Abstract
The analysis of heparan sulfate glycosaminoglycans (HSGAGs) variations in human serum at the disaccharide level has a great potential for disease diagnosis and prognosis. However, the lack of available analytical methodology for the compositional analysis of HSGAGs in human serum remains to be addressed to delineate the possible role of HSGAGs on the onset and/or progression of a disease. In this study, we have developed a method for the in-depth compositional analysis of the 12 heparin/HS-derived disaccharides from human serum using a combination of technologies – fractionation, exhaustive digestion, solid phase extraction and LC-MS/MS. The method exhibits high recovery (72%–110%) and good reproducibility (standard deviation of less than 5%) with a low limit of detection and quantification. Errors from the method validation were within 1.1%. Non-detectable non- or low-sulfated disaccharides in human serum were also detected using the optimized protocol. Further applying this method, the comprehensive analysis of HSGAGs compositions in human sera from female donors showed considerable variations in disaccharide patterns and compositions.
INTRODUCTION
Heparin/heparan sulfate glycosaminoglycans are negatively charged linear polysaccharides that are found on the cell surface and extracellular matrix (ECM). They are the most structurally complex glycosaminoglycans (GAGs) with diverse sulfation patterns and are one of the most informative biopolymers in nature. They are composed of repeating disaccharide units of hexuronic acid linked to glucosamine. The glucosamine may be 6-O-sulfated, N-sulfated, N-acetylated and sporadically 3-O-sulfated, while the hexuronic acid may be 2-O-sulfated. When HSGAGs undergo exhaustive enzymatic digestion, they can generate typically 12 heparin/heparan sulfate (HS)-derived disaccharides including some of the rare 3-O-sulfated disaccharide with modification by differential sulfation patterns1–4.
It is known that HSGAGs have diverse biological functions and are widely involved in many physiological and pathological processes such as blood coagulation and inflammatory response5–6 via interactions with a variety of proteins including growth factors, cytokines and chemokines7–8. The interactions are dependent upon the disaccharide composition and patterns of HSGAGs, which play a significant role in regulating various biological processes. The heterogeneity of HSGAGs is determined by the expression patterns of genes and the presence of various HSGAGs-editing enzymes under different pathological conditions9–13. Inflammatory processes and diseases such as mucopolysaccharidoses, osteoarthritis and myeloma cancer have been reported to correlate to the different disaccharide structures of HSGAGs14–17. Therefore, evaluating the variations (i.e. presence and quantity) of HSGAGs has a great potential for diagnosis and prognosis of diseases.
Human serum or plasma serves as a typical clinical specimen since both are more accessible and convenient for long-term monitoring. It has been demonstrated that the low molecular weight fraction of human serum or plasma provides a rich source for potential biomarkers of diseases generated through enzymatic cleavage18–19. GAGs, such as chondroitin sulfate and HS have been found in human serum or plasma20–21, which carry important biological information. The variations of GAGs in human serum or plasma are also vital for investigating and monitoring disease conditions. For example, the total amount of GAGs in serum was found to be closely related to diabetic pathology22. However, the minute concentration of GAGs in human serum together with the complexity of the serum matrix limits the information on the HSGAGs component in human serum.
The analysis of HSGAGs in human serum is demanding, with the requirements of effective separation, purification and detection. Many efforts have been made to meet these requirements involving various techniques such as Cohn-Oncley fractionation, multiple proteolytic enzyme digestions, ethanol, trichloroacetic acid (TCA) or heating precipitation, dialysis, lipid extraction, chromatographic purification and gel electrophoretic separation23–27. However, to date, none of the above methods have provided for efficient extraction of HSGAGs from human serum. The available methods for the compositional analysis of GAGs in human serum are generally conducted by enzyme or chemical degradation of GAGs into their constituent disaccharide units, followed by quantitative analysis through separation methods using chromatography and electrophoresis. These methods also involve tedious steps to develop appropriate techniques which typically constitute derivatization, isotopic labeling and sulfatase digestion to obtain the baseline separation and identification of isomeric disaccharides28–32.
In this study, a MS-based analytical method for the comprehensive compositional analysis of the 12 heparin/HS-derived disaccharides from human serum, using fractionation, exhaustive enzymatic digestion, solid phase extraction (SPE) and LC-MS/MS has been developed and validated.
EXPERIMENTAL SECTION
Materials
Heparin/heparan sulfate-derived disaccharide standards ΔUA-GlcNAc (IV-A), ΔUA2S-GlcNAc (III-A), ΔUA-GlcNAc6S (II-A), ΔUA2S-GlcNAc6S (I-A), ΔUA2S-GlcNS (III-S), ΔUA-GlcNS6S (II-S), ΔUA2S-GlcNS6S (I-S), ΔUA2S-GlcN6S (I-H), ΔUA-GlcN6S (II-H), ΔUA2S-GlcN (III-H), ΔUA-GlcN (IV-H), and ΔUA2S-GlcNCOEt6S (I-P) were purchased from V-Labs (Covington, LA). ΔUA-GlcNS (IV-S) was obtained from Iduron (Manchester, UK). Heparinase I (heparinase, EC 4.2.2.7), heparinase II (heparitinase II, EC 4.2.2.8), and heparinase III (heparitinase, EC 4.2.2.8) from Flavobacterium heparinum were obtained from Seikagaku Corporation (East Falmouth, MA). Human serum from platelet poor human plasma and human serum from human male AB plasma were procured from Sigma-Aldrich (St Louis, MO). Human sera from female single donors (Hispanic age of 29, African American age of 44 and Caucasian age of 38) were obtained from Innovative Research (Novi, MI).
