Abstract
Osteoblasts are controlled by the individual and combined effects of systemic and local growth regulators. Here we show functional and physical interactions between estradiol (17βE) and Wnt activated pathways in osteoblasts. 17βE increased gene promoter activity by the Wnt pathway transcriptional effector T cell factor (TCF) in an estrogen receptor (ER) dependent way. This occurred independently of its activity through traditional estrogen response elements and was not replicated by androgen receptor activation. 17βE also increased the stimulatory effect of LiCl on TCF activity, LiCl increased the stimulatory effect of 17βE through estrogen response elements, and both were further enhanced by a noncanonical Wnt receptor agonist (WAg) that functions independently of β-catenin stabilization. In contrast to LiCl, WAg increased DNA synthesis and reduced relative collagen synthesis and alkaline phosphatase activity in otherwise untreated or 17βE stimulated cells. In addition, WAg suppressed Runx2, osterix, and alkaline phosphatase mRNA levels, and potently induced osteoprotegerin mRNA, whereas LiCl was ineffective alone and inhibitory in combination with 17βE. A definitive intersection between the 17βE and Wnt pathways occurred at the protein level, where ERα physically associated with TCF-4 independently of its β-catenin binding domain. This interaction required ligand dependent exposure of a TCF binding region that mapped to ERα domain E and was further enhanced by Wnt pathway activation. Our studies reveal highly focused co-regulatory effects between the 17βE and Wnt pathways in osteoblasts that involve activated ERα and TCF-4, and downstream changes in gene expression, osteoblast proliferation, and differentiated cell function.
Keywords: 17βE, estrogen receptor, TCF-4, Runx2, protein interactions, noncanonical effects
1. Introduction
Sex steroids elicit their effects through specific intracellular receptors that drive changes in gene expression in direct and indirect ways. Importantly, the latter are thought to integrate combinatorial events with other regulatory systems, accounting in part for their tissue restricted diversity (Barkhem et al., 2004; Centrella et al., 2004a; Centrella et al., 2004b; Manolagas et al., 2004; Syed et al., 2005; Boonyaratanakornkit and Edwards, 2007). The skeleton is a key sex steroid sensitive tissue. As such, bone loss accelerates when sex steroids levels decline in the elderly or after sex organ ablation (Windahl et al., 2002; Rosen, 2003; McClung, 2005; Sorensen et al., 2007; Windahl et al., 2007). In these situations, an imbalance in bone remodeling occurs in response to a decrease in the otherwise normal constraints on bone resorption imposed by endogenous sex steroids (Ray et al., 1994; Stein and Yang, 1995; McCarthy et al., 1997; Suda et al., 1997; Jilka et al., 2007) and can be restored by sex hormone replacement therapy. Several of the indirect sex steroid sensitive events which are now thought to control bone cell activity include physical and functional interactions between hormone-activated estrogen receptor α (ERα) and the CCAAT enhancer binding protein (C/EBP) and Runx2 transcription factors in osteoblasts. These interactions can limit access of ERα to DNA containing consensus estrogen response elements (ERE) due to an association between the DNA binding domain (DBD) of ERα with C/EBPδ or Runx2, severely limiting direct transcriptional effects by ERα modulating compounds. At the same time, expression of certain C/EBP sensitive target genes including insulin-like growth factor (IGF)-I are also suppressed, whereas certain Runx2 sensitive genes including TGF-β type I receptor are enhanced (McCarthy et al., 1997; McCarthy, 2003; Centrella et al., 2004a; Chang et al., 2005). As a result, these interactions may help to define local growth factor actions in bone in highly focused ways.
In addition to IGF-I and TGF-β, another growth factor system essential for bone growth and remodeling is driven by the several Wnt ligands. One of the better appreciated events induced by Wnt pathway activation is inhibition of GSK3 activity. This results in β-catenin stabilization, its nuclear accumulation, and its stimulatory effect on transcription by members of the lymphoid enhancing factor/T cell factor (TCF) gene family (Arce et al., 2006; Cadigan, 2008a; Cadigan, 2008b; MacDonald et al., 2009). Indeed, expression of canonical Wnt ligands and β-catenin stabilization appear essential for some aspects of Wnt pathway activation in osteoblasts. Still, other Wnt dependent events may require only select Wnt ligands or transcriptional components, driving separate downstream events (Bennett et al., 2005; Gaur et al., 2005; Kulkarni et al., 2005; Tobimatsu et al., 2006; Bergenstock and Partridge, 2007; Chen et al., 2007). In these contexts, osteoblasts express several Wnts that act in complex ways during early and late stages of skeletal formation (Kato et al., 2002; Hoang et al., 2004; Zhang et al., 2004). Transgenic expression or mutation of genes encoding a subset of Wnt ligands may re-direct precursor cells between the osteoblast and adipocyte lineages, regulate the expression of genes associated with osteoblast function, and favor early aspects of osteogenesis (Gong et al., 2001; Bennett et al., 2005; Glass et al., 2005; Takada et al., 2007). Accordingly, targeted deletion of Wnt co-receptor genes LRP-6 during development and LRP-5 post partum are linked with osteopenia and osteoporosis. Moreover, transgenic animals with aberrant expression of native Wnt antagonists also exhibit disrupted skeletal integrity. This too occurs in complex and differentiation dependent ways, suggesting that, despite a clear relationship with bone formation, the Wnt system is naturally limited in highly differentiated osteoblasts (Kato et al., 2002; Vaes et al., 2002; Holmen et al., 2004; van Bezooijen et al., 2004; Glass et al., 2005; Vaes et al., 2005; Mani et al., 2007; Yadav et al., 2008).
Recently we reported that osteoblasts develop independent and co-dependent Wnt pathway responses which relate to their state of differentiation and to their expression of Runx2 (McCarthy and Centrella, 2010). These studies revealed that individual aspects of Wnt pathway activation can drive canonical effects that parallel β-catenin stabilization, and some noncanonical effects through Runx2 activation and its complex formation with TCF-4 in a β-catenin independent way. Given earlier evidence for interactions between ERα and Runx2 (McCarthy, 2003; Juttner and Perry, 2007; McCarthy et al., 2007; Khalid et al., 2008; McCarthy et al., 2008), more recent evidence for interactions between Runx2 and TCF-4 (McCarthy and Centrella, 2010), and the possibility that some ERα modulators might develop their effects in part through Wnt signaling (El-Tanani et al., 2001; Kouzmenko et al., 2004; Kousteni et al., 2007; Sunters et al., 2010; Varea et al., 2010), we asked whether the ERα and Wnt pathways might intersect through any of these components in differentiating osteoblasts. Our results support clear connections among these systems and now reveal novel cross-control between the ERα and Wnt pathways in osteoblasts, involving direct interactions between ERα and TCF transcription factors.
2. Materials and Methods
2.1 Cells
Primary osteoblast-enriched cultures were isolated from parietal bones of 22 day old Sprague-Dawley rat fetuses (Charles River Laboratories), as approved by Yale Institutional Animal Care and Use Committee. Sutures were dissected and cells were released by 5 sequential collagenase digestions. Cells from the last 3 digestions express features of differentiating osteoblasts, including high levels of transcription factor Runx2, parathyroid hormone receptor, type I collagen synthesis, and alkaline phosphatase. They also increase osteocalcin expression in response to dihydroxyvitamin D3, display differential sensitivity to TGF-β, BMP-2, and various prostaglandins (PGs), and form mineralized nodules under conditions that promote long term differentiation in vitro. (Centrella et al., 1987; McCarthy et al., 1988; Centrella et al., 1994; Centrella et al., 1995; Centrella et al., 1996; Ji et al., 1997; Carpenter et al., 1998; Ji et al., 1998). Cells were plated at 4,000/cm2 in Dulbecco’s modified Eagle’s medium supplemented with 100 μg/ml ascorbic acid, nonessential amino acids, antibiotics, and 10% fetal bovine serum, and grown for at least 6 days before transfection or treatments.
2.2 Plasmids
Wnt dependent gene expression was assessed with a firefly luciferase reporter plasmid TOP-Flash containing four TCFRE. ERα dependent gene expression was assessed with a luciferase reporter plasmid driven by an ERE from the frog vitellogenin promoter cloned upstream of a minimal prolactin gene promoter, or a 0.94 kb fragment of the rat oxytocin gene promoter (NCBI reference sequence NC_005102.2) that contains multiple TCFRE and ERE, as revealed by MatInspector (http://www.genomatix.de) analysis. AR dependent gene expression was assessed with luciferase reporter plasmid driven by four consensus ARE cloned upstream of a minimal RSV promoter. Cells were co-transfected with expression plasmids encoding native or mutated ERα, ERβ, or AR. Runx2 transcriptional activation was assessed with expression plasmids encoding full length Runx2 fused to a GAL4 promoter DBD within vector M (M-Runx2), and luciferase reporter plasmid containing five GAL4 REs (5XGAL4) (McCarthy et al., 2000b; McCarthy, 2003; Centrella et al., 2004b; McCarthy et al., 2007). Runx dependent gene expression was assessed with luciferase reporter plasmid SXN1C containing two copies of a Runx RE from the rat TβRI gene promoter cloned into reporter plasmid pGL3-Promoter (Ji et al., 1998; Zawel et al., 1998; Ji et al., 2001; McCarthy, 2003). Endogenous Runx2 levels were reduced with an expression plasmid containing restriction fragments of mouse Runx2 cloned in reversed orientation within vector pSV7d (McCarthy et al., 2000b; Ji et al., 2001; McCarthy, 2003; McCarthy et al., 2007). The ERα DNA-binding domain mutant ERα203/204μ was engineered by site-directed mutagenesis (GeneTailor, Invitrogen) of wild type coding sequence to produce two substitutions in the first zinc finger at residues E203A and G204A, analogous to another well characterized ERα mutant protein (Jakacka et al., 2001). Local Wnt expression was induced with expression plasmids encoding prototypical Wnt1 (McCarthy and Centrella, 2010). Physical interactions between ERα and TCF4 were assessed by co-transfection with expression plasmids encoding M-ERα or its fragments (Chang et al., 2005), a β-catenin independent derivative of TCF4 (Liu et al., 2005) fused to the transactivation domain of Herpes virus protein 16 (VP16) within vector MVN (MVN-TFCt) (McCarthy and Centrella, 2010), and luciferase reporter plasmid 5XGAL4. Functional interactions between ERα and TCF4 were further assessed with expression plasmids encoding untagged proteins and 17βE or Wnt sensitive reporter plasmids.
2.3 Transfections and treatments
Reporter plasmids, expression plasmids, and empty vector plasmids were pre-titrated for optimal expression efficiency and transfected with reagent TransIT LT1 (Mirus). Cell cultures at least 70% confluent (~35,000 cells per cm2) were exposed to 25 ng per cm2 of expression plasmid DNA and/or 37.5 to 75 ng per cm2 of reporter plasmid DNA for 16 h or longer in medium containing 4% charcoal stripped fetal bovine serum. Cells were then treated as indicated in each figure in serum free and phenol red free medium. In studies to determine steroid activity, untreated cells were supplemented with equivalent dilution of ethanol vehicle. In studies to determine Wnt pathway activation, cells not treated with LiCl were supplemented with an equivalent of NaCl, and cells not treated with WAg (Calbiochem) or the GSK3 inhibitor SB 415286 (Sigma) were supplemented with an of equivalent DMSO vehicle. Cells were lysed and extracts were analyzed by comparison to positive and negative control plasmids, and where appropriate, corrected for differences in total protein or 18s rRNA content (McCarthy et al., 2000b; Ji et al., 2001; McCarthy, 2003; McCarthy et al., 2007).
