SUMMARY
The neuronal voltage-dependent sodium channel (Nav1.2), essential for generation and propagation of action potentials, is regulated by calmodulin (CaM) binding to the IQ motif in its α-subunit. A peptide (Nav1.2IQp, KRKQEEVSAIVIQRAYRRYLLKQKVKK) representing the IQ motif had higher affinity for apo CaM than (Ca2+)4-CaM. Association was mediated solely by the C-domain of CaM. A solution structure (2KXW.pdb) of apo 13C,15N-CaM C-domain bound to Nav1.2IQp was determined with NMR. The region of Nav1.2IQp bound to CaM was helical; R1902, an Nav1.2 residue implicated in familial autism, did not contact CaM. The apo C-domain of CaM in this complex shares features of the same domain bound to myosin V IQ motifs (2IX7) and bound to an SK channel peptide (1G4Y) that does not contain an IQ motif. Thermodynamic and structural studies of CaM-Nav1.2IQp interactions show that apo and (Ca2+)4-CaM adopt distinct conformations that both permit tight association with Nav1.2IQp during gating.
Keywords: Allostery, NMR, Titration, Binding, Linkage, Solution Structure
INTRODUCTION
The voltage-dependent sodium channel type II (Nav1.2) is required for propagation of action potentials along unmyelinated axons found primarily in neurons and muscle (Schaller and Caldwell, 2003). Nav1.2 has one α-subunit and one or more β-subunits (Catterall, 2000). The α-subunit has four homologous, pore-forming transmembrane domains, and intracellular domains that control channel gating. Inactivation of Nav1.2 α-subunit involves interactions of the intracellular loop connecting domains III and IV and the C-terminal tail which has been modeled to contain six α-helices (Cormier et al., 2002).
Patch clamp studies showed that deletion of the putative sixth α-helix maintains the channel in a closed, inactivated state (Cormier et al., 2002; Mantegazza et al., 2001). A sequence (residues 1901 to 1927, Nav1.2IQp) within that region contains a classical CaM-binding IQ motif (Mori et al., 2000) (IQxxΦBGΦxxB), where X is any amino acid, Φ is an aromatic residue, and B is Arg or Lys (Figure 1A, B, C). All ten isoforms of NaV contain a single IQ motif that allows channel regulation by CaM (Yu and Catterall, 2003). Many IQ motifs found in myosins, neuronal growth proteins, and ion channels bind preferentially to apo (Ca2+-depleted) CaM (Liu and Storm, 1990; Urbauer et al., 1995). However, this preference is not universal. For example, the IQ motif in CaV 1.2 binds preferentially to (Ca2+)4-CaM (DeMaria et al., 2001).
Figure 1. Features of Voltage-Dependent Sodium Channel IQ Motifs and CaM.
A: Consensus sequence of 10 human voltage-dependent sodium channel IQ motifs. B: Nav1.2IQp sequence modeled as an ideal α-helix. C: Helical wheel projection of IQ motif of Nav1.2IQp displaying its hydrophobic and polar faces. D-G: Four structures of CaM (residues 1-75 blue, 76-80 black, 81-148 red) shown with bound peptide (green) and Ca2+ (yellow spheres) as appropriate. Square icons above ribbon diagrams indicate apo (open circle) or Ca2+-saturated (yellow filled circle) sites in the N-domain (blue) and C-domain (red).
CaM is essential to many eukaryotic signal transduction pathways (Klee et al., 1986). As seen in Figure 1D, it is composed of two domains (N/blue and C/red) that each bind two Ca2+ ions cooperatively. Although the domains are similar in sequence and structure, the C-domain binds Ca2+ 10- to 25-fold more favorably than the N-domain (Klee et al., 1986). Ca2+ binding triggers opening of hydrophobic clefts in each domain of CaM, altering its free energy of association with target proteins (Crivici and Ikura, 1995). The canonical CaM-binding sequence forms a basic, amphipathic, alpha-helix (BAA motif) (Yap et al., 2000) that binds preferentially to (Ca2+)4-CaM forming a compact ellipsoid (Figure 1E shows a BAA motif from CaMKII bound to (Ca2+)4-CaM (1CDM)). There are few available structures of apo CaM bound to targets. In one, two apo CaM molecules are bound to a fragment of myosin V containing two IQ motifs (2IX7; (Houdusse et al., 2006), Figure 1F). The C-domain of CaM adopted a semi-open conformation having interhelical angles smaller than those of (Ca2+)4-CaM. In another structure (1G4Y (Schumacher et al., 2001), Figure 1G), two CaM molecules, each having an apo semi-open C-domain but Ca2+-saturated open N-domain, are bound to two copies of a fragment of the SK channel that does not contain an IQ motif. CaM is widely recognized for extreme molecular plasticity.
To understand the Ca2+-dependent roles of each domain of CaM in binding the IQ motif of NaV1.2, fluorescence spectroscopy and NMR were used to monitor thermodynamic and structural properties of association. We determined that the C-domain of CaM bound Nav1.2IQp with very high affinity (nM Kd values), while the N-domain of CaM interacted weakly with the IQ motif. Nav1.2IQp binding to CaM selectively reduced Ca2+-binding affinity of sites III and IV, without significantly affecting sites I and II. Complexes of Nav1.2IQp with apo CaM (the C-domain alone or full-length CaM) were unperturbed in the presence of elevated NaCl. Heteronuclear NMR studies of apo 13C,15N-CaM76-148 (C-domain alone) bound to Nav1.2IQp were used to determine a solution structure. In it, the C-domain of CaM adopted a semi-open conformation in binding to the primarily helical peptide. Furthermore, although Nav1.2IQp bound tightly to both apo and (Ca2+)4-CaM, it caused unique chemical shift perturbations in each case. This suggests that the binding interface between (Ca2+)4-CaM and Nav1.2IQp is very different from that of apo CaM76-148:Nav1.2IQp, consistent with the physiological function of Ca2+-mediated inactivation Nav1.2. These results are discussed with respect to CaM recognition of IQ motifs in general, and regulation of sodium channels specifically.
RESULTS
CaM Binding to Nav1.2IQp
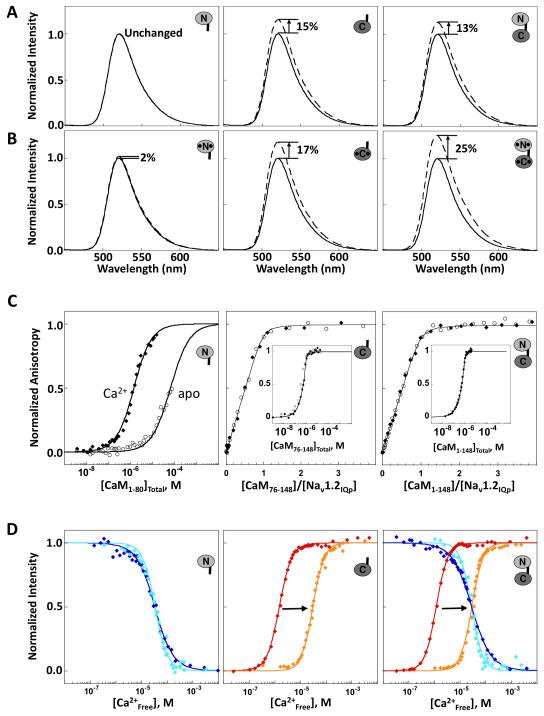
CaM binding to fluoresceinated Nav1.2IQp (Fl-Nav1.2IQp) was monitored by steady-state fluorescence emission to determine domain-specific differences in effects on peak intensity. As shown in Figure 2A for apo CaM, no change in Fl-Nav1.2IQp was observed upon addition of 3 equivalents of CaM1-80 (N-domain). Significant increases in intensity of Fl-Nav1.2IQp were observed after addition of CaM76-148 (C-domain, 15%) or CaM1-148 (full-length, 13%). This domain-specific pattern of change for apo CaM was preserved in 10 mM Ca2+ (Figure 2B), although the increase observed for (Ca2+)4-CaM1-148 (25%) was larger than that for (Ca2+)2-CaM76-148 (17%). The precision in intensity was ~1% for 3 replicate trials.
Figure 2. Domain-specific Interactions of CaM with Nav1.2IQp.
