Abstract
Mitochondria are dynamic organelles that undergo frequent division and fusion, but the molecular mechanisms of these two events are not well understood. Dnm1p, a mitochondria-associated, dynamin-related GTPase was previously shown to mediate mitochondrial fission. Recently, a genome-wide yeast two-hybrid screen identified an uncharacterized protein that interacts with Dnm1p. Cells disrupted in this new gene, which we call NET2, contain a single mitochondrion that consists of a network formed by interconnected tubules, similar to the phenotype of dnm1Δ cells. NET2 encodes a mitochondria-associated protein with a predicted coiled-coil region and six WD-40 repeats. Immunofluorescence microscopy indicates that Net2p is located in distinct, dot-like structures along the mitochondrial surface, many of which colocalize with the Dnm1 protein. Fluorescence and immunoelectron microscopy shows that Dnm1p and Net2p preferentially colocalize at constriction sites along mitochondrial tubules. Our results suggest that Net2p is a new component of the mitochondrial division machinery.
INTRODUCTION
Mitochondria are essential organelles that participate in ATP synthesis, ion homeostasis, cell fate determination, lipid metabolism, and apoptosis (Saraste and Walker, 1982; Tzagoloff, 1983; Attardi and Schatz, 1988; Green and Reed, 1998; Wallace, 1999). To perform these functions, mitochondria can dynamically regulate their number, shape and locations in different eukaryotic cell types (Tzagoloff, 1983; Bereiter-Hahn, 1990; Bereiter-Hahn and Voth, 1994). In growing cells of the yeast Saccharomyces cerevisiae, mitochondria form a branched, tubular reticulum throughout the periphery of the cell (Hoffman and Avers, 1973; Stevens, 1977, 1981). During stationary phase, mitochondria fragment into 30–50 small organelles (Stevens, 1981). Mitochondria in yeast and most other cells are constantly fusing and dividing (Bereiter-Hahn, 1990; Nunnari et al., 1997). Fusion and division are balanced during cell growth so that each cell contains ∼5–10 separate organelles (Stevens, 1977). During meiosis and sporulation of diploid yeast, mitochondria undergo dramatic reorganization utilizing mitochondrial fusion and fission to eventually form four mitochondria, each of which encircles the nuclei of the separate spores (Miyakawa et al., 1984). In yeast, the presence of two GTPases is crucial for successful fusion and division of mitochondria. Fzo1p, an integral protein of the mitochondrial outer membrane, is required for mitochondrial fusion (Hermann et al., 1998; Bleazard et al., 1999; Sesaki and Jensen, 1999) and Dnm1p, a dynamin-related protein, mediates organelle fission (Otsuga et al., 1998; Bleazard et al., 1999; Sesaki and Jensen, 1999).
The first member of the Fzo1 protein family, fuzzy onions, was identified in Drosophila (Hales and Fuller, 1997). Mitochondria in the sperm cells of fuzzy onions mutants fail to fuse their mitochondria and, therefore, accumulate fragmented organelles. fuzzy onions mutants are defective in a mitochondrial transmembrane GTPase, which is required for mitochondrial fusion in sperm. Yeast cells that lack the Fzo1 protein fail to fuse their mitochondria during cell mating (Hermann et al., 1998; Sesaki and Jensen, 1999). Dnm1p was identified as a homologue of mammalian dynamin (Gammie et al., 1995), a protein required to pinch endocytic vesicles from the plasma membrane (De Camilli et al., 1995). Although yeast cells defective in Dnm1p show only a slight defect in endocytosis, they exhibit a striking mitochondrial phenotype. Cells that lack the Dnm1 protein have a single mitochondrion composed of a partially collapsed network of interconnected tubules (Otsuga et al., 1998; Bleazard et al., 1999; Sesaki and Jensen, 1999). Arguing for a direct role in mitochondrial fission, some Dnm1p in the yeast cell appears to be localized at sites of division (Bleazard et al., 1999; Sesaki and Jensen, 1999). Additional evidence that Fzo1p and Dnm1p are critical for fusion and division comes from double mutant studies. In dnm1 mutants, the normal branched, tubular structure of mitochondria is replaced by a single organelle consisting of interconnected tubules (Bleazard et al., 1999; Sesaki and Jensen, 1999). In fzo1 cells, numerous mitochondrial fragments accumulate (Hermann et al., 1998; Bleazard et al., 1999; Sesaki and Jensen, 1999). Surprisingly, the majority of dnm1Δ fzo1Δ double mutants contain several separate tubular mitochondria, reminiscent of wild-type organelles (Sesaki and Jensen, 1999). Thus, the number and shape of mitochondria in yeast cells seems to be controlled by a balance between the events of division and fusion that require Dnm1p and Fzo1p, respectively. In this paper, we present evidence that a new protein, named Net2p, binds to Dnm1p and works with Dnm1p to mediate mitochondrial fission.
MATERIALS AND METHODS
Strains and Relevant Genotypes
Strains used in this study are listed in Table 1. Strains BY4741 and BY4742 were previously described (Brachmann et al., 1998). A yeast strain disrupted in the YJL112w open reading frame (ORF) with kanMX4 (now called net2Δ) was purchased from Research Genetics (Huntsville, AL) and renamed RJ1253. This net2Δ strain was crossed to the dnm1Δ strain RJ1188, and the diploids were sporulated and dissected to generate dnm1Δ net2Δ strain RJ1285. fzo1Δ net2Δ strain RJ1286 was constructed by crossing fzo1Δ strain RJ1232 to RJ1253. Standard yeast media and genetic techniques (Adams et al., 1997) were used.
Table 1.
Strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| BY4741 | MATa leu2 met15 ura3 his3 | Brachmann et al., 1998 |
| BY4742 | MATα leu2 lys2 ura3 his3 | Brachmann et al., 1998 |
| RJ1188 | MATα dnm1::HIS3 trp1 his3 leu2 met15 ura3 | Sesaki and Jensen, 1999 |
| RJ1232 | MATα fzo1::kanMX4 his3 leu2 met15 ura3 trp1 | Sesaki and Jensen, 1999 |
| RJ1253 | MATa net2::kanMX4 his3 leu2 met15 ura3 | |
| RJ1285 | MATa his3 leu2 met15 ura3 trp1 dnm1::HIS3 net2::kanMX4 | This study |
| RJ1286 | MATa his3 leu2 met15 ura3 fzo1::kanMX4 net2::kanMX4 | This study |
| RJ1295 | MATa/MATα fzo1::kanMX4/FZO1 | This study |
| NET2/net2::kanMX4 his3/his3 leu2/leu2 met15/met15 ura3/ura3 | ||
| Y190 (RJ518) | MATa gal4 gal80 his3 trp1 ade2 leu2 URA3::GAL-LACZ LYS2::GAL-HIS3 | Bai and Elledge, 1996 |
| RJ515 | MATa gal4 gal80 his3 trp1 ade2 leu2 URA3::GAL-LACZ LYS2::GAL-HIS3 pTD1 pVA3 | Kerscher et al., 1998 |
Plasmid Construction
pKC2, a CEN-LEU2 plasmid that expresses OM45p-GFP, was constructed as follows. A 1380-base pair (bp) DNA fragment encoding the OM45 ORF and 200 bp of upstream sequences were polymerase chain reaction (PCR) amplified from yeast genomic DNA (Hoffman and Winston, 1987) using oligonucleotides 357 (5′-CCGCTCGAGCATATAATAATTGACAAG-3′) and 358 (5′-TATTGCGGCCGCCGTCCTTTTTCGAGC-3′). This PCR fragment was digested with XhoI and NotI and then inserted into pAA1, a CEN-LEU2 plasmid containing green fluorescent protein (GFP; Sesaki and Jensen, 1999) such that GFP was fused, in frame, to the C terminus of OM45p.
pKC5, a CEN-LEU2 plasmid encoding NET2 with a triple hemagglutinin (HA) epitope fused at the amino terminus, was produced as follows. A 2345-bp DNA fragment containing the NET2-coding sequences and 200 bp downstream were amplified from yeast genomic DNA (Hoffman and Winston, 1987) using oligonucleotides 442 (5′-GCGGGATCCATGTCAGTGAACGACCAAATAAC-3′) and 443 (5′-GCGGGATCCATTTACATTCCAGAACG-3′), digested with BamHI, and inserted into BamHI-cut pJE6. pJE6 contains the triple HA epitope inserted into the NotI site of pJE5 (Emtage and Jensen, 1993). pKC5 encodes the HA-Net2p fusion protein under the control of the TIM23 promoter region.
pKC11, a CEN-URA3 plasmid that expresses the Dnm1p-GFP fusion protein, was constructed by inserting a PvuI fragment containing DNM1-GFP from pHS20 (Sesaki and Jensen, 1999) into PvuI-cut pRS316 (Sikorski and Hieter, 1989).
pKC13, a CEN-URA3 plasmid that expresses the Dnm1p-MYC fusion protein, was constructed as follows. The triple MYC epitope was PCR amplified from an MYC-containing plasmid KB241 (a gift from D. Kornitzer, S. Kron, and G. Fink, Whitehead Institute, Cambridge, MA) using oligonucleotides 487 (5′-GTGCGGCCGCAGAG-GTGAACAAAA-GTTG-3′) and 497 (5′-GAGCGCGGTTAGCATGCCTGCAGGTCGAC-3′), digested with SacII and NotI, and inserted at the C terminus of Dnm1p in SacII/NotI-cut pHS20 (Sesaki and Jensen, 1999), forming pKC12. A SacII/XhoI fragment containing DNM1-MYC from pKC12 was ligated into SacII/XhoI-digested pRS316 (Sikorski and Hieter, 1989) to form pKC13.