Human Serum Fractionation and Digestion
A sample of 20 μL human serum was diluted to 4 mL in 25 mM NH4HCO3 (pH 8.2) in 20% (v/v) acetonitrile (ACN)/H2O and fractionated by a 50 kDa molecular weight cut-off (MWCO) Amicon Ultra-4 centrifugal filter (Millipore, Billerica, MA).33 The sample was centrifuged at 4,000 ×g for 15min to separate the serum components into high- and low-molecular weight (HMW and LMW) serum fractions. The LMW serum fraction was dried in vacuo, resuspended in 50 μL of digestion buffer (20 mM NH4OAc, pH 7.5 and 1 mM Ca(OAc)2) containing 2 mU each of heparinase I, II and III, and incubated at 37 °C for about 20 hrs with gentle agitation.
Solid Phase Extraction (SPE)
The enzyme digestion was spiked with 3 μL 80 mM internal standard I-P solution and purified using a C18+carbon-SPE TopTip (Glygen, Columbia, MD). The TopTip was conditioned twice with 400 μL of 0.1% trifluoroacetic acid (TFA) in 80% (v/v) ACN/H2O, followed by 400 μL Milli-Q water, 3×. The digested sample was diluted to 400 μL with Milli-Q water, applied to TopTip and washed 5× with 400 μL of Milli-Q water. The disaccharides were eluted 3× with 400 μL of 20% (v/v) ACN/H2O and 5× with 400 μL of 0.05% TFA in 40% (v/v) ACN/H2O. All the elution steps were performed by spinning TopTip in centrifuge at 3500 rpm for 1 min. Both fractions were collected, dried in vacuo, redissolved in 60 μL of Milli-Q water before LC-MS/MS analysis.
Liquid Chromatography-Mass Spectrometry Analysis
Mass spectra were acquired using a LTQ 2-D linear ion trap mass spectrometer equipped with an electrospray ionization (ESI) source and directly coupled to an HPLC system (Thermo Electron, San Jose, CA). The SPE purified disaccharide samples were separated using a Hypercarb column (2.1 mm ID × 150 mm, 5 μm) (Thermo Fisher Scientific, Waltham, MA) ran for 40 min with a gradient of 5 mM NH4HCO3/H2O (solvent A) and in 80% ACN/H2O (solvent B) at a flow rate of 200 μL/min. The gradient was programmed as 3.8 min 100% A, linearly changed to 62.5% A in 0.2 min, maintained at 62.5% A for 22 min, modified to 100% A in 0.2 min, and kept at 100% A for 13.8 min. The first 3.8 min was diverted to waste and the remaining 36.2 min was sprayed directly into the mass spectrometer. The analysis was performed in the negative ion mode using a capillary temperature of 250 °C. The instrument was tuned using disaccharide standards for optimal condition before sample analysis. For MS2 experiments, the precursor ions were selected using an isolation width of 3 Da and activated using 18–21% normalized collision energy for 100 ms. Data acquisition and analysis were performed using Xcalibur 2.0 software.
RESULTS AND DISCUSSION
Method Development
An extraction and purification method was developed for the MS-based compositional analysis of heparin/HS-derived disaccharides extracted from human serum. Only a small amount of human serum (20 μL) was needed compared to other methods that require larger amounts of sample (up to 10 mL)34–35. Thus, the method presented here requires much less sample from blood donors and allows for replicate analysis to be made. One of the challenges in the analysis of serum is that it is a complex biological mixture and many high abundant proteins interfere in the analysis of low abundant components such as glycosaminoglycans (GAGs). The majority of proteins identified in human plasma are comprised of albumin, immunoglobulins and transferrins, which account for about 90% of the total protein content.19 All of these proteins have molecular weights larger than 50 kDa. Thus, to address the issue of complexity, the serum sample was first fractionated using a 50 kDa MWCO filter in 25 mM NH4HCO3 (pH 8.2) in 20% (v/v) ACN/H2O, a buffer system that disrupts protein-protein interactions.33 Heparin/HS GAGs contained in the LMW serum fraction were then digested exhaustively by a combination of heparinase I, II and III followed by solid phase extraction. Solid phase extraction as the purification method of the digested GAGs affords an inexpensive, fast, reproducible means of clean-up and there is no concern about dead volume36 in addition to its good selectivity for ionic compounds, specifically in the case of graphitized carbon resin. Lastly, the eluted fractions were analyzed by LC-MS/MS for disaccharide identification and quantification.
