were treated with antimycin (1 μg/ml), oligomycin (3 μg/ml), or CCCP (4.1 μg/ml). In preliminary experiments we verified that these concentrations were sufficient to inhibit the growth of ρ+ cells growing nonfermentatively. For microarray analysis, aliquots of cells were removed 1–2 h after the addition of the inhibitors and compared with untreated (zero time point) controls.
Analysis of genes jointly affected by different mitochondrial perturbations shows a striking degree of overlap between the effects of antimycin treatment and ρo cells (Table 2): 44% (19 genes) of the 43 genes up-regulated in ρo cells are also up-regulated by treatment of ρ+ cells with antimycin. The 19 genes include 10 of the 13 genes mentioned earlier (Table 1A) having a role in peroxisomal function. Moreover, using stringent criteria for designation of an induced gene (detailed in MATERIALS AND METHODS), we found that several additional genes induced by antimycin are also plausibly part of a pathway of up-regulated peroxisomal function. This set includes CTA1 (peroxisomal catalase), FOX2 (peroxisomal β-oxidation protein), POX1 (fatty acyl-CoA oxidase), and IDP3 (peroxisomal NADP-dependent isocitrate dehydrogenase; Table 1B). Altogether, a total of 19% (20 genes) of the 106 genes that are up-regulated in ρo petites or antimycin-treated ρ+ cells either are oleate inducible or have some involvement in peroxisomal metabolism.
In contrast with the similarities in gene induction between ρo and antimycin-treated cells, very few of the up-regulated genes resulting from CCCP or oligomycin treatment were also up-regulated in ρo cells (Table 2A), suggesting that the ρo transcript profile is dominated by the effects of loss of electron transport rather than the loss of mitochondrial ATP synthesis per se. Significant overlap was seen between the genes induced in CCCP and both antimycin and oligomycin (Table 2), but there was no obvious pattern that would suggest a coherent biological role for these genes. Although comparable numbers of genes were up-regulated by each of the inhibitors, a much larger number of genes were down-regulated by oligomycin treatment than by treatment with antimycin or CCCP (Table 2A). Little or no significant overlap was found when considering the genes down-regulated in common by these diverse perturbations (Table 2B).
Peroxisome Proliferation in Respiratory-deficient Cells
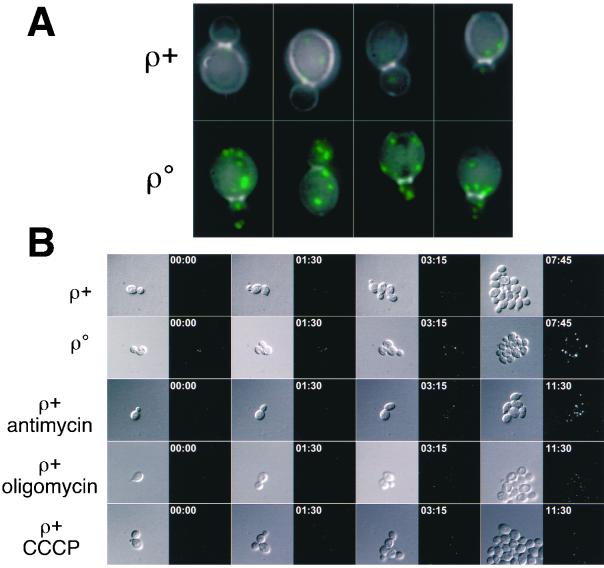
In ρo petites, an intact retrograde pathway is required for oleate induction of peroxisome biogenesis, suggesting a link between mitochondrial dysfunction and peroxisomal activities (Chelstowska and Butow, 1995; Kos et al., 1995). That notion is strengthened by the current experiments, which suggest that peroxisomal activities may play a crucial role in the metabolic remodeling of respiratory-deficient cells. Given that many of the genes up-regulated in ρo petites are known to be induced by oleate, we asked whether enhanced peroxisome proliferation also occurs in those cells. For this purpose, we used a strain expressing a chromosomally integrated fusion gene encoding a derivative of GFP with a peroxisomal matrix-targeting signal, AKL, at its C terminus. This GFP derivative, whose expression is under the control of the constitutive PGK1 promoter, has been shown to target efficiently and exclusively to peroxisomes and is an accurate indicator of peroxisome proliferation induced by exogenous oleic acid (Marshall et al., 1996). The PGK1-GFP-AKL ρ+ strain was converted to a ρo petite by treatment with ethidium bromide.