Yeast Two-Hybrid Experiments
To construct plasmid pOAD-DNM1, a CEN-LEU2 plasmid that carries Dnm1p fused to the activating domain of Gal4p, we first amplified DNM1 sequences from yeast genomic DNA using oligonucleotides 453 (5′-CCACCAAACCCAAAAAAAGAGATCGAATTCCAGCTGACCACCATGGCTAGTTTAGAAGATCTTATTC-3′) and 454 (5′-CATAGATCTCTGCAGGTCGACGGATCCCCGGGAATTGCCATGTTAC-AGAATATTACTAATAAG-3′). Vector pOAD (Uetz et al., 2000) was linearized by digestion with NcoI and treated with phosphatase. We cotransformed pOAD and the DNM1-containing PCR product into yeast strain Y190 to allow homologous recombination to form pOAD-DNM1.
To construct pOBD-NET2, a CEN-TRP1 plasmid carrying NET2 fused to the Gal4p DNA-binding domain, we first amplified NET2 sequences from genomic DNA using oligonucleotides 455 (5′-CCACCAAACCCAAAAAAAGAGATCGAATTCCAGCTGACCACC-ATGTCAGTGAACGACCAAATAAC-3′) and 456 (5′-CATAGATCT-CTGCAGGTCGACGGATCCCCGGGAATTGCCATGTCATACGG-CCCAAATATTTAC-3′). Vector pOBD (Uetz et al., 2000) was digested with EcoRI and NcoI and cotransformed into strain Y190 together with the NET2 PCR product so that pOBD-NET2 was formed by homologous recombination. Y190 cells containing both pOAD-DNM1 and pOBD-NET2 were screened for β-galactosidase activity as described previously (Adams et al., 1997).
Microscopy
Samples were observed using a Axioskop microscope (Carl Zeiss Inc., Thornwood, NY) with a 100× objective. Fluorescence and differential interference contrast (DIC) images were captured with a Photometrics PXL charge-coupled device camera (Roper Industries, Princeton, NJ) using IP Lab software, version 3.2.0 (Signal Analytics, Vienna, VA).
Electron microscopy was performed as previously described (Rieder et al., 1996). Briefly, cells were fixed with 3% glutaraldehyde in 0.1 M sodium cacodylate, pH 7.4, 5 mM calcium chloride, 5 mM magnesium chloride, and 2.5% sucrose for 1 h at 25°C with gentle agitation, spheroplasted, embedded in 2% ultra-low-temperature agarose made in water, cooled, and cut into 1-mm3 pieces. The cells were subsequently postfixed in 1% osmium tetraoxide/1% potassium ferrocyanide in cacodylate buffer (0.1 M cacodylate/5 mM calcium chloride, pH 6.8) at room temperature for 30 min. The blocks of cells were washed four times in water, transferred to 1% thiocarbohydrazide at room temperature for 5 min, washed in water again, transferred to 1% osmium tetraoxide/1% potassium ferrocyanide in cacodylate buffer, pH 6.8, and incubated for 5 min at room temperature. The cells were then washed four times with water, en bloc stained in Kellenberger's uranyl acetate for 2 h, dehydrated through a graded series of ethanol washes, and embedded in Spurr resin. Sections were cut on a Ultracut T ultramicrotome (Leica, Deerfield, IL) and observed on a Philips EM 410 (FEI Co., Peabody, MA).
Immunoelectron microscopy was performed as described by Rieder et al. (1996). Briefly, cells were fixed in suspension for 15 min by adding an equal volume of freshly prepared 8% formaldehyde in phosphate-buffered saline (PBS). Cells were pelleted and resuspended in 4% formaldehyde in PBS and fixed for an additional 18–24 h at 4°C. Cells were then washed briefly in PBS and resuspended in 1% low-temperature–melting agarose. After cooling, the agarose blocks were trimmed into 1-mm3 pieces; infiltrated with 2.3 M sucrose in 20% polyvinylpyrrolidone at pH 7.4 for 2 h, mounted onto cryo-pins, and rapidly frozen in liquid nitrogen. Ultrathin cryosections were cut on a UCT ultramicrotome (Leica, Deerfield, IL) equipped with an FCS cryo-attachment and collected onto Formvar-carbon–coated nickel grids. Grids were washed with PBS containing 2.5% fetal calf serum in 10 mM glycine at pH 7.4, blocked in 10% fetal calf serum for 30 min, and then incubated overnight with a 1:50 dilution of monoclonal antibody to the HA epitope (Santa Cruz Biochemicals, Santa Cruz, CA) and 1:100 dilution of polyclonal rabbit antibody to the MYC epitope (Santa Cruz Biochemicals). After the grids were washed, they were incubated for 2 h in anti-mouse antibody conjugated to 5-nm gold particles and anti-rabbit antibody conjugated to 10-nm gold particles (Jackson Immunoresearch Labs, West Grove, PA). Grids were then washed with PBS, followed by water, and then immersed in a solution of 3.2% polyvinyl alcohol, 0.2% methyl cellulose, and 0.1% uranyl acetate. Grids were examined at 80 kV using the electron microscope.
Indirect Immunofluorescence
To localize the Net2 protein, yeast cells were grown to an OD600 of 0.6–0.7 in synthetic medium containing 2% galactose and the appropriate amino acids. Cells were then fixed with 4% paraformaldehyde (Sigma, St. Louis, MO) for 75 min, spheroplasted with zymolyase 20T (180 μg/ml; ICN, Costa Mesa, CA) and β-glucuronidase (1382.5 U/ml; Sigma) for 1 h at 30°C, then attached to poly-L-lysine (Sigma)-coated glass coverslips, and permeabilized using methanol/acetone as described previously (Harlow and Lane, 1988). Samples were incubated for 30 min with undiluted culture supernatant from 12CA5 cells (Niman et al., 1983). In some experiments cells were also incubated for 30 min with a 1:100 dilution of antiserum to the β subunit of the F1-ATPase, F1β, (a gift from M. Yaffe, University of California, San Diego). Coverslips were washed with PBS supplemented with 1% bovine serum albumin (Calbiochem, La Jolla, CA) and 0.05% Tween-20 (Sigma). Samples were then incubated for 30 min with a 1:200 dilution of fluorescein isothiocyanate (FITC)-coupled goat anti-mouse immunoglobulin G, a 1:500 dilution of rhodamine-conjugated goat anti-rabbit antibodies, or a 1:250 dilution of Cy3-conjugated goat anti-mouse antibodies (all from Boehringer Mannheim, Indianapolis, IN).
Subcellular and Submitochondrial Localization of Net2p and Dnm1p
Net2p and Dnm1p were localized by cellular fractionation essentially as described (Daum et al., 1982) using two strains that each contained a plasmid expressing the protein of interest. net2Δ strain RJ1253 expressed HA-Net2p from pKC5 and dnm1Δ strain RJ1188 contained pHS14, a CEN-LEU2 plasmid with Dnm1p fused to the HA epitope. Each strain was grown to an OD600 of 1.6 in synthetic media containing 2% galactose and supplemented with the appropriate amino acids. Cells were converted to spheroplasts, homogenized, and separated into a mitochondrial pellet and postmitochondrial supernatant by centrifugation at 10,000 × g for 10 min. The mitochondrial pellet was washed in breaking buffer (250 mM sucrose, 1 mM EDTA, and 20 mM HEPES, pH 7.4), supplemented with 1 mM phenylmethylsulfonylfluoride in dimethylsulfoxide (DMSO), 1 mg/ml aprotinin and leupeptin in water, and 1 mM trans-epoxysuccinyl-l-leucylamido(4-guanidino)butane (E-64) in water (all from Sigma).
Proteins were separated by SDS-PAGE (Laemmli, 1970) and then transferred to Immobilon membranes (Haid and Suissa, 1983). HA fusion proteins were identified by incubating membranes with a 1:10,000 dilution of mouse ascites fluid prepared from 12CA5 cells (BABCO, Berkeley, CA). Marker proteins were identified by incubating membranes with 1:10,000 dilutions of antisera against the following proteins: the F1β, Tim23p (Emtage and Jensen, 1993), OM45p (Yaffe et al., 1989), and hexokinase. Immune complexes were detected with horseradish peroxidase-conjugated secondary antibodies (Amersham Pharmacia, Piscataway, NJ) at a 1:10,000 dilution followed by enhanced chemiluminescence (SuperSignal; Pierce, Rockford, IL).