Emission spectra of Fl-Nav1.2IQp in the absence (solid line) and presence (dashed line) of slight excess of (A) apo CaM1-80 (left), apo CaM76-148 (middle), and apo CaM1-148 (right) or (B) Ca2+-saturated CaM. C: Titration of Fl-Nav1.2IQp with apo CaM (open circles) or Ca2+-saturated CaM (filled diamonds). The titrations of apo CaM1-80 were normalized to the maximum signal observed after addition of 10 mM CaCl2 (Newman, 2008). For CaM76-148 (middle), and CaM1-148 (right), binding was stoichiometric and is plotted as a function of the ratio of [CaMtotal]:[peptide], with an inset showing saturation versus [CaMtotal]. D: Equilibrium Ca2+ titrations of CaM1-80 (left), CaM76-148 (middle), and CaM1-148 (right) using intrinsic fluorescence to monitor Ca2+ binding in the absence of peptide [sites I and II (blue) or sites III and IV (red)], or after addition of excess Nav1.2IQp [sites I and II (cyan) and sites III and IV (orange)]. Supported by Tables S1 and S2.
CaM Titration of Nav1.2IQp
The affinities of CaM binding to Fl-Nav1.2IQp were determined using fluorescence anisotropy. Figure 2C shows that the anisotropy of Fl-Nav1.2IQp increased upon binding either apo CaM (open symbols) or Ca2+-saturated CaM (closed symbols). CaM1-80 bound to Nav1.2IQp with low affinity (left panel); the Kd was 1.64 ± 0.65 μM for (Ca2+)2-CaM1-80, and 70.8 ± 5.7 μM for apo CaM1-80. In contrast, both CaM1-148 and CaM76-148 (apo and Ca2+-saturated forms) bound stoichiometrically to Nav1.2IQp, with a Kd ≤10 nM. The titrations were fit to a single-site binding isotherm that accounted for total CaM (Table S1); however, only an upper limit for the Kd can be determined under conditions of limiting CaM (Newman, 2008).
Effect of Nav1.2IQp on Calcium Binding to CaM
To determine how Nav1.2IQp affects the Ca2+-binding properties of each domain of CaM, equilibrium titrations were performed on the isolated domains (CaM1-80, CaM76-148) and compared to the same sites within CaM1-148 (Figure 2D). Changes in intrinsic fluorescence intensity of phenylalanine reported on Ca2+ binding to sites I and II, while changes in tyrosine intensity reported on binding to sites III and IV. Total free energies of binding are listed in Table S2. Titrations are shown in blue (sites I and II) and red (sites III and IV) in the absence of peptide, and shown in cyan (sites I and II) and orange (sites III and IV) in the presence of peptide. Figure 2D shows that Nav1.2IQp had little effect on sites I and II, whether in CaM1-80 (left panel) or CaM1-148 (right panel). However, it selectively reduced the Ca2+ binding affinity of sites III and IV of CaM76-148 (middle) by a factor > 500, and CaM1-148 (right) by a factor > 750 (Table S2).
Residue-Specific Effects of Nav1.2IQp on CaM
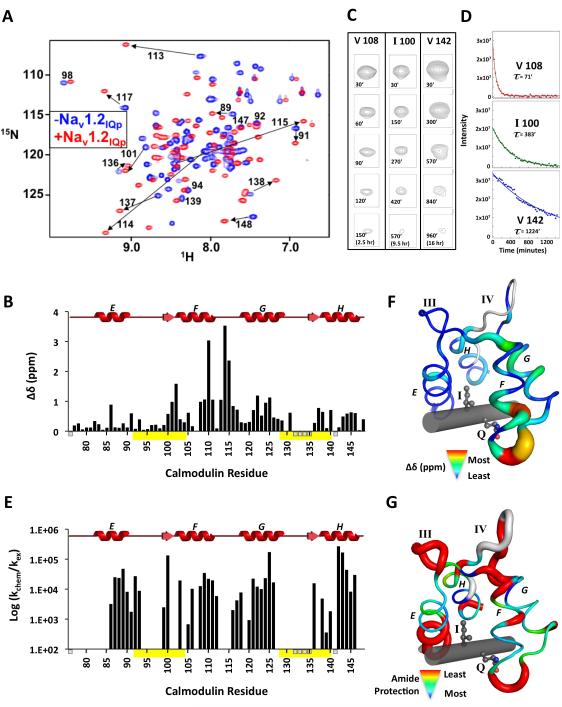
Effects of Nav1.2IQp on apo CaM were also manifested as conformational differences detected in the 15N/1H-HSQC spectra. A spectral overlay of apo CaM1-148 alone and bound to Nav1.2IQp showed that half of the observed amide chemical shifts were nearly identical (Figure S1). An overlay of 15N/1H-HSQC spectra of apo CaM1-80 (N-domain) alone and apo CaM1-148 bound to Nav1.2IQp (Figure S2) showed that resonances unperturbed by peptide binding were attributable to the N-domain. In contrast, resonances corresponding to C-domain residues were dramatically affected by peptide binding. Those in the spectrum of the apo CaM76-148:Nav1.2IQp complex were nearly identical to corresponding C-domain residues in apo CaM1-148:Nav1.2IQp (Figure S3). An overlay of three of these 15N/1H-HSQC spectra in Figure 3A shows that the resonances of (a) apo CaM1-80 alone (green), and (b) apo CaM76-148 bound to Nav1.2IQp (blue) closely resembled the spectrum of (c) apo CaM1-148 bound to Nav1.2IQp (red).
Figure 3. 15N/1H-HSQC Spectra of 13C,15N-CaM.
A: Overlapping peaks in 15N/1H-HSQC spectra of apo CaM1-80 (blue) and apo CaM1-148 with equimolar Nav1.2IQp (red) are boxed by dashed squares, while those overlapped in the apo CaM76-148 (green) and apo CaM1-148 with equimolar Nav1.2IQp (red) are shown by solid circles. The spectra of apo CaM1-80 alone and apo CaM76-148:Nav1.2IQp complex account for > 95% of the peaks of apo CaM1-148-Nav1.2IQp B: Overlay of 15N/1H-HSQC spectra of apo CaM76-148 (green) and (Ca2+)2-CaM76-148 (black), each with equimolar Nav1.2IQp. Cross peaks of apo CaM76-148-Nav1.2IQp are labeled. Supported by Figures S1, S2, S3.
Figure 3B shows the effect of Ca2+ binding. Almost all resonances in the 15 N-HSQC spectrum of (Ca2+)2-CaM76-148 bound to Nav1.2IQp (black) were distinct from those of apo CaM76-148 bound to Nav1.2IQp (green), indicating that most backbone amides have different chemical environments, presumably because they have adopted distinct conformations.
Analysis of 15N/1H-HSQC spectra of apo CaM76-148 ± Nav1.2IQp (Figure 4A) showed that peptide binding caused significant chemical shift perturbations (Δδ) of the CaM backbone amides (δaverage of 0.51 ppm). It also dramatically increased dispersion of 15N/1H-HSQC cross peaks (Figure 4A) by breaking the symmetry of similar chemical environments of homologous residues in the paired EF-hands of apo CaM76-148. Mapping Δδ onto the sequence of CaM76-148 (Figure 4B) indicated that the greatest perturbations were observed in the region between helices F and G. For residues 108-117,Δδaverage was 1.47 ppm. Unassigned amides (for residues 76, 132, 133, 134, 135, and 141) are indicated with short gray bars below the abscissa; yellow bars indicate Ca2+-binding sites III and IV.
Figure 4. 15N/1H-HSQC-Detected Changes in Apo CaM76-148 upon Nav1.2IQp Addition.
A: Overlay of 15N/1H-HSQC spectra of apo CaM76-148 (blue) and apo CaM76-148 bound to Nav1.2IQp (red). B: Quantified backbone amide chemical shift perturbations of apo CaM76-148 upon binding Nav1.2Iqp. Ca2+-binding sites are yellow bars. Unassigned residues (76, 132, 133, 134, 135, 141) are indicated as small bars below the abscissa. C: 15N/1H-HSQC crosspeak intensities at identical contour levels showing residues of apo CaM76-148 bound to Nav1.2IQp. D: Fitted exchange curves and rates for the residues shown in C. E: Bar graph of amide protection factors. Chemical shift perturbations (F) and protection factor magnitudes (G) mapped onto the structure of apo CaM76-148:Nav1.2IQp (unassigned residues are light gray; Nav1.2IQp is a gray rod). Supported by Table S3.
Hydrogen/Deuterium Backbone Amide Exchange
Backbone amide hydrogen/deuterium exchange rates of apo CaM76-148:Nav1.2IQp were determined to identify slowly exchanging amides that are usually located in regions of persistent secondary structure. In the 15N/1H-HSQC spectrum, 37 amide resonances were detected 30’ after addition of D2O to a lyophilized sample. Protection factors (Table S3) were calculated from the decay of cross peak intensities (examples shown in Figure 4C) that were fit to a mono-exponential function (Figure 4D) and corrected for intrinsic exchange rates (Figure 4E) (Bai et al., 1993; Molday et al., 1972). Residues with observable exchange rates were used as H-bond restraints during subsequent structure calculations.