Microscopic examination of log phase cultures of the PGK1-GFP-AKL ρ+ and ρ° strains revealed a dramatic increase in the intensity of peroxisomal profiles, indicative of an increase in peroxisome biogenesis in the ρ° derivative (Figure 4A). When viewed using an intensity range typical for the GFP-AKL fluorescence in ρ° cells, the peroxisomes in ρ+ cells are nearly undetectable (Figure 4A, top row). It should be noted that peroxisomes in ρ+ cells are easily visualized when viewed with a more sensitive intensity scaling function. GFP-AKL foci were completely absent in ρ+ and ρ° derivatives deleted for pex5 (Epstein, Waddle, Hale, Davé, Thornton, Macatee, Garner, and Butow, unpublished results), a peroxin essential for the import of proteins into the peroxisomal matrix (Vanderleij et al., 1993).
Figure 4.
Peroxisome induction in respiratory-deficient cells. (A) Overlay between Calcofluor White staining (gray) and GFP peroxisomal marker (green) in representative ρ+ and ρ° cells. Each image is a 2-D projection of a 3-D image of the entire cell volume. The images were generated with a fixed exposure time and illumination intensity and a camera with a dynamic range exceeding 3 orders of magnitude. (B) Selected time points from 3-D, time-lapse recordings comparing peroxisome biogenesis in respiratory-competent, respiratory-deficient, and drug-treated cells. The DIC images are taken from an equatorial focal plane, whereas the GFP images are 2-D projections showing all peroxisomes in the image volume above a fixed intensity range. Annotation indicates elapsed time in hours:minutes format from start of experiment. Because the drug-treated cells grow slower than untreated ρ+ and ρo cells, a later time point is used for the final images in the drug-treated samples.
Because transcript profiling suggested that only antimycin treatment of ρ+ cells had significant similarities to ρ° cells in terms of the up-regulation of oleate inducible genes (Table 2), we tested whether peroxisome proliferation might also be unique to antimycin treatment. Accordingly, we examined peroxisomal profiles using 3-D time-lapse fluorescence microscopy in living, PGK1-GFP-AKL ρ+ cells treated over a 12-h period with antimycin, oligomycin, or CCCP. The image acquisition parameters were adjusted such that at a constant exposure, excitation level, and intensity scaling ρ+ peroxisomes in untreated cells are barely detectable, whereas those in ρ° cells are bright, distinct foci (compare first to second row in Figure 4B). In agreement with the results of the transcript profiling, we found a striking antimycin-dependent increase in peroxisomal fluorescence ∼3 h after treatment (third image pair, row 3, Figure 4B). The intensity of the peroxisomes in the antimycin-treated ρ+ cells was nearly indistinguishable from that of ρo cells. Moreover, the response was largely specific to antimycin because oligomycin treatment caused only a minor increase in peroxisomal intensity and CCCP-treated cells did not differ from untreated ρ+ cells. Thus, the involvement of enhanced peroxisomal activities in respiratory-deficient cells inferred from transcript profiling (Figures 1 and 2 and Table 1) was borne out by the observation of enhanced peroxisome biogenesis in those cells (Figure 4). The partial inhibition of electron transport activity by oligomycin may account for the slight increase in peroxisome profiles observed in Figure 4, although no induction of peroxisome-related genes was evident in the transcript profiling that was consistently above the stringent cut-off criteria we have applied in these experiments.