RESULTS
YJL112w Encodes a Novel WD-40 Repeat Containing Protein That Interacts with Dnm1p
Dnm1p, a mitochondria-associated GTPase, plays a crucial role in the division of yeast mitochondria (Bleazard et al., 1999; Sesaki and Jensen, 1999). A genome-wide yeast two-hybrid screen (Uetz et al., 2000) showed that the product of ORF YJL112w physically interacted with Dnm1p. We confirmed the yeast two-hybrid interaction between Dnm1p and YJL112w (Figure 1A). Plasmids containing Dnm1p fused to the Gal4p-activating domain (pOAD-DNM1) and the YJL112w ORF fused to the Gal4p DNA-binding domain (pOAD-YJL112w) were constructed and cotransformed into strain Y190, which contains Escherichia coli β-galactosidase under the control of the yeast GAL1 promoter region (GAL1::lacZ; Bai and Elledge, 1996). As shown in Figure 1, Y190 cells that contained both constructs expressed moderate levels of β-galactosidase activity, confirming the interaction between Dnm1p and YJL112w. No β-galactosidase activity was detected in Y190 cells transformed with pOBD-YJL112w, and therefore the pOBD-YJL112w construct was not self-activating. Two proteins known to interact, p53 and the large T antigen (Li and Fields, 1993), produced a positive interaction when expressed as fusion proteins to the Gal4p DNA-binding and the activation domains, respectively (Figure 1A).
Figure 1.
The YJL112w protein physically interacts with Dnm1p, a protein required for mitochondrial division. (A) A yeast two-hybrid assay shows binding between Dnm1p and Net2p. pOAD-DNM1, which expresses Dnm1p fused to the Gal4p DNA-binding domain, and pOBD-YJL112w, which encodes a YJL112w-Gal4p–activating domain fusion, were transformed into the gal1::LACZ strain Y190. As a positive control, Y190 was cotransformed with pVA3, encoding the p53-Gal4p DNA-binding domain fusion, and pTD1, expressing the large T-antigen fused to the Gal4p-activating domain. As a control for self-activation, pOBD-YJL112w was transformed into Y190. β-Galactosidase activity of cells was determined by measuring the hydrolysis of ortho-nitrophenyl-B-d-galactopyranoside. The average of three separate experiments is shown with SD. (B) Predicted domains of Net2p. Analysis by the COILS (Lupas et al., 1991) and WD-repeat (Garcia-Higuera et al., 1996) prediction programs suggest that Net2p contains a coiled-coil region in the amino terminus and six WD-40 repeats in the carboxyl terminus.
As depicted in Figure 1B, sequence analysis predicts that YJL112w is an 80-kDa protein that contains WD-40 repeats in its carboxyl-terminal domain (residues 369–714), and a coiled-coil region in its amino-terminal domain (residues 222- 300).
Cells Disrupted for NET2 Display a Single Mitochondrion Composed of an Interconnected Network of Tubules
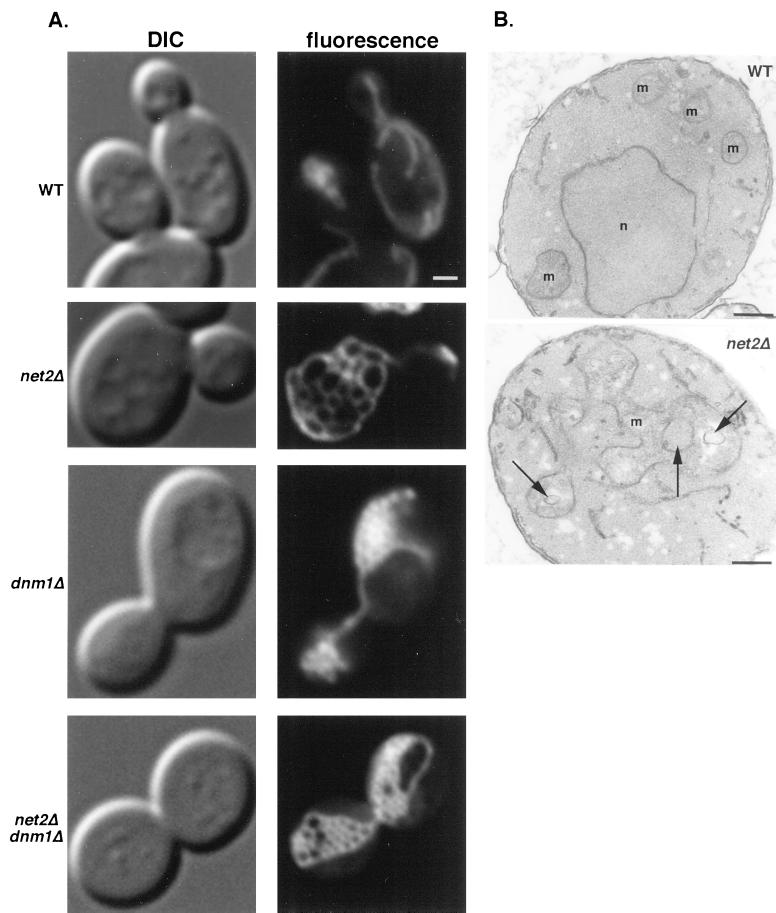
To determine whether YJL112w was involved in mitochondrial division, we examined yeast cells carrying a disruption in this gene (see MATERIALS AND METHODS). Wild-type and disruption strains were transformed with plasmid pHS12 (Sesaki and Jensen, 1999), which labels mitochondria fluorescent green because it expresses a fusion between the mitochondrial targeting signal from the matrix-localized cytochrome oxidase subunit IV (Cox4) protein and the GFP. When examined by fluorescence microscopy, we found that wild-type cells contained 5–10 branched, tubular-shaped mitochondria. In contrast, cells disrupted in YJL112w contained a single organelle consisting of a network of interconnected tubules (Figure 2A). A three-dimensional reconstruction of confocal sections confirmed that one highly branched mitochondrion was present in each net2Δ cell. Thin section electron micrographs (Figure 2B) showed normal cristae structure and were consistent with the idea that YJL112w-disrupted strains contained interconnected mitochondrial tubules. We have named the YJL112w gene NET2 for the complex mitochondrial network observed in the net2 null mutants. Because net2Δ mutants, like dnm1Δ mutants, contain a single mitochondrion per cell, we hypothesized that Net2p, like Dnm1p, is essential for mitochondrial fission.
Figure 2.
net2Δ cells contain a single mitochondrion composed of an interconnected network of tubules. (A) Wild-type strain BY4742 (WT), net2Δ strain RJ1253, dnm1Δ strain RJ1188, and net2Δ dnm1Δ strain 1285, each expressing matrix-targeted GFP from pHS12 (Sesaki and Jensen, 1999), were grown to mid log phase in synthetic medium with 2% galactose and examined by DIC and fluorescence microscopy. Representative images of cells are shown. Bar, 5 μm. (B) Wild-type and net2Δ cells were fixed, embedded, sectioned, and examined by electron microscopy. Arrows indicate spaces that result from the interconnected network of tubules that forms a single mitochondrion in the net2Δ cells. m, mitochondria; n, nucleus. Bar, 0.1 μm.
Although net2Δ mutants contained an interconnected network of tubules similar to that seen in dnm1Δ cells, there were also noticeable differences in the morphology of the organelles seen in the two cell types (Figure 2A). In most dnm1Δ mutants (83 of 100 total cells), the tubular network of mitochondria was collapsed at one or both ends of the organelle, and the mitochondrion was often at the periphery of the cell. The remaining 17 cells were generally more rounded in cell shape and showed larger, more spread-out mitochondrial networks. In contrast to dnm1Δ cells, the majority of net2Δ mutants (86 of 100 total cells) showed completely open mitochondrial networks that spread throughout the cell. The remaining 14 cells had smaller mitochondria with fewer interconnected tubules, but they still exhibited a visible mitochondrial network. To understand the relationship between Net2p and Dnm1p, we examined the phenotype of the net2Δ dnm1Δ double mutant. The mitochondrial morphology resulting from the net2Δ disruption was epistatic to dnm1Δ, because dnm1Δ net2Δ double mutants contained a single mitochondrion with interconnected tubules more similar to that seen in net2Δ cells (Figure 2A).