Solid Phase Extraction of Disaccharides
Graphitized carbon has been shown to be effective in the purification of oligosaccharides extracted from biological matrices.37–41 The SPE clean-up procedure was first optimized and evaluated by comparing three different SPE formats: carbon TopTip, mixed C18 with carbon TopTip (mixed-mode), and separate C18 TopTip and carbon TopTip. The mixed-mode and separate C18 and carbon TopTip formats were observed to have similar efficiency in the clean-up, while the carbon TopTip format showed more contaminant peaks (data not shown). Due to the fact that the mixed-mode SPE format provided a faster and inexpensive means for sample clean-up, the mixed-mode format was used for the succeeding experiments. To achieve maximum recovery, the SPE clean-up procedure was further optimized by examining several elution solvents of different ACN percentages as well as varying the collection volume. The heparin/HS-derived disaccharide IV-H was found to elute in the 20% ACN/H2O fraction and the remaining 11 disaccharides were eluted in the 40% ACN/0.05% TFA fraction. The highest percentage recovery was obtained by collecting 400 μL of 20% ACN/H2O fraction, 3× and 400 μL of 40% ACN/0.05% TFA, 5×.
Compositional Analysis of 12 Disaccharides
An analytical method for the compositional analysis and quantification of commercially pure bovine heparin and heparan sulfate by ESI-MS and MS/MS has been developed by this laboratory1. With this procedure, the digested samples were analyzed directly by ESI-MS without the need for LC separation. The concentrations of non-isomeric disaccharides (I-S, IV-A, I-A, IV-H) and the sum of the concentrations of each set of isomers (III-S/II-S/I-H, IV-S/II-H/III-H, III-A/II-A) can be obtained from the MS1 spectra (full MS scan). The quantification was accomplished by integrating the peak area of the extracted ion chromatograph (XIC) (i.e. m/z 191.5, 378.1, 268.7, 336.3 for the four non-isomeric disaccharides and m/z 247.7, 416.1 and 458.1 for the three sets of isomeric disaccharides, respectively) followed by the calculation of the relative amounts of each isomer by the diagnostic ions in the MS2 spectra. This analytical method was adapted and modified for the compositional analysis of heparin/HS-derived disaccharides from human serum.
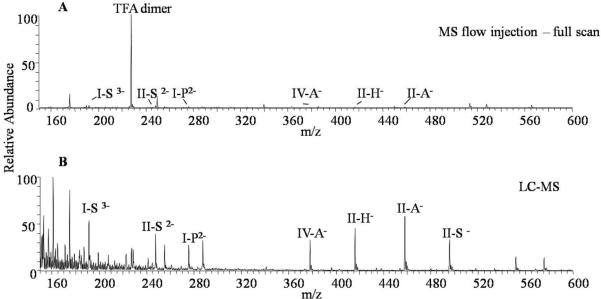
One of the primary issues for the quantitative analysis of heparin/HS-derived disaccharides from biological matrices such as serum or for any quantification of targeted compounds in a complex mixture is the effect of the matrix, which typically affects the signals from the molecules of interest. Particularly for ESI-MS analysis, sample preparation/clean-up is an important factor prior to instrument analysis. For this study, additional online-desalting procedure of the SPE purified disaccharides using graphitized carbon HPLC column was also found to be necessary before ESI-MS/MS analysis. As shown in Figure 1, the sample injected directly into the mass spectrometer displayed a significant TFA suppression of the disaccharide signals in the negative ion mode, while samples analyzed by LC-MS showed a greater improvement in the disaccharide signals and a cleaner mass spectrum.
Figure 1.
Comparison of MS flow injection and LC-MS analysis of a sample of six disaccharide standards (I-S, II-S, IV-A, II-H, II-A and internal standard I-P) in human serum matrix after SPE cleanup: (A) MS full scan spectrum shows a significant TFA dimer ion suppression; (B) LC-MS spectrum shows much better signals of disaccharides.
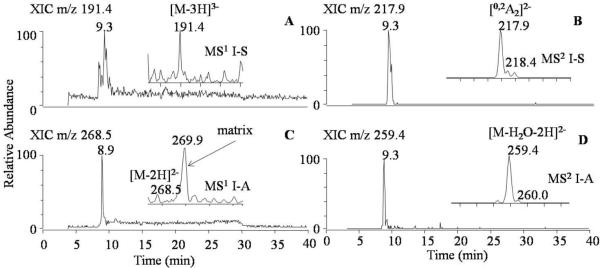
We have developed and improved upon an earlier study for analyzing the 12 common heparin/HS-derived disaccharides1. In the current research effort, we have incorporated LC-MS/MS in an attempt to further optimize this difficult analysis. Additionally, to minimize the effects of human serum matrix, the concentrations of non-isomeric compounds were determined using the diagnostic ions from the MS2 data instead from the MS1 spectra, which effectively improved the quantitative analysis of low abundant disaccharides and provided higher signal-to-noise (S/N). Comparison of the MS1 and MS2 data is shown in Figure 2. The extracted ion chromatograph (XIC) of disaccharide I-S in the MS1 spectrum shows low S/N ratio (Figure 2A), while the MS2 spectrum of I-S (m/z 191.4), diagnostic ion [0,2A2]2- at m/z 217.9 can be quantified more accurately by providing a more defined XIC peak (Figure 2B).42 To further illustrate this point, the MS data for another disaccharide are provided in Figure 2C and 2D. As shown, the XIC of disaccharide I-A is not well distinguished from the matrix peak at m/z 269.9 which partially overlaps with the I-A peak at m/z 268.5 (Figure 2C). On the other hand, for the XIC of the diagnostic ion of I-A (m/z 259.4) in the MS2 data, there is no overlapping peak from the matrix (Figure 2D). Furthermore, some low-abundance disaccharides that were not detected from previous MS1 analysis, such as I-A and IV-H, can now be detected from their MS2 spectra using the characteristic diagnostic ions.