RTG Genes
Because of the importance of the RTG genes in the retrograde response, we next characterized globally the sets of genes whose expression is influenced by one or more of the RTG genes in ρo cells. To this end, we obtained transcript profiles from comparisons of an RTG wild-type ρo strain and rtg1Δ, rtg2Δ, or rtg3Δ ρo derivatives and used cluster analysis to elucidate the sets of genes that are sensitive to the rtg mutations in the ρo background (Figure 5). Figure 5, cluster A, identifies a set of genes that are induced in ρo petites and whose induction depends on signaling via the RTG pathway. The size of this set is limited and, in addition to CIT2, includes CIT3 and PDH1 (discussed above). Figure 5, cluster B, presents additional genes that depend on RTG genes for their expression in ρo cells. Unlike the CIT2 class (Figure 5, cluster A), these genes are not induced in ρo relative to their expression in ρ+ cells. This set includes CIT1, IDH2, and ACO1, which encode enzymes catalyzing the first three steps of the TCA cycle, leading to α-KG and glutamate, a retrograde regulator. These data are in agreement with a previous study showing that expression of these TCA cycle genes becomes dependent on the RTG genes as mitochondrial respiratory function is reduced (Liu and Butow, 1999).
The genes showing enhanced expression in rtgΔ ρo compared with RTG ρo cells are illustrated in Figure 5, cluster C. These genes behave as if they were repressed by RTG gene activity, but this is probably an indirect response to the exacerbated defects suffered by ρo cells when they are also deficient in retrograde signaling. This set of genes includes PUT1 (proline oxidase), CAR1 (arginase), AGP1, and GAP1 (both amino acid carriers), which are all involved in amino acid turnover or transport. The strong, consistent induction of PUT1 and CAR1 suggests that cells are attempting to up-regulate the synthesis of glutamate from nutritionally derived proline and arginine under circumstances in which combined genetic deficiencies preclude synthesis via α-KG derived from the TCA cycle. Although petites are unable to oxidize proline via proline oxidase encoded by PUT1, the synthesis of PUT1 mRNA is nevertheless subject to regulation in petites (Wang and Brandriss, 1987). Finally, Figure 5, cluster D, illustrates genes that are induced in ρo relative to ρ+ but whose expression in ρo cells is nearly independent of the RTG genes. Included in this class are membrane carriers for monocarboxylic (JEN1) and dicarboxylic (DIP5) acids, phosphate (PHO89), and hexose (HXT2). The induction of these genes suggests that the transcriptional response to mitochondrial dysfunction depends on novel retrograde signaling pathways, in addition to those involving RTG1, RTG2, and RTG3.
CONCLUSION
Transcript profiling suggests that respiratory-deficient yeast cells respond to the loss of oxidative phosphorylation by reconfiguring metabolism to increase supplies of acetyl-CoA and OAA from peroxisomal activities and anaplerotic reactions. Because the TCA cycle no longer functions as a cycle in respiratory-deficient cells, stoichiometric amounts of OAA must be available for condensation with acetyl-CoA. This is to ensure that the first three steps of the TCA cycle, which are under control of the RTG genes in cells with compromised mitochondrial function, operate at a rate sufficient to meet cellular demands for glutamate, also a regulator of the retrograde response. Microarray analysis revealed RTG-dependent, retrograde-responsive genes that are likely to function in propionate metabolism. Other retrograde-responsive genes, however, were identified that do not appear to be under control of the RTG genes, suggesting that new pathways will unfold in mitochondria-to-nucleus signaling. Respiratory-competent ρ+ cells treated with different inhibitors of oxidative phosphorylation show the induction of overlapping but nonidentical sets of genes. Of the inhibitors, only antimycin treatment induces many of the same genes induced in ρo petites, a number of which are involved in peroxisomal activities. These array data led to a prediction that respiratory deficiency, but not the loss of mitochondrial ATP synthesis per se, would result in an increase in peroxisome biogenesis. Microscopic examination of cells expressing a derivative of GFP with a peroxisomal matrix-targeting signal confirmed these predictions. Future studies should reveal the similarities or differences in the pathway of peroxisome biogenesis induced by respiratory deficiency and by oleate induction.
Supplementary Material
Table 1A.