2.4 Western blots
After treatments culture medium was removed, and cell layers were washed three times with buffer. Cells were collected by scraping, lysed in 1% Triton X-100 supplemented with protease, phosphatase, and kinase inhibitors, and nuclei were collected by centrifugation. Equal total protein levels from medium, nuclear, and cytoplasmic extracts were fractionated by electrophoresis on SDS 7.5-12% polyacrylamide gradient gels, and blotted onto polyvinylidene fluoride membranes (NEN Life Sciences) along with pre-stained molecular weight markers. Blots were blocked in 5% fat-free powdered milk, probed with a 1:1000 or 1:3000 dilution of primary antibody (Santa Cruz Biotechnologies or BD Transduction Laboratories). Reactive bands were visualized with a 1:2,500 dilution of species specific anti-IgG linked to horseradish peroxidase (Jackson Laboratories, Inc.) and chemiluminescence (Western Lightning, PerkinElmer Life Sciences). Inasmuch as protein levels differ in relative abundance, exposure times or image intensities were varied to produce linear stain to background ratios (Kim et al., 2006; McCarthy and Centrella, 2010).
2.5 mRNA analysis
Total RNA was extracted with acid guanidine monothiocyanate, precipitated with isopropyl alcohol, dissolved in sterile water (Centrella et al., 1995), and purified further using the RNeasy™ Plus Mini Kit (Qiagen). RNA was reverse transcribed with the iScript™ cDNA Synthesis Kit and products were amplified and quantified using specific primers (eurofins mwg/operon; supplemental Table 1) and the iQ™ SYBR® Green Supermix in a CFX96 Real Time polymerase chain reaction (RT-PCR) detection system (Bio-Rad Laboratories, Inc.).
2.6 DNA synthesis
Cells were incubated with 5 μCi/ml [methyl-3H]thymidine (80 Ci/mmol) for the last two hours of culture. Cells were lysed in 0.1 M sodium dodecyl sulfate and 0.1 M sodium hydroxide, collecting the precipitate formed with 10% trichloroacetic acid, and scintillation counting (Centrella et al., 1987; Centrella et al., 1995).
2.7 Collagen synthesis
Cells were incubated with 5 μCi/ml, mole) for the last two hours of culture. Cell layers were lysed by freeze thawing and extracted with 0.5% Triton X-100. Samples were precipitated with cold 10% trichloroacetic acid, and acid insoluble material was collected by centrifugation. Precipitates were extracted with acetone, dried, and rehydrated. [3H-2,3]proline incorporated into collagenase digestible protein (collagen) or noncollagen proteins was determined by differential sensitivity to bacterial collagenase free of nonspecific protease activity. Percent collagen synthesis was calculated by correcting for the 5.4-fold enrichment of proline content in collagen (Centrella et al., 1987; Centrella et al., 1995).
2.8 Alkaline phosphatase activity
Cell layers were lysed by freeze thawing and extracted with 0.5% Triton X-100. Enzyme specific activity was assessed in cell extracts by hydrolysis of p-nitrophenol phosphate, measured at 410 nm after 30 minutes of incubation at 37°C. Data are expressed as pmol of p-nitrophenol released per minute per μg protein (Centrella et al., 1987; Centrella et al., 1995).
2.9 Statistical analysis
Differences were assessed by one way analysis of variance with post hoc analysis when comparing multiple treatments simultaneously, using SigmaStat software (Jandel Corporation), which defaults to Student’s t test when assessing single treatments, from data derived from nine or more replicate samples and three or more independent cell preparations. Western blots were from at least two studies and mRNA levels were from three or more studies. A significant difference was assumed by a P value of <0.05.
3. Results
3.1 Estrogen dependent activation of the Wnt and ERα pathways in osteoblasts
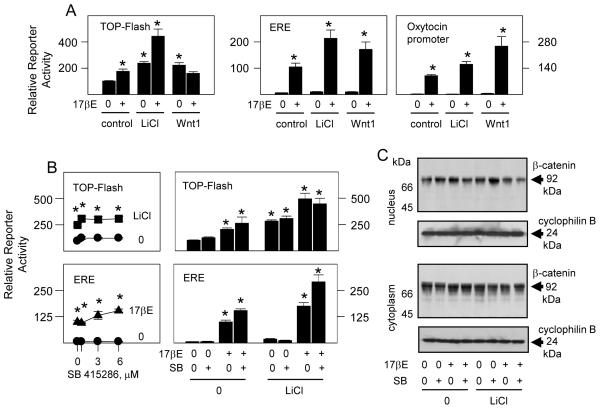
Previous studies in mouse pre-osteoblast cultures showed that the stimulatory effect of LiCl through TCF response elements (TCFRE) with TOP-Flash reporter, a well defined marker system of Wnt pathway induction, is mimicked by some steroid-like compounds including 4-estren-3α,17β-diol (estren), but not by estradiol (17βE) (Kousteni et al., 2007). LiCl, which also readily induces TCF activity in primary cultures of differentiating rat osteoblasts (McCarthy and Centrella, 2010), is similarly active without or with ERα or androgen receptor (AR) expression (Fig. 1A). However, unlike its relative lack of effect in mouse pre-osteoblasts (Kousteni et al., 2007) or in rat osteosarcoma derived cells (Armstrong et al., 2007), 17βE at 10 nM induced significant ERα dependent TCF activity in normal differentiating rat osteoblasts. By contrast, estren, even at 100 nM, or the AR agonist dihydrotestosterone (DHT) at 10 nM, which each fully activate gene expression through AR sensitive RE (ARE) (Centrella et al., 2004b), did not increase TCF activity. Neither LiCl nor 17βE significantly increased gene promoter activity when the TCFRE sites were mutated (FOP-Flash) (Fig. 1B). The amount of 17βE needed to activate TCFRE by comparison to conventional ERE differed by at least two orders of magnitude (Fig. 1C), predicting different mechanisms of action. In agreement with this, the stimulatory effects of 17βE through TCFRE persisted in cells that express ERα with DBD point mutations that severely limit gene activation through ERE (Fig. 1D), whereas estren remained ineffective through either TCFRE or ERE. These findings confirm that TCF activation, in this case by 17βE, does not derive from a direct ERα dependent genomic effect through ERE (Kousteni et al., 2007). Low affinity ligands that bind and partially activate ERα such raloxifene, tamoxifen, and 17α-estradiol, or other agents with estrogen-like effects such as resveratrol, did not induce significant TCF activation. However, the ERα ligand ICI 182780, which at high concentrations can antagonize the stimulatory effect of 17βE in osteoblasts, potently activated TCF dependent gene promoter activity in these cells (left panels, Fig. 1E). Whereas the stimulatory effect of 17βE through ERE was, as expected, suppressed by ICI 182780 (Centrella et al., 2004b), TCF dependent gene expression remained high with both ligands (right panels, Fig. 1E). Thus, TCF can be activated by the Wnt pathway as well as by activated ERα in both pre-osteoblasts and differentiated osteoblasts, albeit with ligand selectivity.
Fig. 1.
Estrogen dependent activation of the Wnt and ERα pathways in osteoblasts. Fetal rat osteoblasts were transfected for 24 hours to express reporter plasmids TOP-Flash with native TCF RE, FOP-Flash with mutated TCF RE, or ERE or ARE driven reporter plasmids in combination with empty vector (no receptor), native ERα or AR, or ERα mutated in its DBD (ERα203/204μ). The cells were treated with vehicle (0), 20 mM LiCl, 0.1 to 10 nM of 17βE, 10 nM of DHT, or 100 nM of estren (En), raloxifene (Ral), tamoxifene (Tam), resveratrol (Res), 17αE, or ICI 128780 (ICI) as indicated, and reporter gene enzyme activity was measured after 24 hours. LiCl significantly increased TOP-Flash activity without or with steroid receptor expression (A,D); 17βE at 10 nM significantly increased TOP-Flash activity with native (A,C,E) or mutated ERα (D); 17βE at 0.1 to 10 nM significantly increased ERE driven reporter only with native ERα (A,C,E); DHT and En significantly increased ARE activity only with AR (A); ICI significantly increased TOP-Flash activity alone or in combination with 17βE, and Ral, Tam, and 17αE significantly increased ERE driven reporter activity (E); (* = P <0.05).
3.2 Canonical Wnt pathway activation enhances ERα activity
In addition to their individual effects shown in Fig. 1, 17βE and LiCl co-enhanced gene expression through TCFRE and ERE, as well as through the oxytocin gene promoter which contains intrinsic TCFRE and ERE that are functional in osteoblasts (McCarthy et al., 2008; McCarthy and Centrella, 2010). Consistent with earlier studies (McCarthy and Centrella, 2010), transgenic expression Wnt1, a canonical Wnt receptor ligand (Huelsken and Behrens, 2002), replicated the response to LiCl treatment through TCFRE. With Wnt1 expression, however, the synergistic effect of 17βE was only evident on gene expression through ERE and the oxytocin gene promoter (Fig. 2A). In an attempt to stabilize β-catenin and drive canonical Wnt pathway activation independently of Wnt1 expression (McCarthy and Centrella, 2010), osteoblasts were treated with SB-415286, a potent GSK3 inhibitor (Coghlan et al., 2000; Smith et al., 2001). As much as 6 μM of SB-415286, a concentration that effectively suppresses GSK3 activity (Coghlan et al., 2000), did not by itself induce significant TCF activity (left panel, Fig. 2B) and did not increase the individual or combined responses to 17βE or LiCl (right panel, Fig. 2B). Similar to LiCl and transgenic Wnt1 expression, however, SB-415286 enhanced the response to 17βE through ERE (lower panels, Fig. 2B). SB-415286 marginally increased nuclear β-catenin levels in untreated osteoblasts, but as with LiCl treatment, this did not persist in 17βE co-treated cells (Fig. 2C). Furthermore, the amounts of nuclear or cytoplasmic GSK3 dependent phospho-β-catenin (at serines 33 and 37 and threonine 41 (Yost et al., 1996); supplemental Fig. 1), could not explain the stimulatory or co-stimulatory effects of 17βE and LiCl. Therefore, the increase in TCF dependent gene expression in response to 17βE did not invariably correlate with changes in nuclear β-catenin and, consistent with evidence in Figs. 1 C and D, did not coincide with the direct effect of 17βE on ERα through ERE.
Fig. 2.