Figure 2.
Comparison of the extracted ion chromatographs (XIC) from the MS1 and MS2 spectra: (A) XIC of I-S3- ([M-3H]3- at m/z 191.4) in MS1 spectrum (shown as an inset); (B) XIC of the diagnostic ion ([0,2A2]2- at m/z 217.9) of I-S in MS2 spectrum (shown as an inset); (C) XIC of I-A2- ([M-2H]2- at m/z 268.5) in MS1 spectrum. The inserted MS1 spectrum shows I-A ion and a matrix peak (m/z 269.9) that partially overlapped with I-A; (D) XIC of the diagnostic ion ([M-H2O-2H]2- at m/z 259.4) of I-A in MS2 spectrum (shown as an inset).
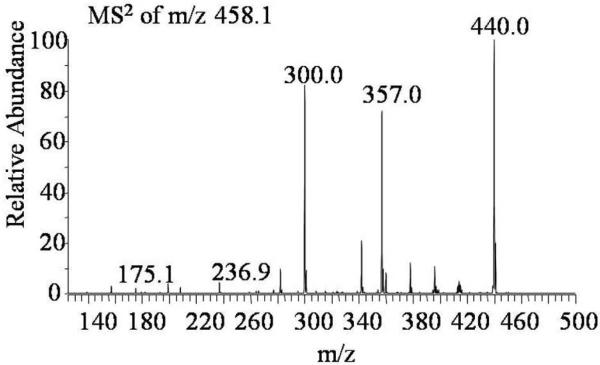
The quantification method for the isomeric disaccharides has been reported2. As an example of that protocol, the quantification of the II-A/III-A isomers (m/z 458.1) is demonstrated here. The quantitative analysis of isomers II-A and III-A are based on the product ions - m/z 357.0 and 236.9, wherein m/z 357.0 corresponds to [0,2A2]− fragment ion from II-A while m/z 236.9 corresponds to [B1]− fragment of III-A (Figure 3). Both m/z 357.0 and 236.9 ions are detected, which suggests that II-A and III-A are present in the digested sample of human serum from the platelet poor human plasma. In sum, the quantitative analysis of all 12 heparin/HS-derived disaccharides from human serum can be accomplished using the protocol described above with high sensitivity and confidence. The 3-O-sulfated isomer of I-S, ΔUA2S→GlcNS3S, a rare disaccharide was also not observed since the MS2 spectrum of m/z 191.5 does not match to the published spectrum of 3-O-sulfated standard.3
Figure 3.
MS2 spectrum of isomeric disaccharides II-A/III-A (m/z 458.1 as the precursor ion) from the digested sample of human serum from platelet poor human plasma. The product ion m/z 357.0 is mostly contributed by [0,2A2]− fragment of II-A and m/z 236.9 corresponds to [B1]− fragment of III-A.
Method Validation
The linearity of the analytical method was determined by spiking the human serum with six different concentrations (from 1 μM to 80 μM) of disaccharide standards and a constant concentration (4 μM) of I-P, as the internal standard (IS), creating a dynamic range of disaccharide-to-IS ratio from 0.25 to 20 (~102). Plotting the peak area ratio of standards and IS, the method demonstrated very good linearity with linear correlation coefficients (R2) ranging from 0.994–0.999. The response factor (R factor) of each of the 12 disaccharides was determined by one-point R factor determination method.1
Moreover, following the equation provided from an earlier publication43, the limit of detection (LOD) and quantification (LOQ) were calculated from the calibration curve as 3.3σ/S and 10σ/S respectively, where σ is the standard deviation of the intercept and S is the slope of the regression line of the calibration curve. Using these equations, the LOD was calculated to be from 3.1 pmol to 17.8 pmol and the LOQ was determined to be from 9.3 pmol to 53.9 pmol (Table 1). The recovery was then determined by spiking a known amount (10 μM each) of the 12 disaccharide standards to the human serum matrix analyzed in triplicate. The concentrations of the disaccharides were compared to that without SPE purification. An internal standard I-P of a constant concentration (4 μM) was also used for the calculations of the percent recoveries for all 12 heparin/HS-derived disaccharide standards. The percent recoveries were determined to range from 72% to 110% with good reproducibility (standard deviation of less than 5%) as shown in Table 1. Also, there was no noticeable desulfation observed during the MS analysis.
