Abstract
The overall structure of a protein-protein complex reflects an intricate arrangement of non-covalent interactions. While intramolecular interactions confer secondary and tertiary structure to individual subunits, intermolecular interactions lead to quaternary structure - the ordered aggregation of separate polypeptide chains into multi-subunit assemblies. The specific ensemble of non-covalent contacts dictates the stability of subunit folds, enforces protein-protein binding specificity, and determines multimer stability. Consequently, non-covalent architecture is likely to play a role in the gas-phase dissociation of these assemblies during tandem mass spectrometry (MS/MS). To further advance the applicability of MS/MS to analytical problems in structural biology, a better understanding of the interplay between the structures and fragmentation behaviors of non-covalent protein complexes is essential. The present work constitutes a systematic study of model protein homodimers (bacteriophage N15 Cro; bacteriophage λ Cro; bacteriophage P22 Arc) with related but divergent structures, both in terms of subunit folds and protein-protein interfaces. Because each of these dimers has a well-characterized structure (solution and / or crystal structure), specific non-covalent features could be correlated with gas-phase disassembly patterns as studied by collision-induced dissociation, surface-induced dissociation, and ion mobility. Of the several respects in which the dimers differed in structure, the presence or absence of intermolecular electrostatic contacts exerted the most significant influence on the gas-phase dissociation behavior. This is attributed to the well-known enhancement of ionic interactions in the absence of bulk solvent. Because salt bridges are general contributors to both intermolecular and intramolecular stability in protein complexes, these observations are broadly applicable to aid in the interpretation or prediction of dissociation spectra for non-covalent protein assemblies.
Introduction
In recent years, mass spectrometry (MS) has developed into an expedient means of examining non-covalent biomolecular ensembles, thus opening new methodological avenues for the advancement of research in structural biology. The emergence of MS into the realm of non-covalent complex analysis is largely attributable to the development of electrospray ionization (ESI)1 and subsequently nanoelectrospray ionization (nano-ESI)2, which are capable of transferring multiply-charged biomolecular complexes from physiologically relevant condensed-phase conditions into the gas phase. This can be accomplished with sufficient delicacy to preserve the specific non-covalent interactions within the native complex. The application of this technology to the study of protein-protein complexes is especially relevant, as an overwhelming majority of proteins carry out their functions as non-covalent constructions of multiple polypeptide chains.3, 4 Since the earliest MS experiments with supramolecular protein assemblies, the development of MS-based methods for probing higher-order protein structure has proceeded apace and has now reached a level of considerable maturity.5–14 Nonetheless, recent reviews not only underscore the promise of MS-based structural biology, but also point out present challenges in the field.15, 16 The need for efficient and informative MS-based methods for determining the subunit topology of quaternary protein assemblies is invariably cited among these current hurdles.
While mass measurement of an intact protein complex to determine oligomeric state or subunit stoichiometry has become relatively straightforward, determination of spatial relationships between constituent subunits remains a significantly more challenging endeavor. One of the more common approaches for probing the structural organization of protein complexes involves partial disruption of non-covalent complexes in the condensed phase using non-native solution conditions (e.g., adjustment of pH, adjustment of ionic strength, or introduction of chaotropes). When appropriate solution conditions are found that produce informative subcomplexes, distinct subunit organizational possibilities can be resolved.17 This approach depends on the distinct binding characteristics of different protein—protein interfaces, which allow stepwise disassembly in solution.
The use of tandem mass spectrometry (MS/MS) to bring about gas-phase dissociation of non-covalent assemblies provides a complement to solution-phase disruption experiments. As discussed throughout the reviews cited above, gas-phase dissociation of non-covalent protein complexes is now reasonably well-understood, albeit in a rather generic sense. At present, relatively little is known regarding the effects of specific non-covalent interactions (or combinations thereof) upon the overall disassembly of protein complexes in vacuo. The totality of non-covalent interactions conferring integrity to protein-protein assemblies usually involves a combination of intramolecular and intermolecular hydrophobic interactions, electrostatic interactions, and hydrogen bonding. Thus, in much the same way that the distinct binding characteristics of different protein—protein interfaces allow stepwise disassembly in solution, there is also the possibility that a specific ensemble of non-covalent contacts imparts a particular dissociation signature during MS/MS.
In this work, dimeric protein complexes with well-established solution and / or crystal structures were studied by collision-induced dissociation (CID), surface-induced dissociation (SID), and ion mobility (IM). Although the dimers of interest are structurally related, each also has a unique combination of non-covalent features with respect to subunit fold and interface structure. When viewed in the context of these known topologies, differences in gas-phase disassembly patterns could be associated with specific elements of non-covalent architecture. This type of understanding is of broad analytical relevance to MS-based efforts in structural biology. The establishment of generally applicable dependencies of gas-phase dissociation behavior upon higher-order structure will provide for more informative interpretation of MS/MS dissociation patterns, while also suggesting the possibility of dissociation pathway prediction for known structures.
Experimental Methods
Dimeric protein complexes
N15 Cro and λ Cro proteins containing C-terminal LEH6 sequence tags were overexpressed in Escherichia coli strain BL21(λDE3) from pET21b-derived plasmids and purified by denaturing Ni-NTA affinity chromatography and size exclusion chromatography as described.18 Arc repressor was overexpressed in Escherichia coli strain BL21(λDE3) from the arc-st11 gene of the plasmid pET800.19 The expressed Arc protein included a C-terminal st11 extension (H6KNQHE) to stabilize the protein against degradation in vivo and to allow for affinity purification. Arc-st11 was purified by Ni2+-NTA affinity chromatography under denaturing conditions essentially as described,20 except that 10 mM imidazole was included in all load and wash steps to increase purity. Following elution in 6 M guanidine-HCl with 0.2 M HOAc, Arc was refolded by dialysis into 1 M ammonium acetate buffer (pH 7.0) for 48 hours at 4°C, with one buffer change after the first 24 hours. Protein concentrations (monomer basis) were determined using UV absorbance at 280 nm. For MS analysis, protein solutions at concentrations of 20–50 µM were buffer exchanged into 100 mM NH4OAc (pH 7.4) using size exclusion spin columns (BioRad Micro Bio-Spin, Hercules, CA, USA), and were then held on ice until the time of analysis. All subsequent analyses were carried out using nano-ESI of this native-like solution.
Mass spectrometry and ion mobility analyses
For each analysis, a 5–10 µL portion of buffer-exchanged protein solution was loaded into a nano-ESI emitter that had been fabricated in-house. The emitters were fashioned from 1.0 mm inner diameter borosilicate capillary tubing, and were pulled to a 1–5 µm inner diameter tip using a Flaming/Brown type micropipette puller (Sutter Instruments Model P-97, Novato, CA, USA). All nano-ESI was performed in static mode through application of a 1.2–1.8 kV potential directly to the solution using a platinum wire. The resulting flow rates were approximately 20–50 nL/min.
