Abstract
We present an efficient pipeline enabling high-throughput analysis of protein structure in solution with small angle X-ray scattering (SAXS). Our SAXS pipeline combines automated sample handling of microliter volumes, temperature and anaerobic control, rapid data collection, data analysis, and couples structural analysis with automated archiving. We subjected 50 representative proteins, mostly from Pyrococcus furiosus, to this pipeline, revealing that 30 were multimeric structures in solution. SAXS analysis allowed us to distinguish aggregated and unfolded proteins, define global structural parameters and oligomeric states for most samples, identify shapes and similar structures for 25 unknown structures, and determine envelopes for 41 proteins. We believe that high throughput SAXS is an enabling technology that may change the way that structural genomics research is done.
Visualizing macromolecular shapes and assemblies that principally determine function is a central challenge for structural molecular biology1. Addressing this challenge requires the capacity to characterize the many complexes and conformations that underlie biological outcomes. Yet, growing metagenomics, proteomics, and bioinformatics contributions are outpacing classical structural biology approaches, creating an increasing structural knowledge gap2,3.
X-ray diffraction and scattering are powerful methods for unraveling structural details and molecular shapes4. Macromolecular X-ray crystallography has been the cornerstone of the structural genomics initiatives5; both crystallography and NMR have provided a deep and broad survey of macromolecular structural properties at high resolution6,7,8. Yet, the stochastic nature of crystallization and the size and time constraints of NMR limit the throughput of these technologies. The application of X-ray scattering in solution, known as small angle X-ray scattering (SAXS), to structural biology has lagged behind crystallography despite its strength in other fields9. However, SAXS use has sharply increased with advances in synchrotron X-ray sources and detectors that improve data quality and reduce the amount of sample required. New algorithms have been developed which can identify accurate shapes and assemblies based upon the scattering data4,10,11. Importantly, SAXS analyses can build upon and be combined with other results to test experimental hypotheses and computational models4.
Though lower in spatial resolution than crystallography or NMR, SAXS offers fundamental advantages for high-throughput structural analyses: structural measurements are carried out in solution, sample preparation is simple, quality global parameters can be obtained for most samples, and SAXS is compatible with and complementary to other biophysical techniques. The ~15 Å spatial resolution of SAXS envelopes is often sufficient to address key biological questions, and several high impact SAXS results have recently been described12–15. Because sample preparation is minimal and data can be rapidly collected and analyzed, SAXS is potentially the highest throughput structural technique. As most macromolecular structures are amenable to SAXS analysis, for example, the structural analysis of all complexes of a metabolic pathway can be considered. In the US alone, the NIH will spend $80 million this year on the Protein Structure Initiative16, which provides structures for 3% to 15% of its targets17, so a cost-effective and efficient means to improve the fraction of protein samples yielding structural information would be very valuable.
Here we report the development of an efficient pipeline enabling robust, broadly applicable, and largely automated SAXS-based structural analyses. Though alternative collection approaches have been reported18,19, we were able to obtain high quality data from small volumes (12 µl) and protein concentrations (~1 mg/ml), with temperature and anaerobic control for sample stability in a modular 96-well format. We subjected 50 proteins, mostly from Pyrococcus furiosus (Pfu), to our pipeline. Our high-throughput SAXS pipeline provided global information (Table 1) for most samples, as well as folding, assembly and three-dimensional envelope information on mono-disperse samples. Such information can be used to judge the amenability of proteins for crystallographic studies and can even be used to infer protein function. Our results demonstrate how automated, high-throughput SAXS can provide a critical enabling technology for producing unique, comprehensive, and complementary solution structural information.
Table1.