Genes induced in respiratory-deficient cells
| Open reading frame | Gene | Fold change in ρ° | Fold change in antimycin | Oleate induction | Function |
|---|---|---|---|---|---|
| YJL037W | 7.1 | 5.2 | Similar and adjacent to YJL038C | ||
| YDR384C | 5.9 | 4.6 | see YCR010C | Similar to YNR002C and YCR010C | |
| YPR002W | PDH1 | 5.8 | 9.43 | Propionate catabolism | |
| YPR001W | CIT3 | 5.2 | 12.7 | Mitochondrial isoform of citrate synthase | |
| YCR005C | CIT2 | 5.1 | 10.0 | + | Peroxisomal citrate synthase |
| YFL011W | HXT10 | 5.0 | 4.5 | High-affinity hexose transporter | |
| YLR348C | DIC1 | 5.0 | 5.3 | Mitochondrial dicarboxylate transporter | |
| YAL054C | ACS1 | 4.2 | 6.2 | + | Inducible acetyl- and propionyl-CoA synthetase |
| YIL057C | 4.2 | 24.6 | + | ||
| YPL265W | DIP5 | 4.2 | 3.2 | Dicarboxylic amino acid permease | |
| YNL036W | NCE103 | 4.0 | Oxidative damage stress response | ||
| YLR307W | CDA1 | 4.0 | Chitin deacetylase | ||
| YOR374W | ALD4 | 3.7 | + | Aldehyde dehydrogenase | |
| YBR296C | PHO89 | 3.6 | Na+ phosphate symporter | ||
| YOR100C | CRC1 | 3.5 | 5.8 | Mitochondrial carnitine carrier | |
| YLR349W | 3.5 | DIC1 reverse internal | |||
| YKL217W | JEN1 | 3.5 | 2.8 | + | Monocarboxylic acid transporter |
| YNL202W | SPS19 | 3.4 | 3.2 | + | Peroxisomal 2,4-dienoyl-CoA reductase |
| YGL254W | FZF1 | 3.4 | Transcription factor; sulfite metabolism | ||
| YJL213W | 3.3 | Similar to arylidialkylphosphatase | |||
| YMR303C | ADH2 | 3.1 | Alcohol dehydrogenase II | ||
| YCR010C | 3.0 | 12.2 | + | Similar to YDR384C and YNR002C | |
| YNR060W | FRE4 | 3.0 | Similar to ferric reductase; electron transport | ||
| YGR260W | TNA1 | 3.0 | Nicotinic acid transporter | ||
| YGL062W | PYC1 | 2.9 | Pyruvate carboxylase | ||
| YNL203C | 2.8 | 3.2 | |||
| YLR089C | 2.6 | Mitochondrial alanine aminotransferase | |||
| YPL147W | PXA1 | 2.6 | 6.3 | + | ABC family long-chain fatty acid transporter |
| YBR105C | VID24 | 2.5 | Peripheral vesicle membrane protein | ||
| YJR148W | BAT2 | 2.5 | Branched-chain amino acid transaminase | ||
| YOL147C | PEX11 | 2.4 | 2.8 | + | Peroxisome biogenesis |
| YER187W | 2.3 | ||||
| YLR004C | 2.3 | Similar to allantoate permease | |||
| YNL012W | SPO1 | 2.3 | Similar to phospholipase B | ||
| YPL281C | ERR2 | 2.2 | Similar to enolase | ||
| YDL174C | DLD1 | 2.2 | Mitochondrial D-lactate dehydrogenase | ||
| YOL119C | 2.2 | Potential monocarboxylate transporter | |||
| YPL018W | CTF19 | 2.2 | 3.2 | Kinetochore protein | |
| YAR068W | 2.2 | ||||
| YMR191W | 2.2 | 3.1 | |||
| YNL279W | 2.2 | ||||
| YOL059W | GPD2 | 2.1 | Glycerol-3-phosphate dehydrogenase (NAD+) | ||
| YBR132C | AGP2 | 2.1 | Carnitine transport across plasma membrane |
Table 1B.