MS and MS/MS studies were carried out using two quadrupole time-of-flight (QTOF) MS instruments: a QTOF2 (Micromass / Waters, Manchester, UK) and a Synapt G2 HDMS (Waters, Manchester, UK). As described in detail elsewhere,21 the QTOF2 instrument was previously modified to include an in-line SID device installed between the RF/DC quadrupole and the hexapole collision cell. In order to accommodate the SID stage, the original collision cell was replaced with a reduced-length version. In this instrument configuration, a broad range of ions or a mass-selected precursor ion could be transmitted through the SID device for MS or MS/MS via CID; alternatively, the DC potentials on SID optics could be adjusted to bring about ion-surface collisions for MS/MS via SID. This design allowed CID and SID to be conducted under otherwise identical conditions on the same instrument. CID was carried out using argon as the collision gas (1 × 10−4 mbar analyzer pressure). The SID target was a gold-coated glass slide (Evaporated Metal Films, Ithaca, NY, USA) covalently modified with a self-assembled monolayer (SAM) of 2-(perfluorodecyl)ethanethiol (FC12). For fluorocarbon SAM preparation, the glass slide was rinsed with ethanol, cleaned using a Boekel UV cleaner (Feasterville, PA, USA), and submerged in an ethanolic solution of 1 mM FC12 for 24 h. The resulting fluorocarbon-modified surface was sonicated five times in fresh portions of ethanol, and was then fitted onto the SID stage.
The Synapt G2 HDMS instrument was equipped with a traveling wave IM cell which allowed drift time measurement of activated precursor ions and CID fragment ions. Precursor ions were subjected to CID within the trapping region stacked ring ion guide (4 mL/min argon flow rate; 3 × 10−2 mbar trap region pressure), and the resulting ion population was then pulsed into the IM cell. Ions were collisionally cooled by helium during injection to the initial chamber of the IM cell (180 mL/min helium flow rate; > 5 mbar helium cell pressure). The remainder of the IM cell was pressurized with nitrogen, which served as the drift gas for separation (90 mL/min nitrogen flow rate; 3 mbar IM cell pressure). The IM DC traveling wave was set to a velocity of 840 m/s with a wave height of 40 V. These conditions were tuned for optimum separation of the dimeric precursor ions and monomeric product ions under study.
Both instruments were operated under conditions specifically chosen to maintain the native structure of protein dimer ions produced by nano-ESI. The cone voltages (QTOF2: 50 V; Synapt G2: 20 V), extractor voltages (QTOF2: 1 V; Synapt G2: 2 V), and z-spray block temperatures (ambient) were all set to ensure mild source conditions. In addition, the pressure in the first differentially pumped region of the instrument was elevated to provide for vibrational relaxation of ions (QTOF2: 5 × 10−3 mbar source chamber pressure; Synapt G2: 6 mbar backing pressure). This was accomplished by adjusting a variable valve (Edwards Speedivalve, West Sussex, UK) placed between the roughing pump and the source region. All DC bias values were also reduced to the minimum practical levels still allowing reasonable ion transmission efficiency. The interested reader is directed to more detailed discussions of methodology for MS and IM analysis of protein complexes.22–24
Results and Discussion
Bacteriophage Cro Dimers as Model Complexes
Bacteriophage Cro proteins comprise a class of homodimeric DNA-binding factors which contribute to the regulation of viral reproduction. Comparison of two orthologous Cro proteins – one from bacteriophage N15, and the other from bacteriophage λ - reveals a unique amalgam of similar and dissimilar structural characteristics. The previously established crystal structures of the N15 Cro and λ Cro dimers are reproduced in Figure 1.25, 26 Although the two complexes have similar molecular weights (N15 Cro, 17.8 kDa; λ Cro, 16.9 kDa), comparable interfacial contact areas (N15 Cro, 1303 Å2; λ Cro, 1354 Å2), and nearly equivalent dimer dissociation constants (N15 Cro, KD = 5 µM; λ Cro, KD = 3 µM),26, 27 in a variety of other respects the dimers are strikingly unalike. The individual subunits of N15 Cro adopt an all α-helical fold and dimerize in a relatively superficial manner likened to “sticky billiard balls,”28 whereby each subunit interacts with only outer surface side chains of the neighboring monomer (Figure 1a). Specifically, a series of interactions between L39 / L39’, V46 / V46’, and Y50 / Y50’ forms a hydrophobic “zipper” motif (Figure 1b). These hydrophobic contacts are known to make major contributions to the dimer stability in solution; indeed, mutations to the “zipper” residues have been found to significantly affect the strength of dimerization (D.J. O’Brien and M.H.J. Cordes, unpublished data). By contrast to N15 Cro, λ Cro subunits take on a mixed α-helix / β-sheet fold, forming a dimer with an intertwined interface that involves the extension of identical strands from each monomer into the central domain of the adjacent subunit (Figure 1c). This interface includes interactions between F58 and the hydrophobic core of the opposite subunit, giving rise to a hydrophobic “ball-and-socket” arrangement. In addition, the λ Cro interface includes a pair of intermolecular electrostatic contacts between E54 / K56’ and E54’ / K56 (Figure 1d). Having well-defined but disparate structures, these dimers provide intriguing exemplars for study of the interplay between subunit structure, interface structure, and gas-phase dissociation behavior in non-covalent protein complexes.
Figure 1.
Previously established crystal structures of the bacteriophage N15 Cro dimer (PDB ID 2hin, from Dubrava et al.26) and the bacteriophage λ Cro dimer (PDB ID 5cro, from Ohlendorf et al.25): (a) N15 Cro overall dimer structure and (b) interface detail, illustrating the homotypic hydrophobic “zipper;” (c) λ Cro overall dimer structure and (d) interface detail, illustrating the hydrophobic “ball-and-socket” arrangement and cross-strand ionic interactions. Individual subunits of the dimers are indicated by distinct coloration. The view in (b) is from the same perspective as in (a), with 90° clockwise rotation about the z-axis. The view in (d) is from above (c), with 45° clockwise rotation about the y-axis.