SAXS characterizations for 34 Pfu samples including structural information.
| Sample | New Structural Results | |||||
|---|---|---|---|---|---|---|
| Class | Gene | Ortholog | RG | Dmax | Assemblies | Envelope |
| Aggregates | PF0418 | ATPase | separable | No | ||
| PF1733 | Cons. hypothetical | inseparable | No | |||
| PF1951 | Aspartate-ammonia ligase | inseparable | No | |||
| Mixtures of Oligomers of Unknown Structure | PF0230 | ArsR transcription regulator | Mostly 2-mer | Yes | ||
| PF0259 | Cons. hypothetical | 26.0 | 84 | Mostly 8-mer | Yes | |
| PF0741 | Thioredoxin-related | 20 | >80 | Mostly 1-mer | Yes | |
| PF1548 | Cons. hypothetical | rings | ~Yes | |||
| PF1605 | Molybdopterin synthase | 21 | >90 | Mostly 1-mer | Yes | |
| Mixtures of Oligomers of Known Structure | PF0094* | Glutaredoxin-like | 28 | 110 | 92% 2-mer/8% 4-mer | Yes |
| PF0380* | Cons. hypothetical | 21 | 125 | 68% 2-mer/32% 1-mer | Yes | |
| PF0939 | Isopropylmalate dehydratase | 23.1 | 82 | 73% 2-mer/27% 1-mer | Yes | |
| PF1909* | Ferredoxin | 13.0 | 38 | 40% 2-mer/60% 1-mer | Yes | |
| Matching PDB Model | PF0863* | Adenylyl cyclase CyaB | 27.4 | 87 | Matching1-mer | Yes |
| PF1061† | Ferredoxin β-grasp fold | 17.7 | 78 | Matching 1-mer | Yes | |
| PF1281* | Superoxide reductase | 22.1 | 80 | Matching 4-mer | Yes | |
| PF1282† | Rubredoxin | 11.0 | 29 | Matching 1-mer | Yes | |
| PF0619 | Cons. hypothetical | 23.1 | 73 | Matching 3-mer | Yes | |
| Crystal Structure of Homolog | PF1026 | Malic enzyme, NAD-binding | 31.8 | 110 | Matching 1-mer | Yes |
| PF1033 | Thioredoxin-like fold | 51.2 | 150 | Matching 10mer | Yes | |
| PF1528 | SNO glutamine amidotransferase | 19.9 | 80 | Matching 1-mer | Yes | |
| PF1674 | Tyrosine/serine phosphatase | 16.7 | 58 | Matching 1-mer | Yes | |
| PF1787 | Acetyl-CoA synthetase | 33.9 | 98 | Novel 3-mer | Yes | |
| Proteins of Unknown Structure | PF0014/0015a | Cons. hypothetical | 55.0 | 165 | >8-mer | Yes |
| PF0553 | Tyrosine phosphatase | 19.2 | 110 | 1-mer | Yes | |
| PF706.1 | Zinc finger | 18.6 | 80 | 1-mer | Unfolded | |
| PF0699 | Conserved | 23.7 | 74 | 2-mer | Yes | |
| PF0715 | NADH oxidase | 23.1 | 96 | 2-mer | Yes | |
| PF0965/0966/0967/0971b | Pyruvate ferredoxin oxidoreductase | 36.9 | 120 | 244kDa | Yes | |
| PF1282/1205c | Nucleotide binding proteina | 24.3 | 95 | 1-mer | Unfolded | |
| PF1291 | Phosphoesterase | 35.6 | 110 | 4-mer | Yes | |
| PF1372 | Cons. hypothetical | 23 | 75 | 4-mer | Yes | |
| PF1911 | Ferredoxin NADP reductase | 30.9 | 101 | 2-mer | Yes | |
| PF1950 | Phosphoribosyl transferase | 25.3 | 100 | 2-mer | Yes | |
| PF2047.1 | Cons. hypothetical | 29.7 | 155 | 1-mer | Unfolded | |
PF number is the Pfu genome ORF30 number; protein names (orthologs) were based on bioinformatics analyses [www.ebi.ac.uk/interpro/]. We divide SAXS samples into three general classes: non-ideal samples, samples with existing atomic structural information and proteins of unknown structures. Non-ideal samples are either aggregated or mixtures of multimers which cannot be modeled with available atomic resolution results. Samples with direct atomic resolution results or from a sequence homolog provide improved resolution even when a mixture is present.
The abbreviation Cons. Hypothetical denotes conserved hypothetical proteins which have unknown function.
Proteins which have been crystallized.
Proteins with models determined from NMR.
PF0014 and 0015 were tandemly expressed in E-Coli. and form a complex
PF0965,PF0966,PF0967 and PF0971 form Pryruvate ferredoxin oxidoreductase and were purified from native biomass.
The PF1282/1205 recombinant fusion protein has the rubredoxin (PF1282) of Pfu as the ‘tag’ to an unrelated putative nucleotide binding protein (PF1205). While the PF1282 portion is folded as evident by its red color (Iron), PF1205 is unfolded as shown below.
RESULTS
High-throughput SAXS data collection platform
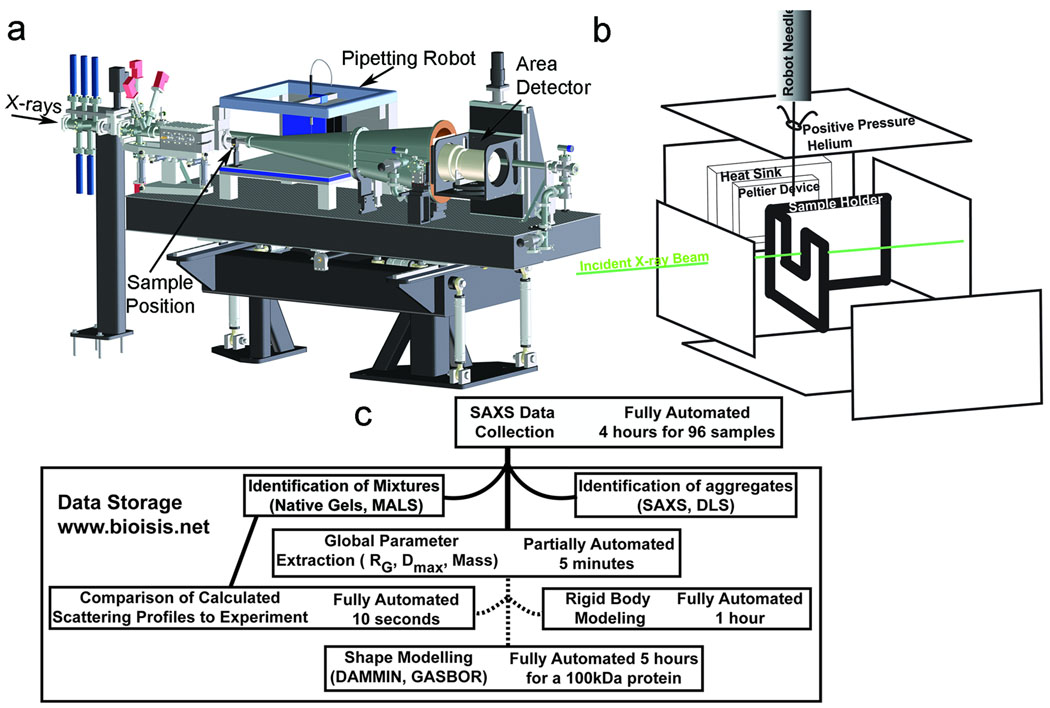
To achieve sufficient X-ray flux for informative scattering with low protein concentration and small volumes, we designed the SIBYLS beamline at the Advanced Light Source. We employ a light path generated by a super-bend20 magnet to provide a 1012 photons/sec flux (1 Å wavelength). The tunable incident wavelength enables rapid adjustment of the q range appropriate for the experiment without changing the sample to detector configuration (q=4π sin(θ /2)/λ where θ is the scattering angle and λ is the wavelength). Scattering is measured on a MAR165® area detector co-axial with the incident beam and 1.5 m from the sample allowing a q range from a minimum of 0.007 Å−1 to a maximum of 4.2 Å−1 (Fig. 1a).