Canonical Wnt pathway activation co-stimulates ERα activity. Fetal rat osteoblasts were transfected for 24 hours to express reporter plasmid TOP-Flash, or ERE driven reporter plasmid, or the oxytocin gene promoter in combination with expression vectors encoding native ERα and Wnt1 (A,B) or with ERα alone (C). The cells were treated with vehicle (0), 10 nM 17βE, 20 mM LiCl, and/or 0.6 to 6 μM of the GSK3 inhibitor SB 415286 (SB) (bottom left panel, B) or 6 μM of SB (bottom right panel, B; C) as indicated, and reporter gene enzyme activity was measured after 24 hours (A,B), or nuclear and cytoplasmic extracts, corrected for total protein content, were probed by Western blot analysis with antibody to β-catenin or cyclophilin B (loading controls) (C). LiCl (A,B) and Wnt 1 expression significantly (A) increased TOP-Flash activity alone, and 17βE significantly increased TOP-Flash activity alone and enhanced the effect of LiCl (A,B). 17βE at 10 nM significantly increased the ERE (A,B) or oxytocin promoter (A) driven reporters alone, and LiCl (A,B) or Wnt1 expression (A) enhanced the effect of 17βE. SB at 6 μM significantly increased the effect of 17βE through ERE in control and LiCl treated cells (B); (* = P <0.05).
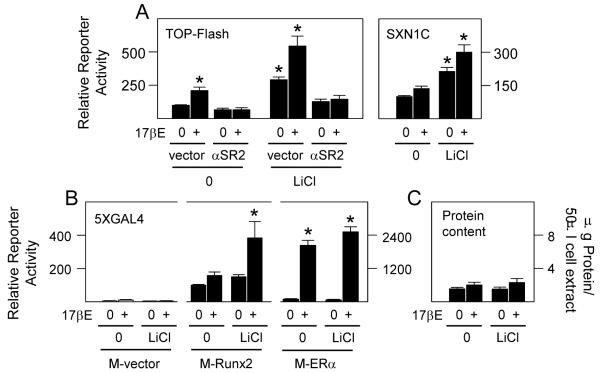
3.3 Influence of Runx2
Previous studies revealed that Wnt pathway dependent activation of TCF is highly limited in differentiating osteoblasts in the absence of endogenous Runx2, which can directly interact with TCF-4 (McCarthy and Centrella, 2010). Similarly, the stimulatory effects of 17βE and LiCl on TCF activity were suppressed when Runx2 levels were silenced (left panel, Fig. 3A). In addition, the gene promoter fragment SXN1C (right panel, Fig. 3A), which is driven by endogenous Runx2 (Chang et al., 1998; Ji et al., 1998; Ji et al., 2001; McCarthy, 2003), and transgenic GAL4-DBD tagged Runx2 (M-Runx2) dependent gene transactivation through GAL4 RE (McCarthy, 2003) (center four lanes, Fig. 3B), were potently co-induced by 17βE and LiCl. These results were unlikely due to nonspecific effects by LiCl on either Runx2, ERα, or reporter protein stability since LiCl alone did not significantly increase basal reporter enzyme activity in empty vector transfected cells (M-vector), through M-Runx2, or through GAL4-DBD tagged ERα (M-ERα) (Fig. 3B), and it did not increase total protein content in control or 17βE induced cultures (Fig. 3C). Thus, whereas the ERα and Wnt pathways coalesce in differentiating osteoblasts without an apparent involvement by β-catenin, in select cases this intersection can occur in a Runx2 restricted way.
Fig. 3.
Runx2 is required for efficient Wnt pathway activation in ERα activated osteoblasts. Fetal rat osteoblasts were transfected for 24 hours to express reporter plasmids TOP-Flash, the Runx sensitive reporter SXN1C, or 5XGAL4 in combination with expression plasmid encoding native ERα (A), vector pSV7d or pSV7d encoding Runx2 in antisense orientation (αSR2) (Ji et al., 2001; McCarthy, 2003; McCarthy and Centrella, 2010), empty vector M driving the expression of GAL4DBD or vector M fused to full length Runx2 (M-Runx2; center panel B) or ERα (M-ERα, right panel B) as indicated, or with expression vector encoding native ERα (C). The cells were treated with vehicle (0), 10 nM 17βE and/or 20 mM LiCl as indicated, and reporter gene enzyme activity was measured after 24 hours. 17βE significantly increased reporter gene expression through TOP-Flash, SXN1C, and 5XGAL4 in combination with M-Runx2 or M-ERα (A,B); LiCl significantly increased reporter gene expression through TOP-Flash, SXN1C, and 5XGAL4 in combination with M-ERα (A,B); 17βE and/or LiCl together significantly co-increased reporter gene expression through TOP-Flash, SXN1C, and 5XGAL4 in combination with M-Runx2 (A,B); αSR2 expression significantly suppressed TOP-Flash activity in 17βE and/or LiCl activated cells (left panel, A); (* = P <0.05).
3.4 Further ERα dependent co-activation in combination with a noncanonical Wnt pathway agonist
We then tested the possibility that 17βE interacted with a noncanonical Wnt pathway, using the synthetic noncanonical agonist (WAg), 2-amino-4-[3,4-(methylenedioxy) benzyl-amino]-6-(3-methoxy phenyl) pyrimidine (Liu et al., 2005; McCarthy and Centrella, 2010). 17βE enhanced the stimulatory effect of WAg through TCFRE in osteoblasts, as well as its synergistic interaction with canonical Wnt signaling by LiCl (upper panel, Fig. 4A). WAg also increased 17βE induced gene expression through ERE by ERα (lower panel, Fig. 4A). This interaction also occurred with ERβ, although basal gene promoter activity was higher with ERβ expression (supplemental Fig. 2). In contrast, activation of AR by DHT failed to enhance TCF activity in LiCl plus WAg activated osteoblasts (upper left panel, Fig. 4B). Moreover, co-treatment with LiCl plus WAg failed to induce AR activity through ARE, although together these agents increased the stimulatory effect of DHT (lower left panel, Fig. 4B). This effect was modest, however, relative to ERα activation (lower right panel, Fig. 4B). Notably, the stimulatory effect of LiCl plus WAg on TCF activity in the absence of sex steroid treatment was attenuated in osteoblasts that express AR by comparison to those that express ERα (upper panels, Fig. 4B), revealing that AR can arrest TCF activity and may thus distinguish some estrogen and androgen dependent effects. Consistent with earlier evidence (McCarthy and Centrella, 2010), nuclear β-catenin levels increased in response to LiCl but appeared largely unchanged in WAg activated osteoblasts without or with LiCl co-treatment (Fig. 4C). Similarly, the relative increase in nuclear β-catenin that occurred with 17βE treatment (here and Fig. 2C) did not persist in Wnt pathway co-activated cells where their co-stimulatory effects on gene expression were maximal (left panels, Fig. 4D). By contrast to β-catenin, TCF-4 is predominantly nuclear in osteoblasts, but it is also relatively enriched in 17βE treated cells (center panels, Fig. 4D). Whereas Wnt pathway activation increased ERα dependent gene expression, it did not significantly enhance ERα stability or its relative distribution between the nuclei and the cytoplasm (right panels, Fig. 4D). There was also an apparent decline in nuclear TCF-4 and ERα in response to Wnt pathway activation, which may relate in part to correction for a 1.65-fold enrichment in total nuclear protein as a result of treatment. Even so, 17βE also enhanced the relative amounts of nuclear TCF-4 and ERα in Wnt pathway activated osteoblasts. Therefore, 17βE may promote the assembly of these and other relevant proteins in the nucleus, and contribute to in part to the potent stimulatory effects that occur through TCFRE and ERE when the Wnt and ERα pathways are co-activated.
Fig. 4.
Further ERα dependent co-activation in combination with a noncanonical Wnt pathway agonist. Fetal rat osteoblasts were transfected for 24 hours to express reporter plasmids TOP-Flash, or ERE or ARE driven reporter plasmids in combination with expression plasmids encoding native ERα, ERβ, or AR (A,B), or native ERα alone (C,D). The cells were treated with vehicle (0), 20 mM LiCl and/or 10 μM WAg alone or in combination with 10 nM of 17βE or 10 nM of DHT as indicated, and reporter gene enzyme activity was measured after 24 hours (A,B), or nuclear and cytoplasmic extracts, corrected for total protein content, were probed by Western blot analysis with antibody to β-catenin (C,D), TCF-4, ERα (D), or cyclophilin B (loading controls) (C,D), as shown. LiCl and WAg together significantly increased reporter gene expression through TOP-Flash and ERE by comparison to either agent alone; LiCl plus WAg significantly increased the stimulatory effect of 17βE on TOP-Flash and ERE activity in ERα and ERβ transfected cells (A,B); and LiCl plus WAg significantly increased the stimulatory effect of DHT on ARE activity in AR transfected cells (B); (* = P <0.05).
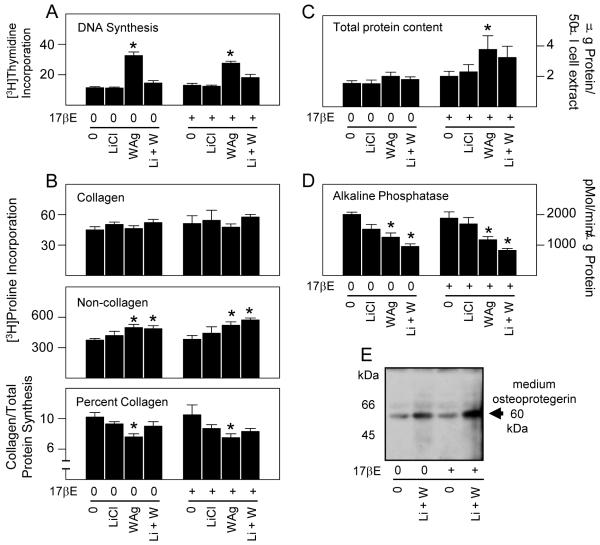
3.5 ERα activation does not modify basal or Wnt dependent effects on osteoblast differentiation
After 24 hours of treatment, LiCl did not directly affect DNA synthesis but suppressed the mitogenic effect of WAg (Fig. 5A). Whereas WAg stimulated DNA synthesis, it did not increase the rate of new collagen synthesis. However, by comparison to its stimulatory effect on noncollagenous protein synthesis, the percent of newly synthesized collagen was suppressed (Fig. 5B). In this short time frame WAg increased total cellular protein content (Fig. 5C), whereas LiCl and WAg, individually and together, reduced alkaline phosphatase specific activity (Fig. 5D). In addition, Wnt pathway activation increased the relative expression of osteoprotegerin, a potent inhibitor of RANK activation and osteoclast activity secreted by osteoblasts into the culture medium (Fig. 5E). Despite these somewhat rapid effects, 17βE did not modify the metabolic response of osteoblasts to Wnt pathway activation in this time frame (Figs. 5A, B, D, E). Similarly, total protein content remained stable or modestly increased in 17βE induced cells (Fig. 5C), consistent with its overall effect on newly synthesized noncollagenous protein. Thus, short term exposure to WAg, which can activate the Wnt pathway in noncanonical ways in differentiating osteoblasts (McCarthy and Centrella, 2010), increases replication and limits some early aspects of differentiation, whereas these same events are relatively insensitive to 17βE in this time frame.