Structure of Apo CaM76-148:Nav1.2IQp
Structural statistics of the apo CaM76-148:Nav1.2IQp complex are in Table 1. This structure was determined based on 109 dihedral angular restraints and 1865 unambiguous distance restraints comprised of 457 intra-residue, 298 short, 300 medium, 331 long, 245 intra-peptide, 188 intermolecular, and 46 hydrogen bonds. The restraints are distributed evenly throughout the protein, with a slightly higher number observed in α-helical and β-sheet regions than in the empty Ca2+-binding sites III and IV. Figure 5A shows a superposition of the 20 lowest energy structures of the apo CaM76-148:Nav1.2IQp complex that best satisfied the experimental restraints. The interhelical angles calculated by UCSF Chimera (Pettersen et al., 2004) between helices E-F and G-H of these structures were 77.4° ± 1.7 and 78.0° ± 2.3 respectively. The apo CaM76-148:Nav1.2IQp complex was well defined, with an ensemble RMSD of 0.31 ± 0.05 Å (Table 1). Figure S4 shows a Ramachandran plot of the final averaged structure.
Table 1.
Structural statistics and root-mean-square deviation for 20 structures of apo CaM76-148:Nav1.2IQp complex
| Structural statisticsa | <SA> | |
|---|---|---|
| RMSD from experimental distance restraints (Å)b | ||
| All (1865) | 0.008 ± 0.001 | 0.007 |
| CaM intra-residue (457) | 0.007 ± 0.002 | 0.005 |
| CaM sequential (298) | 0.005 ± 0.004 | 0.003 |
| CaM medium range (300) | 0.009 ± 0.002 | 0.008 |
| CaM long range (331) | 0.006 ± 0.001 | 0.005 |
| Intra-peptide (245) | 0.005 ± 0.002 | 0.003 |
| CaM-peptide intermolecular (188) | 0.012 ± 0.002 | 0.011 |
| hydrogen bond (46) | 0.016 ± 0.001 | 0.021 |
| RMSD from experimental torsional angle restraints (deg)c | ||
|
| ||
| Φ and Ψ angles (109) | 0.3 ± 0.03 | 0.2 |
| Ramachandran Statisticsd | ||
|
| ||
| Residues in most favored regions | 80.2% | |
| Residues in additional allowed regions | 17.6% | |
| Residues in generously allowed regions | 2.2% | |
| Residues in disallowed regions | 0.0% | |
| CNS Potential Energy (kcal/mol) | ||
|
| ||
| Etot | 89 ± 4.8 | 80 |
| Ebond | 5 ± 0.5 | 1 |
| Eang | 65 ± 2.9 | 62 |
| Eimp | 5 ± 0.6 | 4 |
| Erepel | 6 ± 1.2 | 6 |
| Enoe | 6 ± 1.2 | 5 |
| Ecdih | 1 ± 0.1 | 1 |
| Cartesian coordinate RMSD (Å) | N, Ca, and C’ | all heavy |
|
| ||
| <SA> vs. e | 0.31 ± 0.05 | 0.95 ± 0.11 |
Where <SA> is the ensemble of 20 NMR-derived solution structures of CaM/peptide; is the mean atomic structure; is the energy-minimized average structure. The CNS Frepel function was used to simulate van der Waals interactions using a force constant of 4.0 kcal/mol Å−4 with the atomic radii set to 0.8 times their CHARMM values.
Distance restraints were employed with a square-well potential (Fnoe = 50 kcal/mol Å−2). Hydrogen bonds were given bounds of 1.8-2.4 Å (H-O) and 2.7-3.3 Å (N-O). No distance restraint was violated by more than 0.3 Å in any of the final structures.
Torsional restraints were applied with values derived from an analysis of the C’, N, Ca, Ha, and Cb chemical shifts using the TALOS program. Force constant of 200 kcal mol−1 rad−2 was applied for all torsional restraints.
Ramachandran statistics calculated with ProCheck. http://www.ebi.ac.uk/thornton-srv/software/PROCHECK/
RMSD for CaM residues 80-128 and 134-146 and peptide residues 1905-1920.
Figure 5. Solution Structure of Apo CaM76-148:Nav1.2IQp.
A: Ensemble of 20 lowest energy structures of apo CaM76-148 (black) bound to Nav1.2IQp (red) B: Hydrophobic interaction interfaces of 20 lowest energy structures in 2KXW. Apo CaM76-148 and residues 1904-1922 of Nav1.2IQp (green backbone) with selected interfacial hydrophobic residues shown in sticks. C: Vacuum electrostatic potentials mapped on the surface apo CaM76-148 and Nav1.2IQp. Surfaces are positive (blue), negative (red) or hydrophobic (white). Supported by Figure S4 and Movies S1 and S2.
Interface Between Nav1.2IQp and Apo CaM76-148
The hydrophobic interface of apo CaM76-148:Nav1.2IQp was well defined as judged by the constrained positions of interacting residues (Figure 5B). The buried solvent-accessible hydrophobic surface calculated using GETAREA (Fraczkiewicz and Braun, 1998) was 1393 Å2 (751 Å2 Nav1.2IQp, 642 Å2 apo CaM76-148). In apo CaM76-148, hydrophobic residues A88, V91, F92, L112, and M145 accounted for 43% of its buried surface, while in Nav1.2IQp, hydrophobic residues V1911, I1912, Y1916, Y1919, and L1920 accounted for 53.5% of its buried surface area. To demonstrate how well residues at the interface were constrained, a superposition of the 20 lowest energy structures was rotated by 2° increments in PyMol and captured with ImageJ. Movie S1 shows an overlay of apo CaM76-148 models (wireframe) bound to Nav1.2IQp (cartoon and sticks). Movie S2 shows the apo CaM76-148 models with all sidechains, followed by models showing only hydrophobic sidechains located at the interface with Nav1.2IQp.
Figure 5C shows vacuum electrostatic potentials for coordinates of apo CaM76-148 and Nav1.2IQp corresponding to their conformation in the complex as calculated using Pymol (Schrödinger, LLC.). As expected, favorable electrostatic interactions were observed between negatively charged apo CaM76-148 and positively charged Nav1.2IQp., and the interface is dominated by hydrophobic surface.
Figure 6A shows all contacts between apo CaM76-148 and Nav1.2IQp that are • 4.5 Å (tabulated by by Contacts of Structural Units (CSU) (Sobolev et al., 1999)). Interactions of the IQ residues (I1912, Q1913) with apo CaM76-148 were identified on the basis of numerous NOEs detected in (a) the 3D 13C-edited and 15N-edited NOESY spectra and (b) the 13C-edited and 12C,14N-filtered 3D NOESY spectra. Figures 6B and 6C show NOEs for selected residues of apo CaM76-148. As seen in the CSU analysis, I1912 inserted into the shallow hydrophobic pocket of semi-open apo CaM76-148 (Figure 6D) while Q1913 formed hydrogen bonds with backbone atoms L112 and E114 in the turn connecting helices F and G (Figure 6E). A comparison of CaM sequences from 102 species (Ataman et al., 2007) showed that residues E114 and G113 were identical in all, while L112 was 89.2% identical.
Figure 6. Analysis of apo CaM76-148:Nav1.2IQp Binding Interface.
A: Residues of apo CaM76-148 within 4.5 Å of each residue of Nav1.2IQp in 2KXW are listed; four peptide residues (I1912, Q1913, Y1916, Y1919) made • 5 contacts (highlighted in red). 5 contacts (highlighted in red). B: Slices of NOESY spectra showing intermolecular NOEs observed between I1912 of Nav1.2IQp and apo CaM76-148. Slices from the 3D 13C-edited NOESY spectra are in blue boxes, while slices from 3D 13C-edited and 12C,14N-filtered NOESY spectrum are in red boxes. C: Top panel shows Q1913 carboxamide proton shifts in the 2D 12C,14N-filtered NOESY spectrum, and lower panels show slices of 15N-edited NOESY spectra showing NOEs between Nav1.2IQp Q1913 NεH2 and 7 loop backbone amides in apo CaM76-148. D: Location of apo CaM76-148 residues shown in panel B relative to I1912. E: Location of apo CaM76-148 residues shown in panel C relative to Q1913. Dashed lines show hydrogen bonds between the carboxamide of Q1913 and backbone atoms of L112 and E114.