Figure 1.
High-throughput SAXS pipeline. (a) Configuration of the SAXS endstation shows X-ray beam path, sample position, pipetting robot, and area detector. (b) Schematic of the sample area showing how the sample is loaded by the robot into a temperature-controlled cell. Positive helium pressure reduces air scatter and oxidative damage. (c) SAXS analysis tree for rapid and robust data processing and analysis. Proteins are first categorized as aggregated (using either the scattering curve itself or dynamic light scattering (DLS)), mixtures (based on native gel electrophoresis or multi-angle light scattering (MALS)), or mono-disperse samples. For monodisperse samples, SAXS data next defines global solution structural parameters radius of gyration, maximum dimension, and calculated mass. Sequence-based homology search discovers existing structures that can be used to analyze both mixtures and monodisperse samples. Approximate time scales are noted in each step. Perl scripts are used to collect information and begin processes for dashed paths. Both primary data and derived shapes are stored at the BioIsis internet accessible utility.
To transfer 96-well plate samples to the SAXS sample cell, we implemented a Hamilton® pipetting robot. Both sample cell and the 96-well plate are temperature controlled with the sample plate sealed by a pierceable aluminum sheet. The robot needle transfers samples to the helium-filled sample holder (Fig. 1b), providing an anaerobic environment with low X-ray scattering cross-section; reducing background.
Protocol for high-throughput SAXS data analysis
For efficient analysis of data quality and information, we developed a SAXS analysis tree (Fig 1c). We automated the program data flow with Perl scripts (Supplementary Software) for the ATSAS21 program suite similar to those recently reported22. Output is standardized for automated incorporation into our database. Job scheduling is also automated on computer clusters. Data analysis begins with defining global sample parameters and comparisons of experimental and calculated scattering curves where prior structural information exists. To test the scattering information, we employ two different molecular envelope determination programs: DAMMIN23 and GASBOR10, which determine a compact envelope by minimizing differences between experimental and calculated scattering.
Ten independent DAMMIN runs are spawned by default once data enters the system. Mass is estimated using half the Porod volume9 calculated from q < 0.25 Å−1. For most samples, we found this estimate sufficient to identify oligomeric state. When ambiguous, mass was estimated by the extrapolated intensity at zero scattering angle9,24. The time required to traverse the analysis tree is size dependent: 40 minutes for a 20 kDa protein to 1.5 days for a 500 kDa complex run in parallel with other proteins. With current computational resources, our throughput exceeds 20 proteins per week for a full analysis; >1,000 macromolecules could be analyzed per year.
Automated data storage and quality control
To aid communication of our results as well as for promoting objective quality assessment, testing of newly available atomic resolution models and SAXS algorithm development, we created the web accessible database Bioisis (Biologically integrated structures in solution: www.bioisis.net). A powerful aspect of SAXS data collection is the ability to characterize macromolecules in many solution conditions. In the Bioisis database all experimental details are saved and associated with each sample. Database functionality has been enhanced for Pfu, Sulfolobus solfataricus and Halobacterium salinarum, including gene annotations and a search engine for gene number or a key word in the annotation.
Testing prototypical sample sets
To test whether SAXS can provide proteomic-scale information, we analyzed protein targets from 2 sources: 34 recombinantly expressed Pfu samples with a 9 amino-acid His-tag (Table 1) plus 16 Joint Center for Structural Genomics (JCSG) targets with 19 amino-acid His-tags (Supplementary Table 1). We focus here on the results from Pfu samples where 29 of the 34 proteins had failed to crystallize despite systematic efforts. These are labeled by open reading frame (ORF), and are prototypic of gene products providing sequences for current structural genomics efforts.
To aid analysis, we divided samples into three general classes (Table 1): non-ideal proteins (aggregated or mixed assembly states), proteins with existing structural information (either directly or from a sequence homolog) and proteins with unknown structures. We first characterized the samples by non-denaturing gel electrophoresis and light scattering. Non-ideal samples exhibit mixtures of states or aggregation that restricts SAXS analyses (Fig. 2). Proteins with existing structural information (from themselves or sequence homologs) allow higher resolution analyses. Proteins of unknown structure are monodispersed with no or incomplete structural homology. For these novel protein structures, we show that SAXS not only provides shape and assembly information, but also identifies similar known structures based upon direct comparisons of experimental scattering with that calculated from known structures.
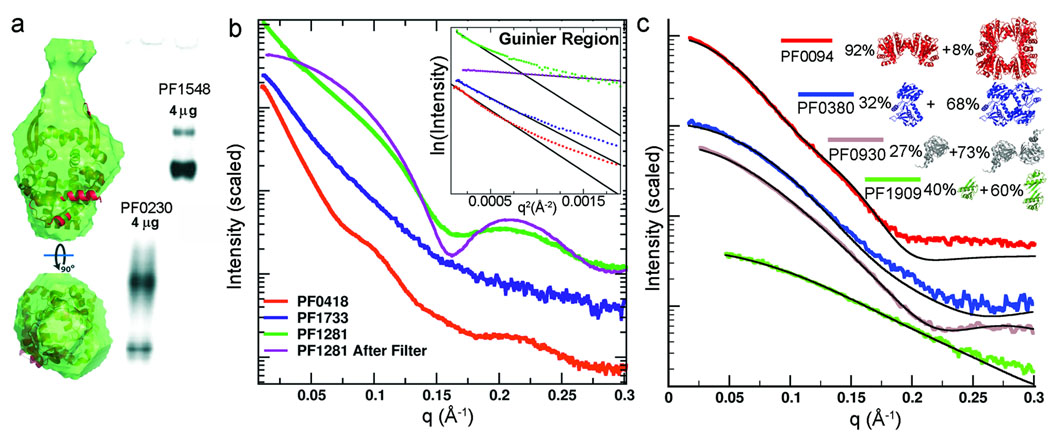
Figure 2.