Fig. 5.
ERα activation does not modify the basal or immediate Wnt dependent effects on osteoblast differentiation. Fetal rat osteoblasts were transfected for 24 hours to express native ERα. The cells were treated for 24 hours with vehicle (0), 20 mM LiCl and/or 10 μM WAg alone or in combination with 10 nM of 17βE as indicated. DNA synthesis measured by pulse labeling with [3H]thymidine during the last 2 hours of treatment, and assaying acid insoluble [3H]thymidine incorporation (A). Collagen and noncellagen protein synthesis was measured by pulse labeling with [3H]proline during the last 2 hours of treatment, and assaying acid insoluble [3H]proline incorporation into collagenase digestible and collagenase insensitive cell extracts. Collagen/total protein synthesis was calculated by correcting for the 5.4 fold relative enrichment of proline in collagen (Peterkofsky and Diegelmann, 1971; Centrella et al., 1992) (B). Total protein content was measured with Bradford reagent (Bradford, 1976) (C). Alkaline phosphatase specific activity was measured by p-nitrophenol phosphate hydrolysis and correcting for protein content (Centrella et al., 1995) (D). Osteoprotegerin secretion was determined in conditioned medium extracts (McCarthy et al., 1994), corrected for total protein content, by Western blot (E). WAg significantly increased DNA synthesis rate (A), and suppressed percent collagen synthesis (B) and alkaline phosphatase activity (D) in vehicle or 17βE treated cells, and significantly increased total protein content in 17βE treated cells (C); (* = P <0.05).
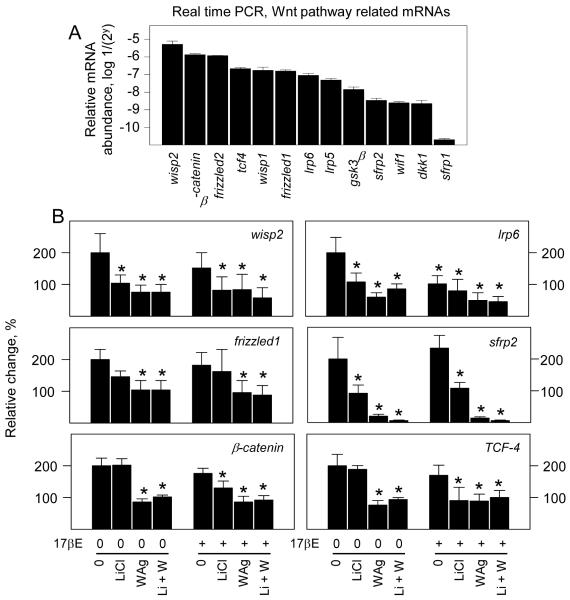
3.6 ERα activation selectively alters canonical Wnt pathway dependent effects on mRNA expression by osteoblasts
To address the possibility that the ERα and Wnt pathways intersected in ways not yet apparent on basal aspects of differentiated osteoblast function, steady state mRNA levels of several genes associated with osteoblast activity and Wnt pathway activation were measured by real time (RT)-PCR. Of the several osteoblast related gene products that we measured, the most abundant were mRNAs encoding the α1 chain of type I collagen (α1A1) and alkaline phosphatase, which differed by 2 orders of magnitude from each other. Other less well expressed but still highly abundant mRNAs were those encoding the bone remodeling regulators osteocalcin and osteoprotegerin, and the osteoblast enriched transcription factors Runx2 and osterix (Fig. 6A). Treatment with vehicle, LiCl, WAg, or 17βE in any combination had no effect on 18s rRNA levels (upper left panel, Fig. 6B) or osteocalcin mRNA (not shown) after 24 hours of exposure, indicating equivalent RNA extraction and recovery, and a lack of generalized or nonspecific stimulatory or inhibitory effects in this time period. Analogous to the relatively high and stable levels of new collagen protein synthesis, mRNA encoding α1A1 differed by 1.5 to 2-fold in response to 17βE or Wnt pathway activators, but no effects were significant. By contrast, and in agreement with its effect on alkaline phosphatase activity, WAg rapidly suppressed alkaline phosphatase mRNA alone and in combination with LiCl. WAg also suppressed mRNA encoding Runx2 and osterix. By comparison, WAg strongly enhanced osteoprotegerin mRNA. WAg therefore rapidly down regulated the mRNA levels of some genes associated with osteoblast differentiation and increased the mRNA encoding a powerful inhibitor of osteoclast-dependent bone resorption. None of these mRNAs was significantly changed by LiCl alone. With the exception of a 40% reduction in alkaline phosphatase mRNA, exposure to 17βE by itself did not significantly affect the levels of these several gene products. However, other than α1A1 chain of type I collagen and osteoprotegerin, which, of all the mRNAs examined, were not suppressed by WAg, co-treatment with 17βE was inhibitory in combination with LiCl (Fig. 6B). Thus, there appears to be separate sets of genes whose expression parallel individual Wnt pathways, some of which are revealed by 17βE treatment.
Fig. 6.
Wnt pathway activation selectively regulates differentiation related mRNA expression by osteoblasts. Fetal rat osteoblasts were transfected for 24 hours to express native ERα. The cells were treated for 24 hours with vehicle (0), 20 mM LiCl and/or 10 μM WAg alone or in combination with 10 nM of 17βE as indicated, and mRNA levels were measured by RT-PCR. *s designate significant differences from untreated cells; (* = P <0.05).
Steady state levels of mRNA encoding a panel of Wnt signaling components were also readily measured. Of these, all but mRNA encoding secreted frizzled related protein 1 (SFRP1), a decoy binding component that among other functions, can limit Wnt signaling through cell membrane receptors (Bovolenta et al., 2008; MacDonald et al., 2009) were highly expressed within three orders of magnitude relative to each other (Fig. 7A) and to several osteoblast related genes (Fig 6A). No significant variations were evident in mRNAs encoding Wnt receptor components Frizzled 2 or LRP5, the Wnt1 inducible signaling pathway protein WISP1, GSK3β, Wnt binding protein WIF1, the Wnt signaling inhibitor Dickkopf1 (DKK1) or SFRP1 within 24 hours of treatment (not shown). Treatment with LiCl suppressed the levels of WISP2, LRP6, and SFRP2 mRNAs, and treatment with WAg suppressed these as well as β-catenin, TCF-4, and Frizzled 1 mRNAs. 17βE alone suppressed LRP6 mRNA and, in combination with LiCl, suppressed β-catenin and TCF-4 mRNAs (Fig. 7B). Therefore, the effect of 17βE on Wnt pathway activation cannot be explained by rapid increases the expression of either Wnt receptors or these several regulatory components of TCF dependent gene expression.
Fig. 7.
ERα activation selectively alters canonical Wnt pathway dependent effects on mRNA expression by osteoblasts. Fetal rat osteoblasts were transfected for 24 hours to express native ERα. The cells were treated for 24 hours with vehicle (0), 20 mM LiCl and/or 10 μM WAg alone or in combination with 10 nM of 17βE as indicated, and mRNA levels were measured by RT-PCR. *s designate significant differences from untreated cells; (* = P <0.05).
3.7 ERα physically associates with TCF-4
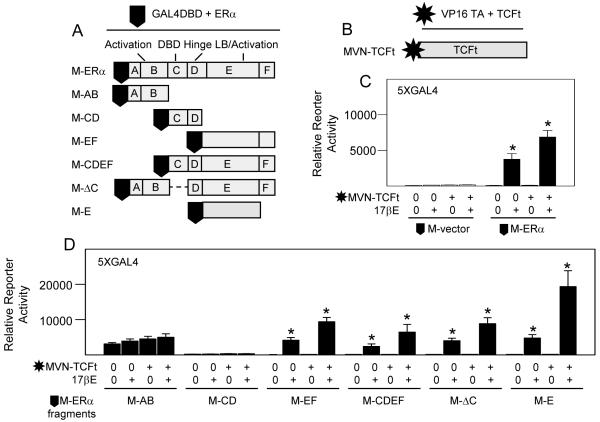
In the absence of evidence supporting a functional role for β-catenin or an increase in the expression of mRNAs encoding several principal regulators of Wnt pathway activation in response to 17βE, we hypothesized that the rapid, potent and bifunctional transcriptional effects that occur by 17βE and Wnt pathway activation might rather derive from molecular interactions between ERα and TCF-4 proteins. This was assessed in osteoblasts by a two-hybrid gene expression assay comprising GAL4-DBD tagged ERα and a large fragment of TCF-4 (TCFt) fused to the herpes virus protein 16 gene transactivation domain (M-ERα and MVN-TCFt, Figs. 8A,B) in combination with reporter plasmid 5XGAL4. The ERα and TCFt fusion constructs included highly efficient nuclear localization sequences to ensure appropriate nuclear accumulation. Importantly, TCFt lacked its amino terminal β-catenin binding domain to test our hypothesis that β-catenin is dispensable for functional interactions between ERα and TCFs. Predictably, empty M-vector (no ERα) generated no gene activation without or with 17βE treatment, either alone or in combination with MVN-TCFt (left four lanes, Fig. 8C). However, co-expression of MVN-TCFt significantly increased M-ERα dependent gene activation in 17βE treated osteoblasts (right four lanes, Fig. 8C), revealing a physical interaction between these proteins that required ligand dependent activation of ERα. To determine possible TCF binding domains on ERα, osteoblasts were transfected with GAL4-DBD tagged ERα truncation fragments which correspond to several of its well defined functional regions, including: its ligand independent gene transactivation domain [M-AB]; its hinge region required for homodimerization with another molecule of activated ERα and its DBD required for binding to ERE [M-CD]; its ligand binding and ligand dependent gene transactivation domains [M-EF]; domain E alone [M-E]; or ERα lacking only its own DBD [M-ΔC] to eliminate possible compromising downstream effects by activation of gene expression through ERE. As with intact M-ERα, all regions except the ligand independent transactivation domain AB were inactive in the absence of 17βE. Notably, MVN-TCFt significantly interacted with all ERα fragments which contained domain E in 17βE treated cells, and highly significant interaction occurred with ERα domain E alone (Fig. 8D).
Fig. 8.
ERα physically associates with TCF-4. Fetal rat osteoblasts were transfected for 24 hours to express reporter plasmid 5XGAL4 in combination with expression plasmids encoding empty vector M driving the expression of GAL4DBD or vector M fused to full length ERα or the ERα fragments indicated (A), and/or empty vector MVN driving the expression of the herpes virus protein 16 transactivation domain or MVN fused to transcription factor TCF-4 lacking its β-catenin binding domain (TCFt) (B). The cells were treated vehicle (0) or 10 nM 17βE and reporter gene enzyme activity was measured after 24 hours. 17βE significantly increased reporter gene expression through 5XGAL4 and MVN-TCTt significantly enhanced ERα dependent gene expression in 17βE treated cells (C). ERα fragment AB significantly enhanced reporter gene expression through 5XGAL4 relative to empty vector M but was not further increased by 17βE or MVN-TCFt co-expression, whereas all ERα fragments retaining domain E were significantly activated by 17βE and further enhanced by MVN-TCFt co-expression (D); (* = P <0.05).