Mitochondrial Shape in net2Δ Mutants Does Not Depend on the Actin Cytoskeleton
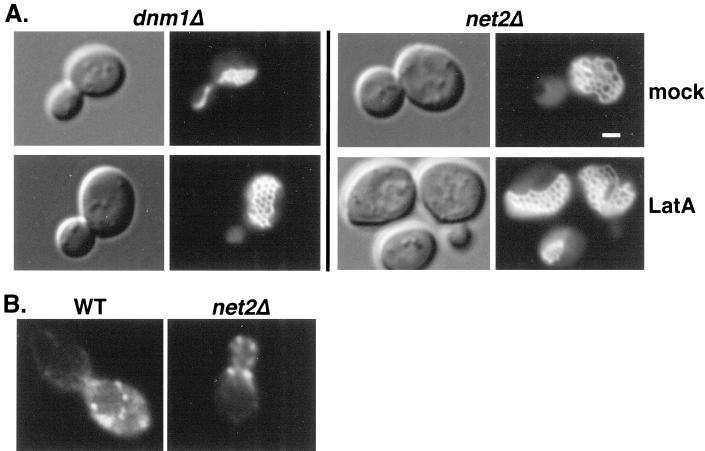
Yeast mitochondria are proposed to interact with the actin cytoskeleton (Lazzarino et al., 1994), and disruption of actin filaments by treating cells with latrunculin A causes mitochondria to fragment (Boldogh et al., 1998). This latrunculin A-induced fragmentation of mitochondria depended on Dnm1p function. In contrast to wild-type cells, dnm1Δ cells treated with latrunculin A did not fragment and remained as a single mitochondrial network (Jensen et al., 2000). However, instead of the partially collapsed structures seen in untreated cells, we found that latrunculin A-treated dnm1Δ mutants contained completely open mitochondrial networks, suggesting that the maintenance of partially collapsed mitochondrial networks in dnm1Δ mutants depended on an interaction between mitochondria and the actin cytoskeleton (A. Aiken Hobbs, unpublished results; Figure 3A). Phalloidin staining confirmed that actin cables and patches were disorganized in drug-treated cells. In contrast, the mitochondrial network seen in net2Δ mutants was unchanged by latrunculin A treatment (Figure 3A). Fully open tubular networks are seen in net2Δ cells with or without drug treatment. These results raised the possibility that net2Δ mutants were defective in mitochondria binding to actin.
Figure 3.
net2Δ cells display a mitochondrial network similar to dnm1Δ cells in which the actin cytoskeleton has been disrupted. (A) When treated with latrunculin A, dnm1Δ cells exhibit a mitochondrial morphology nearly identical to net2Δ cells. Strains RJ 1188 (dnm1Δ) and RJ 1253 (net2Δ), both expressing matrix-targeted GFP from pHS12, were grown to OD600 of 0.7 and then incubated with 250 μM latrunculin A in DMSO (LatA) or an equal volume of DMSO (mock) for 45 min at 24°C. Cells were then examined by DIC and fluorescence microscopy. Representative images are shown. Bar, 3 μm. (B) Actin patches and cables appear normal in net2Δ cells. Wild-type and net2Δ cells were fixed and stained with Alexa594-phalloidin (Molecular Probes, Eugene, OR) to visualize the actin cytoskeleton (Adams and Pringle, 1991). Representative fluorescent images are shown.
To determine whether Net2p was directly required for the organization of the actin cytoskeleton, we stained WT and net2Δ cells with fluorescent phalloidin and found that the distribution of actin cables and patches appeared identical in wild-type and net2Δ cells (Figure 3B). We therefore concluded that Net2p was not an integral part of the actin cytoskeleton but may bind mitochondria and actin.
fzo1Δ net2Δ Cells Contain Tubular, Nonfragmented Mitochondria
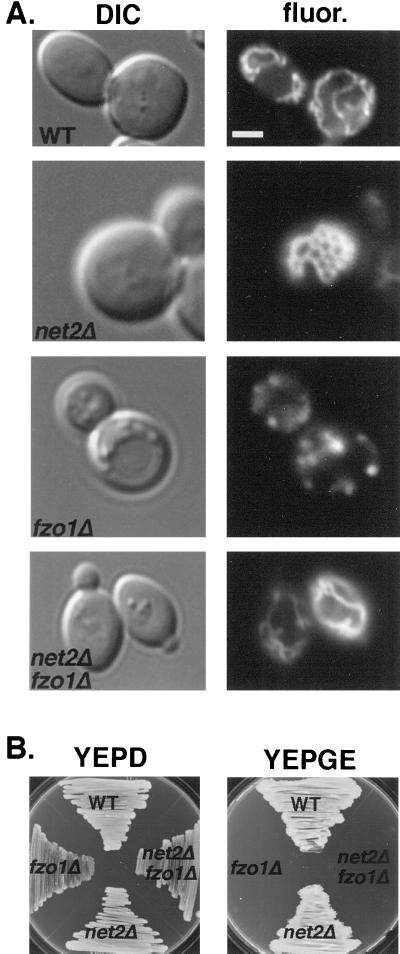
Mitochondrial division and fusion have been proposed to antagonistically regulate mitochondrial number and shape (Sesaki and Jensen, 1999). In fzo1Δ mutants, which are defective in mitochondrial fusion, ongoing division produces numerous mitochondrial fragments (Figure 4A). In dnm1Δ mutants, fusion without division produces a single organelle (Figure 2A). In dnm1Δ fzo1Δ double mutants, the number and shape of mitochondria is almost wild type (Bleazard et al., 1999; Sesaki and Jensen, 1999). Because Net2p and Dnm1p have similar phenotypes, we tested the idea that Net2p is required for mitochondrial fission by constructing net2Δ fzo1Δ double mutants and examining their mitochondrial morphology. The double mutant cells contained several normal tubule-shaped mitochondria similar to wild-type cells (Figure 4A). These seemingly normal tubules in net2Δ fzo1Δ mutants were not due to restored fusion activity, because the double mutant was still defective in the process of mitochondrial fusion normally seen after cell mating (K. Cerveny, unpublished observations). These results showed that disruption of NET2 suppresses the mitochondrial fragmentation phenotype seen in fzo1Δ mutants and suggested that Dnm1p and Net2p are both required for mitochondrial division.
Figure 4.
Deletion of NET2 suppresses fragmentation of mitochondria seen in fzo1Δ cells but does not prevent loss of mitochondrial DNA in fzo1Δ cells. (A) fzo1Δ net2Δ cells contain normal, tubular mitochondria. Wild-type (BY4742) and fzo1Δ strain RJ1232, net2Δ strain RJ1253, and fzo1Δ net2Δ strain RJ1286, all expressing OM45p-GFP from pKC2, were grown to mid log phase and viewed by DIC and fluorescence (fluor.) microscopy. Representative images of cells are shown. Bar, 5 μm. (B) Deletion of NET2 in fzo1Δ cells does not suppress the loss of mtDNA in fzo1Δ cells. NET2/net2Δ fzo1Δ/FZO1 (strain RJ1295) cells were sporulated, and a representative tetratype is depicted growing on medium containing either dextrose (YEPD) or glycerol plus ethanol (YEPGE) as the carbon source.
fzo1Δ mutants rapidly lose mitochondrial DNA (mtDNA), and this phenotype can be suppressed by inactivating Dnm1p (Bleazard et al., 1999; Sesaki and Jensen, 1999). To determine whether net2Δ also suppresses the loss of mtDNA in fzo1Δ mutants, we crossed net2Δ (strain RJ1253) to fzo1Δ (strain RJ1232) and analyzed the meiotic products (Figure 4B). All eight net2Δ fzo1Δ double mutants examined failed to grow on a nonfermentable carbon source (YEPGE), indicating that net2Δ did not suppress the loss of mtDNA. Direct examination of net2Δ fzo1Δ cells with the fluorescent DNA stain, 4′,6′-diamindino-2-pheylindole, confirmed that none of these double mutants contained mtDNA. Although it appears that disruption of NET2 differs from dnm1 deletions, with regard to suppressing the mtDNA loss of fzo1Δ cells, it is important to note that not all dnm1Δ fzo1Δ double mutants maintain mtDNA. Only ∼25% of dnm1Δ fzo1Δ cells produced in genetic crosses contain mtDNA (H. Sesaki, unpublished observations). Whether a small number of net2Δ fzo1Δ segregants will retain mtDNA awaits further studies.
Net2p Is Located in Punctate Structures on the Mitochondrial Surface
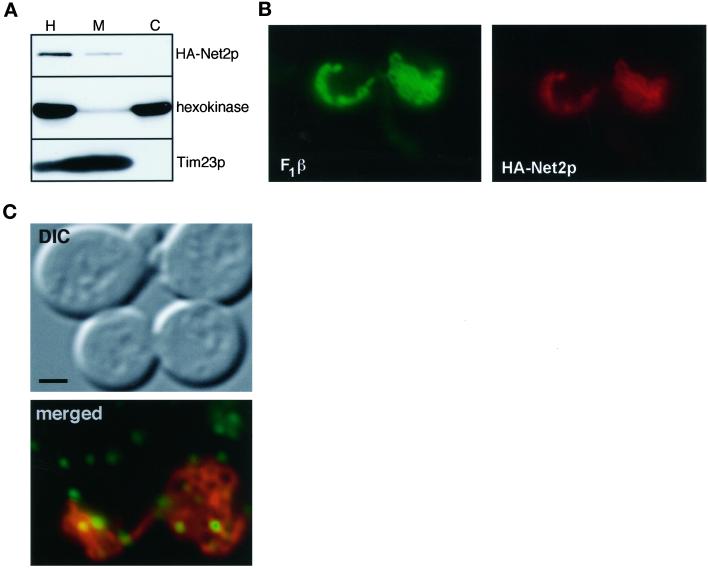
Because the Net2 protein appeared to play a role in mitochondrial division, we asked where Net2p was located in the yeast cell. We fused the HA epitope of influenza HA (Field et al., 1988) to the amino terminus of Net2p and expressed this construct in net2Δ cells. Normal mitochondrial morphology was seen in these cells, indicating that the HA-Net2p fusion protein was functional (see Figure 6). Cells that expressed HA-Net2p were homogenized and separated into a mitochondrial fraction and a crude cytosolic pellet by centrifugation at 10,000 × g for 10 min (Figure 5A). Western blots showed that the HA-Net2 protein was found in the mitochondrial pellet along with Tim23p, a mitochondrial protein (Emtage and Jensen, 1993). We therefore concluded that HA-Net2p is a mitochondrial protein. In contrast, a Dnm1p-HA fusion protein (Sesaki and Jensen, 1999) cofractionated with hexokinase in the cytosol (Figure 5B). Previous studies showed that Dnm1p associated with the mitochondria (Otsuga et al., 1998). In light of these data, it is important to note that the Dnm1p-HA fusion protein fully complemented the DNM1 deletion (Sesaki and Jensen, 1999). Even when we loaded five times more protein in the mitochondrial lane for the Dnm1p-HA samples as compared with the HA-Net2p samples, no Dnm1p was found with the mitochondrial pellet.