SAXS analysis provides feedback on challenging samples that are polydisperse or inhomogeneous. (a) PF0230 and PF1548 were mixtures by native gel electrophoresis. Overlaying the SAXS-predicted PF0230 envelope with a close homolog (PDB 2CWE) revealed consistency to the homolog dimer with additional density indicating a larger species. (b) SAXS results directly discerned aggregation based on low angle Guinier regions (insert) for three protein samples PF0418 (red), PF1733 (blue) and PF1281 (green). Features (oscillations) in the SAXS scattering curve for PF0418 and PF1281 suggest that small adjustments in sample preparation may yield workable data, e.g. PF1281 was markedly improved after passing through a filter (purple). (c) Probable multimers may be identified when atomic resolution results are available of the protein or a homolog. Here, multimers in crystal lattices (PF0094 homolog PDB 1J08, PF0380 PDB 1VK1, PF0930 homolog PDB 1V7L, and PF1090 PDB 1SJ1) are used to identify a best fit to the SAXS data.
Non-Ideal protein samples can guide sample improvements
Samples are non-ideal when light scattering or other techniques suggest aggregation or mixed states (Table 1). Native gel electrophoresis showed that PF0230 and PF1548 were mixed oligomeric species (Fig. 2a). Results for all SAXS derived parameters on mixtures are electron number and population weighted averages of parameters determined from each component individually. Algorithms for envelope determination assume homogeneous solutions, so interpretations must take any mixed state into account. The PF0230 envelope (Fig. 2a), for example, is overlaid on the proposed biological unit from a homolog crystal structure. The lower portion fits the dimer; yet, the extension is probably an average of dimers mixed with larger oligomers. For PF1548, gel filtration analysis and forward scattering indicate large multimeric assemblies. Reconstructed envelopes suggest rings with a propensity to stack.
Mixed oligomeric or aggregate samples produce scattering curves dominated at the smallest angles by the largest particles, which can confound subsequent analysis. However, SAXS probes structural details at and below 15 Å, so such samples may generate useful information if interpreted cautiously as SAXS is additive. Three proteins, PF0418, PF1281 and PF1733, were aggregated based on a small or non-existent Guinier region in the measured q space (Fig. 2b). This metric identifies particles with RG > 75 Å and a Dmax (longest dimension across the molecule) of at least 340 Å (larger than a ribosome). Scattering curve oscillations beyond q > 0.1 Å −1 with such a large RG indicate ordered and population-wide correlations on a much smaller length scale (e.g. data from PF0418 and PF1281). The absence of such oscillations (e.g. PF1733) typifies a heterogeneous assembly population with a substantial fraction having large dimensions.
Reversible aggregation is identified from scattering curve changes as a function of concentration, a metric that may guide crystallization experiments. JCSG samples were prepared for crystallography and concentrated to 23 mg/ml on average. We find that such high concentrations (> 5 mg/ml) often cause artificial multimerization states and aggregation. These concentrations may increase crystal nucleation but also heterogeneity, adversely affecting SAXS and other analyses. Aggregated samples whose scattering shows oscillations are often salvageable by removing aggregates. For example, passing samples through a 100kDa filter yielded scattering characteristic of a monodisperse solution for PF1281 (Fig. 2b). Given the scattering features observable for PF0418, interpretable SAXS results would likely be obtained with additional sample preparation, such as filtration. Six JCSG samples were rescued in this manner (Supplementary Table 1).
Homologous structures improve resolution
To take full-advantage of scattering information, it is important to identify and employ additional information when available4. An initial step in our analysis tree is the application of sequence analysis to identify any known detailed structures for samples. Atomic models were available for 7 Pfu samples. Seven others had sequence homology to proteins of comparable size with an existing structure in the Protein Data Bank (PDB) (Table 1). Existing detailed structures allowed comparisons of measured scattering curves with those calculated from atomic models.
In four cases (Table 1, Fig. 2c), non-denaturing gel electrophoresis showed multimeric forms and the data could be fit as a mixture of assemblies found in crystallographic lattices. SAXS data can identify which multimers are relevant even when mixtures are present. For example PF0094 fit multimers found in a homolog better than those found in its own determined crystal lattice.
Six samples had SAXS curves that matched those calculated from single multimeric states suggested by PDB structures (Fig. 3b). PF1281 was initially aggregated based upon the SAXS results, but a homogenous solution was obtained after spin column filtration just prior to data collection (Fig. 2b).