Calcium Binding to Apo CaM1-148:Nav1.2IQp Complex
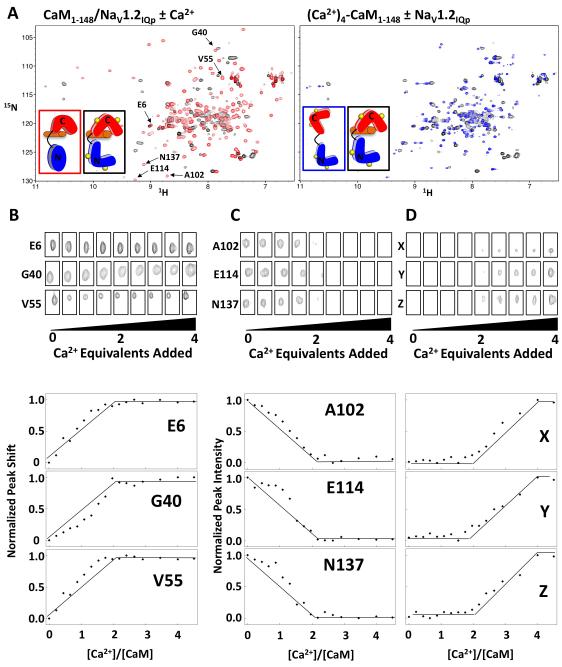
Figure 7A shows that apo CaM1-148 bound to Nav1.2IQp underwent a structural transition upon Ca2+ binding. Figure 7A (left panel) shows the overlay of 15N/1H-HSQC spectra of apo CaM1-148 (red) and (Ca2+)4-CaM1-148 (black) bound to Nav1.2IQp, indicating that Ca2+ caused significant change in the chemical environment of apo CaM1-148 resonances within the complex. For reference, Figure 7A (right panel) compares (Ca2+)4-CaM1-148 alone and after addition of Nav1.2IQp. Figures 7B, C, and D show the 15N/1H-HSQC cross peaks for selected residues of the N- and C-domains of apo CaM1-148 when bound to Nav1.2IQp at increasing [Ca2+]. The N-domain of apo CaM1-148 was in fast exchange over the course of the Ca2+ titration; representative resonances are shown in Figure 7B. Plots of the normalized Ca2+-dependent change in chemical shifts indicated that the N-domain titrated almost completely between 0 and 2 Ca2+ equivalents.
Figure 7. Ca2+ binding to Apo CaM1-148:Nav1.2IQp.
A: 15N/1H-HSQC spectral overlays of apo (red) and (Ca2+)4-CaM1-148 (black) when bound to Nav1.2IQp (left panel) and (Ca2+)4-CaM1-148 alone (blue) and bound (black) to Nav1.2IQp (right panel). B: 15N/1H-HSQC cross peak intensity and position as a function of added Ca2+ equivalents for selected N-domain residues (E6, G40, V55) of apo CaM1-148, selected C-domain residues (A102, E114, N137) of CaM1-148, and three unassigned C-domain residues (X, Y, Z) of CaM1-148 when Ca2+-saturated. The quantified change in chemical shift perturbations as a function of added Ca2+ equivalents for these residues are plotted in the lower panels. Supported by Figure S6.
Observation of Ca2+-induced changes in peak position and intensity of the C-domain of apo CaM1-148 bound to Nav1.2IQp (Figure 7C) showed that peaks were in intermediate or slow exchange. Thus, changes in peak intensities were used to determine relative populations over the course of the titration. These peaks broadened beyond the limit of detection after addition of 2 equivalents of Ca2+ (Figure 7C) as has been seen for Ca2+ binding to melittin-bound CaM (Newman, 2008). New peaks that appeared at the midpoint of the Ca2+ titration and increased through the addition of 4 equivalents of Ca2+ (see Figure 7D) are likely to correspond to residues in the C-domain of (Ca2+)4-CaM1-148 based on correlations with effects of Nav1.2IQp on Ca2+-binding shown in Figure 2. These data indicated that, when bound to Nav1.2IQp, each domain of CaM1-148 binds 2 Ca2+, and sites I and II have a slightly more favorable Ca2+-binding affinity than sites III and IV.
Effect of Ca2+ on CaM1-148:Nav1.2IQp
As shown in Figure 2C, apo CaM1-148 bound to Fl-Nav1.2IQp stoichiometrically at a 1:1 ratio, with binding mediated by the C-domain of CaM. If Ca2+ binding to CaM1-148 induced association of the N-domain with Nav1.2IQp, then the structure would become more compact. To test this, Ca2+ was added to apo CaM1-148:Fl-Nav1.2IQp (Figure S5A). The hydrodynamic behavior of the complex was monitored by fluorescence anisotropy, but no change was observed. Evaluated with the NMR spectra in Figures 3 and 7, this suggests that the Ca2+-saturated N-domain is free to bind elsewhere on Nav1.2 or to another target protein.
Effect of Na+ upon Nav1.2IQp Binding to CaM
Proteins in the plasma membrane experience large fluctuations in Na+ concentration during Nav1.2 gating (Venosa, 1974) which might affect the affinity of Nav1.2IQp for CaM. To test this, Fl-Nav1.2IQp was saturated with apo CaM76-148 or apo CaM1-148 and then titrated with NaCl in a matching buffer over a range from 0 to 650 mM (Figure S5B). The observed anisotropy of Nav1.2IQp was constant, indicating that NaCl has a negligible effect on Nav1.2IQp dissociation from CaM.
DISCUSSION
Nav1.2 contains a CaM-binding IQ motif involved in channel inactivation which is necessary for controlling Na+ balance. Thermodynamic studies and the structure of apo semi-open CaM76-148 bound to Nav1.2IQp reported here provide insights regarding the roles of residues that are conserved in IQ motifs in ion channels, and differences between the two domains of CaM. They provides a foundation for further studies of Ca2+-mediated switching of channel activity.
Domain-Specific Binding of CaM to Nav1.2IQp
The energetics of CaM binding to Nav1.2IQp reflect large differences between the two domains of CaM that would permit major regulatory differences. In the complex, apo CaM76-148 did not contact R1902, a residue implicated in familial autism (R1902C is known to interfere with binding of (Ca2+)4-CaM (Weiss et al., 2003)). This leaves open the question of how (Ca2+)2-CaM76-148 interacts with R1902, and whether the mutation may exert its influence via effects on the tertiary structure of the channel. The CaM C-domain is necessary and sufficient for binding Nav1.2IQp. Its high affinity (nM Kd) for CaM would promote constitutive binding to Nav1.2 independent of intracellular [Ca2+]. Strong, Ca2+-independent binding is similar to behavior observed for Nav1.4 in electrophysiological and FRET studies by Tomaselli and coworkers who showed that CaM remained bound to the IQ motif region as Nav1.4 adopted multiple conformations in response to voltage changes and Ca2+ (Biswas et al., 2008). The weak (μM) affinity of CaM1-80 for Nav1.2IQp and residue-specific NMR studies showing that Nav1.2IQp caused barely detectable perturbations of the N-domain (either as a fragment or within full-length CaM) both argue that the N-domain of CaM does not bind Nav1.2IQp in a manner that is physiologically significant.
Effect of Nav1.2IQp on Ca2+ Binding
Unlike kinases that increase the Ca2+-binding affinity of both domains of CaM by interacting favorably with the open conformation of each domain, binding of Nav1.2IQp selectively lowered the overall Ca2+-binding affinity of CaM by 4.03 kcal/mol (Figure 2D). This decrease was attributable almost entirely to interactions of the C-domain of CaM (••G2 of 3.89 kcal/mol; Table S1). Binding of Nav1.2IQp imposed tertiary constraints that lowered the probability of Ca2+ binding to sites III and IV. The effects of Nav1.2IQp on •G2 for Ca2+ binding to the individual domains of CaM was additive (i.e., similar that observed in CaM1-148), indicating that interdomain interactions in CaM1-148 were minimal (Table S1). This was consistent with domain-specific effects seen with fluorescence in Figure 2A, B, C and with NMR in Figures 3, S1, S2, and S3.
Semi-Open Apo C-domain Binds IQ Motifs
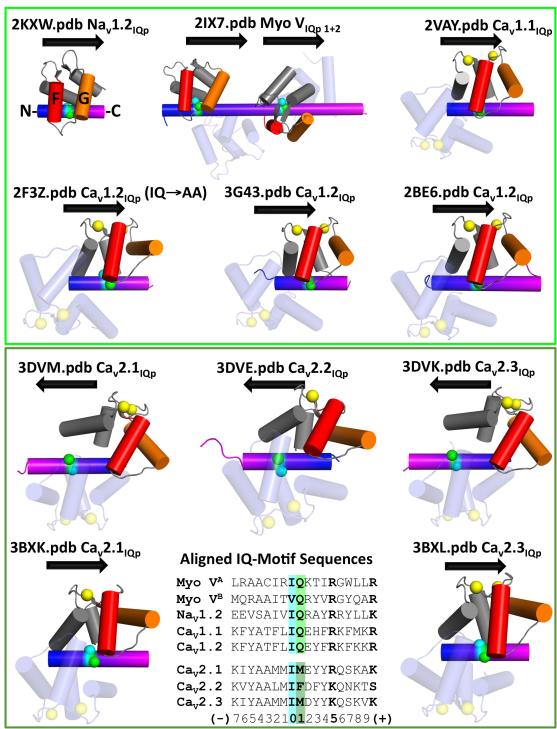
Figure 8A shows a structural comparison of (a) apo CaM76-148 bound to Nav1.2IQp (determined in this study) to (b) the C-domains of two apo CaM1-148 molecules bound to two neighboring IQ motifs found in myosin V (2IX7), (c) apo CaM-like proteins bound to IQ motifs of MYO2P (1M46, 1M45, 1N2D) and (d) ELC bound to myosin heavy chain (3JVT). In all of these, each set of paired EF-hands of the C-domain adopts a semi-open conformation when bound to its respective IQ motif. Moreover, the residues located at positions “0”/Ile and “1”/Gln of the IQ motif interact with a similar subset of residues in the apo C-domain.