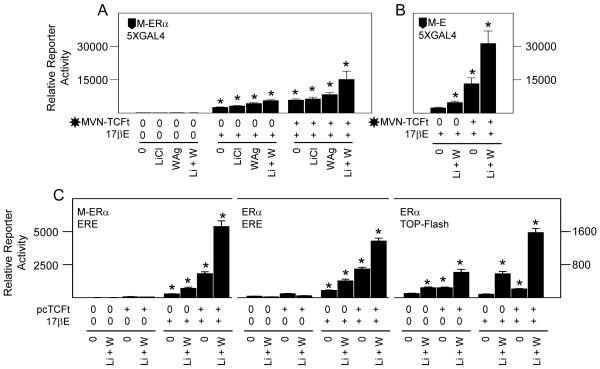
3.8 Wnt pathway activation enhances physical and functional interactions between ERα and TCF-4
In agreement with their combined effects on ligand activated ERα through ERE (Fig. 4A), LiCl plus WAg enhanced M-ERα dependent gene transactivation in 17βE treated cell cultures, and this was further increased by MVN-TCFt expression (Fig. 9A). An analogous and highly potent co-induction also occurred with the M-fusion protein containing ERα domain E alone (Fig. 9B). To eliminate possible contributions by the GAL4 DBD on ERα or by the herpes virus protein 16 gene transactivation domain on TCFt to functional interactions between the parental proteins, the cells were instead transfected with M-ERα and the ERE driven reporter plasmid, or with untagged ERα and either the ERE or TCFRE driven reporter plasmids, alone or in combination with an expression vector encoding untagged, β-catenin independent, TCFt. Regarding ERE, gene expression again required 17βE and was further enhanced by LiCl plus WAg. Moreover, this effect was potently increased by transgenic TCFt expression (left and middle panels, Fig. 9C). Regarding TCFRE, the stimulatory effect of LiCl plus WAg was also enhanced by transgenic TCFt expression, and their combined effect was further increased by 17βE (right panel, Fig. 9C). Therefore, endogenous (Fig. 4) and transgenic (Figs. 8 and 9) Wnt pathway activated transcription factors interact with ERα. A physical association between TCF-4, which is expressed at the mRNA and protein levels by differentiating osteoblasts (McCarthy and Centrella, 2010), with ERα principally involves ERα domain E. This can occur in a β-catenin independent manner in these cells since LiCl by itself has no effect on ERα activation (Figs. 1 and 2) and, consistent with the lack of response to the GSK3 inhibitor SB-415286 (Fig. 2B), functional gene transactivation through TCFRE is potentiated in combination with transgenic TCFt that lacks β-catenin binding potential.
Fig. 9.
Wnt pathway activation enhances physical and functional interactions between ERα and TCF-4. Fetal rat osteoblasts were transfected for 24 hours to express reporter plasmids 5XGAL4, ERE driven reporter plasmid, or TOP-Flash reporter plasmid in combination with expression plasmids encoding vector M driving the expression of GAL4DBD fused to full length ERα (A, left panel C), ERα fragment E (B), vector MVN driving the expression of the herpes virus protein 16 transactivation domain or MVN fused to β-catenin independent TCF-4 (A,B), or β-catenin independent TCF-4 driven by expression plasmid pcDNA3 (pcTCFt) (C). The cells were treated vehicle (0), 20 mM LiCl and/or 10 μM WAg alone or in combination with 10 nM of 17βE as indicated, and reporter gene enzyme activity was measured after 24 hours. 17βE significantly increased reporter gene expression through 5XGAL4 (A,B), ERE, or TOP-Flash (C); LiCl plus WAg significantly increased the effect of 17βE; MVN-TCTt expression significantly enhanced ERα (A) or ERα fragment E (B) dependent gene expression in 17βE treated cells through 5XGAL4 (A,B), and pcTCFt expression significantly enhanced ERα activity in 17βE treated cells through ERE or TOP-Flash reporters (C); (* = P <0.05).
Discussion
Estrogens have clear positive and negative, direct and combinatorial, effects on gene expression and biochemical activity, by which they can integrate the effects of local growth factor systems in osteoblasts (Centrella et al., 2004a). Of these, components of the Wnt growth factor pathway are now considered requisite during earlier and later aspects of bone formation and remodeling (Milat and Ng, 2009). Like the many regulatory systems that modulate bone growth and repair, the Wnt pathway cooperates, coordinates, or antagonizes the response to other local and systemic bone growth regulators (Lian et al., 2006; Bergenstock and Partridge, 2007; Kugimiya et al., 2007; Reinhold and Naski, 2007; Milat and Ng, 2009; Sunters et al., 2010). We recently reported that differentiating osteoblasts react in both β-catenin dependent (so-called canonical) and β-catenin independent noncanonical ways to Wnt pathway restricted activators. Moreover, by comparison to fibroblasts, Wnt pathway activation is highly dependent on the essential osteogenic transcription factor Runx2 (McCarthy and Centrella, 2010). We and others demonstrated that the Wnt pathway targeted transcription factor TCF-4 physically associates with Runx2 (Reinhold and Naski, 2007; McCarthy and Centrella, 2010). We extended this to show that their interaction increases Runx2 activity, which is enhanced by biochemical co-activation of Runx2 and the Wnt pathway independently of β-catenin (McCarthy and Centrella, 2010).
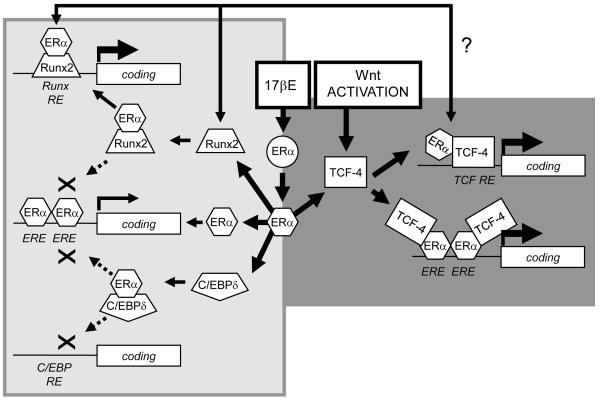
In the present study we further show that the 17βE and Wnt induced pathways are linked in osteoblasts. We find physical and functional interactions between ERα and TCF-4 that require hormone dependent ERα activation. The co-stimulatory effect of 17βE and Wnt pathway activation occurs without preferential nuclear re-localization of ERα, which presides in both the cytoplasm and the nucleus even in the absence of ligand (Htun et al., 1999), or the involvement of β-catenin stabilization or binding. The intersection between the 17βE and Wnt pathways is at least in part cross-inductive, and differs notably from the physical and functional interactions that occur between ERα and either Runx2 or C/EBPδ. In the latter instances, hormone dependent complex formation with ERα enhances Runx2 activity but reduces nuclear C/EBPδ levels and C/EBPδ dependent gene expression. Domain mapping studies reveal that Runx2 and C/EBPδ each associate with the ERα DBD and accordingly suppress ERα dependent gene expression (McCarthy, 2003; Chang et al., 2005). By contrast, our current studies show that TCF-4 binds hormone activated ERα principally within ERα domain E in parallel with greater ERα activity in Wnt pathway co-induced cells. Thus, the combinatorial effects of ERα activation with Runx2, C/EBPδ, and TCF4 are functionally distinct and focused in part through different ERα domains. Although the 17βE and Wnt pathways are each highly influenced by Runx2 expression, it remains unclear at this point if they are distinct or involve complex interactions by Runx2 with ERα (McCarthy, 2003), C/EBPδ (McCarthy et al., 2000b), or TCF-4 (McCarthy and Centrella, 2010). It remains important to emphasize that each of these signaling systems exhibits both feed-forward and also counter-regulatory or compensatory responses that may be temporally and spacially regulated. For example, high levels of Runx2 feed back to suppress ERα activity (McCarthy, 2003), and high levels of C/EBPδ feed back to suppress Runx2 activity (McCarthy et al., 2000b). As shown in these studies, noncanonical Wnt pathway activation can potently suppress new Runx2 mRNA expression, and 17βE drives or supports decreases in the steady state levels of mRNAs encoding the Wnt receptor component LRP6 and TCF-4. Nevertheless, the stimulatory effect of 17βE on Wnt pathway activation does not appear directly related to rapid changes in mRNAs encoding well characterized Wnt receptors, or to the multiple downstream regulatory or transcriptional components of TCF dependent gene expression that we measured. Still, future studies may reveal that other Wnt pathway related components or TCF gene family members may contribute these overall effects. A model related to some of these interactions is shown in Fig. 10.
Fig. 10.
Transcription factor restricted functional interactions with ERα in osteoblasts. A model based on results from these studies proposes that, in differentiating osteoblasts, 17βE activated ERα enhances gene expression through TCF response elements (TCF RE), and Wnt pathway induction enhances the transcriptional effect of 17βE activated ERα through ERE response elements in a β-catenin independent way through a functional complex formation between ERα and TCF-4 (right half of figure). These co-stimulatory events contrast with earlier studies revealing that an interaction between ERα and Runx2 that enhances Runx2 dependent gene expression through Runx response elements (Runx RE) and suppresses ERα dependent gene expression through ERE (McCarthy, 2003); and between ERα and C/EBP that co-suppresses gene expression by ERα through ERE and C/EBP through C/EBP response elements (C/EBP RE) (Chang et al., 2005) (left half of figure). Perhaps imposed on this, but not detailed in this figure, and the physical and functional interactions between TCF-4 and Runx2 (McCarthy and Centrella, 2010), and between Runx2 and C/EBP (McCarthy et al., 2000b), and the interactions between these factors by the osteoblast growth factor TGF-β and/or IGF-I pathways (McCarthy et al., 1997; McCarthy et al., 2000a; Centrella et al., 2004a; McCarthy and Centrella, 2010). The importance of changes in specific downstream genes regulated by the individual or combined effects of ERα and TCF activation and the mechanisms by which they occur, now just initiated here (Figs. 6,7), await further studies.
Our current results in osteoblasts both concur and also differ in part with those in other tissue derived cells. Previous studies in a noninvasive rat mammary epithelial cell line predicted an intersection between ERα and TCFs by the use of complex expression, sequestration, and 17βE induced reporter plasmid systems, and by physical interactions between ERs and TCFs synthesized in reticulocyte lysates (El-Tanani et al., 2001). In that cell context, 17βE enhanced downstream gene expression by TCF-1 through the osteopontin gene promoter independently of canonical ERE, whereas endogenous TCF-4 appeared inhibitory, even though both TCFs bound ERα or ERβ. In agreement with those studies, we detected a significant albeit relatively attenuated response to 17βE in cells that express ERβ versus ERα and the involvement of the E domain (also called AF2) within ERα. In contrast, however, we found a positive functional interaction with TCF-4 in osteoblasts. Our current results may thus differ in part from those findings due to cell lineage differences. Surprisingly, the mammary epithelial cell studies offered no evidence for the importance of this interaction on Wnt pathway activation (El-Tanani et al., 2001). In addition, while we found minimal evidence for interactions between the androgen/AR and Wnt pathways in osteoblasts, earlier studies in prostate cancer cells showed cross-control between these systems, engendered by competition between the DBDs of AR and TCF for β-catenin and by TCF-4 dependent suppression of AR activity (Chesire and Isaacs, 2002). Thus, these results also differ distinctly from the ERα and TCF-4 interactions that we found with regard to both steroid receptor domain interactions and to the influence of β-catenin. Still, they may help to explain our finding that AR expression can potently limit TCF dependent gene expression in osteoblasts when they are co-stimulated by LiCl and WAg.