Table 1.
Recovery, reproducibility, limit of detection (LOD) and limit of quantification (LOQ) of the method.
| DSCa | m/z | Recovery (%)b Mean ± SDc | LODd (pmol) | LOQd (pmol) | |
|---|---|---|---|---|---|
| IV-A | D0A0 | 378.11− | 102 ± 4 | 11.9 | 36.1 |
| III-A | D2A0 | 458.11− | 99 ± 2 | 11.6 | 35.3 |
| II-A | D0A6 | 458.11− | 101 ± 1 | 3.1 | 9.3 |
| I-A | D2A6 | 268.72− | 96 ± 2 | 4.9 | 14.8 |
| IV-S | D0S0 | 416.11− | 110 ± 4 | 8.8 | 26.6 |
| III-S | D2S0 | 247.72− | 72 ± 2 | 10.1 | 30.7 |
| II-S | D0S6 | 247.72− | 95 ± 3 | 6.7 | 20.3 |
| I-S | D2S6 | 191.53− | 79 ± 3 | 12.9 | 39.1 |
| I-H | D2H6 | 247.72− | 96 ± 5 | 9.8 | 29.6 |
| II-H | D0H6 | 416.11− | 99 ± 2 | 7.1 | 21.5 |
| III-H | D2H0 | 416.11− | 80 ± 3 | 17.8 | 53.9 |
| IV-H | D0H0 | 336.31− | 98 ± 4 | 10.2 | 31.0 |
DSC, the disaccharide structure code.4
The recovery of the purification method was determined by comparing disaccharide standards in human serum matrix with and without SPE purification.
SD, standard deviation of the mean.
LOD and LOQ was calculated as 3.3σ/S and 10σ/S respectively using calibration curve, where σ is the standard deviation of the intercept and S is the slope of the regression line of calibration curve.
The compositional analysis method was further verified by analyzing a mock mixture that consists of 10 μM each of the 12 heparin/HS-derived disaccharide standards in the human serum matrix following the protocol described above. Since the mock mixture is composed of the same amount of disaccharides, each disaccharide is calculated to have a theoretical composition of 8.3%. By comparing these values to those of the experimentally determined percent compositions, the percent errors were calculated to be within 1.1% (Table 2). These data demonstrates that our method for compositional analysis in human serum has adequate sensitivity and recovery compared to other method for quantitative analysis of disaccharides in complex biological matrix.44–46 Before every sample analysis, a mock mixture is analyzed to serve as a validation step and to monitor the accuracy of the method even when the analysis is performed on different days.
Table 2.
Method validation using serum spiked with a mock mixture of twelve disaccharide standardsa.
| m/z | Theo. (%) | Expt'l (%) | Diff. (%) | |
|---|---|---|---|---|
| IV-A | 378.11− | 8.3 | 8.3 | 0.0 |
| III-A | 458.11− | 8.3 | 9.2 | 0.9 |
| II-A | 458.11− | 8.3 | 8.6 | 0.3 |
| I-A | 268.72− | 8.3 | 7.5 | −0.8 |
| IV-S | 416.11− | 8.3 | 8.1 | −0.2 |
| III-S | 247.72− | 8.3 | 8.2 | −0.1 |
| II-S | 247.72− | 8.3 | 8.3 | 0.0 |
| I-S | 191.53− | 8.3 | 8.3 | 0.0 |
| I-H | 247.72− | 8.3 | 8.8 | 0.5 |
| II-H | 416.11− | 8.3 | 8.4 | 0.1 |
| III-H | 416.11− | 8.3 | 9.2 | 0.9 |
| IV-H | 336.31− | 8.3 | 7.2 | −1.1 |
The purification and compositional analysis method was validated using serum matrix spiked with a mock mixture of 10 μM each of 12 heparin/HS-derived disaccharide standards.
Analysis of Two Commercial Human Sera
The method was then applied to analyze the composition of disaccharides extracted from two commercially available human sera - human serum from platelet poor human plasma (platelet poor) and from male AB human plasma (male AB). Three aliquots from each of the human sera were fractionated, digested, purified using SPE and analyzed by LC-MS/MS. To ensure the accuracy of the compositional analysis, the disaccharide standards used for R-factor determination were purified and analyzed on the same day employing the same tuning profile. In order to ensure accuracy of extraction from human sera, and also to guarantee that no bias occurs during extraction, we also performed a control experiment in which heparan sulfate was doped into human serum. The data for this experiment is provided in Supplementary Material Table S-1. These data clearly show that our extraction method and efficiency is both accurate and precise.2 Table 3 lists the calculated compositions of each of the 12 disaccharides averaged from three preparations (n=3) and their standard deviations. These two sera have several abundant disaccharides in common (i.e. IV-A, III-A, II-A, IV-S, III-S, and II-S). The major differences between these two sera were observed in the compositions of I-S and I-A disaccharides, which were much higher in the platelet poor sample. While the remaining disaccharides IV-A, IV-S, II-A, III-A, II-S, III-S and I-S were observed to be in good agreement with previous reports1,2,35,44,47. However, some low-abundance disaccharides, which have not been observed in the earlier reports such as I-A were also detected and quantified using this method.