Collision-Induced Dissociation of Bacteriophage Cro Dimers
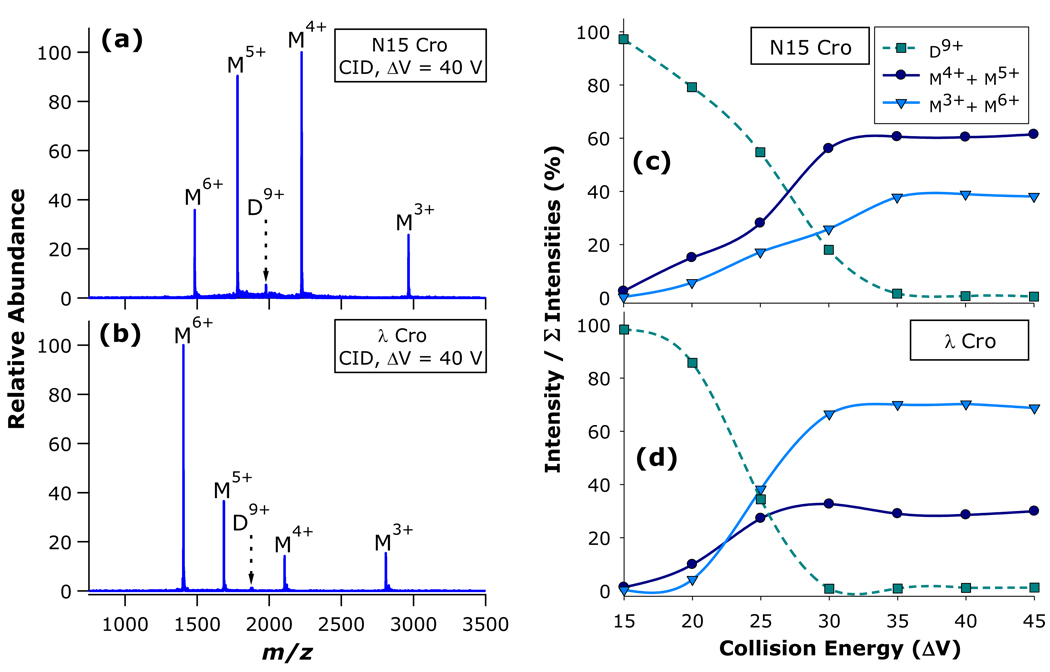
When nanosprayed from native-like solution conditions, both Cro protein preparations exhibited monomers and dimers in proportions approximately consistent with expectations based on their KD values and solution concentrations (Figures S1 and S2 in Supporting Information). The CID characteristics for N15 Cro and λ Cro dimers are summarized in Figure 2, with pronounced differences in the dissociation spectra of the two dimers being immediately apparent. The results demonstrate that each dimer can access two routes to dissociation: one pathway resulting in a symmetric division of charge with respect to mass, which produces monomer ions M4+ and M5+ from the precursor dimer D9+; and a second channel leading to asymmetric division of charge with respect to mass and yielding M3+ and M6+ from D9+. At the same collision energy (ΔV = 40 V), the N15 Cro D9+ precursor ion dissociates predominantly through the charge-symmetric pathway (Figure 2a), whereas the λ Cro D9+ precursor ion dissociates mainly through the charge-asymmetric channel (Figure 2b). The fractional intensities of the precursor ion (D9+), the charge-symmetric complementary monomer pair (M4++M5+), and the charge-asymmetric complementary monomer pair (M3++M6+) were also determined as a function of collision energy for both N15 Cro (Figure 2c) and λ Cro (Figure 2d). These energy-resolved CID experiments confirm that the preferential dissociation of each Cro dimer along either the symmetric (N15 Cro) or asymmetric (λ Cro) channel is maintained over a range of collision energies.
Figure 2.
CID of N15 Cro and λ Cro dimers: CID mass spectra of (a) N15 Cro and (b) λ Cro; and energy-resolved CID curves for (c) N15 Cro and (d) λ Cro. Dimers (D) and monomers (M) are labelled with their corresponding charge states.
The proportion of product ions arising from the two dissociation channels appears to be closely tied to structure. These initial results suggest that the fraction of monomers yielded through symmetric versus asymmetric pathways hinges on the non-covalent architecture of the precursor dimers; furthermore, the dissociation pathway has implications for the conformation of monomer product ions. According to the generally accepted model of non-covalent protein complex dissociation, which is now well-established on both experimental29–35 and theoretical36, 37 grounds, multi-step activation of a protein complex during CID generally leads to gradual unfolding of one subunit. As the subunit takes on a more extended structure, proton transfer to this newly exposed surface area results in the unfolded subunit gaining a number of charges that is disproportionate with respect to mass, but proportionate with respect to the increased surface area. In accord with this mechanism, the vast majority of non-covalent protein complexes dissociate with asymmetric division of charge amongst the product ions. In this respect, the dissociation of λ Cro better harmonizes with established behavior, while dissociation of N15 Cro mainly through the symmetric pathway is rather unusual. Notably, under certain circumstances charge-symmetric CID of protein complexes has been observed. For example, symmetric charge partitioning has been noticed for dimers electrosprayed from non-native conditions (in which case the constituent monomers were presumably denatured prior to activation)38; for dimers with subunits prevented from unfolding due to covalent constraints (i.e., disulfide bridging, chemical cross-linking);32 for a tetrameric complex having a dimer-of-dimers arrangement where the monomer-monomer interface was significantly more stable than the dimer-dimer interface;39 and for complexes with charge states deliberately reduced to much lower than typical for native nano-ESI of protein complexes.40 None of these circumstances explains the distinct dissociation behaviors of N15 Cro and λ Cro, as neither contains intra-subunit disulfide bonds or cross-links, both have only a single subunit interface, and both were interrogated as the 9+ charge state (a typical charge state for protein complexes in the molecular weight range of these dimers when nanosprayed from native-like conditions). More recently, it has been posited that atypical dissociation patterns may be partly driven by Coulombic instability; if so, a protein complex with a relatively high number of charges per unit surface area might be expected to dissociate through pathways other than the unfolding / monomer release model.41 While appealing, this hypothesis does not seem a likely explanation for the difference in dissociation of N15 Cro and λ Cro, as the charge densities of the D9+ precursor ions differ by only 3.8%: 9 z / 7743 Å2 = 1.162 × 10−3 z/Å2 for N15 Cro; 9 z / 8105 Å2 = 1.110 × 10−3 z/Å2 for λ Cro (surface areas from Dubrava et al.26).
