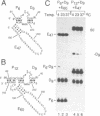
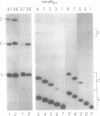
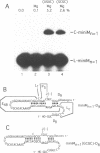
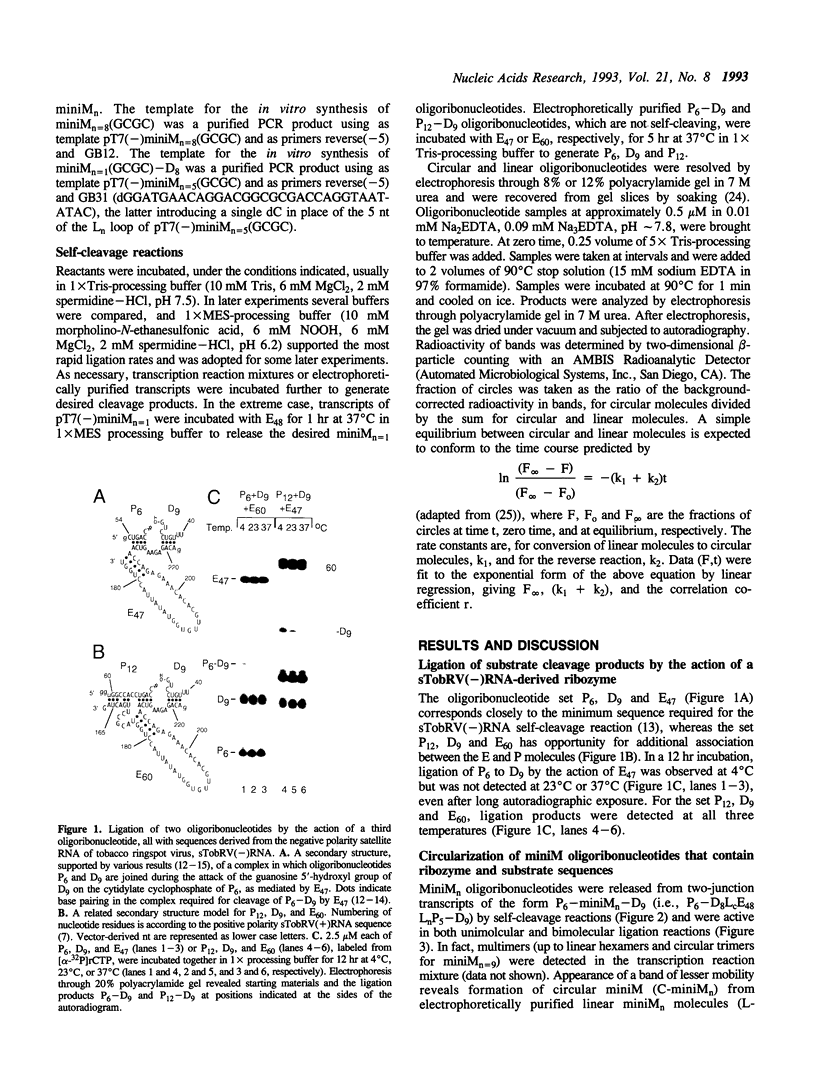
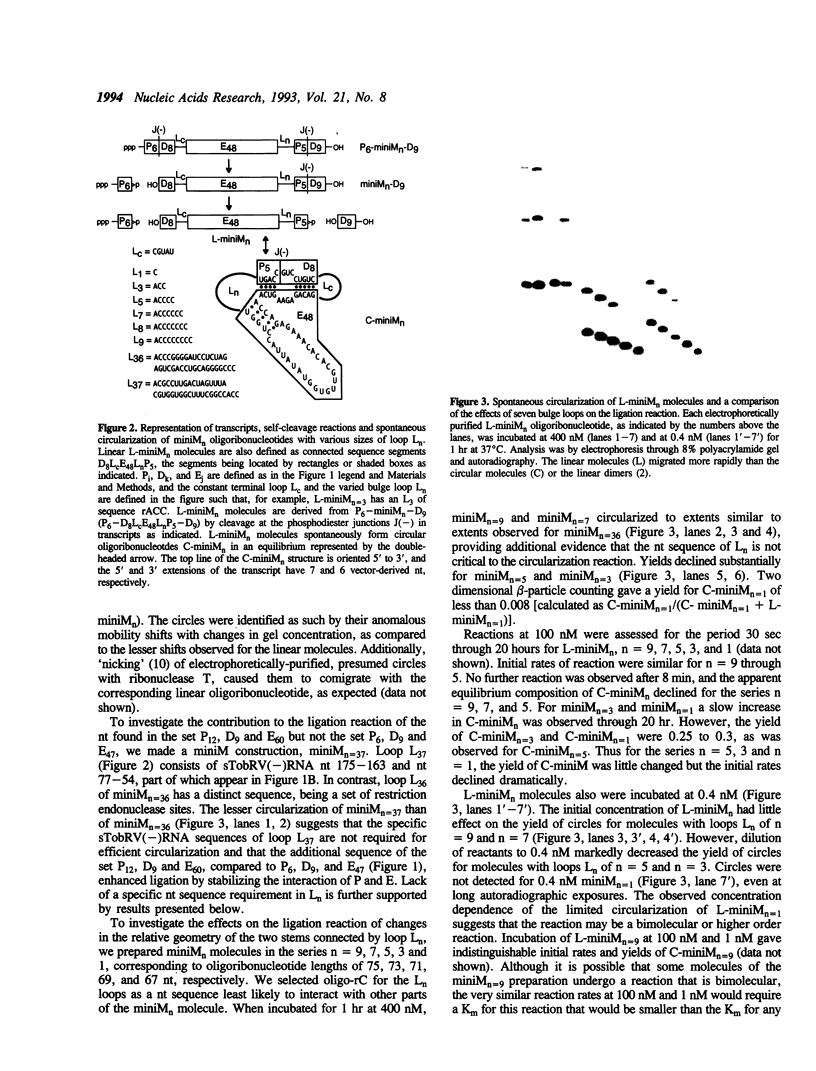
Abstract
The less abundant polarity of the satellite RNA of tobacco ringspot virus, designated sTobRV(-)RNA, contains a ribozyme and its substrate. We demonstrate that the ribozyme can catalyze the ligation of substrate cleavage products and that oligoribonucleotides, termed 'mini-monomers' and containing little more than covalently attached ribozyme and substrate cleavage products, circularized spontaneously, efficiently and reversibly. The kinetics of ligation and cleavage of one such mini-monomer was consistent with a simple unimolecular reaction at some temperatures. Evidence suggests that the circular ligation product includes a 5 bp stem that is connected to a 4 bp stem by a bulge loop. Reduction of the bulge loop to one nt is expected to place the 4 and 5 bp helices in a nearly coaxial, rather than an angled or parallel, orientation. Such molecules did not circularize in a unimolecular reaction but did when incubated with second, trans-acting oligoribonucleotides that had either the original or a substituted 4 bp helix. These results suggest that a bulge loop that is too small prevents formation of geometry essential for unimolecular ligation. We suggest the term 'paperclip' to represent the arrangement of RNA strands in the region of sTobRV(-)RNA that participates in the cleavage and ligation reactions.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bruening G., Passmore B. K., van Tol H., Buzayan J. M., Feldstein P. A. Replication of a plant virus satellite RNA: evidence favors transcription of circular templates of both polarities. Mol Plant Microbe Interact. 1991 May-Jun;4(3):219–225. doi: 10.1094/mpmi-4-219. [DOI] [PubMed] [Google Scholar]
- Doudna J. A., Szostak J. W. RNA-catalysed synthesis of complementary-strand RNA. Nature. 1989 Jun 15;339(6225):519–522. doi: 10.1038/339519a0. [DOI] [PubMed] [Google Scholar]