Table 3.
The comprehensive compositional analysis of the 12 heparin/HS-derived disaccharides extracted from human sera from platelet poor human plasma (n=3) and human serum from male AB plasma (n=3).
| m/z | Platelet Poora(%) Mean ± SDc | Male ABb(%) Mean ± SDc | |
|---|---|---|---|
| IV-A | 378.11− | 37.2 ± 0.2 | 52.9 ± 1.7 |
| III-A | 458.11− | 1.5 ± 0.5 | 1.3 ± 0.1 |
| II-A | 458.11− | 14.3 ± 1.0 | 11.2 ± 1.4 |
| I-A | 268.72− | 3.2 ± 0.5 | 0.3 ± 0.1 |
| IV-S | 416.11− | 14.6 ± 2.0 | 12.2 ± 1.5 |
| III-S | 247.72− | 8.6 ± 0.4 | 10.5 ± 2.1 |
| II-S | 247.72− | 12.1 ± 2.1 | 8.0 ± 0.4 |
| I-S | 191.53− | 5.9 ± 0.6 | 0.5 ± 0.2 |
| I-H | 247.72− | 0.8 ± 0.1 | 2.0 ± 0.4 |
| II-H | 416.11− | 0.6 ± 0.2 | 0.0 ± 0.0 |
| III-H | 416.11− | 0.7 ± 0.2 | 0.5 ± 0.2 |
| IV-H | 336.31− | 0.4 ± 0.1 | 0.7 ± 0.1 |
Platelet poor, human serum from platelet poor human plasma
Male AB, human serum from male AB plasma
SD, standard deviation of the mean
Analysis of Donor Sera
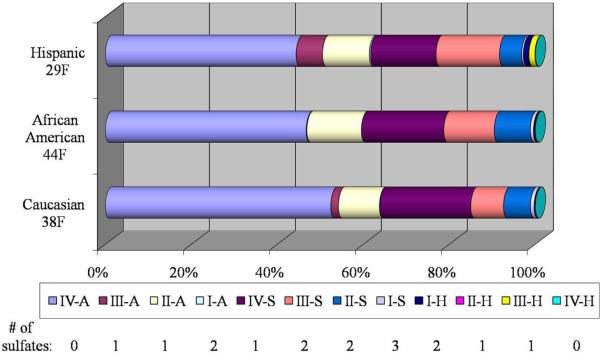
The human serum samples were from three female single donors - Hispanic (29 years old), African American (44 years old) and Caucasian (38 years old). These three human sera were plotted according to the compositions of the 12 heparin/HS-derived disaccharides as shown in Figure 4. The results showed that the most abundant disaccharides were IV-A, II-A, IV-S, III-S and II-S. As illustrated in Figure 4, the three sera show dramatic differences in the compositions of disaccharides, such as III-A and I-H. These differences may be due to various factors which cannot be determined at this point. However, these data clearly indicate that our method is sensitive to variations in disaccharide profiles from human sera and has a great potential in probing for the changes of HS compositions with various factors such as ethnicities and backgrounds.
Figure 4.
Bar graph of the compositions of the 12 heparin/HS-derived disaccharides from three single donor sera, 38 years old Caucasian female (Caucasian 38F), 44 years old African American female (African American 44F) and 29 years old Hispanic female (Hispanic 29F). The 12 disaccharides are shown in different colors as legend list below and the length of each color represents the percentage composition of the represented disaccharide. The number of sulfate groups in each 12 disaccharide is listed under the color legend accordingly.
CONCLUSIONS
An analytical method for the compositional analysis of the 12 heparin/HS-derived disaccharides extracted from human serum was developed, validated, and applied in analyzing two pooled human sera and three single donor sera. The method developed consists of a combination of techniques including fractionation, exhaustive digestion, purification and LC-MS/MS, showing low LOD and LOQ down to picomole level with a good accuracy and precision. Furthermore, the effects from biological matrix can be reduced significantly. The comprehensive compositional analysis of human sera from various single donors suggests that this protocol can be applied to probe for the differences in the compositions of heparin/HS-derived disaccharides in human serum from patients, which has significant potential in biomarker discovery, disease diagnosis and prognosis.
Supplementary Material
ACKNOWLEDGMENTS
Financial support for this research is provided by the National Institutes of Health (Grant No. GM 047356).