Ion Mobility Analysis of Bacteriophage Cro Dimer Dissociation
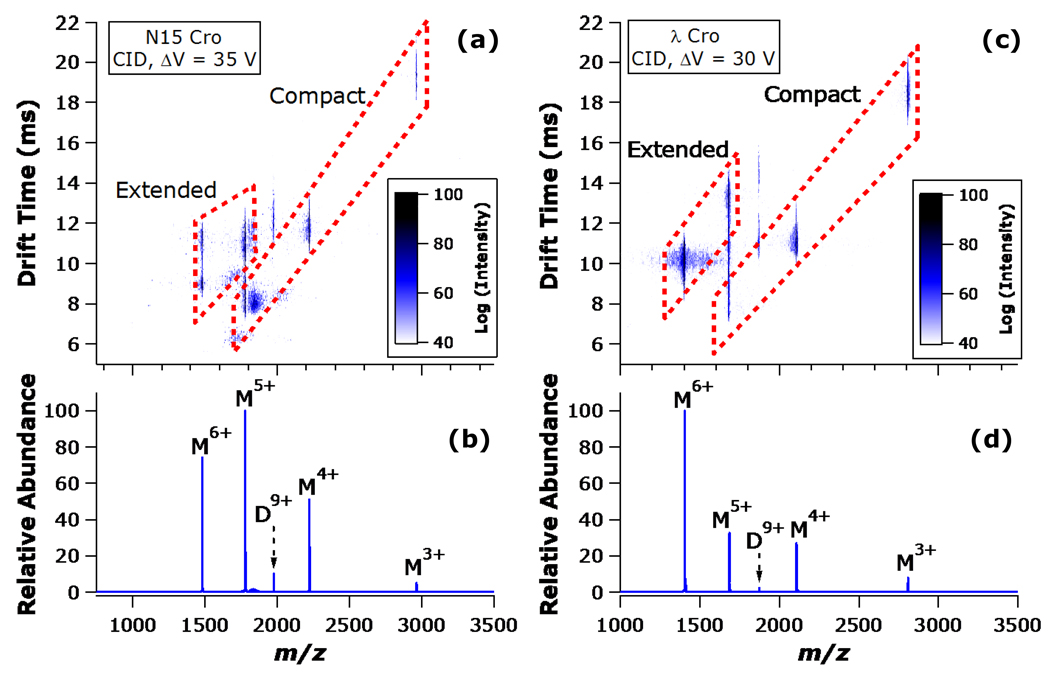
In order to more directly address the question of whether the difference between N15 Cro and λ Cro dissociation is related to subunit unfolding (or lack thereof), IM analysis was applied in conjunction with CID. Figure 3 summarizes the results from CID of N15 Cro and λ Cro D9+ precursors, followed by IM separation of the resulting monomeric products. As expected, the monomers having lower charge states (M3+, M4+) appear to retain a relatively compact conformation, whereas the monomers with the higher charge states (M5+, M6+) assume a mixture of compact and extended states. In traveling wave IM, drift times scale as the inverse square of ion mobility.42 Because ion mobility is directly proportional to charge, it also follows that drift times should scale as the inverse square of charge state in the limiting case that collisional cross section remains unchanged. The structures assigned as compact in Figure 3 have drift times that scale with the inverse square of charge state, as expected for a family of relatively folded monomers differing in charge but not shape (all compact conformers have drift times falling within 10% of drift times predicted based on inverse square scaling with charge state). Those conformers assigned as extended in Figure 3 deviate markedly from prediction and take on longer drift times despite their higher charge states (as expected for structures having larger collisional cross sections). Moreover, the ratio of these folded and unfolded conformations is consistent with the observed dissociation symmetry. Specifically, IM analysis confirms that the charge-symmetric dissociation channel of N15 Cro produces a significant proportion of folded monomeric product. The M5+ monomer consists of a roughly equal mixture of folded and unfolded states, while the M6+ monomer takes on two distinct unfolded states, with the more unfolded of the two being approximately half as abundant as the less extended form. (Figure 3a, and the corresponding mass spectrum in Figure 3b). By contrast, the λ Cro M5+ ion is detected primarily (approximately 80%) in an extended conformation while the M6+ ion is observed solely in an extended conformation (Figure 3c, and the corresponding mass spectrum in Figure 3d).
Figure 3.
IM of N15 Cro dimer and λ Cro dimer CID products: (a) N15 Cro two-dimensional IM heat map and (b) corresponding CID mass spectrum; (c) λ Cro two-dimensional IM heat map and (d) corresponding CID mass spectrum. The heat map intensity scales are inset (logarithmic scales are used for clarity). Dimers (D) and monomers (M) are labelled with their corresponding charge states. The dashed boxes highlight the transition from relatively compact structures at the lower charge states (drift times decrease with increasing charge), to relatively extended structures at the higher charge states (drift times increase despite increasing charge).
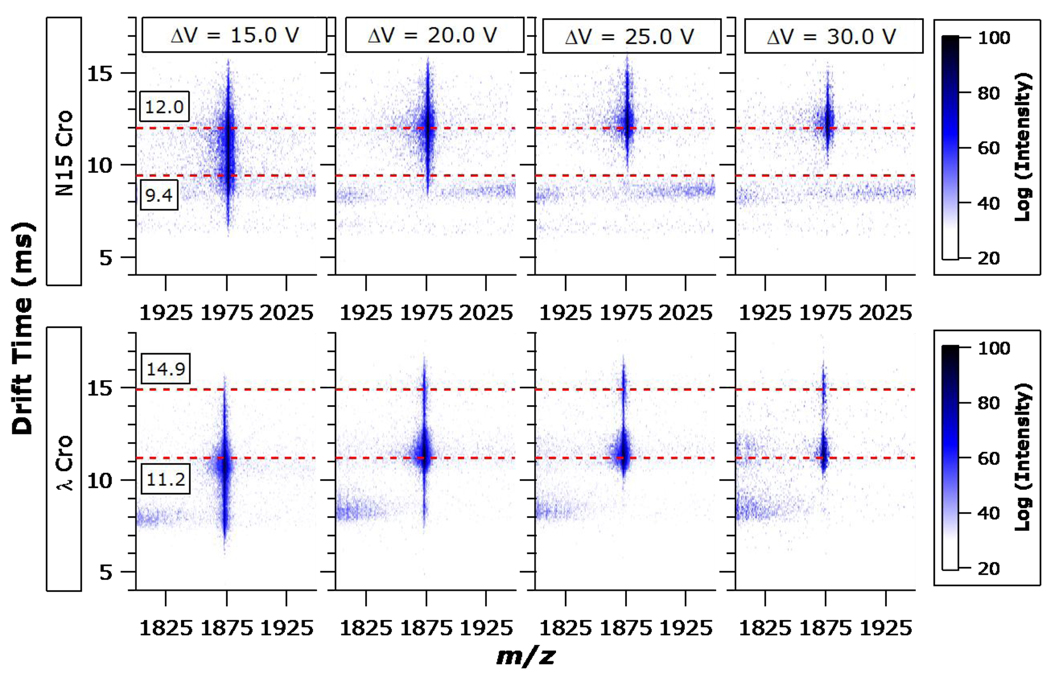
As illustrated in Figure 4, subjecting the N15 Cro and λ Cro dimers to increasing collision energies and monitoring the drift time of the activated precursors provides additional insights on their dissociation behaviors. In the case of N15 Cro, the D9+ ion initially has an arrival time distribution (ATD) with maxima at 9.4 ms and 12.0 ms. With increasing collision energy, the activated N15 Cro dimers preferentially anneal to a preexisting structure that had contributed to the initial ATD but do not appear to give rise to new unfolded states prior to disociation. At ΔV = 30 V, D9+ of N15 Cro has taken on an ATD with a single maxiumum centered around a drift time of 12.0 ms. In the case of λ Cro, the initial ATD with a major peak centered at a drift time of 11.2 ms gradually gives rise to a second, distinct maximum centered at 14.9 ms drift time. This provides evidence for energy-resolved unfolding of the λ Cro D9+ ion into a distinct, extended state prior to dissociation. The relatively low abundance of the extended λ Cro conformer may be a consequence of facile dissociation of the dimer once one subunit has become unfolded. By contrast, the N15 Cro dimer undergoes little or no unfolding prior to dissociation and merely converts to one of the two initial conformations when energized.