Oleate-inducible genes up-regulated uniquely after both 1 and 2 h of antimycin treatment
| Open reading frame | Gene | Fold change in antimycin | Function |
|---|---|---|---|
| YFL014W | HSP12 | 13.2 | 12-kDa heat shock protein |
| YKR097W | PCK1 | 8.7 | Phosphoenolpyruvate carboxykinase |
| YGL205W | POX1 | 7.8 | Fatty acyl-CoA oxidase |
| YDR256C | CTA1 | 5.2 | Peroxisomal catalase A |
| YKR009C | FOX2 | 4.0 | Peroxisomal beta-oxidation protein |
| YOL052CA | DDR2 | 3.1 | Induced by DNA damage and diverse stress |
| YNL009W | IDP3 | 2.8 | Peroxisomal NADP-dependent IDH |
Genes induced in ρo petite and antimycin treated cells. (A) All genes induced in ρo petites by at least 2-fold in both early and late logarithmic phase cultures. The average of the early and late log phase transcriptional responses is shown. Respiratory competent ρ+ cells were treated with 1 μg/ml of antimycin. The average response to antimycin treatment is also shown for the subset of genes that were affected by antimycin treatment after both 1 and 2 h of treatment. Functional descriptions were adapted from YPD Proteome and SGD data bases (Costanzo et al., 2000; Cherry et al., 1998). (B) All oleate induced genes (Kal et al., 1999) induced uniquely after both 1 and 2 h of antimycin treatment. The averages of responses after 1 and 2 h of treatment are shown.
ACKNOWLEDGMENTS
We thank T. Fonden for oligo design, K. Kupfer for helpful discussions of numerical methods, D. Middelman for software support, and J. Goodman for GFP-AKL strains. We also thank M. Eisen and G. Sherlock for clustering software and V. Iyer and J. DeRisi for advice on microarrays. Finally, we are grateful to M. McCammon for many helpful discussions. This work was supported by National Institutes of Health grants GM22525 and CA77811 and a Robert A. Welch Grant (I-0642) to R.A.B. C.B.E. was supported in part by a Postdoctoral Fellowship (AG05781) from the National Institutes of Health.
Footnotes
Online version of this article contains video material and is available at www.molbiolcell.org.
REFERENCES
- Baysal BE, Ferrell RE, Willett-Brozick JE, Lawrence EC, Myssiorek D, Bosch A, van der Mey A, Taschner PE, Rubinstein WS, Myers EN, Richard C W, 3rd, Cornelisse CJ, Devilee P, Devlin B. Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science. 2000;287:848–851. doi: 10.1126/science.287.5454.848. [DOI] [PubMed] [Google Scholar]
- Bork P, Sander C, Valencia A. An ATPase. domain common to prokaryotic cell cycle proteins, sugar kinases, actin and hsp70 heat shock proteins. Proc Natl Acad Sci USA. 1992;89:7290–7294. doi: 10.1073/pnas.89.16.7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li JC, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Chelstowska A, Butow RA. RTG genes in yeast that function in communication between mitochondria and the nucleus are also required for expression of genes encoding peroxisomal proteins. J Biol Chem. 1995;270:18141–18146. doi: 10.1074/jbc.270.30.18141. [DOI] [PubMed] [Google Scholar]
- Cherry JM, Adler C, Ball C, Chervitz SA, Dwight SS, Hester ET, Jia Y, Juvik G, Roe T, Schroeder M, Weng S, Botstein D. SGD: Saccharomyces genome database. Nucleic Acids Res. 1998;26:73–79. doi: 10.1093/nar/26.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortopassi GA, Wong A. Mitochondria in organismal aging and degeneration. Biochim Biophys Acta. 1999;1410:183–193. doi: 10.1016/s0005-2728(98)00166-2. [DOI] [PubMed] [Google Scholar]
- Costanzo MC, Hogan JD, Cusick ME, Davis BP, Fancher AM, Hodges PE, Kondu P, Lengieza C, Lew-Smith JE, Lingner C, Roberg-Perez KJ, Tillberg M, Brooks JE, Garrels JI. The yeast proteome database (YPD) and Caenorhabditis elegans proteome database (WormPD): comprehensive resources for the organization and comparison of model organism protein information. Nucleic Acids Res. 2000;28:73–76. doi: 10.1093/nar/28.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein, C.B., Hale, W, IV, and Butow, R.A. (2000). Numerical methods for handling uncertainty in microarray data: an example analyzing perturbed mitochondrial function in yeast. Methods Cell Biol. 66 (in press). [DOI] [PubMed]