REFERENCES
- (1).Saad OM, Leary JA. Anal. Chem. 2003;75:2985–2995. doi: 10.1021/ac0340455. [DOI] [PubMed] [Google Scholar]
- (2).Saad OM, Ebel H, Uchimura K, Rosen SD, Bertozzi CR, Leary JA. Glycobiology. 2005;15:818–826. doi: 10.1093/glycob/cwi064. [DOI] [PubMed] [Google Scholar]
- (3).Meissen JK, Sweeney MD, Girardi M, Lawrence R, Esko JD, Leary JA. J. Am. Soc. Mass Spectrom. 2009;20:652–657. doi: 10.1016/j.jasms.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Lawrence R, Lu H, Rosenberg RD, Esko JD, Zhang L. Nat. Methods. 2008;5:291–292. doi: 10.1038/nmeth0408-291. [DOI] [PubMed] [Google Scholar]
- (5).Varki A, Cummings RD, Esko JD. Essentials of Glycobiology. 2nd ed. Vol. 16. Cold Spring Harbor Laboratory Press; New York: 2008. [PubMed] [Google Scholar]
- (6).Lortat-Jacob H, Grosdidier A, Imberty A. Proc. Natl. Acad. Sci. U.S.A. 2001;99:1299–1234. doi: 10.1073/pnas.032497699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Gandhi NS, Mancera RL. Chem. Biol. Drug Des. 2008;72:455–482. doi: 10.1111/j.1747-0285.2008.00741.x. [DOI] [PubMed] [Google Scholar]
- (8).Sasisekharan R, Raman R, Prabhakar V. Annu. Rev. Biomed. Eng. 2006;8:181–231. doi: 10.1146/annurev.bioeng.8.061505.095745. [DOI] [PubMed] [Google Scholar]
- (9).Lindahl U, Kusche-Gullberg M, Kjelle L. J. Biol. Chem. 1998;273:24979–24982. doi: 10.1074/jbc.273.39.24979. [DOI] [PubMed] [Google Scholar]
- (10).Nakato H, Kimata K. Biochim. Biophys. Acta. 2002;1573:312–318. doi: 10.1016/s0304-4165(02)00398-7. [DOI] [PubMed] [Google Scholar]
- (11).Gesslbauer B, Rek A, Falsone F, Rajkovic E, Kungl AJ. Proteomics. 2007;7:2870–2880. doi: 10.1002/pmic.200700176. [DOI] [PubMed] [Google Scholar]
- (12).Bengtsson J, Eriksson I, Kjelle L. Biochemistry. 2003;42:2110–2115. doi: 10.1021/bi026928g. [DOI] [PubMed] [Google Scholar]
- (13).Sasisekharan R, Shriver Z, Venkataraman G, Narayanasami U. Nat. Rev. Cancer. 2002;2:521–528. doi: 10.1038/nrc842. [DOI] [PubMed] [Google Scholar]
- (14).Wilson PJ, Morris CP, Anson DS, Occhiodoro T, Bielicki J, Clements PR, Hopwood JJ. Proc. Natl. Acad. Sci.U.S.A. 1990;87:8531–8535. doi: 10.1073/pnas.87.21.8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Otsuki S, Taniguchi N, Grogan SP, D'Lima D, Kinoshita M, Lotz M. Arthritis Res. Ther. 2008;10:R61. doi: 10.1186/ar2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Carter NM, Ali S, Kirby JA. J. Cell Sci. 2003;116:3591–3600. doi: 10.1242/jcs.00662. [DOI] [PubMed] [Google Scholar]
- (17).Yang Y, MacLeod V, Dai Y, Khotskaya-Sample Y, Shriver Z, Venkataraman G, Sasisekharan R, Naggi A, Torri G, Casu B, Vlodavsky I, Suva LJ, Epstein J, Yaccoby S, S. JD, Jr, Barlogie B, Sanderson RD. Blood. 2007;110:2041–2048. doi: 10.1182/blood-2007-04-082495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Anderson NL, Polanski M, Pieper R, Gatlin T, Tirumalai RS, Conrads TP, Veenstra TD, Adkins JN, Pounds JG, Fagan R, Lobley A. Mol. Cell. Proteomics. 2004;3:311–326. doi: 10.1074/mcp.M300127-MCP200. [DOI] [PubMed] [Google Scholar]
- (19).Adkins JN, Varnum SM, Auberry KJ, Moore RJ, Angell NH, Smith RD, Springer DL, Pounds JG. Mol. Cell. Proteomics. 2002;1:947–955. doi: 10.1074/mcp.m200066-mcp200. [DOI] [PubMed] [Google Scholar]
- (20).Lamari FN, Theocharis AD, Asimakopoulou AP, Malavaki CJ, Karamanos NK. Biomed. Chromatogr. 2006;20:539–550. doi: 10.1002/bmc.669. [DOI] [PubMed] [Google Scholar]
- (21).Staprans I, Felts JM. J. Clin. Invest. 1985;76:1984–1991. doi: 10.1172/JCI112198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Komosinska-Vassev K, Olczyk K, Koźma EM, Olczyk P, Wisowski G, Winsz-Szczotka K. Clin. Chem. Lab. Med. 2005;43:924–929. doi: 10.1515/CCLM.2005.158. [DOI] [PubMed] [Google Scholar]