Figure 4.
IM of N15 Cro (upper row) and λ Cro (lower row) residual precursor dimers (D9+) following activation at different collision energies (columns). The heat map intensity scales are inset (logarithmic scales are used for clarity). Dashed lines at drift times of 9.4 ms and 12.0 ms are provided in the upper row; dashed lines at drift times of 11.2 ms and 14.9 ms are provided in the lower row.
Surface-Induced Dissociation of Bacteriophage Cro Dimers
As discussed above, CID of protein complexes generally yields charge-asymmetric product ions due to unfolding and proton transfer prior to subunit loss. This laboratory has previously demonstrated that, by contrast, SID yields more charge-symmetric product ions for a variety of protein complexes including dimers,43 tetramers and pentamers,44 dodecamers,45, 46 and most recently a hexameric complex containing three distinct types of subunits.47 These additional dissociation pathways arise as a consequence of the one-step activation of SID, which provides more efficient conversion of kinetic energy to internal energy owing to the large effective mass of the surface. Because surface collision allows high energy deposition within a short timeframe, sufficient kinetic shift is achieved to allow subunit dissociation on a timescale faster than that of protein unfolding. SID activation is thus markedly different from CID activation in a QTOF, which deposits much smaller increments of energy in a stepwise manner. During beam-type CID, the first collision provides the highest conversion of kinetic to vibrational energy, with each subsequent collision depositing successively lower amounts of energy.
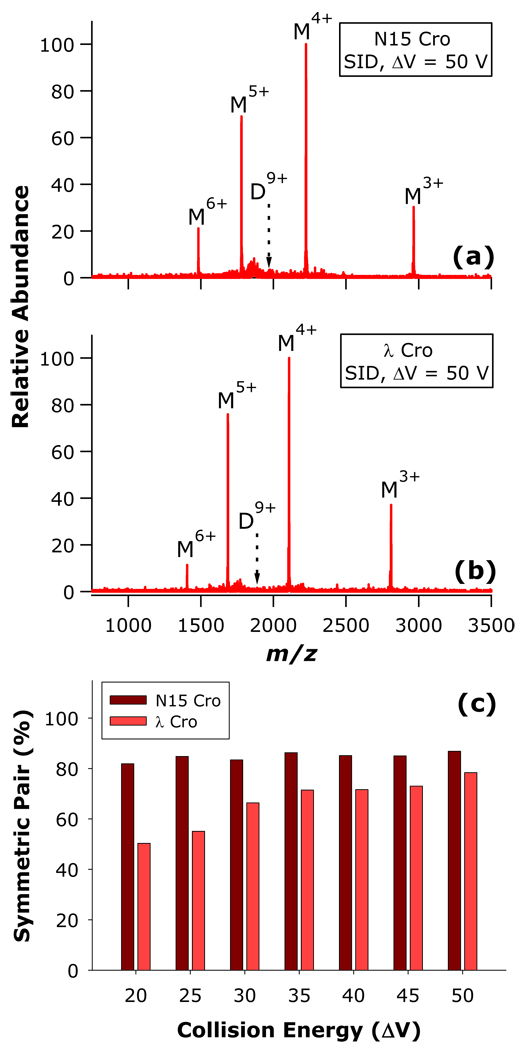
Consistent with this prior experience, SID of both N15 Cro and λ Cro brings about dissociation mainly through the charge-symmetric pathway, with both dimers dissociating to a higher proportion of the M4++M5+ complementary pair relative to CID (Figure 5a,b). While ΔV = 50 V SID spectra of D9+ from N15 Cro and λ Cro are very similar in appearance, energy-resolved SID demonstrates that λ Cro requires greater collision energy to achieve a proportion of symmetric monomer pair comparable to that of N15 Cro (Figure 5c). This is consistent with other results presented herein which indicate that the barrier towards symmetric dissociation is much higher for λ Cro than N15 Cro. Interestingly, this data also furnishes evidence that SID is able to access charge-symmetric dissociation products which are presumably relatively folded despite the intermingling of the subunits composing the λ Cro dimer.
Figure 5.
SID of N15 Cro and λ Cro dimers: SID mass spectra for (a) N15 Cro and (b) λ Cro; and (c) proportion of charge-symmetric monomer pair as a function of collision energy.
Although the SID energy dependence of N15 Cro dissociation symmetry is not apparent in Figure 5c, it is speculated that the onset for high-symmetry dissociation lies at lower SID collision energies than can be accessed using the current SID device (the lowest practical collision energy for protein complexes is ca. ΔV = 20 V). Attempts were made to access lower energy SID pathways by substitution of the fluorocarbon-modified collision surface with a hydrocarbon-modified surface (using 1-hexadecanethiol) which has a characteristically lower energy transfer efficiency.48–50 Even when applying the minumum possible collision ΔV and using the hydrocarbon-modified surface, N15 Cro dissociation produces a similar proportion of the charge-symmetric complementary pair as was seen with the fluorocarbon-modified surface (data not shown). This is consistent with the “sticky billiard balls” topology of the N15 Cro dimer, and with the prevalence of the charge-symmetric dissociation products yielded by this dimer upon CID.