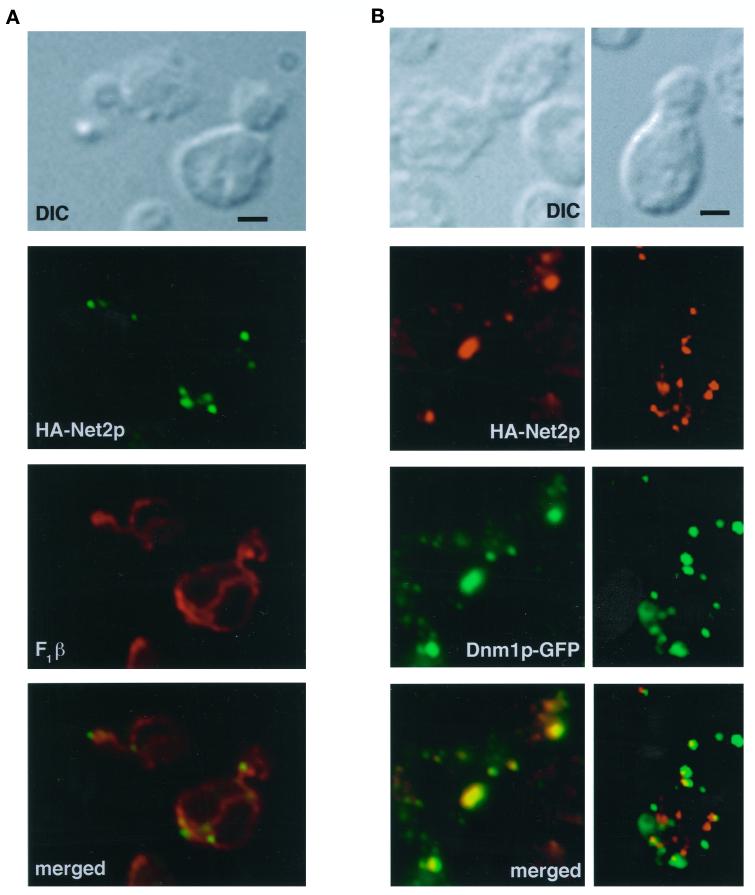
Figure 6.
Net2p forms punctate structures that colocalize with mitochondria and with Dnm1p. (A) net2Δ cells expressing HA-Net2p from pKC5 were fixed, permeabilized, and then incubated with antibodies to the HA epitope and the mitochondrial protein, F1β. Immune complexes were detected with either rhodamine-conjugated secondary antibodies (F1β) or FITC-linked antibodies (HA-Net2p), and cells were examined by DIC and fluorescence microscopy. Single and merged images are shown. Bar, 5 μm. (B) Most of Net2p-HA colocalizes with Dnm1p-GFP. net2Δ dnm1Δ cells (RJ1285) that expressed HA-Net2p from pKC5 and Dnm1p-GFP from pKC11 were fixed, permeabilized, and incubated with HA antibodies. Cells were then incubated with CY3-conjugated goat anti-mouse immunoglobulin G. Representative images from the green (Dnm1p-GFP) and red (HA-Net2p) channels, as well as merged images, are shown. Bar, 5 μm.
Figure 5.
Net2p is peripherally associated with the mitochondrial outer membrane. (A) Net2p cofractionates with mitochondria. net2Δ cells expressing HA-Net2p from pKC5 were subjected to cell fractionation. Aliquots of homogenate (H), cytosol (C), and mitochondria (M) representing equivalent numbers of cells were subjected to SDS-PAGE and analyzed by Western blotting with antibodies to the HA-epitope (HA-Net2p), hexokinase, and Tim23p. (B) Dnm1p is found in the cytosol after cell fractionation. dnm1Δ cells expressing Dnm1p-HA from pHS14 (Sesaki and Jensen, 1999) were treated and analyzed as in A except that 100 μg of total protein was loaded per lane. (C) Net2p is a peripheral membrane protein. Mitochondria (150 μg) from net2Δ cells expressing HA-Net2p from pKC5 were resuspended to a concentration of 1 mg/ml total protein in either 0.1 M Na2CO3, pH 11, or 1 M NaCl and then centrifuged at 100,000 × g for 60 min. Equal amounts of pellets (P) and supernatants (S) were subjected to SDS-PAGE and analyzed by Western blotting with antibodies to the HA epitope (HA-Net2p), the β-subunit of the F1-ATPase (F1β), and Tim23p. (D) Net2p associates with the mitochondrial outer membrane. mitochondria (150 μg, 1 mg/ml) from cells expressing HA-Net2p were incubated either in the presence (+) or absence (−) of trypsin (150 μg/ml) on ice for 20 min in breaking buffer (0.625 M sucrose in 20 mM HEPES, pH 7.4). Lima bean trypsin inhibitor (Sigma) was then added to a concentration of 2 mg/ml. Mitochondria were pelleted and subjected to SDS-PAGE and analyzed by Western blotting with antibodies directed against the HA epitope (HA-Net2p), OM45p, and Tim23p.
We found that Net2p is peripherally associated with the mitochondrial outer membrane. As shown in Figure 5C, mitochondria isolated from HA-Net2p-expressing cells were treated with either salt or alkali and separated into a pellet and supernatant by centrifugation. Western blots showed that HA-Net2p pelleted with mitochondria after treatment with sodium chloride. However, after treatment with sodium carbonate, pH 11, HA-Net2p was found in the supernatant along with a peripheral membrane protein, the β-subunit of the F1-ATPase (F1β). We concluded that HA-Net2p associated tightly with mitochondrial membranes. When HA-Net2p-containing mitochondria were incubated with trypsin, all of the HA-Net2 protein was digested, similarly to OM45p, a mitochondrial outer membrane protein that faces the cytosol (Figure 5D; Yaffe et al., 1989). In contrast, Tim23p, which contains a protease-sensitive domain that faces the intermembrane space (Ryan et al., 1998), was protected from digestion by the outer membrane. These results indicated that Net2p associates with the cytosolic face of the mitochondrial outer membrane.
Immunofluorescence studies localized Net2p to punctate structures on the mitochondrial surface (Figure 6A). net2Δ cells that expressed HA-Net2p were fixed, permeabilized, and double labeled using antibodies to the HA epitope (Figure 6, green) and antibodies against the mitochondrial F1β protein (Figure 6, red). In contrast to F1β, which showed uniform staining of mitochondrial tubules, virtually all of the HA-Net2p associated with the mitochondria in dot-like structures. Three-dimensional reconstructions of merged F1β (red) and HA-Net2p (green) confocal images confirmed the mitochondrial location of the HA-Net2p dots (K. Cerveny, unpublished observations). On average, we observed between 7 and 16 HA-Net2p dots per yeast cell. Generally the dots appeared to be distributed along the tubules and were often found at branch points in the mitochondrial network (see Figure 6A). Interestingly, of 25 budded cells examined, all 25 cells had at least one Net2p-containing dot in the bud neck (Figure 6A), suggesting that the bud neck is one place where mitochondrial division always occurs.