In contrast to the influence of the Wnt pathway on ERα activity, the regulatory effect of 17βE on TCF dependent gene expression does not require direct ERα dependent gene activation through its own DBD. Even so, ligand induced activation of ERα was clearly an essential component of the interaction between ERα and TCF-4. However, only two of the several ERα ligands in our test panel, 17βE and the purported ERα antagonist ICI 182780, enhanced TCF dependent gene expression. Specific conformational changes in ERα in response to ligand binding may therefore be necessary for its association with TCF-4, presumably to reconfigure and properly present ERα domain E, and may only occur with certain ligands. In addition, they may involve Runx2 which can be transcriptionally co-activated by either 17βE (McCarthy, 2003) or ICI 182780 (supplemental Fig. 3). Thus, some ER agonists or antagonists may cause previously unrecognized or unanticipated responses that involve interactions with other transcriptional components like the TCF, Runx, or C/EBP transcription factors, and account in part for their specificity in various tissues where individual binding partners have restricted expression or biological effects.
Finally, of the several biochemical responses or mRNA levels that we examined, we found highly focused effects by 17βE, alone or in combination with canonical or noncanonical Wnt pathway activation, within the 24 hour treatment interval that we employed. In most instances, neither 17βE nor canonical Wnt signaling by LiCl directly affected general osteoblast activity in this relatively short time frame, whereas β-catenin independent Wnt pathway activation by WAg enhanced DNA synthesis and limited the expression of differentiated cell function. By and large, 17βE also had minimal direct effects on the panels of mRNAs that we examined. In some instances, however, it suppressed steady state mRNA levels in combination with canonical Wnt signaling, and maintained or enforced the response to WAg. Curiously, Wnt pathway activation suppressed Runx2 mRNA as well some mRNAs thought to be under direct control of Runx2, even while it directly enhanced Runx2 transcriptional activity (these studies and (McCarthy and Centrella, 2010)), confirming that in many instances the expression of Runx2 dependent genes are controlled post-transcriptionally by effects on the activation potential of pre-existing Runx2 protein, on its association with other proteins, or on its stability (Chang et al., 1998; Tintut et al., 1999; McCarthy et al., 2000b; Selvamurugan et al., 2000; McCarthy, 2003; Selvamurugan et al., 2004; McCarthy et al., 2007). Even so, our results verify that Runx2 competent cells drive or prime osteoblasts to respond to Wnt signaling (McCarthy and Centrella, 2010) and predict the possible involvement of one or more, still unknown, Runx2 sensitive genes.
In summary, our studies reveal cross-control between metabolic pathways activated by 17βE and Wnt signaling in differentiated osteoblasts replete for Runx2 expression. They further demonstrate selective interactions by 17βE with so-called canonical and noncanonical Wnt pathways at the biochemical level, and define a functional ERα domain restricted interaction with TCF-4 which can occur without the involvement of β-catenin. We extended the importance of this interaction to show that it requires 17βE activation of ERα, is enhanced by full Wnt pathway activation, is functionally consequential on both ERα and TCF dependent gene expression, and occurs within a cell where both pathways have clear physiological importance. These interactions may account for pathway selective effects on gene expression and biological activity in bone by circulating 17βE, and perhaps tissue restricted effects by some local (McCarthy et al., 2008) or pharmacological SERMs that can activate ERα, and thereby sustain skeletal integrity.
Supplementary Material
Acknowledgements
We are grateful to Drs. Stuart Adler (Washington Univ., St. Louis, MO), Chawnshang Chang (University of Rochester, NY), Ronald M. Evans, Peter G. Schultz, and Jun Liu (Scripps Research Institute, CA), John Wysolmerski (Yale University), Yoshiaki Ito (National University of Singapore), Ivan J. Sadowski (University of British Columbia, Canada), and Jan-Ake Gustafsson (Karolinska Institute, Sweden) for some of the vectors, reporter, or expression plasmids; and to Drs. Joseph A. Madri, and Dianne Duffey (Yale University), for some of the antibodies used in these studies. As always, we are indebted to Dr. Madri for helpful comments and advice during manuscript preparation.
Abbreviations
- DBD
DNA binding domain
- GSK
glycogen synthase kinase
- RE
response element
- RT-PCR
real time polymerase chain reaction
- Runx
runt homology domain transcription factor
- TCF
T cell factor
- WAg
Wnt pathway agonist
Footnotes
Disclosure statement: The authors have nothing to disclose.
Supported by NIAMS award AR39201 (MC and TLM), RSDP/UCSF-K12-HD000849 (CBK), and Yale Department of Surgery.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arce L, Yokoyama NN, Waterman ML. Diversity of LEF/TCF action in development and disease. Oncogene. 2006;25:7492–504. doi: 10.1038/sj.onc.1210056. [DOI] [PubMed] [Google Scholar]
- Armstrong VJ, Muzylak M, Sunters A, Zaman G, Saxon LK, Price JS, Lanyon LE. Wnt/beta-catenin signaling is a component of osteoblastic bone cell early responses to load-bearing and requires estrogen receptor alpha. J Biol Chem. 2007;282:20715–27. doi: 10.1074/jbc.M703224200. [DOI] [PubMed] [Google Scholar]
- Barkhem T, Nilsson S, Gustafsson JA. Molecular mechanisms, physiological consequences and pharmacological implications of estrogen receptor action. Am J Pharmacogenomics. 2004;4:19–28. doi: 10.2165/00129785-200404010-00003. [DOI] [PubMed] [Google Scholar]
- Bennett CN, Longo KA, Wright WS, Suva LJ, Lane TF, Hankenson KD, MacDougald OA. Regulation of osteoblastogenesis and bone mass by Wnt10b. Proc Natl Acad Sci U S A. 2005;102:3324–9. doi: 10.1073/pnas.0408742102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergenstock MK, Partridge NC. Parathyroid hormone stimulation of noncanonical Wnt signaling in bone. Ann N Y Acad Sci. 2007;1116:354–9. doi: 10.1196/annals.1402.047. [DOI] [PubMed] [Google Scholar]
- Boonyaratanakornkit V, Edwards DP. Receptor mechanisms mediating non-genomic actions of sex steroids. Semin Reprod Med. 2007;25:139–53. doi: 10.1055/s-2007-973427. [DOI] [PubMed] [Google Scholar]
- Bovolenta P, Esteve P, Ruiz JM, Cisneros E, Lopez-Rios J. Beyond Wnt inhibition: new functions of secreted Frizzled-related proteins in development and disease. J Cell Sci. 2008;121:737–46. doi: 10.1242/jcs.026096. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Cadigan KM. Wnt-beta-catenin signaling. Curr Biol. 2008a;18:R943–7. doi: 10.1016/j.cub.2008.08.017. [DOI] [PubMed] [Google Scholar]
- Cadigan KM. Wnt/beta-catenin signaling: turning the switch. Dev Cell. 2008b;14:322–3. doi: 10.1016/j.devcel.2008.02.006. [DOI] [PubMed] [Google Scholar]
- Carpenter TO, Moltz KC, Ellis B, Andreoli M, McCarthy TL, Centrella M, Bryan D, Gundberg CM. Osteocalcin production in primary osteoblast cultures derived from normal and Hyp mice. Endocrinology. 1998;139:35–43. doi: 10.1210/endo.139.1.5677. [DOI] [PubMed] [Google Scholar]
- Centrella M, Casinghino S, Gundberg C, McCarthy TL, Wozney J, Rosen V. Changes in bone morphogenetic protein sensitivity relative to differentiation in fetal rat bone cell cultures. Ann N Y Acad Sci. 1996;785:224–6. doi: 10.1111/j.1749-6632.1996.tb56267.x. [DOI] [PubMed] [Google Scholar]
- Centrella M, Casinghino S, Ignotz R, McCarthy TL. Multiple regulatory effects by transforming growth factor-beta on type I collagen levels in osteoblast-enriched cultures from fetal rat bone. Endocrinology. 1992;131:2863–72. doi: 10.1210/endo.131.6.1446624. [DOI] [PubMed] [Google Scholar]
- Centrella M, Casinghino S, Kim J, Pham T, Rosen V, Wozney J, McCarthy TL. Independent changes in type I and type II receptors for transforming growth factor beta induced by bone morphogenetic protein 2 parallel expression of the osteoblast phenotype. Mol Cell Biol. 1995;15:3273–81. doi: 10.1128/mcb.15.6.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centrella M, Casinghino S, McCarthy TL. Differential actions of prostaglandins in separate cell populations from fetal rat bone. Endocrinology. 1994;135:1611–20. doi: 10.1210/endo.135.4.7925124. [DOI] [PubMed] [Google Scholar]
- Centrella M, Christakos S, McCarthy TL. Skeletal hormones and the C/EBP and Runx transcription factors: interactions that integrate and redefine gene expression. Gene. 2004a;342:13–24. doi: 10.1016/j.gene.2004.06.036. [DOI] [PubMed] [Google Scholar]
- Centrella M, McCarthy TL, Canalis E. Transforming growth factor beta is a bifunctional regulator of replication and collagen synthesis in osteoblast-enriched cell cultures from fetal rat bone. J Biol Chem. 1987;262:2869–74. [PubMed] [Google Scholar]
- Centrella M, McCarthy TL, Chang W, Labaree DC, Hochberg RB. Estren (4-Estren-3alpha,17beta-diol) is a prohormone that regulates both androgenic and estrogenic transcriptional effects through the androgen receptor. Mol Endo. 2004b;18:1120–1130. doi: 10.1210/me.2003-0491. [DOI] [PubMed] [Google Scholar]