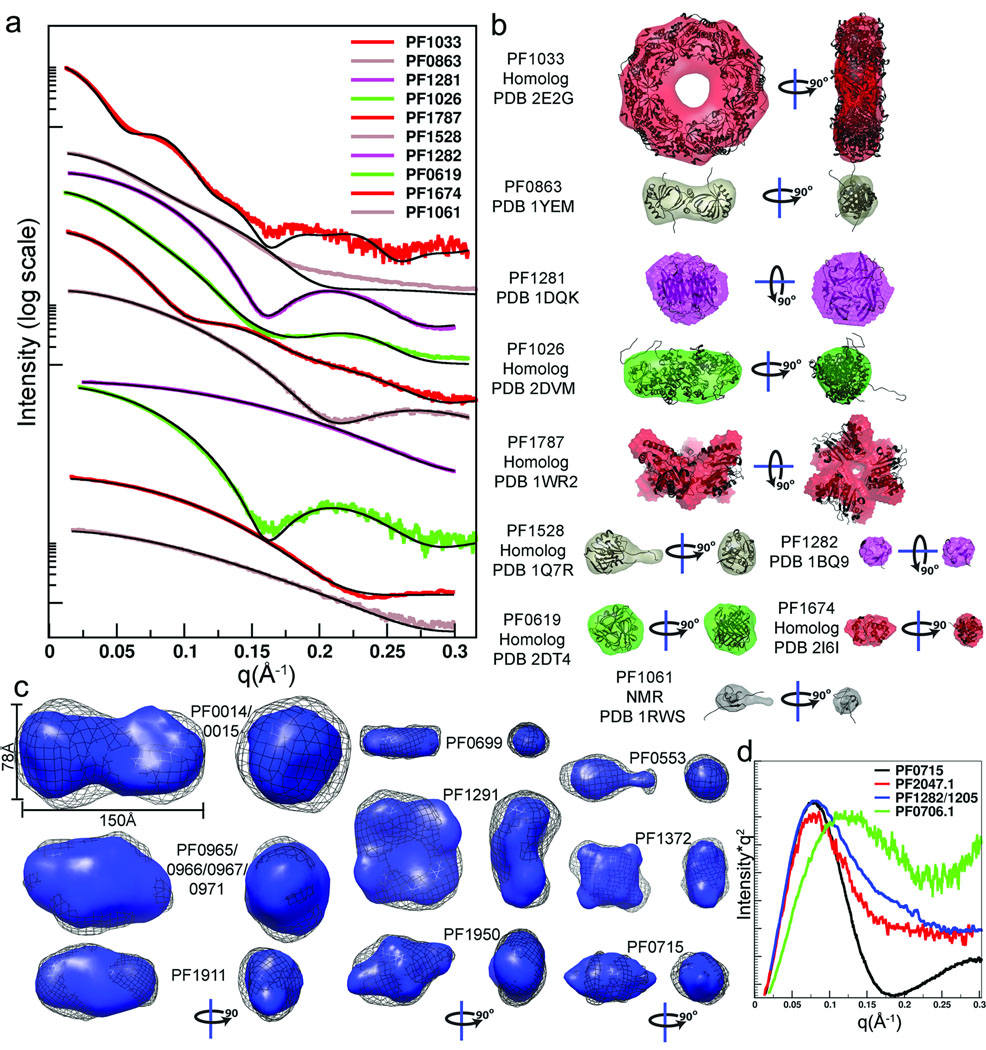
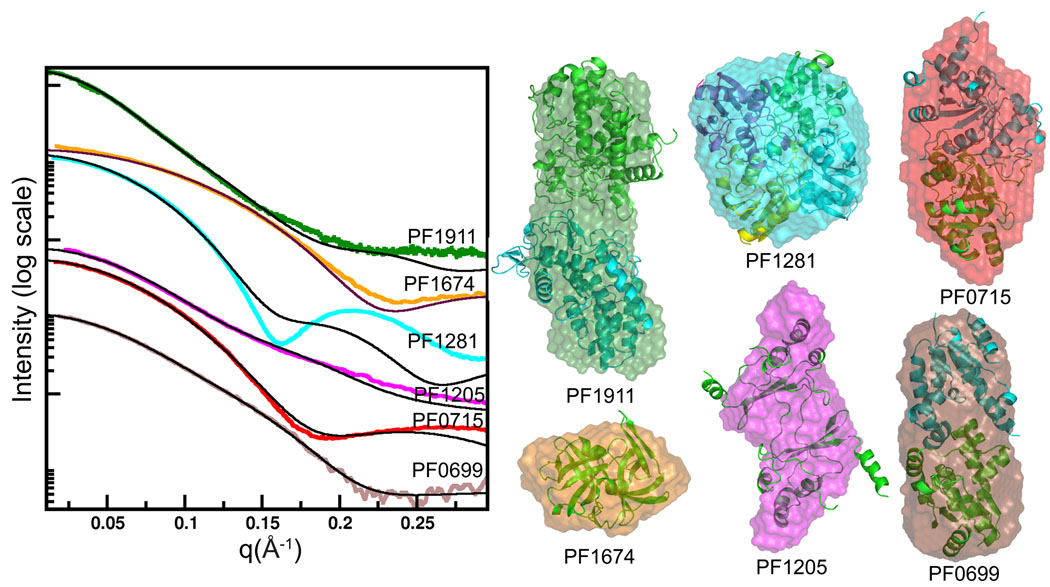
Figure 3.
SAXS provides accurate shape and assembly in solution for most samples. (a) For the ten proteins with structural homologs or existing structures, the experimental scattering data (colors) were compared with the scattering curve calculated for the matching structure (black). (b) For monodisperse samples, the envelope determinations (colored as in a) were overlaid with the existing structures (ribbons). All monomeric units had a seven amino-acid His-tag attached. (c) For the 9 proteins with no pre-existing structural information, envelope predictions from two independent programs were compared and generally agree. The DAMMIN results (black mesh) were generated without symmetry. The GASBOR results used 2-fold symmetry for PF0014/0015, PF0965/0966/0967/0971, PF1911 (dimer), PF00716 (dimer), PF0699 (dimer) and PF1950 (dimer). Four-fold symmetry was imposed on tetrameric PF1291 and PF1372. (d) Plotting the SAXS data as I*q2 vs. q (Kratky plot) highlights proteins with large unfolded regions. The Kratky plot of PF0715 is shown for comparison of a folded protein and shows characteristic parabolic behavior at wide angles. In contrast PF0706.1, PF2047.1, and PF1282/1205 have SAXS data consistent with unfolded regions as reflected in the non-parabolic wide-angle properties.
PF1674 matches the scattering profile calculated from the monomeric state of a distant homolog (Table 1). In contrast, PF1787 did not fit the monomer scattering profile, nor any multimer in the crystal structure of a homolog with 55% sequence identity (PDB 1WR2). We applied rigid body modeling of three subunits and found a best fit to the experimental data, supporting the reconstructed envelope with 3-fold symmetry (Fig. 4).