Net2p and Dnm1p Colocalize in Yeast Cells but Associate with Mitochondria Independently of Each Other
Previous studies showed that Dnm1p localizes to the mitochondrial surface in large, punctate structures (Bleazard et al., 1999; Sesaki and Jensen, 1999), similar to HA-Net2p. Because yeast two-hybrid analysis suggested that Dnm1p and Net2p physically interact (Uetz et al., 2000; Figure 1), we asked whether these proteins also coaligned in vivo. When yeast cells that expressed HA-Net2p and a Dnm1p-GFP fusion proteins were examined by immunofluorescence microscopy, both Net2p and Dnm1p were found in similar dot-like structures on the mitochondrial surface (Figure 6B). When red and green images were merged, a variable number of dot-like structures contained both Dnm1p and Net2p. For example, in one cell, 9 of 11 punctate dots contained both proteins (Figure 6B, left). In another cell, 7 of 14 dots contained Dnm1p and Net2p (Figure 6B, right). Quantitation of 30 total cells showed that colocalization ranged from 50 to 90%. However, when 25 budded cells were examined, every punctate structure found in the bud neck always contained both Dnm1p and Net2p. Specifically, of 32 HA-Net2p dots located in the bud necks, all colocalized with Dnm1p-GFP. We concluded that Dnm1p and Net2p physically associate on the mitochondrial surface, and we hypothesized that the Dnm1p-Net2p interaction is dynamic.
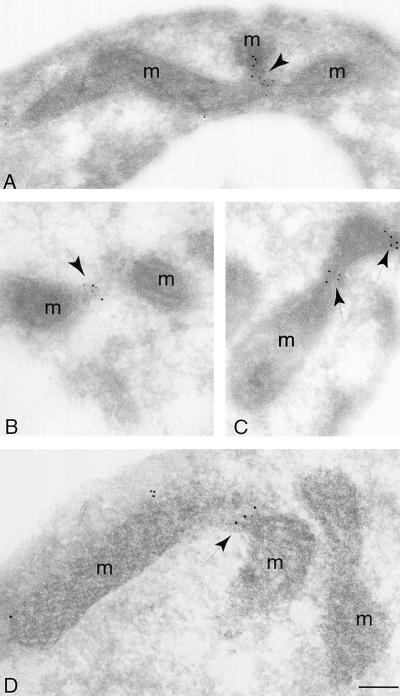
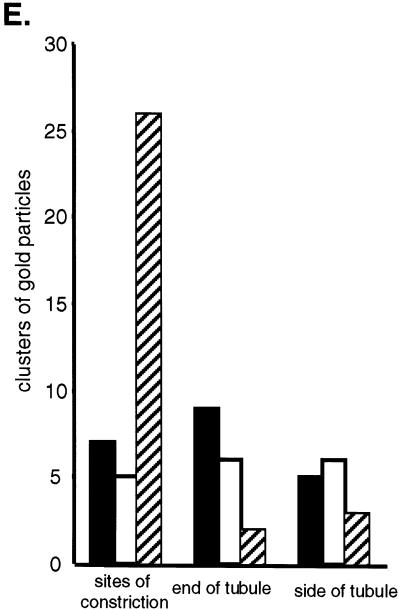
Net2p and Dnm1p appear to associate at sites of active mitochondrial division. We examined mitochondria in cells expressing HA-Net2p and Dnm1p-MYC fusion proteins by immunogold electron microscopy. To quantitate the association of Net2p and Dnm1p and their relationship to mitochondrial constriction sites, a total of 69 clusters of gold particles containing Dnm1p, Net2p, or both Dnm1p and Net2p were counted. Mitochondrial constrictions appear to represent sites of ongoing division. For example, Figures 7, A and C, shows intermediate constrictions, and Figure 7B is an example of a mitochondrial tubule that appears almost completely divided. When both Net2p and Dnm1p were found together, they were located at constrictions in the mitochondrial tubule nearly 84% of the time (Figure 7E). In contrast, only 30% of the clusters containing either Net2p (n = 21) or Dnm1p (n = 17) were found at mitochondrial constrictions (Figure 7E). We found that the number of clusters containing only Net2p or Dnm1p were evenly distributed between the ends of mitochondria and the sides of tubules (Figure 7E). Our results therefore suggest that Net2p and Dnm1p together act to catalyze mitochondrial fission.
Figure 7.
Net2p and Dnm1p colocalize at constriction sites along mitochondrial tubules. (A–D) Immunoelectron micrographs show colocalization of HA-Net2p and Dnm1p-MYC at sites of mitochondrial division. net2Δ dnm1Δ cells (RJ1285) expressing HA-Net2p from pKC5 and Dnm1p-MYC from pKC13 were fixed, embedded, and frozen. Cryosections were incubated with mouse antibodies to the HA epitope and rabbit antibodies to the MYC epitope, followed by incubation with anti-mouse antibodies conjugated to 5-nm gold particles (HA-Net2p) and anti-rabbit antibodies linked to 10-nm gold particles (Dnm1p-MYC). After the sections were stained, they were examined by electron microscopy. Arrows indicate clusters of gold particles at mitochondrial constrictions. m, mitochondria. Bar, 0.5 μm. (E) Quantitation of the immunogold electron micrographs show that the majority of Net2p-Dnm1p-containing complexes are found at sites of mitochondrial division. A total of 69 clusters of gold particles containing Net2p (black bars), Dnm1p (white bars) or both Net2p and Dnm1p (hatched bars) were counted and grouped based on their locations along the mitochondria.
We found that Dnm1p does not require Net2p for its mitochondrial association. In net2Δ cells that expressed Dnm1p-GFP, we found that Dnm1p was localized in punctate dots on the mitochondrial tubules, virtually identical in size and number to those seen in wild-type cells (compare Figures 6B and 8C). Similarly, Net2p localization to mitochondria did not depend on Dnm1p function. When dnm1Δ cells that expressed HA-Net2p were fractionated by differential centrifugation, HA-Net2p pelleted with the mitochondrial fraction (Figure 8B). However, we found that localization of Net2p to punctate dots on mitochondria required Dnm1p. When dnm1Δ cells expressing HA-Net2p were examined, we found Net2p evenly distributed on the mitochondrial surface (Figure 8A). Our results suggest that recruitment of Net2p into dot-like structures requires Dnm1p function, but binding to mitochondria is independent of the Dnm1 protein.
Figure 8.
Net2p and Dnm1p do not recruit each other to the mitochondrial outer membrane. (A) Location of Net2p in dnm1Δ cells. dnm1Δ cells, strain RJ1188, expressing HA-Net2p from pKC5 were grown to OD600 of 0.8 in synthetic medium with 2% galactose and subjected to cell fractionation. Aliquots of homogenate (H), cytosol (C), and mitochondria (M) representing equivalent numbers of cells were subjected to SDS-PAGE and analyzed by Western blotting with antibodies against the HA epitope (HA-Net2p), hexokinase, and Tim23p. (B) Formation of punctate structures containing Net2p requires Dnm1p. net2Δ dnm1Δ cells (RJ1285), expressing HA-Net2p from pKC5, were fixed, permeabilzed, and then incubated with antibodies to the HA epitope and the mitochondrial F1β protein. Immune complexes were visualized with FITC-conjugated antibodies (F1β) and CY3-linked antibodies (HA-Net2p). Representative images from the green (F1β) and red (HA-Net2p) channels are shown. (C) Localization of Dnm1p in net2Δ cells. net2Δ strain RJ1253, constitutively expressing Dnm1p-GFP from pHS20 (Sesaki and Jensen, 1999), was transformed with pHS51, which expresses Cox4-RFP under the control of the galactose-inducible GAL1 promoter. Cells were grown in media with 2% raffinose to an OD600 of 0.3 and then resuspended in synthetic medium containing 2% galactose and 2% sucrose for 2.5 h to induce expression of matrix-targeted RFP. Cells were then examined by DIC and fluorescence microscopy. A representative merged image from the red (cox4-RFP mitochondria) and green (Dnm1p-GFP) is shown. Bar, 5 μm. Note that the upper two cells are not expressing Cox4-RFP.
DISCUSSION
Our results indicate that Net2p is a new protein required for the division of mitochondria in yeast. For example, net2Δ cells have a single mitochondrion composed of interconnected tubules instead of the 5–10 separate tubules seen in wild-type cells. This phenotype is very similar to that seen in cells lacking Dnm1p, a protein previously shown to be required for mitochondrial scission (Bleazard et al., 1999; Sesaki and Jensen, 1999). In addition, disruption of NET2 suppresses the fragmentation of mitochondria in fzo1Δ cells. net2Δ fzo1Δ double mutants contain tubule-shaped mitochondria very similar to those seen in wild-type cells. Similar results were seen previously in dnm1 fzo1 double mutants (Sesaki and Jensen, 1999). These results indicate that both net2 and dnm1 mutants are defective in division of mitochondria, an activity that is antagonistic to fusion. We speculate that mitochondrial number and shape is normally controlled by a balance between division, mediated by Net2p and Dnm1p, and fusion, mediated by Fzo1p. The physical interaction between Net2p and Dnm1p provides additional evidence that Net2p plays a role in mitochondrial division.
NET2 encodes a novel 80-kDa protein that is predicted to contain six WD-40 repeats in its carboxyl terminus and an amino terminal coiled-coil region. The WD-40 repeats are predicted to form a β-propeller tertiary structure, which has been implicated in a wide variety of protein-protein interactions (Garcia-Higuera et al., 1996). Coiled-coil motifs have also been found to mediate the association between many different proteins (Lupas, 1996). Because the Net2 protein interacts with Dnm1p in the yeast two-hybrid assay, we are currently testing which domain of Net2p is required for Dnm1p binding. It is tempting to speculate that the amino-terminal coiled-coil region of Net2p may interact with the GTPase effector domain of Dnm1p, which also appears to contain a coiled-coil motif.