Figure 8. Conserved Features of Apo Semi-Open EF-Hand Domains and Model for CaM:NaV1.2 Binding to Recognition Motifs.
A: Structural comparison of apo CaM76-148 (red):Nav1.2IQp (green rod) complex (2KXW) with the C-domains of apo CaM and apo CaM-like proteins (gray) bound to canonical IQ motifs (2IX7, 1M45, 1M46, 1ND2, and 3JVT). I and Q residues of all IQ motifs are sticks; Cα of residue 113 of CaM is a red sphere. B: Structural comparison of apo CaM76-148 (red):Nav1.2IQp (green rod) complex with the apo C-domain of (Ca2+)2-CaM1-148 (orange) bound to SKp (cyan). I1912 and Q1913 (Nav1.2IQp) and L428 and N426 (SKp) are ball-and-stick; CaM residue 113 shown as red or orange sphere. Sequences of Nav1.2IQp and SKp are aligned structurally based on the hydrophobic interaction of I1912 and L428 and carboxamide-containing Q1913 and N426. C: Two alternative polarities for peptides binding to the C-domain of CaM; NF-GC depicts the N-terminus (blue) closest to helix F (red), while CF-GN depicts the C-terminus (magenta) closest to helix F and farthest from helix G (orange). D: Relative positions of the hydrophobic (cyan spheres) and carboxamide (green spheres) residues. The blue-to-magenta color gradient indicates NF-GC polarity. E: Proposed model of CaM1-148 interacting with Nav1.2. The IQ motif interacts solely with the semi-open C-domain of CaM. The N-domain is in a closed conformation that may interact with an unidentified site X elsewhere on Nav1.2. Under Ca2+-saturating conditions, the C-domain remains bound to Nav1.2IQp; the open N-domain may bind to site Y (potentially distinct from site X). Supported by Figure S6.
At position “0”, a hydrophobic (typically branched) residue contacts hydrophobic sidechains of the semi-open C-domain, burying what would otherwise be solvent-exposed hydrophobic surface. Conservation of Gln at position “1” of canonical IQ motifs preserves hydrogen bonding interactions between its carboxamide and the backbone of CaM residues located in the turn connecting α-helices F and G (Figure 6E). As observed in the 15N/1H-HSQC data (Figure 4A and 4D), residues located in this turn exhibited the largest chemical shift perturbation upon peptide binding. Examination of amide hydrogen exchange protection factors mapped onto the structure (Figure 4F) suggests that this turn is solvent-exposed. Therefore, the H-bonds involved in this turn may be different from those observed in classic α-helices and β-sheets.
Tomaselli and coworkers, studying NaV1.4, showed that residues I and Q are required for gating, and that substitution by alanine eliminated the proximity of CaM to the C-terminus of Nav1.4 under all conditions (e.g., resting and high Ca2+) (Biswas et al., 2008). The interactions observed between the IQ residues of Nav1.2IQp and apo CaM could not be satisfied by alanine substitutions. With Ala at position “0”, there would be less of a penalty for solvent exposure relative to Ile, Leu, Met, or Phe at that position. With Ala at position “1”, there would be no carboxamide interaction between the sodium channel and atoms in the backbone of the loop between α-helices F and G of CaM.
Semi-Open Apo C-Domain Binding to a Non-IQ motif
An apo domain of CaM can bind to a sequence without an IQ motif, as seen in the 1G4Y structure of partially apo CaM bound to a peptide (SKp, gating domain residues 413-488) derived from the small conductance potassium channel (SK Channel) (Schumacher et al., 2001). A superposition of apo CaM76-148:Nav1.2IQp and the apo C-domain of 1G4Y showed that both adopt the semi-open conformation (Figure 8B).
SKp shares a key feature seen in the IQ motif-containing peptides bound to apo CaM, CaM-like proteins, and ELC. Figures 8A and 8B show that a carboxamide-containing side chain (Asn in SKp vs. Gln in IQ motifs) forms hydrogen bonds with backbone residues between the F and G helices of CaM. This conserved hydrogen-bonding network influences the position of the IQ motif or SKp helix relative to the apo C-domain. Another similarity was apparent in the conserved hydrophobic interaction between I1912 (Nav1.2IQp) or L428 (SKp) and the core of the apo C-domain (Figure 8B), even though these residues differ in their position in the primary sequence relative to the carboxamide side chain (Q1913 in Nav1.2IQp or N426 in SKp).
As depicted in Figure 8C, both SKp and Nav1.2IQp adopted the NF-GC orientation. where F and G refer to the helices of CaM between sites III and IV, and N and C refer to the termini of the CaM-binding sequence. In Nav1.2IQp, the conserved hydrophobic residue (I1912) that inserts into the core of apo CaM76-148 precedes the conserved carboxamide-containing residue (Q1913). Although in SKp, L428 (the residue homologous to I1912) is located 2 residues distal to the carboxamide-containing residue (N426), the relative angular positions of IQ and N-L are the same (Figure 8D). This illustrates how peptides can recognize the semi-open conformation of apo C-domain regardless of whether they contain an IQ motif.
Orientations of IQ motif Binding to CaM
If the contacts seen in Figure 8A were maintained by all apo CaM-binding motifs, then all canonical IQ motifs would bind to the semi-open C-domain of apo CaM in the NF-GC orientation. At this time, there are too few deposited structures of apo CaM bound to IQ motifs to test this prediction. Figure 9 (upper panel) depicts available structures of apo CaM and (Ca2+)4-CaM bound to peptides containing canonical IQ motifs derived from indicated target proteins. In these cases, position “0” is occupied by Ile in all structures except that of myosin VB (which has Val), and position “1” is only occupied by Gln. The orientation of peptide binding to each of this is NF-GC (N- and C- termini of peptide blue and magenta respectively), as depicted by rightward arrows.
Figure 9. Orientation of Canonical and Non-Canonical IQ Motifs Bound to CaM.
CaM is bound to peptides containing canonical (upper panel) or non-canonical (lower panel) IQ motifs. Each ribbon diagram is labeled with PDB ID and degree of Ca2+ binding (yellow spheres). In CaM, helix F is red, and G is orange. Helical polarity of each IQ motif is indicated by a color gradient from N- (blue) to C-terminus (magenta) and corresponding arrow. Residues at positions 0 (cyan) and 1 (green) are indicated by spheres. In the lower panel, sequences of binding motifs are aligned according to positions 0 and 1.
We infer that apo CaM binds to Nav1.2IQp and other canonical IQ motifs in the NF-GC orientation because of the Gln located at position “1”. The simplest model for Ca2+-mediated regulation of NaV1.2 is that the NF-GC orientation is also used by (Ca2+)4-CaM when binding Nav1.2, because NF-GC has been observed in all structures of (Ca2+)4-CaM bound to canonical IQ-motifs from other channels. Adoption of the opposite (CF-GN) orientation by (Ca2+)4-CaM would require the C-domain of apo CaM to release from the IQ motif upon Ca2+ influx, and re-associate when Ca2+-saturated.
Both orientations of (Ca2+)4-CaM have been observed (Figure 9, lower panel) in complexes with peptides that contain non-canonical IQ motifs (e.g., sequences having Met or Phe at position “1”, rather than Gln). Structures of (Ca2+)4-CaM bound to fragments with these sites from Cav2.1, Cav2.2, and Cav2.3 adopted alternative orientations in different studies. Different peptide lengths of Cav2.1 and Cav2.3, and distinct crystallization conditions were used in these studies, which may account for the structural differences (Kim et al., 2008; Mori et al., 2008). At this time, there are no available structures of apo CaM bound to a CaV IQ-motif.