Bacteriophage P22 Arc Dimer Structure and Dissociation
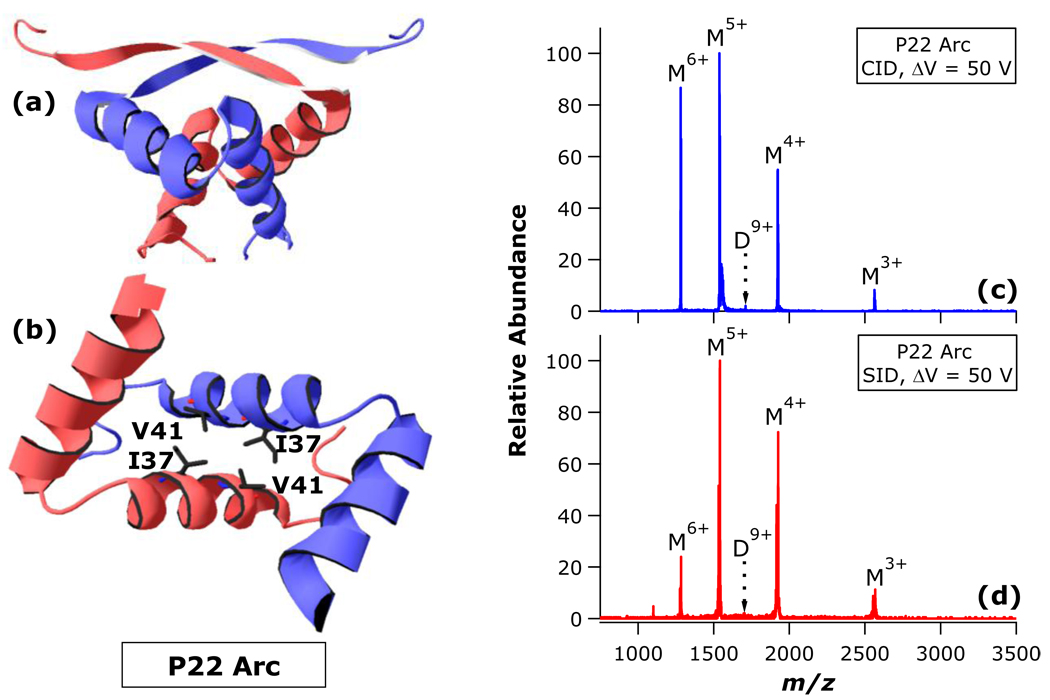
Having established that N15 Cro dissociation and λ Cro dissociation differ in charge symmetry due to differences in the degree of gas-phase unfolding prior to dissociation, we next sought to determine which structural differences are most closely linked to the differences in dissociation behavior. As discussed above, perhaps the two most obvious differences between N15 Cro and λ Cro dimer structures are (1) the “sticky billiard balls” versus intertwined interfaces, respectively, and (2) the presence of inter-subunit salt bridging in the λ Cro interface, but not the N15 Cro interface. In order to resolve which of these structural disparities is most influential in gas-phase disassembly, the dissociation behavior of a third model homodimer was explored. The previously determined solution structure of the bacteriophage P22 Arc repressor is presented in Figures 6a,b.51 Like the Cro dimers, the 15.4 kDa P22 Arc dimer is a DNA-binding regulatory factor. The dimer assumes an intertwined interface similar in some respects to λ Cro, but with an even greater degree of inter-subunit convolution (Figure 6a). The monomers are interfaced through a core tetrad of hydrophobic side chains (V41, I37, V41’, I37’; Figure 6b), with other regions of hydrophobic interaction stabilizing the dimer as well. While the presence of electrostatic interactions across the Arc interface is a formal possibility (and is seen in some subunit pairings of Arc in crystal structures), to the best of our knowledge none have been clearly demonstrated in the solution structure.
Figure 6.
Previously established solution structure (PDB ID 1arr, from Bonvin et al.51) and MS/MS behavior of bacteriophage P22 Arc dimer: (a) P22 Arc overall dimer structure and (b) interface detail, illustrating the hydrophobic core; (c) CID and (d) SID of P22 Arc dimer. Individual subunits of the dimer are indicated by distinct coloration. The view in (b) is from above (a), with 90° counterclockwise rotation about the y-axis. In (b), the β-sheets have been removed for clarity. In (c) and (d), dimers (D) and monomers (M) are labelled with their corresponding charge states.
As anticipated, native nano-ESI of P22 Arc produced a mixture of dimer and monomer signals (Figure S3 in Supporting Information). In a fashion similar to N15 Cro, CID of P22 Arc D9+ results in monomers with unusually high charge symmetry (Figure 6c) over a range of collision energies (Figure S4 in Supporting Information). Furthermore, SID again produces a higher proportion of the charge-symmetric complementary monomer pair (Figure 6d). Overall, P22 Arc dissociation behavior more closely parallels that of N15 Cro despite the fact that P22 Arc has an intertwined configuration of subunits that bears more similarity to λ Cro. These observations suggest that the presence of an intertwined interface does not make a decisive contribution to the dissociation characteristics of these dimers. Thus, it is proposed that the interfacial electrostatic interactions of λ Cro contribute enhanced interface stability in the gas phase, and thus the barrier for subunit unfolding is reached before the barrier for subunit loss. This model is consistent with the asymmetric CID pattern of λ Cro, and is also in concurrence with many examples of enhanced ionic interactions in vacuo.5, 52–57 These results also suggest that interfaces lacking salt bridges are more likely to allow complex dissociation to occur with minimal subunit unfolding and thus high charge symmetry, as observed for N15 Cro.
Conclusions
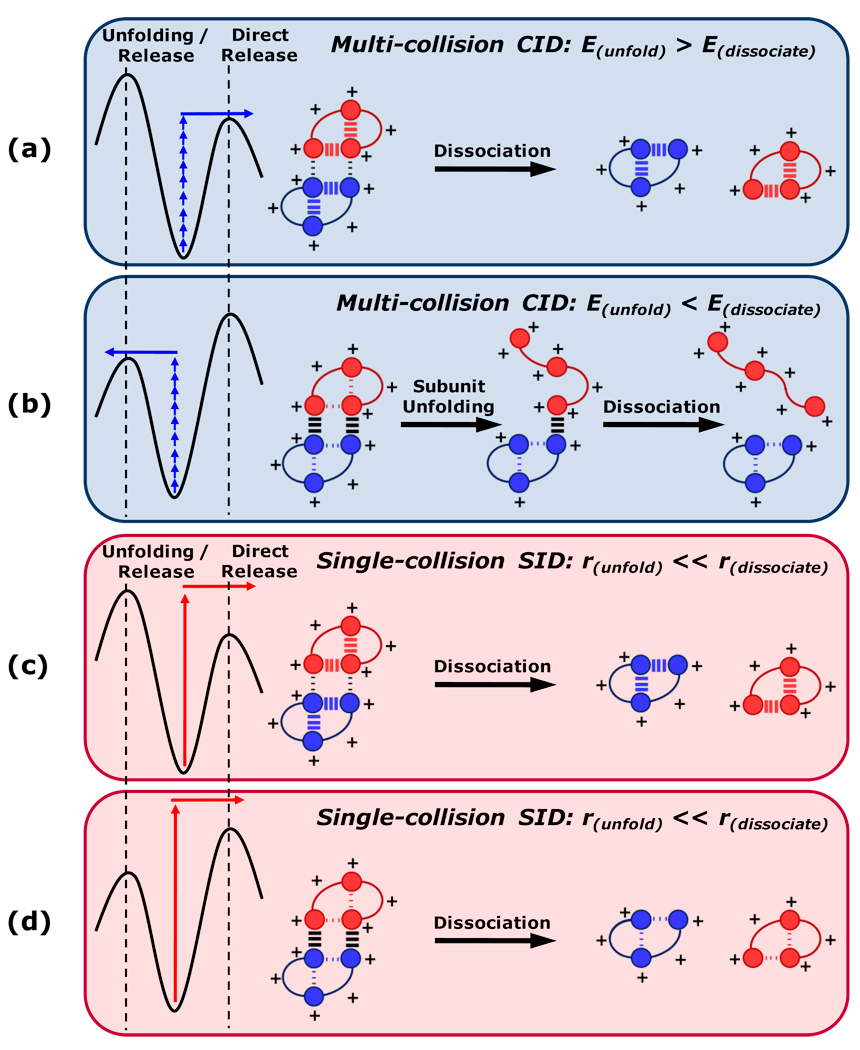
With the benefit of related homodimers having well-characterized but divergent structures, the effects of specific structural features on gas-phase disassembly pathways have been evaluated. The over-arching findings of this study are summarized graphically in Figure 7. The internal energy deposition typical of CID leads to fragments with charge states and structures that are strongly influenced by the relative gas-phase stability of subunit folds and interfacial contacts. In multi-collision CID, only the lowest-energy dissociation pathways of non-covalent protein complexes are accessed. Thus, if the activation barrier towards subunit unfolding is greater than that of subunit release, dissociation proceeds largely according to a charge-symmetric pathway and produces a significant proportion of relatively folded product ions (Figure 7a). Conversely, if the activation barrier for subunit unfolding is accessed prior to subunit release, the majority of resulting product ions are asymmetrically charged, and those with high charge states assume extended conformations (Figure 7b). Therefore, the lowest-energy dissociation pathway may lead to either symmetric or asymmetric dissociation depending on the relative gas-phase stabilities of intramolecular and intermolecular non-covalent interactions. Electrostatic contacts have strong influence over which outcome is observed, being particularly stable in the gas phase due to the absence of solvent dielectric. It is therefore proposed that the presence or absence of interfacial salt bridges is a strong determinant of whether unfolding / release or direct release is more favorable. Surface collision provides access to dissociation pathways other than those of lowest energy, and moreover deposits sufficient internal energy to bring about protein complex dissociation more rapidly than large-scale structural rearrangement of subunits. Thus, regardless of the relative energy requirements of subunit unfolding and direct dissociation, the kinetic barrier towards subunit unfolding allows SID to yield symmetrically charged products (Figure 7c,d). This being said, the proportion of charge-symmetric product ions exhibits clear SID energy dependence when the subunit interface involves ionic interactions. Importantly, the schemes shown in Figure 7 are intended to explain only the major dissociation pathway in each circumstance; certainly, multiple dissociation channels are accessed to varying degrees in each instance. Because the structural characteristics studied here are ubiquitous features of non-covalent protein complexes, these observations have far-reaching, general implications for the interpretation and prediction of gas-phase dissociation patterns for non-covalent protein complexes.