Immunofluorescence studies indicate that both Net2p and Dnm1p are located in punctate structures on the surface of mitochondrial tubules. We found that a variable number of these dot-like complexes contain both Dnm1p and Net2p. In some cells, we found that ∼50% of the dots contained Net2p and Dnm1p, whereas in other cells, more than 90% of the dot-like structures contained both proteins. The remaining dot-like structures contained either Net2p or Dnm1p alone. Immmunogold electron microscopy also indicates that the punctate dots contain Net2p, Dnm1p, or both Net2p and Dnm1p. Our results suggest that the Net2p-Dnm1p interaction is dynamic. We propose that structures containing both Net2p and Dnm1p are active in mitochondrial division, whereas those that contain only Net2p or Dnm1p are pre- or postdivision complexes. Supporting this idea, immunogold staining showed that sites of mitochondrial constriction are enriched in complexes containing both Net2p and Dnm1p. We also found that in the neck region of budded cells Dnm1p and Net2p were always associated, and at least one punctate dot in the bud neck contained both Net2p and Dnm1p. Because the bud neck is the site of cytokinesis, it is plausible that cells activate division at this site to ensure the segregation of mitochondria to both mother and daughter cells.
While this paper was in review, two reports were published also describing the isolation and characterization of Net2p, called either Mdv1p (Tieu and Nunnari, 2000) or Gag3p (Fekkes et al., 2000). Both papers similarly concluded that Net2p/Mdv1p/Gag3p is a mitochondrially associated, Dnm1p-interacting protein required for division. However, Tieu and Nunnari (2000) concluded that all of the punctate structures on the mitochondrial surface contained both Mdv1p and Dnm1p. They did not observe dots that contained only Mdv1p or only Dnm1p. It is possible that the results of the two papers differ because our microscopy was done after cells were fixed, whereas Tieu and Nunnari examined live cells. Alternatively, the GFP-Mdv1p construct used by Tieu and Nunnari was overproduced by expression from the GAL1 promoter, possibly altering the distribution of Mdv1p. Clearly, the interaction between Dnm1p and Net2p/Mdv1p/Gag3p requires additional study.
We found that localization of Net2p to mitochondria does not require Dnm1p but that the punctate distribution of Net2p is dependent on Dnm1p function. Cell fractionations show that Net2p remains associated with mitochondria. However, in dnm1Δ cells Net2p is evenly distributed along mitochondrial tubules and not in punctate dot-like structures. In cell fractionations, the Net2p protein remains on the mitochondrial surface, whereas Dnm1p is found in the postmitochondrial supernatant. In intact cells, however, most of a Dnm1p-GFP fusion protein and all of HA-Net2p fusion is located in punctate structures on the mitochondrial surface. These results indicate that the association of Dnm1p with mitochondria is more labile than that of Net2p. They also raise the possibility that Net2p plays a role in anchoring Dnm1p onto the mitochondrial surface. Arguing against this idea, we found that Dnm1-GFP remains associated with mitochondria in net2Δ cells. Therefore, Dnm1p associates with the mitochondrial outer membrane by a mechanism independent of Net2p. Although Dnm1p-GFP-containing dots are located on the mitochondrial tubules in net2Δ cells, division does not occur in the absence of the Net2p protein. Recently, a mitochondrial outer membrane protein, called Fis1p, was shown to mediate mitochondrial fission (Mozdy et al. 2000). In fis1 mutants, only some of Net2p and none of the Dnm1 protein remained bound to mitochondria, suggesting that Fis1p plays a role in recruiting both proteins to the mitochondrial surface (Mozdy et al. 2000).
Our results also suggest that the actin cytoskeleton is somehow involved in mitochondrial division. Yeast mitochondria appear to interact with the actin cytoskeleton (Drubin et al., 1993; Lazzarino et al., 1994; Smith et al., 1995). Although this association has been implicated in mitochondrial shape and movement, it is also likely that an actin-mitochondria interaction plays a role in division and fusion. Disruption of the actin network with latrunculin A causes mitochondria to fragment (Boldogh et al., 1998), and this fragmentation requires Dnm1p (Jensen et al., 2000; see Figure 3). Our results raise the possibility that Net2p/Mdv1p/Gag3p mediates a connection between the division machinery and actin. In dnm1Δ mutants, mitochondrial networks are partially collapsed, but treatment of dnm1Δ cells with latrunculin A results in completely open networks. Thus, the collapsed networks seen in untreated cells appear to result from an interaction with actin. In contrast, net2Δ networks are fully open even in the absence of latrunculin A treatment, suggesting that the association of mitochondria with actin requires Net2p/Mdv1p/Gag3p. Experiments to test this possibility directly are in progress. Recently, Ochoa et al., (2000) found a functional link between dynamin and the actin cytoskeleton and proposed that actin assists in endocytic vesicle formation. They suggested that an actin cytoskeletal scaffold forms around the neck of endocytic vesicles in a dynamin-dependent manner and provides the force for membrane fission (Ochoa et al., 2000). The role of the actin cytoskeleton in yeast mitochondrial division awaits further studies.
Several possible models explain how Net2p functions with Dnm1p to divide mitochondria. For example, Net2p and Dnm1p may both act directly to pinch mitochondria. Dnm1p is a homologue of dynamin that has been proposed to function as a mechanochemical pinchase (Hinshaw and Schmid, 1995). Dnm1p and Net2p may function together to constrict and pinch mitochondrial membranes. Supporting a direct role for both Dnm1p and Net2p, we found that overexpression of Dnm1p in net2Δ mutants does not rescue the mitochondrial division defect, and multicopy plasmids containing NET2 do not restore fission in dnm1Δ cells. In a second model, Net2p divides mitochondria after activation by Dnm1p. Dynamin has been proposed to regulate the endocytic fission machinery (Sever et al., 1999), such as the lipid-modifying endophilin protein (Schmidt et al., 1999). By analogy, Dnm1p may stimulate activity of Net2p and other members of the mitochondrial fission machinery. Alternatively, Net2p may regulate the activity of Dnm1p. For instance, Net2p may enhance the GTPase activity of Dnm1p or stimulate the exchange of GDP for GTP. Clearly, additional studies are needed to elucidate the mechanisms by which Dnm1p and Net2p mediate mitochondrial fission. Furthermore, because both Dnm1p and Net2p are associated with the mitochondrial outer membrane, it will be interesting to determine whether scission of the mitochondrial inner membrane requires machinery separate from that required for outer membrane division.
ACKNOWLEDGMENTS
We thank Hiromi Sesaki, Kathy Wilson, Carolyn Machamer, Naresh Sepuri, Alyson Aiken Hobbs, Matthew Youngman, and Cory Dunn for productive discussions and critical comments on the manuscript. We also thank Michael Yaffe for antiserum to F1β, Ben Glick for the RFP variant, Jef Boeke for strains, and Stan Fields for the pOAD and pOBD plasmids. This work was supported by grant R01-GM54021 from the United States Public Health Service to R.E.J. and in part by National Institutes of Health training grant 2T32-GM07445 to K.L.C.