Role of the N-domain of CaM1-148 in Nav1.2 Recognition
The N-domains of both apo and (Ca2+)4-CaM1-148 do not interact with Nav1.2IQp as shown by titrations monitored by steady-state fluorescence (Figures 2A and 2B), fluorescence anisotropy (Figure 2C), and additive 15N/1H-HSQC spectra (Figures 3A, S2). However, studies presented here do not rule out that it may interact elsewhere on NaV1.2, as suggested schematically in Figure 8E by site X (for the apo closed N-domain) and site Y (for the Ca2+-saturated open N-domain). The interaction of apo N-domain might mimic that seen in a complex of CaM bound to a fragment of the anthrax edema factor (Drum et al., 2002) which involves a helix-helix interaction rather than an interface with the cleft of CaM.
Previous studies (Mori et al., 2000) showed that Nav1.2 contains a second CaM-binding motif comprised of residues 1913-1938 (Nav1.2BAA-1913-1938) that binds to (Ca2+)4-CaM, but not apo CaM. This sequence has characteristics of a classic BAA-motif that preferentially binds the open conformation of Ca2+-saturated CaM. Thus, the N-domain could bind to a site within Nav1.21913-1938 while the C-domain remains tethered to Nav1.2IQp (Figure 8E). To test this model, Fl-Nav1.2IQp was titrated with apo CaM1-148 and monitored by fluorescence anisotropy (Figure S6A) to assure saturation. When Nav1.21913-1938 was added in 6-fold excess, it competed for Nav1.2IQp; there was no evidence for simultaneous binding of both Nav1.2 2IQp and Nav1.21913-1938 to (Ca2+)4-CaM. This suggests that a binding site for Ca2+-saturated N-domain may be located elsewhere in Nav1.2 or another protein (shown schematically as site Y in Figure 8E). For example, the intracellular linker between domains III and IV of other sodium channels binds (Ca2+)4-CaM (Sarhan et al., 2009; Shah et al., 2006). Alternatively, the conformation of these residues within Nav1.2 may not be fully represented by a peptide.
Conclusion
The studies reported here provide the first structure of apo CaM76-148:Nav1.2IQp. The structural interface and the binding energy for association of Nav1.2IQp and apo CaM are mediated by the C-domain alone. This implies that a preferred binding site for the N-domain of CaM must lie elsewhere. In the apo CaM76-148:Nav1.2IQp structure, CaM adopts a semi-open conformation similar to the C-domain of apo CaM alone (Swindells and Ikura, 1996), CaM-like proteins (Terrak et al., 2003), essential light chain (ELC) (Swindells and Ikura, 1996), apo CaM when bound to IQ motifs in myosin (Houdusse et al., 2006) and the apo C-domain of CaM bound to the SK channel (Schumacher et al., 2001). At the CaM-peptide interface in apo CaM76-148:Nav1.2IQp, the highest number of noncovalent interactions was observed between apo CaM76-148 and 4 residues (I1912, Q1913, Y1916, and Y1919) of Nav1.2IQp. These sidechains are strongly conserved in IQ motifs found in NaV isoforms, with the exception of the Y1919 which is a histidine in four of the ten isoforms (Figure 1A). There are now multiple structures of (Ca2+)4-CaM bound to peptides representing IQ motifs of ion channels, but there are no cases in which the corresponding complex of apo CaM bound to the same peptide is available. Discovering how Ca2+-mediated switching of CaM regulates NaV1.2 will require future analysis of calcium-saturated CaM76-148:Nav1.2IQp, studies of apo CaM binding to Nav1.2IQp having substitutions that mimic mutations known to alter gating properties, and dissection of interactions of CaM with larger fragments of the intracellular C-terminal sequence of the channel.
EXPERIMENTAL PROCEDURES
Calmodulin
Isotopes were obtained from Cambridge Isotope Laboratories (Andover, MA). CaM over-expression was IPTG-induced in E. coli BL21(DE3) cells containing a recombinant pT7-7 vector expressing full-length (residues 1-148), N-domain (1-80), or C-domain (76-148) of Paramecium CaM (C. Kung, University of Wisconsin, Madison, WI) which is 88% identical to mammalian CaM. A genetic screen of Paramecia demonstrated that CaM mutations directly affected Na+ channel function and chemotactic behavior (Kung et al., 1992). 15N-labeled proteins were over-expressed in minimal media (2 g/L unlabeled glucose as carbon source, 1 g/L 15NH4Cl as sole nitrogen source). 13C,15N-labeled proteins were produced using 2 g/L 13C-glucose as sole carbon source and 1 g/L 15NH4Cl as sole nitrogen source. CaM was purified as described previously (Putkey et al., 1985). Proteins were 97-99% pure as judged by silver-stained SDS-PAGE gels; concentration was determined by UV spectroscopy of CaM native at pH 7.4 or denatured with NaOH.
Peptides
GenScript Corporation (Scotch Plains, NJ) synthesized peptides representing residues 1901-1927 of the Nav1.2 α-subunit (KRKQEEVSAIVIQRAYRRYLLKQKVKK, 3.36 kDa) with (Fl-Nav1.2IQp) or without (Nav1.2IQp) fluorescein at the N-terminus. The W.M. Keck Biotechnology Resource Center (New Haven, CT) synthesized Nav1.2BAA-1913-1938 representing residues 1913 to 1938 of the Nav1.2 α-subunit (QRAYRRYLLKQKVKKVSSIYKKDKGK).
Emission Spectra
Emission spectra (λex 430 nm, bandpasses 2 nm (excitation), 10 nm (emission)) of Fl-Nav1.2IQp (1 μM) ± 3 μM CaM1-148, CaM1-80 or CaM76-148 in 50 mM HEPES, 100 mM KCl, 5 mM NTA, 50 μM EGTA, and 1 mM MgCl2 (pH 7.4) were collected at 22 °C with a PTI-QM4 Fluorimeter (PTI, Birmingham, NJ). Apo samples were Ca2+-saturated by addition of CaCl2 in matching buffer to a final concentration of 10 mM. Spectra were normalized to λmax of Fl-Nav1.2IQp alone and corrected for dilution.
Equilibrium Calcium Titrations
Ca2+ titrations of sites I and II (N-domain) and sites III and IV (C-domain) of CaM alone (6 μM) or with 9 μM Nav1.2IQp (50% excess) were monitored at 22 °C with a PTI-QM4 Fluorimeter (Photon Technology International, Birmingham, NJ) and analyzed as described previously (Theoharis et al., 2008).
Nav1.2IQp Titration of CaM1-148
15N/1H-HSQC spectroscopy monitored a titration of Nav1.2IQp into apo 15N-CaM1-148 (400 μM) in 10 mM D4-imidazole, 100 mM KCl, 50 μM D16-EDTA, 0.01% NaN3, pH 6.8. Upon saturation of apo CaM1-148 with Nav1.2IQp, CaCl2 was added to a final concentration of 5 mM. Nav1.2IQp and Ca2+ saturation of CaM1-148 were confirmed by plateaus in the intensity of peaks corresponding to either the Nav1.2IQp:apo CaM1-148 or Nav1.2IQp:(Ca2+)4-CaM1-148.
Preparation of 13C,15N-CaM76-148:Nav1.2IQp Complex
A 1.5 mM 1:1 complex of 12C,14N-Na 1.2IQp:13C, 15N-apo CaM76-148 was prepared in 10 mM D4-imidazole, 100 mM KCl, 50 μM D16-EDTA, 0.01% NaN3, pH 6.8. Nitrogen-based experiments were conducted in 90% H20 / 10% D20, while carbon-based experiments were conducted in 100% D2O. All spectra, with the exception of amide exchange, were collected on samples in Shigemi (Allison Park, PA) microscale NMR tubes whose magnetic susceptibility was matched to D2O.
15N/1H-HSQC Monitored Amide Exchange of 15N-CaM76-148:Nav1.2IQp Complex
The 12C,14N-Nav1.2IQp:13C, 15N-apo CaM76-148 complex (described above) was lyophilized in a Speed-Vac Model VG-5 (Savant). The complex was re-suspended in 99.96% D2O (Cambridge Isotope Laboratories, Andover, MA). HMQC spectra prior to lyophilization in H2O were identical to those taken in D2O. The pH of the sample was not adjusted after addition of D2O. The sample was placed immediately in a Bruker 500 MHz Avance II spectrometer; data acquisition began within 5 min. 15N/1H-HSQC spectra were acquired at intervals of 30 min for 23 h. Spectra were processed with NMRPipe (Delaglio et al., 1995). Individual peak intensities were determined with Sparky (Goddard and Kneller). Using the Solver function in Microsoft Excel, peak intensities were fit to Equation 1.
| (1) |
where I is intensity at time t, I0 is initial intensity, t0 is time zero, τ is the apparent decay lifetime, and b represents intensity at the asymptote.