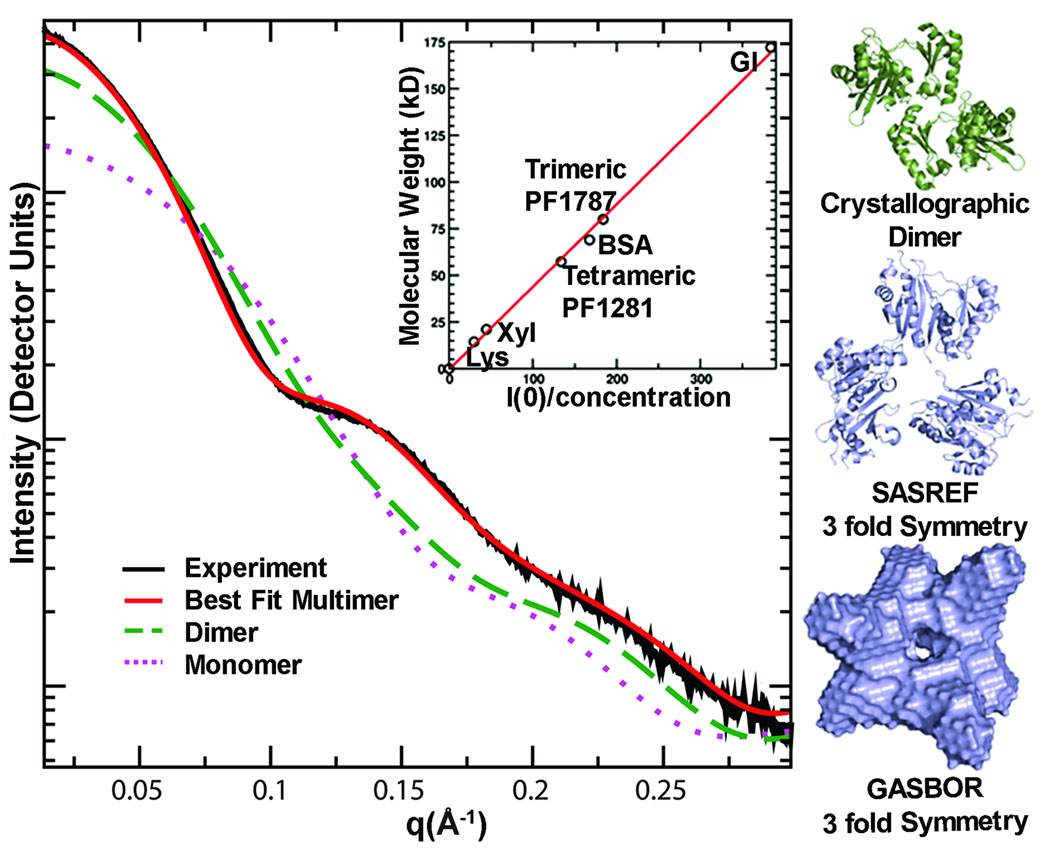
Figure 4.
SAXS determines accurate assembly state in solution, as shown for acetyl-CoA synthetase subunit (PF1787). The experimental scattering curve for PF1787 (black) is shown with calculated scattering curves for monomeric (magenta dots) and dimeric (green dashes) atomic resolution structures of homologs. The best fit (red) to the experimental SAXS data is calculated from a 3-fold symmetric trimer derived from a monomeric homologue (PDB 1WR2). The trimeric form of PF1787 was confirmed using I(0), the extrapolated intensity at 0 scattering angle, normalized for concentration (inset). Proteins standards lysozyme (Lys), xylanase (Xyl), PF1281, bovine serum albumin (BSA) and glucose isomerase (GI) were used to place the data on a relative scale. Relevant structures from analysis of PF1787 are shown on the right. The crystallographic dimer (green) is a flexibly-linked 2-domain protein. Models with 3-fold symmetry enforced (blue) match the SAXS results.
Visualizing novel assemblies and envelopes
Nine of the Pfu proteins (and 3 of the JCSG targets) that we analyzed by SAXS were novel with no known atomic models of sequence homologs of similar length. Assumptions of monodispersity rely on single bands from native gel electrophoresis. We determined shape and assembly from scattering curves (Fig. 3d). To test envelope consistency, models from both DAMMIN without enforced symmetry and GASBOR with appropriate symmetry were compared. The shapes generated by these independent approaches are consistent with one another. GASBOR results contour shapes with greater detail. For three proteins, a Kratky plot25 indicated significant unfolded regions (Fig. 3d). Envelopes were generated for each and those for PF2047.1 and the PF1205/1282 fusion reveal a compact region. However, conformationally heterogeneous samples yield envelopes representing the average shape.
DISCUSSION
Macromolecular information is encoded in shape and assembly, so methods to bridge the growing gap between structural information and highly productive genomic and proteomic advances are needed. Structural genomics efforts have greatly increased the throughput of protein structure analysis (http://sg.pdb.org/), but even with the best efforts, up to ~85 to ~97% of samples cannot be easily characterized by crystallography17. In contrast, our SAXS pipeline yielded solution structural information for 31 of 34 Pfu samples and 10 of 16 JCSG targets, for a success rate of 82%, whereas crystallography efforts only characterized 7 of 34 Pfu targets (21%), typical of structural genomics efforts. Furthermore, SAXS provides superior accuracy for solution conformation and assembly, complementing higher resolution methods such as crystallography and NMR.
SAXS data can have direct implications for determining biological functions as well as for guiding crystallization and biochemical characterizations. For example, a fusion protein created to aid PF1205 purification by adding rubredoxin (PF1282), was purified and soluble, but as indicated by SAXS analyses PF1282/1205 lacked structure and would be unlikely to crystallize. Comparison of SAXS data to those calculated from known structures may guide molecular replacement efforts and identify novel folds (Fig. 5). Our scattering curve from PF0699 matched remarkably well to a scattering profile calculated from a solved structure in the PDB (see DARA26). PF0699 is a conserved hypothetical protein that was matched to E. coli shikimate kinase I (PDB 1KAG27), which acts in the chorismate biosynthesis pathway. Pfu has this pathway involving a known shikimate kinase (PF1694), so an analogous protein (PF0699) is intriguing. We have also identified promising functional leads for PF0715 and PF1911. Improvements in identifying structural homologs using calculated profiles from existing structures are expected. For example, the solved crystal structure of superoxide reductase PF1281 (1DQE) is DARA’s second ranked tetramer with a higher score given to PDB 1JTK. Yet, comparison of the scattering curves over a wider q range immediately highlights the superior fit of the correct structure (Fig. 3a and 5). Similarly, for conserved hypothetical protein PF1674, its homolog was the 25th ranked structure, while structures with poorer overall fit to the data were ranked higher. This is likely the result of overweighting low-resolution features. An additional limitation is the small number of solved crystal structures, especially for very small and very large proteins.