Figure 7.
Schematic summary of predominant dimer dissociation behaviors in several possible scenarios. Individual subunits of the hypothetical dimers are indicated by distinct coloration, while domains participating in non-covalent interactions are symbolized by circles. Inter-subunit and intra-subunit interactions are indicated by hash marks between domains, with the weights of the marks indicating the relative gas-phase stabilities of the interactions. Potential energy diagrams for each scenario are shown at left. E, energy; r, rate.
Supplementary Material
Acknowledgements
This research was supported by NSF DBI (award number 0923551 to V.H.W.), NIH (award number GM066806 to M.H.J.C.), Science Foundation Arizona Graduate Research Fellowships awarded to A.E.B and K.L.H., and an NSF Graduate Research Fellowship awarded to A.E.B.
Footnotes
Supporting Information Available
Native mass spectra of N15 Cro (Figure S1), λ Cro (Figure S2), and P22 Arc (Figure S3); and energy-resolved collision-induced dissociation of P22 Arc (Figure S4). This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Science. 1989;246:64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 2.Wilm M, Mann M. Analytical Chemistry. 1996;68:1–8. doi: 10.1021/ac9509519. [DOI] [PubMed] [Google Scholar]
- 3.Goodsell DS, Olson AJ. Annual Review of Biophysics and Biomolecular Structure. 2000;29:105–153. doi: 10.1146/annurev.biophys.29.1.105. [DOI] [PubMed] [Google Scholar]
- 4.Shen HB, Chou KC. Journal of Proteome Research. 2009;8:1577–1584. doi: 10.1021/pr800957q. [DOI] [PubMed] [Google Scholar]
- 5.Loo JA. Mass Spectrometry Reviews. 1997;16:1–23. doi: 10.1002/(SICI)1098-2787(1997)16:1<1::AID-MAS1>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 6.Rostom AA, Robinson CV. Current Opinion in Structural Biology. 1999;9:135–141. doi: 10.1016/s0959-440x(99)80018-9. [DOI] [PubMed] [Google Scholar]
- 7.Heck AJR, van den Heuvel RHH. Mass Spectrometry Reviews. 2004;23:368–389. doi: 10.1002/mas.10081. [DOI] [PubMed] [Google Scholar]
- 8.van den Heuvel RHH, Heck AJR. Current Opinion in Chemical Biology. 2004;8:519–526. doi: 10.1016/j.cbpa.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 9.Benesch JL, Ruotolo BT, Simmons DA, Robinson CV. Chemical Reviews. 2007;107:3544–3567. doi: 10.1021/cr068289b. [DOI] [PubMed] [Google Scholar]
- 10.Sharon M, Robinson CV. Annual Review of Biochemistry. 2007;76:167–193. doi: 10.1146/annurev.biochem.76.061005.090816. [DOI] [PubMed] [Google Scholar]
- 11.Wyttenbach T, Bowers MT. Annual Review of Physical Chemistry. 2007;58:511–533. doi: 10.1146/annurev.physchem.58.032806.104515. [DOI] [PubMed] [Google Scholar]
- 12.Morton VL, Stockley PG, Stonehouse NJ, Ashcroft AE. Mass Spectrometry Reviews. 2008;27:575–595. doi: 10.1002/mas.20176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taverner T, Hernandez H, Sharon M, Ruotolo BT, Matak-Vinkovic D, Devos D, Russell RB, Robinson CV. Accounts of Chemical Research. 2008;41:617–627. doi: 10.1021/ar700218q. [DOI] [PubMed] [Google Scholar]
- 14.Uetrecht C, Rose RJ, Duijn Ev, Lorenzen K, Heck AJR. Chemical Society Reviews. 2010;39:1633–1655. doi: 10.1039/b914002f. [DOI] [PubMed] [Google Scholar]
- 15.Sharon M. Journal of the American Society for Mass Spectrometry. 2010;21:487–500. doi: 10.1016/j.jasms.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 16.van Duijn E. Journal of the American Society for Mass Spectrometry. 2010;21:971–978. doi: 10.1016/j.jasms.2009.12.010. [DOI] [PubMed] [Google Scholar]
- 17.Levy ED, Erba EB, Robinson CV, Teichmann SA. Nature. 2008;453:1262–1265. doi: 10.1038/nature06942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roessler CG, Hall BM, Anderson WJ, Ingram WM, Roberts SA, Montfort WR, Cordes MH. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:2343–2348. doi: 10.1073/pnas.0711589105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Milla ME, Brown BM, Sauer RT. Nature Structural Biology. 1994;1:518–523. doi: 10.1038/nsb0894-518. [DOI] [PubMed] [Google Scholar]
- 20.Milla ME, Brown BM, Sauer RT. Protein Science. 1993;2:2198–2205. doi: 10.1002/pro.5560021219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galhena AS, Dagan S, Jones CM, Beardsley RL, Wysocki VH. Analytical Chemistry. 2008;80:1425–1436. doi: 10.1021/ac701782q. [DOI] [PubMed] [Google Scholar]
- 22.Hernandez H, Robinson CV. Nature Protocols. 2007;2:715–726. doi: 10.1038/nprot.2007.73. [DOI] [PubMed] [Google Scholar]
- 23.Heck AJR. Nature Methods. 2008;5:927–933. doi: 10.1038/nmeth.1265. [DOI] [PubMed] [Google Scholar]
- 24.Ruotolo BT, Benesch JLP, Sandercock AM, Hyung S-J, Robinson CV. Nature Protocols. 2008;3:1139–1152. doi: 10.1038/nprot.2008.78. [DOI] [PubMed] [Google Scholar]
- 25.Ohlendorf DH, Tronrud DE, Matthews BW. Journal of Molecular Biology. 1998;280:129–136. doi: 10.1006/jmbi.1998.1849. [DOI] [PubMed] [Google Scholar]