Structure Determination
NMR spectra were collected at 25 °C on either a Bruker Avance II 500 MHz or cryoprobe-equipped 800 MHz spectrometer. The 1H, 15N, and 13C resonances of the apo CaM76-148 backbone were assigned using triple resonance experiments (HNCA, HN(CO)CA, HNCACB, HN(CO)CACB, HNCO, and HN(CA)CO) (Yamazaki et al., 1994) with uniformly 13C,15N-labeled apo CaM76-148 bound to unlabeled Nav1.2IQp. 1Hα resonances were assigned from an 15N-edited TOCSY spectrum using a uniformly 15N-labeled protein (Clore and Gronenborn, 1994) and from HA(CACO)NH experiments using a uniformly 15N and 13C-labeled sample. Side chains were assigned from 3D H(CCO)NH-TOCSY, C(CO)NH-TOCSY, HCCH-TOCSY, 15N-edited TOCSY, and 15N or 13C edited NOESY spectra (Clore and Gronenborn, 1994; Fesik and Zuiderweg, 1988).
CaM was 13C, 15N-labeled but Nav1.2IQp had natural abundance isotopes. We used isotope-filtered 2D TOCSY and NOESY experiments with mixing times of 26 to 46 ms (Gemmecker et al., 1992; Ikura and Bax, 1992; Otting and Wuthrich, 1989) to assign the unlabeled Nav1.2IQp. Isotope-filters for 13C and 15N were employed twice (once preceding t1 and once preceding t2) to effectively suppress the signals derived from 13C-attached protons and 15N-attached protons. As a result, 2D spectra for protons located on the unlabeled peptide were obtained; thus, only intrapeptide signals were observed in these doubly isotope-filtered spectra. To assign intermolecular NOEs between the unlabeled peptide and 13C,15N-CaM, we used 13C-edited and 14N,12C-filtered 3D NOESY experiments with mixing times of 80 to 120 ms as described previously (Burgering et al., 1993). Representative intrapeptide and intermolecular NOEs of the CaM/peptide complex were shown in Figure 6. All spectra were processed with NMRPipe (Delaglio et al., 1995) and analyzed with Sparky (Goddard and Kneller).
Apo CaM76-148 :Nav1.2IQp Structure Calculations
Structures of the complex were generated using a torsion-angle molecular dynamics protocol (Stein et al., 1997) with CNS (Brunger et al., 1998). Structure calculations employed 1819 NMR-derived distance restraints from the analysis of 3D 15N- and 13C-resolved NOESY spectra acquired with a mixing time of 120 ms (Fesik and Zuiderweg, 1988). The NOE-derived distance restraints were given upper bounds of 3.0, 4.0, 5.0, and 6.0 Å based upon measured NOE intensities. From an analysis of amide exchange rates measured from a series of 15N/1H-HSQC spectra recorded after the addition of D2O, 46 hydrogen bonds for the α-helices were included in the structural calculations. In addition, 109 Φ and angular restraints derived from an analysis of C, N, Cα, Hα, and Cβ chemical shifts using the TALOS program (Cornilescu et al., 1999) were included in the structural calculations. A square-well potential was employed to constrain the NMR-derived distance restraints with FNOE set to 150 and 50 kcal mol−1 Å−2 during stages of high temperature and slow-cooling torsion angle dynamics and final stage of conjugate gradient minimization, respectively. Force constants of 100 and 200 kcal mol−1 rad−2 were applied to all torsional restraints during the stage of high temperature torsion angle dynamics and the rest stages of structural calculations, respectively.
Quantification of Chemical Shifts
To determine the change in chemical shift upon Nav1.2IQp binding to apo CaM76-148, or Ca2+ binding to apo CaM1-148, net perturbations in both the 1H and 15N dimensions were quantified using the modified Pythagorean theorem in Equation 2.
| (2) |
Here, Δδ refers to the linear displacement of a specific resonance peak from its initial position in the reference spectrum.
CaM Binding to Nav1.2IQp
Binding of CaM to Fl-Nav1.2IQp (50 mM HEPES, 1 mM MgCl2, 5 mM NTA, 50 μM EGTA, pH 7.4) was monitored by fluorescence anisotropy at 22 °C using a Fluorolog 3 (Jobin Yvon, Horiba) fluorimeter. A slight excess of CaM was added (reaching a 1.2:1 ratio of CaM:Nav1.2IQp). The complex was then titrated with a matching buffer that contained 5 M NaCl. Changes in the anisotropy of Fl-Nav1.2IQp were monitored and analyzed numerically as described previously (Theoharis et al., 2008).
Accession Numbers
The coordinates for this structure were deposited in May, 2010 with the PDB code of 2KXW. A preliminary report of this structure was presented at the 2010 Annual Biophysical Meeting (Feldkamp, Yu, and Shea, 2010).
Note Added in Proof
During revision of this article, Chagot and Chazin deposited and reported a structure (2L53) of apo CaM1-148 bound to a peptide representing the IQ motif of NaV1.5 (E1901-L1927) with a short N-terminal tag (GPGS) (Chagot and Chazin, 2011). Superposition of 2KXW and 2L53 using pairs of 68 Cα atoms located in the well defined secondary structure elements showed a RMSD of 1.2 Å. In both 2KXW and 2L53, the apo C-domain adopts the semi-open tertiary conformation. The peptides bound in the same orientation and location despite differences in their sequences. The IQ residues in both peptides interact with almost identical sets of residues on CaM. Slight differences are observed in the two calcium binding loops which are very mobile in the absence of bound calcium. The orientation of the sidechain of Y1916 in our structure is different from that of the corresponding residue F1912 in 2L53, probably reflecting the slight differences in the sequences of both CaM and peptides used in these studies.
Supplementary Material
Acknowledgments
Supported by the National Institutes of Health RO1 GM 57001 to M.A.S., and by the Roy J. Carver Charitable Trust Grant 01-224.
Abbreviations
- NOE
Nuclear Overhauser effect
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ataman ZA, Gakhar L, Sorensen BR, Hell JW, Shea MA. The NMDA Receptor NR1 C1 Region Bound to Calmodulin: Structural Insights into Functional Differences between Homologous Domains. Structure. 2007;15:1603–1617. doi: 10.1016/j.str.2007.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S, Deschenes I, Disilvestre D, Tian Y, Halperin VL, Tomaselli GF. Calmodulin regulation of Nav1.4 current: role of binding to the carboxyl terminus. J Gen Physiol. 2008;131:197–209. doi: 10.1085/jgp.200709863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges N, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. 1998;D54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Burgering M, Boelens R, Kaptein R. Observation of intersubunit NOEs in a dimeric P22 Mnt repressor mutant by a time-shared [15N,13C] double half-filter technique. J. Biomol. NMR. 1993;3:709–714. [Google Scholar]
- Catterall AC. Structure and regulation of voltage-gated Ca2+-channels. Annu.Rev.Cell Dev.Biol. 2000;16:521–555. doi: 10.1146/annurev.cellbio.16.1.521. [DOI] [PubMed] [Google Scholar]
- Chagot B, Chazin WJ. Solution NMR Structure of Apo-Calmodulin in Complex with the IQ Motif of Human Cardiac Sodium Channel NaV1.5. Journal of Molecular Biology. 2011;406:106–119. doi: 10.1016/j.jmb.2010.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clore GM, Gronenborn AM. Multidimensional heteronuclear magnetic resonance of proteins. Meths. Enzymol. 1994;239:349–363. doi: 10.1016/s0076-6879(94)39013-4. [DOI] [PubMed] [Google Scholar]
- Cormier JW, Rivolta I, Tateyama M, Yang AS, Kass RS. Secondary Structure of the Human Cardiac Na+ Channel C Terminus. Journal of Biological Chemistry. 2002;277:9233–9241. doi: 10.1074/jbc.M110204200. [DOI] [PubMed] [Google Scholar]
- Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- Crivici A, Ikura M. Molecular and Structural Basis of Target Recognition by Calmodulin. Annual Review of Biophysics and Biomolecular Structure. 1995;24:85–116. doi: 10.1146/annurev.bb.24.060195.000505. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- DeMaria CD, Soong TW, Alseikhan BA, Alvania RS, Yue DT. Calmodulin bifurcates the local Ca2+ signal that modulates P/Q-type Ca2+ channels. Nature. 2001;411:484–489. doi: 10.1038/35078091. [DOI] [PubMed] [Google Scholar]