- Fedor M. J., Uhlenbeck O. C. Substrate sequence effects on "hammerhead" RNA catalytic efficiency. Proc Natl Acad Sci U S A. 1990 Mar;87(5):1668–1672. doi: 10.1073/pnas.87.5.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldstein P. A., Buzayan J. M., Bruening G. Two sequences participating in the autolytic processing of satellite tobacco ringspot virus complementary RNA. Gene. 1989 Oct 15;82(1):53–61. doi: 10.1016/0378-1119(89)90029-2. [DOI] [PubMed] [Google Scholar]
- Feldstein P. A., Buzayan J. M., van Tol H., deBear J., Gough G. R., Gilham P. T., Bruening G. Specific association between an endoribonucleolytic sequence from a satellite RNA and a substrate analogue containing a 2'-5' phosphodiester. Proc Natl Acad Sci U S A. 1990 Apr;87(7):2623–2627. doi: 10.1073/pnas.87.7.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster A. C., Davies C., Sheldon C. C., Jeffries A. C., Symons R. H. Self-cleaving viroid and newt RNAs may only be active as dimers. Nature. 1988 Jul 21;334(6179):265–267. doi: 10.1038/334265a0. [DOI] [PubMed] [Google Scholar]
- Forster A. C., Symons R. H. Self-cleavage of plus and minus RNAs of a virusoid and a structural model for the active sites. Cell. 1987 Apr 24;49(2):211–220. doi: 10.1016/0092-8674(87)90562-9. [DOI] [PubMed] [Google Scholar]
- Gish G., Eckstein F. DNA and RNA sequence determination based on phosphorothioate chemistry. Science. 1988 Jun 10;240(4858):1520–1522. doi: 10.1126/science.2453926. [DOI] [PubMed] [Google Scholar]
- Hampel A., Tritz R., Hicks M., Cruz P. 'Hairpin' catalytic RNA model: evidence for helices and sequence requirement for substrate RNA. Nucleic Acids Res. 1990 Jan 25;18(2):299–304. doi: 10.1093/nar/18.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampel A., Tritz R. RNA catalytic properties of the minimum (-)sTRSV sequence. Biochemistry. 1989 Jun 13;28(12):4929–4933. doi: 10.1021/bi00438a002. [DOI] [PubMed] [Google Scholar]
- Haseloff J., Gerlach W. L. Sequences required for self-catalysed cleavage of the satellite RNA of tobacco ringspot virus. Gene. 1989 Oct 15;82(1):43–52. doi: 10.1016/0378-1119(89)90028-0. [DOI] [PubMed] [Google Scholar]
- Haseloff J., Gerlach W. L. Simple RNA enzymes with new and highly specific endoribonuclease activities. Nature. 1988 Aug 18;334(6183):585–591. doi: 10.1038/334585a0. [DOI] [PubMed] [Google Scholar]