- 26.Dubrava MS, Ingram WM, Roberts SA, Weichsel A, Montfort WR, Cordes MHJ. Protein Science. 2008;17:803–812. doi: 10.1110/ps.073330808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.LeFevre KR, Cordes MH. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:2345–2350. doi: 10.1073/pnas.0537925100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bennett MJ, Schlunegger MP, Eisenberg D. Protein Science. 1995;4:2455–2468. doi: 10.1002/pro.5560041202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwartz BL, Bruce JE, Anderson GA, Hofstadler SA, Rockwood AL, Smith RD, Chilkoti A, Stayton PS. Journal of the American Society for Mass Spectrometry. 1995;6:459–465. doi: 10.1016/1044-0305(95)00191-F. [DOI] [PubMed] [Google Scholar]
- 30.Versluis C, van der Staaij A, Stokvis E, Heck AJR, de Craene B. Journal of the American Society for Mass Spectrometry. 2001;12:329–336. doi: 10.1016/S1044-0305(00)00227-0. [DOI] [PubMed] [Google Scholar]
- 31.Felitsyn N, Kitova EN, Klassen JS. Analytical Chemistry. 2001;73:4647–4661. doi: 10.1021/ac0103975. [DOI] [PubMed] [Google Scholar]
- 32.Jurchen JC, Williams ER. Journal of the American Chemical Society. 2003;125:2817–2826. doi: 10.1021/ja0211508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jurchen JC, Garcia DE, Williams ER. Journal of the American Society for Mass Spectrometry. 2004;15:1408–1415. doi: 10.1016/j.jasms.2004.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benesch JLP, Aquilina JA, Ruotolo BT, Sobott F, Robinson CV. Chemistry and Biology. 2006;13:597–605. doi: 10.1016/j.chembiol.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 35.Ruotolo BT, Hyung SJ, Robinson PM, Giles K, Bateman RH, Robinson CV. Angewandte Chemie International Edition. 2007;46:8001–8004. doi: 10.1002/anie.200702161. [DOI] [PubMed] [Google Scholar]
- 36.Wanasundara SN, Thachuk M. Journal of the American Society for Mass Spectrometry. 2007;18:2242–2253. doi: 10.1016/j.jasms.2007.09.022. [DOI] [PubMed] [Google Scholar]
- 37.Wanasundara SN, Thachuk M. The Journal of Physical Chemistry A. 2009;113:3814–3821. doi: 10.1021/jp8094227. [DOI] [PubMed] [Google Scholar]
- 38.Smith RD, Light-Wahl KJ, Winger BE, Loo JA. Organic Mass Spectrometry. 1992;27:811–821. [Google Scholar]
- 39.van den Heuvel RHH, van Duijn E, Mazon H, Synowsky SA, Lorenzen K, Versluis C, Brouns SJJ, Langridge D, van der Oost J, Hoyes J, Heck AJR. Analytical Chemistry. 2006;78:7473–7483. doi: 10.1021/ac061039a. [DOI] [PubMed] [Google Scholar]
- 40.Pagel K, Hyung S-J, Ruotolo BT, Robinson CV. Analytical Chemistry. 2010;82:5363–5372. doi: 10.1021/ac101121r. [DOI] [PubMed] [Google Scholar]
- 41.Erba EB, Ruotolo BT, Barsky D, Robinson CV. Analytical Chemistry. 2010;82:9702–9710. doi: 10.1021/ac101778e. [DOI] [PubMed] [Google Scholar]
- 42.Shvartsburg AA, Smith RD. Analytical Chemistry. 2008;80:9689–9699. doi: 10.1021/ac8016295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones CM, Beardsley RL, Galhena AS, Dagan S, Cheng G, Wysocki VH. Journal of the American Chemical Society. 2006;128:15044–15045. doi: 10.1021/ja064586m. [DOI] [PubMed] [Google Scholar]
- 44.Beardsley RL, Jones CM, Galhena AS, Wysocki VH. Analytical Chemistry. 2009;81:1347–1356. doi: 10.1021/ac801883k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wysocki VH, Joyce KE, Jones CM, Beardsley RL. Journal of the American Society for Mass Spectrometry. 2008;19:190–208. doi: 10.1016/j.jasms.2007.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wysocki VH, Jones CM, Galhena AS, Blackwell AE. Journal of the American Society for Mass Spectrometry. 2008;19:903–913. doi: 10.1016/j.jasms.2008.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blackwell AE, Dodds ED, Bandarian V, Wysocki VH. Analytical Chemistry. 2011 doi: 10.1021/ac200452b. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vekey K, Somogyi A, Wysocki VH. Journal of Mass Spectrometry. 1995;30:212–217. [Google Scholar]
- 49.Smith DL, Wysocki VH, Colorado R, Shmakova OE, Graupe M, Lee TR. Langmuir. 2002;18:3895–3902. [Google Scholar]
- 50.Laskin J, Futrell JH. Journal of Chemical Physics. 2003;119:3413–3420. [Google Scholar]
- 51.Bonvin AM, Vis H, Breg JN, Burgering MJ, Boelens R, Kaptein R. Journal of Molecular Biology. 1994;236:328–341. doi: 10.1006/jmbi.1994.1138. [DOI] [PubMed] [Google Scholar]
- 52.Loo JA. International Journal of Mass Spectrometry. 2000;200:175–186. [Google Scholar]
- 53.Daniel JM, Friess SD, Rajagopalan S, Wendt S, Zenobi R. International Journal of Mass Spectrometry. 2002;216:1–27. [Google Scholar]
- 54.Breuker K, McLafferty FW. Proceedings of the National Academy of Sciences. 2008;105:18145–18152. doi: 10.1073/pnas.0807005105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yin S, Xie Y, Loo JA. Journal of the American Society for Mass Spectrometry. 2008;19:1199–1208. doi: 10.1016/j.jasms.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yin S, Loo JA. Journal of the American Society for Mass Spectrometry. 2010;21:899–907. doi: 10.1016/j.jasms.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 57.Breuker K, Brüschweiler S, Tollinger M. Angewandte Chemie International Edition. 2011;50:873–877. doi: 10.1002/anie.201005112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.