- Green DR, Reed JC. Mitochondria and apoptosis. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- Heddi A, Stepien G, Benke PJ, Wallace DC. Coordinate induction of energy gene expression in tissues of mitochondrial disease patients. J Biol Chem. 1999;274:22968–22976. doi: 10.1074/jbc.274.33.22968. [DOI] [PubMed] [Google Scholar]
- Horswill AR, Escalante-Semerena JC. Propionate catabolism in Salmonella typhimurium LT2: two divergently transcribed units comprise the prp locus at 8.5 centisomes, prpR encodes a member of the sigma-54 family of activators, and the prpBCDE genes constitute an operon. J Bacteriol. 1997;179:928–940. doi: 10.1128/jb.179.3.928-940.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, Armour CD, Bennett HA, Coffey E, Dai H, He YD, Kidd MJ, King AM, Meyer MR, Slade D, Lum PY, Stepaniants SB, Shoemaker DD, Gachotte D, Chakraburtty K, Simon J, Bard M, Friend SH. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- Jia Y, Rothermel B, Thornton J, Butow RA. A basic helix-loop-helix zipper transcription complex functions in a signaling pathway from mitochondria to the nucleus. Mol Cell Biol. 1997;17:1110–1117. doi: 10.1128/mcb.17.3.1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kal AJ, van Zonneveld AJ, Benes V, vandenBerg M, Koerkamp MG, Albermann K, Strack N, Ruijter JM, Richter A, Dujon B, Ansorge W, Tabak HF. Dynamics of gene expression revealed by comparison of serial analysis of gene expression transcript profiles from yeast grown on two different carbon sources. Mol Biol Cell. 1999;10:1859–1872. doi: 10.1091/mbc.10.6.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchman PA, Kim S, Lai CY, Jazwinski SM. Interorganelle signaling is a determinant of longevity in Saccharomyces cerevisiae. Genetics. 1999;152:179–190. doi: 10.1093/genetics/152.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohrer K, Domdey H. Preparation of high molecular weight RNA. Methods Enzymol. 1991;194:398–405. doi: 10.1016/0076-6879(91)94030-g. [DOI] [PubMed] [Google Scholar]
- Kos W, Kal AJ, Vanwilpe S, Tabak HF. Expression of genes encoding peroxisomal proteins in Saccharomyces cerevisiae is regulated by different circuits of transcriptional control. Biochim Biophys Acta. 1995;1264:79–86. doi: 10.1016/0167-4781(95)00127-3. [DOI] [PubMed] [Google Scholar]
- Kunau W-H, Bühne A, Moreno del la Garza M, Kionka M, Mateblowski M, Shultz-Borchard U, Thieringer R. Comparative enzymology of β-oxidation. Biochem Soc Trans. 1988;16:418–420. doi: 10.1042/bst0160418. [DOI] [PubMed] [Google Scholar]
- Liao X, Butow RA. RTG1 and RTG2: two yeast genes required for a novel path of communication from mitochondria to the nucleus. Cell. 1993;72:61–71. doi: 10.1016/0092-8674(93)90050-z. [DOI] [PubMed] [Google Scholar]
- Liao XS, Small WC, Srere PA, Butow RA. Intramitochondrial functions regulate nonmitochondrial citrate synthase (CIT2) expression in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:38–46. doi: 10.1128/mcb.11.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Butow RA. A transcriptional switch in the expression of yeast tricarboxylic acid cycle genes in response to a reduction or loss of respiratory function. Mol Cell Biol. 1999;19:6720–6728. doi: 10.1128/mcb.19.10.6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall PA, Dyer JM, Quick ME, Goodman JM. Redox-sensitive homodimerization of Pex11p: a proposed mechanism to regulate peroxisomal division. J Cell Biol. 1996;135:123–137. doi: 10.1083/jcb.135.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh VS, Morgan MM, Scott R, Clements LS, Butow RA. The mitochondrial genotype can influence nuclear gene expression in yeast. Science. 1987;235:576–580. doi: 10.1126/science.3027892. [DOI] [PubMed] [Google Scholar]