- Herschlag D., Cech T. R. Catalysis of RNA cleavage by the Tetrahymena thermophila ribozyme. 1. Kinetic description of the reaction of an RNA substrate complementary to the active site. Biochemistry. 1990 Nov 6;29(44):10159–10171. doi: 10.1021/bi00496a003. [DOI] [PubMed] [Google Scholar]
- Kaper J. M., Tousignant M. E., Steger G. Nucleotide sequence predicts circularity and self-cleavage of 300-ribonucleotide satellite of arabis mosaic virus. Biochem Biophys Res Commun. 1988 Jul 15;154(1):318–325. doi: 10.1016/0006-291x(88)90687-0. [DOI] [PubMed] [Google Scholar]
- Prody G. A., Bakos J. T., Buzayan J. M., Schneider I. R., Bruening G. Autolytic processing of dimeric plant virus satellite RNA. Science. 1986 Mar 28;231(4745):1577–1580. doi: 10.1126/science.231.4745.1577. [DOI] [PubMed] [Google Scholar]
- Rubino L., Tousignant M. E., Steger G., Kaper J. M. Nucleotide sequence and structural analysis of two satellite RNAs associated with chicory yellow mottle virus. J Gen Virol. 1990 Sep;71(Pt 9):1897–1903. doi: 10.1099/0022-1317-71-9-1897. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saville B. J., Collins R. A. A site-specific self-cleavage reaction performed by a novel RNA in Neurospora mitochondria. Cell. 1990 May 18;61(4):685–696. doi: 10.1016/0092-8674(90)90480-3. [DOI] [PubMed] [Google Scholar]
- Saville B. J., Collins R. A. RNA-mediated ligation of self-cleavage products of a Neurospora mitochondrial plasmid transcript. Proc Natl Acad Sci U S A. 1991 Oct 1;88(19):8826–8830. doi: 10.1073/pnas.88.19.8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons R. H. Self-cleavage of RNA in the replication of small pathogens of plants and animals. Trends Biochem Sci. 1989 Nov;14(11):445–450. doi: 10.1016/0968-0004(89)90103-5. [DOI] [PubMed] [Google Scholar]
- Symons R. H. Small catalytic RNAs. Annu Rev Biochem. 1992;61:641–671. doi: 10.1146/annurev.bi.61.070192.003233. [DOI] [PubMed] [Google Scholar]
- Van Tol H., Buzayan J. M., Bruening G. Evidence for spontaneous circle formation in the replication of the satellite RNA of tobacco ringspot virus. Virology. 1991 Jan;180(1):23–30. doi: 10.1016/0042-6822(91)90005-v. [DOI] [PubMed] [Google Scholar]