REFERENCES
- Adams A, Gottschling D, Kaiser C, Stearns T. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. [Google Scholar]
- Adams AE, Pringle JR. Staining of actin with fluorochrome-conjugated phalloidin. Methods Enzymol. 1991;194:729–731. doi: 10.1016/0076-6879(91)94054-g. [DOI] [PubMed] [Google Scholar]
- Attardi G, Schatz G. Biogenesis of mitochondria. Annu Rev Cell Biol. 1988;4:289–333. doi: 10.1146/annurev.cb.04.110188.001445. [DOI] [PubMed] [Google Scholar]
- Bai C, Elledge SJ. Gene identification using the yeast two-hybrid system. Methods Enzymol. 1996;273:331–347. doi: 10.1016/s0076-6879(96)73029-x. [DOI] [PubMed] [Google Scholar]
- Bereiter-Hahn J. Behavior of mitochondria in the living cell. Int Rev Cytol. 1990;122:1–62. doi: 10.1016/s0074-7696(08)61205-x. [DOI] [PubMed] [Google Scholar]
- Bereiter-Hahn J, Voth M. Dynamics of mitochondria in living cells: shape changes, dislocations, fusion, and fission of mitochondria. Microsc Res Tech. 1994;27:198–219. doi: 10.1002/jemt.1070270303. [DOI] [PubMed] [Google Scholar]
- Bleazard W, McCaffery JM, King EJ, Bale S, Mozdy A, Tieu Q, Nunnari J, Shaw JM. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat Cell Biol. 1999;1:298–304. doi: 10.1038/13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldogh I, Vojtov N, Karmon S, Pon LA. Interaction between mitochondria and the actin cytoskeleton in budding yeast requires two integral mitochondrial outer membrane proteins, Mmm1p and Mdm10p. J Cell Biol. 1998;141:1371–1381. doi: 10.1083/jcb.141.6.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains from Saccharomyces cerevisiae 288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Daum G, Böhni PC, Schatz G. Import of proteins into mitochondria: cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J Cell Biol. 1982;257:13028–13033. [PubMed] [Google Scholar]
- De Camilli P, Takei K, McPherson PS. The function of dynamin in endocytosis. Curr Opin Neurobiol. 1995;5:559–565. doi: 10.1016/0959-4388(95)80059-x. [DOI] [PubMed] [Google Scholar]
- Drubin DG, Jones HD, Wertman KF. Actin structure and function: roles in mitochondrial organization and morphogenesis in budding yeast and identification of the phalloidin-binding site. Mol Biol Cell. 1993;4:1277–1294. doi: 10.1091/mbc.4.12.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emtage JL, Jensen RE. MAS6 encodes an essential inner membrane component of the yeast mitochondrial protein import pathway. J Cell Biol. 1993;122:1003–1012. doi: 10.1083/jcb.122.5.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekkes P, Shepard KA, Yaffe MP. Gag3p, an outer membrane protein required for fission of mitochondrial tubules. J Cell Biol. 2000;151:333–340. doi: 10.1083/jcb.151.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field J, Nikawa J, Broek D, MacDonald B, Rodgers L, Wilson IA, Lerner RA, Wigler M. Purification of a RAS-responsive adenylyl cyclase complex from Saccharomyces cerevisiae by use of an epitope addition method. Mol Cell Biol. 1988;8:2159–2165. doi: 10.1128/mcb.8.5.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammie AE, Kurihara LJ, Vallee RB, Rose MD. DNM1, a dynamin-related gene, participates in endosomal trafficking in yeast. J Cell Biol. 1995;130:553–566. doi: 10.1083/jcb.130.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Higuera I, Fenoglio J, Li Y, Lewis C, Panchenko MP, Reiner O, Smith TF, Neer EJ. Folding of proteins with WD-repeats: comparison of six members of the WD-repeat superfamily to the G protein beta subunit. Biochemistry. 1996;35:13985–13994. doi: 10.1021/bi9612879. [DOI] [PubMed] [Google Scholar]
- Green DR, Reed JC. Mitochondria and apoptosis. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- Haid A, Suissa M. Immunochemical identification of membrane proteins after sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Methods Enzymol. 1983;96:192–205. doi: 10.1016/s0076-6879(83)96017-2. [DOI] [PubMed] [Google Scholar]
- Hales KG, Fuller MT. Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell. 1997;90:121–129. doi: 10.1016/s0092-8674(00)80319-0. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- Hermann GJ, Thatcher JW, Mills JP, Hales KG, Fuller MT, Nunnari J, Shaw JM. Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J Cell Biol. 1998;143:359–373. doi: 10.1083/jcb.143.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinshaw JE, Schmid SL. Dynamin self-assembles into rings suggesting a mechanism for coated vesicle budding. Nature. 1995;374:190–192. doi: 10.1038/374190a0. [DOI] [PubMed] [Google Scholar]
- Hoffman CS, Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation into Escherichia coli. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- Hoffman HP, Avers CJ. Mitochondrion of yeast: ultrastructural evidence for one giant, branched organelle per cell. Science. 1973;181:749–751. doi: 10.1126/science.181.4101.749. [DOI] [PubMed] [Google Scholar]
- Jensen RE, Aiken Hobbs AE, Cerveny K, Sesaki H. Yeast mitochondrial dynamics: fusion, division, segregation and shape. Microsc Res Tech. 2000;51:573–583. doi: 10.1002/1097-0029(20001215)51:6<573::AID-JEMT7>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lazzarino DA, Boldogh I, Smith MG, Rosand J, Pon LA. Yeast mitochondria contain ATP-sensitive, reversible actin-binding activity. Mol Biol Cell. 1994;5:807–818. doi: 10.1091/mbc.5.7.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Fields S. Identification of mutations in p53 that affect its binding to SV40 large T antigen by using the yeast two-hybrid system. FASEB J. 1993;7:957–963. doi: 10.1096/fasebj.7.10.8344494. [DOI] [PubMed] [Google Scholar]
- Lupas A. Prediction and analysis of coiled-coil structures. Methods Enzymol. 1996;266:513–525. doi: 10.1016/s0076-6879(96)66032-7. [DOI] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Miyakawa I, Aoi H, Sando N, Kuroiwa T. Fluorescence microscopic studies of mitochondrial nucleoids during meiosis and sporulation in the yeast Saccharomyces cerevisiae. J Cell Sci. 1984;66:21–38. doi: 10.1242/jcs.66.1.21. [DOI] [PubMed] [Google Scholar]
- Mozdy AD, McCaffery JM, Shaw JM. Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J Cell Biol. 2000;151:367–380. doi: 10.1083/jcb.151.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niman HL, Houghten RA, Walker LE, Reisfeld RA, Wilson IA, Hogle JM, Lerner RA. Generation of protein-reactive antibodies by short peptides is an event of high frequency: implications for the structural basis of immune recognition. Proc Natl Acad Sci USA. 1983;80:4949–4953. doi: 10.1073/pnas.80.16.4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnari J, Marshall WF, Straight A, Murray A, Sedat JW, Walter P. Mitochondrial transmission during mating in Saccharomyces cerevisiae is determined by mitochondrial fusion and fission and the intramitochondrial segregation of mitochondrial DNA. Mol Biol Cell. 1997;8:1233–1242. doi: 10.1091/mbc.8.7.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochoa GC, Slepnev VI, Neff L, Ringstad N, Takei K, Daniell L, Kim W, Cao H, McNiven M, Baron R, De Camilli P. A functional link between dynamin and the actin cytoskeleton at podosomes. J Cell Biol. 2000;150:377–389. doi: 10.1083/jcb.150.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuga D, Keegan BR, Brisch E, Thatcher JW, Hermann GJ, Bleazard W, Shaw JM. The dynamin-related GTPase, Dnm1p, controls mitochondrial morphology in yeast. J Cell Biol. 1998;143:333–349. doi: 10.1083/jcb.143.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder SE, Banta LM, Kohrer K, McCaffery JM, Emr SD. Multilamellar endosome-like compartment accumulates in the yeast vps28 vacuolar protein sorting mutant. Mol Biol Cell. 1996;7:985–999. doi: 10.1091/mbc.7.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan KR, Leung RS, Jensen RE. Characterization of the mitochondrial inner membrane translocase complex: the Tim23p hydrophobic domain interacts with Tim17p but not with other Tim23p molecules. Mol Cell Biol. 1998;18:178–187. doi: 10.1128/mcb.18.1.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraste M, Walker JE. Internal sequence repeats and the path of polypeptide in mitochondrial ADP/ATP translocase. FEBS Lett. 1982;144:250–254. doi: 10.1016/0014-5793(82)80648-0. [DOI] [PubMed] [Google Scholar]
- Schmidt A, Wolde M, Thiele C, Fest W, Kratzin H, Podtelejnikov AV, Witke W, Huttner WB, Soling HD. Endophilin I mediates synaptic vesicle formation by transfer of arachidonate to lysophosphatidic acid. Nature. 1999;401:133–141. doi: 10.1038/43613. [DOI] [PubMed] [Google Scholar]
- Sesaki H, Jensen RE. Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape. J Cell Biol. 1999;147:699–706. doi: 10.1083/jcb.147.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sever S, Muhlberg AB, Schmid SL. Impairment of dynamin's GAP domain stimulates receptor-mediated endocytosis. Nature. 1999;398:481–486. doi: 10.1038/19024. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MG, Simon VR, O'Sullivan H, Pon LA. Organelle-cytoskeletal interactions: actin mutations inhibit meiosis-dependent mitochondrial rearrangement in the budding yeast Saccharomyces cerevisiae. Mol Biol Cell. 1995;6:1381–1396. doi: 10.1091/mbc.6.10.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens B. Mitochondrial structure. In: Strathern JN, Jones EW, Broach JR, editors. The Molecular Biology of the Yeast Saccharomyces, Life cycle and Inheritance. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1981. pp. 471–504. [Google Scholar]
- Stevens BJ. Variation in number and volume of the mitochondria in yeast according to growth conditions: a study based on serial sectioning and computer graphics reconstitution. Biol Cell. 1977;28:37–56. [Google Scholar]
- Tieu Q, Nunnari J. Mdv1p is a WD repeat protein that interacts with the dynamin-related GTPase, dnm1p, to trigger mitochondrial division. J Cell Biol. 2000;151:353–366. doi: 10.1083/jcb.151.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzagoloff A. Mitochondria. New York: Plenum Press; 1983. [Google Scholar]
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg JM. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- Wallace DC. Mitochondrial diseases in man and mouse. Science. 1999;283:1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- Yaffe MP, Jensen RE, Guido EC. The major 45-kDa protein of the yeast mitochondrial outer membrane is not essential for cell growth or mitochondrial function. J Biol Chem. 1989;264:21091–21096. [PubMed] [Google Scholar]