Figure 5.
SAXS defines accurate shape and assembly in solution for unknown structures and can uncover unsuspected structural similarity. Experimental scattering curves for proteins with no known structural homolog (left, color) were compared with calculated scattering (black curves on left) from PDB structures identified by DARA26, a database of scattering curves calculated from the PDB database. Results from the shape reconstruction program GASBOR (colored envelopes) are overlaid onto the structures identified by DARA (ribbon models, right). In addition, PF1674 and PF1281 with known structures show a limitation in the DARA search (see text) and the need for better comparative algorithms.
Symmetry provides powerful constraints on SAXS reconstructions, so our observation that a surprising 60% of samples formed multimers bodes well for accurate reconstructions. Our SAXS results indicate a trimeric assembly for PF1787 (Fig. 4): a flexibly-linked, two-domain protein, which is one of two acetyl CoA synthetase (ACS) subunits. ACS generates ATP, CoASH and acetate and was purified from Pfu biomass as a heteromeric complex (PF1781 and PF1540) with an α2β2 stoichiometry28. How the trimeric solution structure of PF1787 acts in this ACS reaction can now be experimentally investigated.
The JCSG protein set allowed testing a 19-residue His-Tag. His-tags (on average ~8% of the proteins) increase Dmax, and add significant shape heterogeneity, resulting in lower resolution. The disordered His-tags are also asymmetric, making symmetry in envelope calculations less valid although the core is symmetric. Yet, tags may be modeled if core atomic models are available (Supplementary Figure 1).
A serious stated challenge to current structural genomics efforts is the absence of a clear path for a more comprehensive characterization of proteins including their biologically relevant complexes and conformations29. Our high-throughput SAXS pipeline can deal with complexes and conformations in solution, can rapidly evaluate numerous physiological conditions and ligand interactions, characterizes proteins with unstructured regions, and identifies structural similarities without requiring sequence homology. In general, SAXS can provide solution structural information at resolutions often sufficient for functional insights into how these proteins work in the context of their pathways and networks. Whereas crystallography provides precision by high-resolution structures, it does not guarantee accuracy of conformational and assembly state under physiological conditions as well as SAXS. We anticipate that high-throughput SAXS may therefore help address bottlenecks in current structural genomics efforts and aid fundamental research in proteomics and systems biology.
METHODS
SAXS data collection
All SAXS data collection was performed at the SIBYLS beamline, an international user facility. An application for experiments is accessible at www.bl1231.als.lbl.gov. The data collection strategy has been designed to minimize errors due to instrumentation, radiation damage and concentration dependant phenomenon. The strategy applied depended on available stock concentration and size of the protein. SAXS data were collected on 3 serial dilutions of each sample preparation starting at a maximum 10 and a minimum of 1 mg/ml. Sample loading for data collection for each protein proceeded in the following order, lowest concentration, middle concentration, highest concentration followed by a final buffer measurement. The sample cell was washed between protein solutions using a mild detergent soak for 1 minute followed by 3 rinses with buffer solution. The subtraction of buffer collected before the sample was compared to buffer collected after each sample to insure the subtraction process was not subject to instrument variations. Data was collected from two short and one long X-ray exposure for each protein sample. The short exposures were compared against one another to identify whether significant radiation damage occurred on this time scale. The beam size at the sample is 4×1mm and converges at the detector to a 100×100µm spot. The large beam size at the sample spreads radiation over the entire sample greatly reducing radiation damage. Concentrations were compared against one another to determine whether concentration dependant structure factors contributed to the data. In two cases minor concentration dependence was observed and corrected by extrapolating behavior to zero concentration9 using code developed in house. A final scattering curve used for analysis was created for each sample (Supplementary Figure 2).
For most samples only 1 Å X-rays were used. Short exposures were 0.5 seconds while the long exposures were 5 seconds. However for proteins with long dimensions such as PF1548, 1.5 Å wavelength was also used to better define the maximum distances. Short and long exposures were 4 and 40 seconds respectively. All data were collected at room temperature.
SAXS data analysis
For global parameter (Table1, Supplementary Tables 1 and 2) and pair distribution (Supplementary Figure 3) extraction, we used PRIMUS21. X-ray scattering curves calculated from atomic models by CRYSOL31 were compared to observed. Molecular envelopes were generated by both DAMMIN23 and GASBOR10. Mass was estimated from the Porod volume and by the extrapolated intensity at zero q based upon three standards collected in the same experimental settings24. GASBOR requires the number of residues. Mixtures of proteins with known structures were analyzed with OLIGOMER21. SASREF32 was used for rigid body docking.
Leveraging the protein structure database
BLAST33 was used to identify homologs with PDB structures. To test SAXS identification of similar structures, we used the web utility DARA26 (Database for rapid protein characterization) to rank agreement between experimental data and scattering curves (q < 0.15 Å−1) calculated from PDB structures. Stored scattering profiles calculated from PDB atomic coordinates were scanned to match profiles to experimental data.
Sample Preparation
Expression clones
The PF0015-PF0014 co-expression pET24d Bam plasmid consisted of His-tagged PF0015 with in-frame TEV-site between the His-tag and the protein N-terminus, followed by non-tagged PF0014, while the pET24d Bam expression plasmid for PF1205 included Pfu rubredoxin fused in-frame between the His-tag and PF1205. The remaining His-tagged recombinant proteins had previously been prepared by an X-ray crystallographic structural genomics effort and were cloned in the expression plasmid, pET24d Bam34. The expression clones for SOR, Rd and Fd have been previously described35–37 and were used for the production of native (non-tagged) recombinant protein.