- Pronk JT, van der Linden-Beuman A, Verduyn C, Scheffers WA, van Dijken JP. Propionate metabolism in Saccharomyces cerevisiae: implications for the metabolon hypothesis. Microbiology. 1994;140:717–722. doi: 10.1099/00221287-140-4-717. [DOI] [PubMed] [Google Scholar]
- Rothermel BA, Shyjan AW, Etheredge JL, Butow RA. Transactivation by Rtg1p, a basic helix-loop-helix protein that functions in communication between mitochondria and the nucleus in yeast. J Biol Chem. 1995;49:29476–29482. doi: 10.1074/jbc.270.49.29476. [DOI] [PubMed] [Google Scholar]
- Sekito T, Thornton J, Butow RA. Mitochondria-to-nuclear signaling is regulated by the subcellular localization of the transcription factors Rtg1p and Rtg3p. Mol Biol Cell. 2000;11:2103–2115. doi: 10.1091/mbc.11.6.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoneczny S, Chelstowska A, Rytka A. Study of the coinduction by fatty acids of catalse A and acyl CoA oxidase in standard and mutant S. cerevisiae strains. Eur J Biochem. 1988;174:297–302. doi: 10.1111/j.1432-1033.1988.tb14097.x. [DOI] [PubMed] [Google Scholar]
- Tabuchii T, Serizawa N, Uchiyama H. A novel pathway for the partial oxidation of propionyl-CoA to pyruvate via seven-carbon tricarboxylic acids in yeasts. Agric Biol Chem. 1974;38:2571–2572. [Google Scholar]
- Textor S, Wendisch VF, De Graaf AA, Muller U, Linder MI, Linder D, Buckel W. Propionate oxidation in Escherichia coli: evidence for operation of a methylcitrate cycle in bacteria. Arch Microbiol. 1997;168:428–436. doi: 10.1007/s002030050518. [DOI] [PubMed] [Google Scholar]
- van den Berg MA, de Jong-Gubbels P, Kortland CJ, van Dijken JP, Pronk JT, Steensma HY. The two acetyl-coenzyme A synthetases of Saccharomyces cerevisiae differ with respect to kinetic properties and transcriptional regulation. J Biol Chem. 1996;271:28953–28959. doi: 10.1074/jbc.271.46.28953. [DOI] [PubMed] [Google Scholar]
- van Roermund CW, Hettema EH, van den Berg M, Tabak HF, Wanders RJ. Molecular characterization of carnitine-dependent transport of acetyl-CoA from peroxisomes to mitochondria in Saccharomyces cerevisiae and identification of a plasma membrane carnitine transporter, Agp2p. EMBO J. 1999;18:5843–5852. doi: 10.1093/emboj/18.21.5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderleij I, Franse MM, Elgersma Y, Distel B, Tabak HF. PAS10 is a tetratricopeptide-repeat protein that is essential for the import of most matrix proteins into peroxisomes of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1993;90:11782–11786. doi: 10.1073/pnas.90.24.11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenhuis M, Mateblowski M, Kunau W, Harder W. Proliferation of microbodies in Saccharomyces cerevisiae. Yeast. 1987;3:77–84. doi: 10.1002/yea.320030204. [DOI] [PubMed] [Google Scholar]
- Verduyn C, Postma E, Scheffers WA, van Dijken JP. Energetics of Saccharomyces cerevisiae in anaerobic glucose-limited chemostat cultures. J Gen Microbiol. 1990;136:405–412. doi: 10.1099/00221287-136-3-405. [DOI] [PubMed] [Google Scholar]
- Waddle JA, Karpova TS, Waterston RH, Cooper JA. J. Cell Biol, 1996;132:861–870. doi: 10.1083/jcb.132.5.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC. Mitochondrial diseases in man and mouse. Science. 1999;283:1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- Wang SS, Brandriss MC. Proline utilization in Saccharomyces cerevisiae: sequence, regulation, and mitochondrial localization of the PUT1 gene product. Mol Cell Biol. 1987;7:4431–4440. doi: 10.1128/mcb.7.12.4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.