- Drum CL, Yan S, Bard J, Shen Y, LU D, Soelaiman S, Grabarek Z, Bohm A, Tang W. Structural basis for the activation of anthrax adenylyl cyclase exotoxin by calmodulin. Nature. 2002;415:396–402. doi: 10.1038/415396a. [DOI] [PubMed] [Google Scholar]
- Feldkamp MD, Yu L, Shea MA. Calmodulin Regulation of the Neuronal Voltage-Dependent Sodium Channel. Biophysical Journal. 2010;98:310a. [Google Scholar]
- Fesik SW, Zuiderweg ERP. Heteronuclear three-dimensional NMR spectroscopy. A strategy for the simplification of homonuclear two-dimensional NMR spectra. J. Magn. Reson. 1988;78:588–593. [Google Scholar]
- Fraczkiewicz R, Braun W. Exact and Efficient Analytical Calculation of the Accessible Surface Areas and Their Gradients for Macromolecules. J. Comp. Chem. 1998;19:319–333. [Google Scholar]
- Gemmecker G, Olejniczak ET, Fesik SW. An improved method for selectively observing protons attached to 12C in the presence of 1H-13C spin pairs. J. Magn. Reson. 1992;96:199–204. [Google Scholar]
- Goddard TD, Kneller DG. SPARKY. University of California; San Francisco: [Google Scholar]
- Houdusse A, Gaucher JF, Krementsova E, Mui S, Trybus KM, Cohen C. Crystal structure of apo-calmodulin bound to the first two IQ motifs of myosin V reveals essential recognition features. Proc Natl Acad Sci U S A. 2006;103:19326–19331. doi: 10.1073/pnas.0609436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikura M, Bax A. Isotope-filtered 2D NMR of a protein-peptide complex: study of a skeletal muscle myosin light chain kinase fragment bound to calmodulin. J. Am. Chem. Soc. 1992;114:2433–2440. [Google Scholar]
- Kim EY, Rumpf CH, Fujiwara Y, Cooley ES, Van Petegem F, Minor DL., Jr. Structures of CaV2 Ca2+/CaM-IQ domain complexes reveal binding modes that underlie calcium-dependent inactivation and facilitation. Structure. 2008;16:1455–1467. doi: 10.1016/j.str.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klee CB, Newton DL, Ni WC, Haiech J. Regulation of the calcium signal by calmodulin. In: Evered D, Whelan J, editors. Calcium and the Cell. John Wiley & Sons; Chichester, New York: 1986. pp. 162–182. [DOI] [PubMed] [Google Scholar]
- Kung C, Preston RR, Maley ME, Ling K-Y, Kanabrocki JA, Seavey BR, Saimi Y. In vivo Paramecium mutants show that calmodulin orchestrates membrane responses to stimuli. Cell Calcium. 1992;13:413–425. doi: 10.1016/0143-4160(92)90054-v. [DOI] [PubMed] [Google Scholar]
- Liu Y, Storm DR. Regulation of free calmodulin levels by neuromodulin: Neuron growth and regeneration. TIPS. 1990;11:107–111. doi: 10.1016/0165-6147(90)90195-e. [DOI] [PubMed] [Google Scholar]
- Mantegazza M, Yu FH, Catterall WA, Scheuer T. Role of the C-terminal domain in inactivation of brain and cardiac sodium channel. Proceedings of the National Academy of Science. 2001;98:15348–15353. doi: 10.1073/pnas.211563298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molday RS, Englander SW, Kallen RG. Primary structure effects on peptide group hydrogen exchange. Biochemistry. 1972;11:150–158. doi: 10.1021/bi00752a003. [DOI] [PubMed] [Google Scholar]
- Mori M, Konno T, Ozawa T, Murata M, Imoto K, Nagayama K. Novel Interaction of the Voltage-Dependent Sodium Channel (VDSC) with Calmodulin: Does VDSC Acquire Calmodulin-Mediated Ca2+-Sensitivity? Biochemistry. 2000;39:1316–1323. doi: 10.1021/bi9912600. [DOI] [PubMed] [Google Scholar]
- Mori MX, Kooi C.W. Vander, Leahy DJ, Yue DT. Crystal structure of the CaV2 IQ domain in complex with Ca2+/calmodulin: high-resolution mechanistic implications for channel regulation by Ca2+ Structure. 2008;16:607–620. doi: 10.1016/j.str.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman RA, Van Scyoc WS, Sorensen BR, Jaren OR, Shea MA. Interdomain cooperativity of calmodulin to melittin preferentially increases calcium affinity of sites I and II. Proteins: Structure, Function, and Bioinformatics. 2008;71:1792–1812. doi: 10.1002/prot.21861. [DOI] [PubMed] [Google Scholar]
- Otting G, Wuthrich K. Extended heteronuclear editing of 2D 1H NMR spectra of isotope-labeled proteins, using the X(omega1, omega2) double half filter. J. Magn. Reson. 1989;85:586–594. [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Putkey JA, Slaughter GR, Means AR. Bacterial expression and characterization of proteins derived from the chicken calmodulin cDNA and a calmodulin processed gene. Journal of Biological Chemistry. 1985;260:4704–4712. [PubMed] [Google Scholar]
- Sarhan MF, Van Petegem F, Ahern CA. A double tyrosine motif in the cardiac sodium channel domain III-IV linker couples calcium-dependent calmodulin binding to inactivation gating. J Biol Chem. 2009;284:33265–33274. doi: 10.1074/jbc.M109.052910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller KL, Caldwell JH. Expression and distribution of voltage-gated sodium channels in the cerebellum. Cerebellum. 2003;2:2–9. doi: 10.1080/14734220309424. [DOI] [PubMed] [Google Scholar]
- Schumacher MA, Rivard AF, Bachinger HP, Adelman JP. Structure of the gating domain of a Ca2+-activated K+ channel complexed with Ca2+/calmodulin. Nature. 2001;410:1120–1124. doi: 10.1038/35074145. [DOI] [PubMed] [Google Scholar]
- Shah VN, Wingo TL, Weiss KL, Williams CK, Balser JR, Chazin WJ. Calcium-dependent regulation of the voltage-gated sodium channel hH1: intrinsic and extrinsic sensors use a common molecular switch. Proc Natl Acad Sci U S A. 2006;103:3592–3597. doi: 10.1073/pnas.0507397103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolev V, Sorokine A, Prilusky J, Abola EE, Edelman M. Automated analysis of interatomic contacts in proteins. Bioinformatics. 1999;15:327–332. doi: 10.1093/bioinformatics/15.4.327. [DOI] [PubMed] [Google Scholar]
- Stein EG, Rice LM, Brünger AT. Torsion-Angle Molecular Dynamics as a New Efficient Tool for NMR Structure Calculation. J. Magn. Reson. 1997;124:154–164. doi: 10.1006/jmre.1996.1027. [DOI] [PubMed] [Google Scholar]
- Swindells MB, Ikura M. Pre-formation of the semi-open conformation by the apo-calmodulin C-terminal domain and implications for binding IQ-motifs. Nature Structural Biology. 1996;3:501–504. doi: 10.1038/nsb0696-501. [DOI] [PubMed] [Google Scholar]
- Terrak M, Wu G, Stafford WF, Lu RC, Dominguez R. Two distinct myosin light chain structures are induced by specific variations within the bound IQ motifs - functional implications. European Molecular Biology Organization Journal. 2003;22:362–371. doi: 10.1093/emboj/cdg058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theoharis NT, Sorensen BR, Theisen-Toupal J, Shea MA. The Neuronal Voltage-Dependent Sodium Channel Type II IQ Motif Lowers the Calcium Affinity of the C-Domain of Calmodulin. Biochemistry. 2008;47:112–123. doi: 10.1021/bi7013129. [DOI] [PubMed] [Google Scholar]
- Urbauer JL, Short JH, Dow LK, Wand AJ. Structural Analysis of a Novel Interaction by Calmodulin: High Affinity Binding of a Peptide in the Absence of Calcium. Biochemistry. 1995;34:8099–8109. doi: 10.1021/bi00025a016. [DOI] [PubMed] [Google Scholar]
- Venosa RA. Inward movement of sodium ions in resting and stimulated frog′s sartorius muscle. J Physiol. 1974;241:155–173. doi: 10.1113/jphysiol.1974.sp010646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss LA, Escayg A, Kearney JA, Trudeau M, MacDonald BT, Mori M, Reichert J, Buxbaum JD, Meisler MH. Sodium channels SCN1A, SCN2A and SCN3A in familial autism. Molecular Psychistry. 2003;8:186–194. doi: 10.1038/sj.mp.4001241. [DOI] [PubMed] [Google Scholar]
- Yamazaki T, Lee W, Arrowsmith CH, Muhandiram DR, Kay LE. A suite of triple-resonance NMR experiments for the backbone assignment of 15N, 13C, 2H-labeled proteins with high sensitivity. J. Am. Chem. Soc. 1994;116:11655–11666. [Google Scholar]
- Yap KL, Kim J, Truong K, Sherman M, Yuan T, Ikura M. Calmodulin Target Database. Journal of Structural and Functional Genomics. 2000;1:8–14. doi: 10.1023/a:1011320027914. [DOI] [PubMed] [Google Scholar]
- Yu FH, Catterall WA. Overview of the voltage-gated sodium channel family. Genome Biology. 2003;4:207.201–207.207. doi: 10.1186/gb-2003-4-3-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.