- Chang DJ, Ji C, Kim KK, Casinghino S, McCarthy TL, Centrella M. Reduction in transforming growth factor beta receptor I expression and transcription factor CBFa1 on bone cells by glucocorticoid. J Biol Chem. 1998;273:4892–6. doi: 10.1074/jbc.273.9.4892. [DOI] [PubMed] [Google Scholar]
- Chang W, Parra M, Centrella M, McCarthy TL. Interactions between CCAAT enhancer binding protein delta and estrogen receptor alpha control insulin-like growth factor I (igf1) and estrogen receptor-dependent gene expression in osteoblasts. Gene. 2005;345:225–35. doi: 10.1016/j.gene.2004.11.017. [DOI] [PubMed] [Google Scholar]
- Chen Y, Whetstone HC, Youn A, Nadesan P, Chow EC, Lin AC, Alman BA. Beta-catenin signaling pathway is crucial for bone morphogenetic protein 2 to induce new bone formation. J Biol Chem. 2007;282:526–33. doi: 10.1074/jbc.M602700200. [DOI] [PubMed] [Google Scholar]
- Chesire DR, Isaacs WB. Ligand-dependent inhibition of beta-catenin/TCF signaling by androgen receptor. Oncogene. 2002;21:8453–69. doi: 10.1038/sj.onc.1206049. [DOI] [PubMed] [Google Scholar]
- Coghlan MP, Culbert AA, Cross DA, Corcoran SL, Yates JW, Pearce NJ, Rausch OL, Murphy GJ, Carter PS, Roxbee Cox L, Mills D, Brown MJ, Haigh D, Ward RW, Smith DG, Murray KJ, Reith AD, Holder JC. Selective small molecule inhibitors of glycogen synthase kinase-3 modulate glycogen metabolism and gene transcription. Chem Biol. 2000;7:793–803. doi: 10.1016/s1074-5521(00)00025-9. [DOI] [PubMed] [Google Scholar]
- El-Tanani M, Fernig DG, Barraclough R, Green C, Rudland P. Differential modulation of transcriptional activity of estrogen receptors by direct protein-protein interactions with the T cell factor family of transcription factors. J Biol Chem. 2001;276:41675–82. doi: 10.1074/jbc.M103966200. [DOI] [PubMed] [Google Scholar]
- Gaur T, Lengner CJ, Hovhannisyan H, Bhat RA, Bodine PV, Komm BS, Javed A, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Canonical WNT signaling promotes osteogenesis by directly stimulating Runx2 gene expression. J Biol Chem. 2005;280:33132–40. doi: 10.1074/jbc.M500608200. [DOI] [PubMed] [Google Scholar]
- Glass DA, 2nd, Bialek P, Ahn JD, Starbuck M, Patel MS, Clevers H, Taketo MM, Long F, McMahon AP, Lang RA, Karsenty G. Canonical Wnt signaling in differentiated osteoblasts controls osteoclast differentiation. Dev Cell. 2005;8:751–64. doi: 10.1016/j.devcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- Gong Y, Slee RB, Fukai N, Rawadi G, Roman-Roman S, Reginato AM, Wang H, Cundy T, Glorieux FH, Lev D, Zacharin M, Oexle K, Marcelino J, Suwairi W, Heeger S, Sabatakos G, Apte S, Adkins WN, Allgrove J, Arslan-Kirchner M, Batch JA, Beighton P, Black GC, Boles RG, Boon LM, Borrone C, Brunner HG, Carle GF, Dallapiccola B, De Paepe A, Floege B, Halfhide ML, Hall B, Hennekam RC, Hirose T, Jans A, Juppner H, Kim CA, Keppler-Noreuil K, Kohlschuetter A, LaCombe D, Lambert M, Lemyre E, Letteboer T, Peltonen L, Ramesar RS, Romanengo M, Somer H, Steichen-Gersdorf E, Steinmann B, Sullivan B, Superti-Furga A, Swoboda W, van den Boogaard MJ, Van Hul W, Vikkula M, Votruba M, Zabel B, Garcia T, Baron R, Olsen BR, Warman ML. LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell. 2001;107:513–23. doi: 10.1016/s0092-8674(01)00571-2. [DOI] [PubMed] [Google Scholar]
- Hoang BH, Kubo T, Healey JH, Sowers R, Mazza B, Yang R, Huvos AG, Meyers PA, Gorlick R. Expression of LDL receptor-related protein 5 (LRP5) as a novel marker for disease progression in high-grade osteosarcoma. Int J Cancer. 2004;109:106–11. doi: 10.1002/ijc.11677. [DOI] [PubMed] [Google Scholar]
- Holmen SL, Giambernardi TA, Zylstra CR, Buckner-Berghuis BD, Resau JH, Hess JF, Glatt V, Bouxsein ML, Ai M, Warman ML, Williams BO. Decreased BMD and limb deformities in mice carrying mutations in both Lrp5 and Lrp6. J Bone Miner Res. 2004;19:2033–40. doi: 10.1359/JBMR.040907. [DOI] [PubMed] [Google Scholar]
- Htun H, Holth LT, Walker D, Davie JR, Hager GL. Direct visualization of the human estrogen receptor alpha reveals a role for ligand in the nuclear distribution of the receptor. Mol Biol Cell. 1999;10:471–86. doi: 10.1091/mbc.10.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsken J, Behrens J. The Wnt signalling pathway. J Cell Sci. 2002;115:3977–8. doi: 10.1242/jcs.00089. [DOI] [PubMed] [Google Scholar]
- Jakacka M, Ito M, Weiss J, Chien PY, Gehm BD, Jameson JL. Estrogen receptor binding to DNA is not required for its activity through the nonclassical AP1 pathway. J Biol Chem. 2001;276:13615–21. doi: 10.1074/jbc.M008384200. [DOI] [PubMed] [Google Scholar]
- Ji C, Casinghino S, Chang DJ, Chen Y, Javed A, Ito Y, Hiebert SW, Lian JB, Stein GS, McCarthy TL, Centrella M. CBFa(AML/PEBP2)-related elements in the TGF-beta type I receptor promoter and expression with osteoblast differentiation. J Cell Biochem. 1998;69:353–63. [PubMed] [Google Scholar]
- Ji C, Casinghino S, McCarthy TL, Centrella M. Multiple and essential Sp1 binding sites in the promoter for transforming growth factor-beta type I receptor. J Biol Chem. 1997;272:21260–7. doi: 10.1074/jbc.272.34.21260. [DOI] [PubMed] [Google Scholar]
- Ji C, Eickelberg O, McCarthy TL, Centrella M. Control and counter-control of TGF-beta activity through FAST and Runx (CBFa) transcriptional elements in osteoblasts. Endocrinology. 2001;142:3873–9. doi: 10.1210/endo.142.9.8399. [DOI] [PubMed] [Google Scholar]
- Jilka RL, Weinstein RS, Parfitt AM, Manolagas SC. Quantifying osteoblast and osteocyte apoptosis: challenges and rewards. J Bone Miner Res. 2007;22:1492–501. doi: 10.1359/jbmr.070518. [DOI] [PubMed] [Google Scholar]
- Juttner KV, Perry MJ. High-dose estrogen-induced osteogenesis is decreased in aged RUNX2(+/−) mice. Bone. 2007;41:25–32. doi: 10.1016/j.bone.2007.03.012. [DOI] [PubMed] [Google Scholar]
- Kato M, Patel MS, Levasseur R, Lobov I, Chang BH, Glass DA, 2nd, Hartmann C, Li L, Hwang TH, Brayton CF, Lang RA, Karsenty G, Chan L. Cbfa1-independent decrease in osteoblast proliferation, osteopenia, and persistent embryonic eye vascularization in mice deficient in Lrp5, a Wnt coreceptor. J Cell Biol. 2002;157:303–14. doi: 10.1083/jcb.200201089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalid O, Baniwal SK, Purcell DJ, Leclerc N, Gabet Y, Stallcup MR, Coetzee GA, Frenkel B. Modulation of Runx2 activity by Estrogen Receptor {alpha}: Implications for Osteoporosis and Breast Cancer. Endocrinology. 2008 doi: 10.1210/en.2008-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KK, Ji C, Chang W, Wells RG, Gundberg CM, McCarthy TL, Centrella M. Repetitive exposure to TGF-beta suppresses TGF-beta type I receptor expression by differentiated osteoblasts. Gene. 2006;379:175–84. doi: 10.1016/j.gene.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Kousteni S, Almeida M, Han L, Bellido T, Jilka RL, Manolagas SC. Induction of osteoblast differentiation by selective activation of kinase-mediated actions of the estrogen receptor. Mol Cell Biol. 2007;27:1516–30. doi: 10.1128/MCB.01550-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzmenko AP, Takeyama K, Ito S, Furutani T, Sawatsubashi S, Maki A, Suzuki E, Kawasaki Y, Akiyama T, Tabata T, Kato S. Wnt/beta-catenin and estrogen signaling converge in vivo. J Biol Chem. 2004;279:40255–8. doi: 10.1074/jbc.C400331200. [DOI] [PubMed] [Google Scholar]
- Kugimiya F, Kawaguchi H, Ohba S, Kawamura N, Hirata M, Chikuda H, Azuma Y, Woodgett JR, Nakamura K, Chung UI. GSK-3beta controls osteogenesis through regulating Runx2 activity. PLoS One. 2007;2:e837. doi: 10.1371/journal.pone.0000837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni NH, Halladay DL, Miles RR, Gilbert LM, Frolik CA, Galvin RJ, Martin TJ, Gillespie MT, Onyia JE. Effects of parathyroid hormone on Wnt signaling pathway in bone. J Cell Biochem. 2005;95:1178–90. doi: 10.1002/jcb.20506. [DOI] [PubMed] [Google Scholar]
- Lian JB, Stein GS, Javed A, van Wijnen AJ, Stein JL, Montecino M, Hassan MQ, Gaur T, Lengner CJ, Young DW. Networks and hubs for the transcriptional control of osteoblastogenesis. Rev Endocr Metab Disord. 2006;7:1–16. doi: 10.1007/s11154-006-9001-5. [DOI] [PubMed] [Google Scholar]
- Liu J, Wu X, Mitchell B, Kintner C, Ding S, Schultz PG. A small-molecule agonist of the Wnt signaling pathway. Angew Chem Int Ed Engl. 2005;44:1987–90. doi: 10.1002/anie.200462552. [DOI] [PubMed] [Google Scholar]
- MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani A, Radhakrishnan J, Wang H, Mani A, Mani MA, Nelson-Williams C, Carew KS, Mane S, Najmabadi H, Wu D, Lifton RP. LRP6 mutation in a family with early coronary disease and metabolic risk factors. Science. 2007;315:1278–82. doi: 10.1126/science.1136370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolagas SC, Kousteni S, Chen JR, Schuller M, Plotkin L, Bellido T. Kinase-mediated transcription, activators of nongenotropic estrogen-like signaling (ANGELS), and osteoporosis: a different perspective on the HRT dilemma. Kidney Int Suppl. 2004:S41–9. doi: 10.1111/j.1523-1755.2004.09107.x. [DOI] [PubMed] [Google Scholar]
- McCarthy TL, Casinghino S, Centrella M, Canalis E. Complex pattern of insulin-like growth factor binding protein expression in primary rat osteoblast enriched cultures: regulation by prostaglandin E2, growth hormone, and the insulin-like growth factors. Journal of Cellular Physiology. 1994;160:163–75. doi: 10.1002/jcp.1041600119. [DOI] [PubMed] [Google Scholar]