- (23).Green AA, Hughes WL. Methods Enzymol. 1955;1:67–90. [Google Scholar]
- (24).Ambrosius M, Kleesiek K, Götting C. J. Chromatogr., A. 2008;1201:54–60. doi: 10.1016/j.chroma.2008.06.007. [DOI] [PubMed] [Google Scholar]
- (25).Volpi N, Cusmano M, Venturelli T. Biochim. Biophys. Acta. 1995;1243:49–58. doi: 10.1016/0304-4165(94)00123-f. [DOI] [PubMed] [Google Scholar]
- (26).Hitchcock AM, Yates KE, Shortkroff S, Costello CE, Zaia J. Glycobiology. 2006;17:25–35. doi: 10.1093/glycob/cwl046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Zhang F, Sun P, Muñoz E, Chi L, Sakai S, Toida T, Zhang H, Mousa S, Linhardt RJ. Anal. Biochem. 2006;353:284–286. doi: 10.1016/j.ab.2006.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Ambrosius M, Kleesiek K, Götting C. J. Chromatogr., A. 2008;1201:54–60. doi: 10.1016/j.chroma.2008.06.007. [DOI] [PubMed] [Google Scholar]
- (29).Lamari FN, Militsopoulou M, Mitropoulou TN, Hjerpe A, Karamanos NK. Biomed. Chromatogr. 2002;16:95–102. doi: 10.1002/bmc.144. [DOI] [PubMed] [Google Scholar]
- (30).Ziegler A, Zaia J. J. Chromatogr., B: Anal. Technol. Biomed. Life Sci. 2006;837:76–86. doi: 10.1016/j.jchromb.2006.04.013. [DOI] [PubMed] [Google Scholar]
- (31).Lawrence R, Olson SK, Steele RE, Wang L, Warrior R, Cummings RD, Esko JD. The J. Biol. Chem. 2008;283:33674–33684. doi: 10.1074/jbc.M804288200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Turnbull JE. Methods Mol. Biol. 2001;171:129–139. doi: 10.1385/1-59259-209-0:129. [DOI] [PubMed] [Google Scholar]
- (33).Tirumalai RS, Chan KC, Prieto DA, Issaq HJ, Conrads TP, Veenstra TD. Mol. Cell. Proteomics. 2003;2:1096–1103. doi: 10.1074/mcp.M300031-MCP200. [DOI] [PubMed] [Google Scholar]
- (34).Komosińska-Vassev K, Olczyk K, Koźma EM, Olczyk P, Wisowski G, Winsz-Szczotka K. Clin. Chem. Lab. Med. 2005;43:924–929. doi: 10.1515/CCLM.2005.158. [DOI] [PubMed] [Google Scholar]
- (35).Lu H, Mcdowell LM, Studelska DR, Zhang L. Glycobiology. 2010;2:13–28. [PMC free article] [PubMed] [Google Scholar]
- (36).Aitken A. Proteomics Protoc. Handb. 2005:307–309. [Google Scholar]
- (37).Packer NH, Lawson MA, Jardine DR, Redmond JW. Glycoconjugate J. 1998;15:737–747. doi: 10.1023/a:1006983125913. [DOI] [PubMed] [Google Scholar]
- (38).Ruhaak LR, Deelder AM, Wuhrer M. Anal. Bioanal. Chem. 2009;394:163–174. doi: 10.1007/s00216-009-2664-5. [DOI] [PubMed] [Google Scholar]
- (39).Estrella RP, Whitelock JM, Packer NH, Karlsson NG. Anal. Chem. 2007;79:3597–3606. doi: 10.1021/ac0622227. [DOI] [PubMed] [Google Scholar]
- (40).Leiserowitz GS, Lebrilla C, Miyamoto S, An HJ, Duong H, Kirmiz C, Li B, Liu H, Lam KS. Int. J. Gynecol. Cance. 2008;18:470–475. doi: 10.1111/j.1525-1438.2007.01028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Pereira L. J. Liq. Chromatogr. Relat. Technol. 2008;31:1687–1731. [Google Scholar]
- (42).Costello CE. Glycoconjugate J. 1988;5:397–409. [Google Scholar]
- (43).Rolim A, Oishi T, Maciel CPM, Zague V, Pinto CASO, Kaneko TM, Consiglieri VO, Velasco MVR. Int. J. Pharm. 2006;308:107–114. doi: 10.1016/j.ijpharm.2005.10.031. [DOI] [PubMed] [Google Scholar]
- (44).Oguma T, Tomatsu S, Montano AM, Okazaki O. Anal. Biochem. 2007;368:79–86. doi: 10.1016/j.ab.2007.05.016. [DOI] [PubMed] [Google Scholar]
- (45).Karlsson NG, Schulz BL, Packer NH, Whitelock JM. J. Chromatogr., B. 2005;824:139–147. doi: 10.1016/j.jchromb.2005.07.014. [DOI] [PubMed] [Google Scholar]
- (46).Upreti VV, Khurana M, Cox DS, Eddington ND. J. Chromatogr., B. 2006;831:156–162. doi: 10.1016/j.jchromb.2005.11.047. [DOI] [PubMed] [Google Scholar]
- (47).Shi X, Zaia J. J. Biol. Chem. 2009;284:11806–11814. doi: 10.1074/jbc.M809637200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.