- McCarthy TL, Centrella M. Novel links among Wnt and TGF-beta signaling and Runx2. Mol Endocrinol. 2010;24:587–97. doi: 10.1210/me.2009-0379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy TL, Centrella M, Canalis E. Further biochemical and molecular characterization of primary rat parietal bone cell cultures. J Bone Miner Res. 1988;3:401–8. doi: 10.1002/jbmr.5650030406. [DOI] [PubMed] [Google Scholar]
- McCarthy TL, Chang W-Z, Liu Y, Centrella M. Runx2 integrates estrogen activity in osteoblasts. J Biol Chem. 2003;278:43121–43129. doi: 10.1074/jbc.M306531200. [DOI] [PubMed] [Google Scholar]
- McCarthy TL, Clough ME, Gundberg CM, Centrella M. Expression of an estrogen receptor agonist in differentiating osteoblast cultures. Proc Natl Acad Sci U S A. 2008;105:7022–7. doi: 10.1073/pnas.0800085105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy TL, Hochberg RB, Labaree DC, Centrella M. 3-ketosteroid reductase activity and expression by fetal rat osteoblasts. J Biol Chem. 2007;282:34003–12. doi: 10.1074/jbc.M707502200. [DOI] [PubMed] [Google Scholar]
- McCarthy TL, Ji C, Centrella M. Links among growth factors, hormones, and nuclear factors with essential roles in bone formation. Crit Rev Oral Biol Med. 2000a;11:409–22. doi: 10.1177/10454411000110040201. [DOI] [PubMed] [Google Scholar]
- McCarthy TL, Ji C, Chen Y, Kim KK, Imagawa M, Ito Y, Centrella M. Runt domain factor (Runx)-dependent effects on CCAAT/ enhancer-binding protein delta expression and activity in osteoblasts. J Biol Chem. 2000b;275:21746–53. doi: 10.1074/jbc.M002291200. [DOI] [PubMed] [Google Scholar]
- McCarthy TL, Ji C, Shu H, Casinghino S, Crothers K, Rotwein P, Centrella M. 17beta-estradiol potently suppresses cAMP-induced insulin-like growth factor-I gene activation in primary rat osteoblast cultures. J Biol Chem. 1997;272:18132–9. doi: 10.1074/jbc.272.29.18132. [DOI] [PubMed] [Google Scholar]
- McClung MR. The relationship between bone mineral density and fracture risk. Curr Osteoporos Rep. 2005;3:57–63. doi: 10.1007/s11914-005-0005-y. [DOI] [PubMed] [Google Scholar]
- Milat F, Ng KW. Is Wnt signalling the final common pathway leading to bone formation? Mol Cell Endocrinol. 2009 doi: 10.1016/j.mce.2009.06.002. [DOI] [PubMed] [Google Scholar]
- Peterkofsky B, Diegelmann R. Use of a mixture of proteinase-free collagenases for the specific assay of radioactive collagen in the presence of other proteins. Biochemistry. 1971;10:988–94. doi: 10.1021/bi00782a009. [DOI] [PubMed] [Google Scholar]
- Ray A, Prefontaine KE, Ray P. Down-modulation of interleukin-6 gene expression by 17 beta-estradiol in the absence of high affinity DNA binding by the estrogen receptor. J Biol Chem. 1994;269:12940–6. [PubMed] [Google Scholar]
- Reinhold MI, Naski MC. Direct interactions of Runx2 and canonical Wnt signaling induce FGF18. J Biol Chem. 2007;282:3653–63. doi: 10.1074/jbc.M608995200. [DOI] [PubMed] [Google Scholar]
- Rosen CJ. The cellular and clinical parameters of anabolic therapy for osteoporosis. Crit Rev Eukaryot Gene Expr. 2003;13:25–38. doi: 10.1615/critreveukaryotgeneexpr.v13.i1.30. [DOI] [PubMed] [Google Scholar]
- Selvamurugan N, Kwok S, Partridge NC. Smad3 Interacts with JunB and Cbfa1/Runx2 for Transforming Growth Factor-{beta}1-stimulated Collagenase-3 Expression in Human Breast Cancer Cells. J. Biol. Chem. 2004;279:27764–27773. doi: 10.1074/jbc.M312870200. [DOI] [PubMed] [Google Scholar]
- Selvamurugan N, Pulumati MR, Tyson DR, Partridge NC. Parathyroid hormone regulation of the rat collagenase-3 promoter by protein kinase A-dependent transactivation of core binding factor alpha1. J Biol Chem. 2000;275:5037–42. doi: 10.1074/jbc.275.7.5037. [DOI] [PubMed] [Google Scholar]
- Smith DG, Buffet M, Fenwick AE, Haigh D, Ife RJ, Saunders M, Slingsby BP, Stacey R, Ward RW. 3-Anilino-4-arylmaleimides: potent and selective inhibitors of glycogen synthase kinase-3 (GSK-3) Bioorg Med Chem Lett. 2001;11:635–9. doi: 10.1016/s0960-894x(00)00721-6. [DOI] [PubMed] [Google Scholar]
- Sorensen MG, Henriksen K, Schaller S, Karsdal MA. Biochemical markers in preclinical models of osteoporosis. Biomarkers. 2007;12:266–86. doi: 10.1080/13547500601070842. [DOI] [PubMed] [Google Scholar]
- Stein B, Yang MX. Repression of the interleukin-6 promoter by estrogen receptor is mediated by NF-kappa B and C/EBP beta. Mol Cell Biol. 1995;15:4971–9. doi: 10.1128/mcb.15.9.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suda T, Nakamura I, Jimi E, Takahashi N. Regulation of osteoclast function. J Bone Miner Res. 1997;12:869–79. doi: 10.1359/jbmr.1997.12.6.869. [DOI] [PubMed] [Google Scholar]
- Sunters A, Armstrong VJ, Zaman G, Kypta RM, Kawano Y, Lanyon LE, Price JS. Mechano-transduction in osteoblastic cells involves strain-regulated estrogen receptor alpha-mediated control of insulin-like growth factor (IGF) I receptor sensitivity to Ambient IGF, leading to phosphatidylinositol 3-kinase/AKT-dependent Wnt/LRP5 receptor-independent activation of beta-catenin signaling. J Biol Chem. 2010;285:8743–58. doi: 10.1074/jbc.M109.027086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed FA, Modder UI, Fraser DG, Spelsberg TC, Rosen CJ, Krust A, Chambon P, Jameson JL, Khosla S. Skeletal effects of estrogen are mediated by opposing actions of classical and nonclassical estrogen receptor pathways. J Bone Miner Res. 2005;20:1992–2001. doi: 10.1359/JBMR.050713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada I, Mihara M, Suzawa M, Ohtake F, Kobayashi S, Igarashi M, Youn MY, Takeyama K, Nakamura T, Mezaki Y, Takezawa S, Yogiashi Y, Kitagawa H, Yamada G, Takada S, Minami Y, Shibuya H, Matsumoto K, Kato S. A histone lysine methyltransferase activated by non-canonical Wnt signalling suppresses PPAR-gamma transactivation. Nat Cell Biol. 2007;9:1273–85. doi: 10.1038/ncb1647. [DOI] [PubMed] [Google Scholar]
- Tintut Y, Parhami F, Le V, Karsenty G, Demer LL. Inhibition of osteoblast-specific transcription factor Cbfa1 by the cAMP pathway in osteoblastic cells. Ubiquitin/proteasome-dependent regulation. J Biol Chem. 1999;274:28875–9. doi: 10.1074/jbc.274.41.28875. [DOI] [PubMed] [Google Scholar]
- Tobimatsu T, Kaji H, Sowa H, Naito J, Canaff L, Hendy GN, Sugimoto T, Chihara K. Parathyroid hormone increases beta-catenin levels through Smad3 in mouse osteoblastic cells. Endocrinology. 2006;147:2583–90. doi: 10.1210/en.2005-1627. [DOI] [PubMed] [Google Scholar]
- Vaes BL, Dechering KJ, Feijen A, Hendriks JM, Lefevre C, Mummery CL, Olijve W, van Zoelen EJ, Steegenga WT. Comprehensive microarray analysis of bone morphogenetic protein 2-induced osteoblast differentiation resulting in the identification of novel markers for bone development. J Bone Miner Res. 2002;17:2106–18. doi: 10.1359/jbmr.2002.17.12.2106. [DOI] [PubMed] [Google Scholar]
- Vaes BL, Dechering KJ, van Someren EP, Hendriks JM, van de Ven CJ, Feijen A, Mummery CL, Reinders MJ, Olijve W, van Zoelen EJ, Steegenga WT. Microarray analysis reveals expression regulation of Wnt antagonists in differentiating osteoblasts. Bone. 2005;36:803–11. doi: 10.1016/j.bone.2005.02.001. [DOI] [PubMed] [Google Scholar]
- van Bezooijen RL, Roelen BA, Visser A, van der Wee-Pals L, de Wilt E, Karperien M, Hamersma H, Papapoulos SE, ten Dijke P, Lowik CW. Sclerostin is an osteocyte-expressed negative regulator of bone formation, but not a classical BMP antagonist. J Exp Med. 2004;199:805–14. doi: 10.1084/jem.20031454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varea O, Arevalo MA, Garrido JJ, Garcia-Segura LM, Wandosell F, Mendez P. Interaction of estrogen receptors with insulin-like growth factor-I and Wnt signaling in the nervous system. Steroids. 2010;75:565–9. doi: 10.1016/j.steroids.2009.09.006. [DOI] [PubMed] [Google Scholar]
- Windahl SH, Andersson G, Gustafsson JA. Elucidation of estrogen receptor function in bone with the use of mouse models. Trends Endocrinol Metab. 2002;13:195–200. doi: 10.1016/s1043-2760(02)00594-5. [DOI] [PubMed] [Google Scholar]
- Windahl SH, Lagerquist MK, Andersson N, Jochems C, Kallkopf A, Hakansson C, Inzunza J, Gustafsson JA, van der Saag PT, Carlsten H, Pettersson K, Ohlsson C. Identification of target cells for the genomic effects of estrogens in bone. Endocrinology. 2007;148:5688–95. doi: 10.1210/en.2007-0508. [DOI] [PubMed] [Google Scholar]
- Yadav VK, Ryu JH, Suda N, Tanaka KF, Gingrich JA, Schutz G, Glorieux FH, Chiang CY, Zajac JD, Insogna KL, Mann JJ, Hen R, Ducy P, Karsenty G. Lrp5 controls bone formation by inhibiting serotonin synthesis in the duodenum. Cell. 2008;135:825–37. doi: 10.1016/j.cell.2008.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yost C, Torres M, Miller JR, Huang E, Kimelman D, Moon RT. The axis-inducing activity, stability, and subcellular distribution of beta-catenin is regulated in Xenopus embryos by glycogen synthase kinase 3. Genes Dev. 1996;10:1443–54. doi: 10.1101/gad.10.12.1443. [DOI] [PubMed] [Google Scholar]
- Zawel L, Dai JL, Buckhaults P, Zhou S, Kinzler KW, Vogelstein B, Kern SE. Human Smad3 and Smad4 are sequence-specific transcription activators. Mol Cell. 1998;1:611–7. doi: 10.1016/s1097-2765(00)80061-1. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang Y, Li X, Zhang J, Mao J, Li Z, Zheng J, Li L, Harris S, Wu D. The LRP5 high-bone-mass G171V mutation disrupts LRP5 interaction with Mesd. Mol Cell Biol. 2004;24:4677–84. doi: 10.1128/MCB.24.11.4677-4684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.