Expression in E. coli and purification
All the His-tagged proteins were produced in the E.coli strain, BL21 Star DE3 pRIL (Stratagene) as the host. The His-tagged recombinant proteins were purified according to the high-throughput protocols established for Pfu protein production38. In brief, cells from 1-liter induced cultures were lysed and heated at 80°C for 30 min to precipitate E. coli proteins, cooled to 4°C, and then clarified by centrifugation (40,000 ×g). The clarified supernatant was applied to a 5 ml His-trap Ni affinity column (5 ml) using an AKTA explorer (GE Healthcare, Piscataway, NJ). The column was washed with 5 column volumes (CV) of 20 mM phosphate buffer, pH 7.0, containing 500 mM NaCl, 10 mM imidazole, 5% (vol/vol) glycerol, and 2 mM dithiothreitol. The absorbed protein was eluted with a gradient of 0 to 500 mM imidazole over 20 CV. The major protein peak was collected and concentrated to 10 ml by ultrafiltration (Millipore, Bedford, MA), diluted 15-fold in 20 mM Tris buffer, pH 8.0, containing 5% (vol/vol) glycerol and 2 mM dithiothreitol, and then applied to a column (5 ml) of Q Sepharose (GE Healthcare). The column was washed with 5 CV of the same buffer, and the bound proteins were eluted with a 0 to 1 M NaCl gradient over 20 CV. The major protein peak was concentrated to 5 ml and applied to a 16/60 column size exclusion column of Superdex 75 or Superdex 200 (for PF0015-PF0014) (GE Healthcare) equilibrated with the same Tris buffer. The major protein peak from this column was collected and concentrated to a volume of ~1 ml by ultrafiltration. Samples were buffer exchanged into 20 mM Tris pH 8.0, 300 mM NaCl and 2 mM DTT for SAXS analysis. Recombinant, native (untagged) rubredoxin (PF1282, Rd), superoxide reductase (PF1281, SOR) and ferredoxin (PF1909, Fd) were expressed and purified as described previously 39–41.
Analytical procedures
Protein concentrations were estimated using the Biuret protein assay 42. SDS-PAGE and Native-PAGE analysis of protein samples were done using 4–20% gradient gels (Criterion gel system, Biorad) and run according to the manufacturer’s instructions.
Supplementary Material
ACKNOWLEDGEMENTS
This research is part of the MAGGIE project supported by the Department of Energy (DE-FG0207ER64326) and benefited from allocation of supercomputer time at NERSC. Support for advancement of SAXS technologies at the Lawrence Berkeley National Laboratory SIBYLS beamline of the ALS came from the DOE program Integrated Diffraction Analysis Technologies (IDAT) under Contract DE-AC02-05CH11231 with the U.S. Department of Energy. We thank Ian Wilson and Mark Knuth from the Scripps Research Institute for their Joint Center for Structural Genomics samples.
Footnotes
Author Contributions G.L.H., J.A.T., M.H, S.C., and K.A.F. designed the SIBYLS beamline for high-throughput. G.L.H., J.A.T. and M.W.W.A. wrote the manuscript. A.L.M., F.L.P., F.E.J., S.E.T., R.P.R., R.C.H., and G.L.H. prepared samples for data collection. G.L.H and S.E.T collected SAXS data. M.H and G.L.H. wrote code for analysis. R.P.R. designed www.bioisis.net. S.J. prepared PF0380 and PF2047.1. B.D.D. prepared PF0014/0015. J.W.S. prepared PF1787.
References
- 1.Robinson CV, Sali A, Baumeister W. The molecular sociology of the cell. Nature. 2007;450:973. doi: 10.1038/nature06523. [DOI] [PubMed] [Google Scholar]
- 2.Green BD, Keller M. Capturing the uncultivated majority. Curr. Opin. Biotechnol. 2006;17:236. doi: 10.1016/j.copbio.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 3.Wilmes P, Bond PL. Metaproteomics: studying functional gene expression in microbial ecosystems. Trends Microbiol. 2006;14:92. doi: 10.1016/j.tim.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 4.Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q. Rev. Biophys. 2007;40:191. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- 5.Fox BG, et al. Structural genomics: from genes to structures with valuable materials and many questions in between. Nat. Methods. 2008;5:129. doi: 10.1038/nmeth0208-129. [DOI] [PubMed] [Google Scholar]
- 6.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 7.Berman HM, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sigrist CJ, et al. PROSITE: a documented database using patterns and profiles as motif descriptors. Brief Bioinform. 2002;3:265. doi: 10.1093/bib/3.3.265. [DOI] [PubMed] [Google Scholar]
- 9.Glatter O, Kratky O. Small Angle X-ray Scattering. London: Academic Press; 1982. [Google Scholar]
- 10.Svergun DI, Petoukhov MV, Koch MH. Determination of domain structure of proteins from X-ray solution scattering. Biophys. J. 2001;76:2879. doi: 10.1016/S0006-3495(01)76260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chacon P, Diaz JF, Moran F, Andreu JM. Reconstruction of protein form with X-ray solution scattering and a genetic algorithm. J. Mol. Biol. 2000;299:1289. doi: 10.1006/jmbi.2000.3784. [DOI] [PubMed] [Google Scholar]
- 12.Yamagata A, Tainer JA. Hexameric structures of the archaeal secretion ATPase GspE and implications for a universal secretion mechanism. Embo Journal. 2007;26:878. doi: 10.1038/sj.emboj.7601544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krukenberg KA, et al. Multiple conformations of E. coli Hsp90 in solution: insights into the conformational dynamics of Hsp90. Structure. 2008;16:755. doi: 10.1016/j.str.2008.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pascal JM, et al. A flexible interface between DNA ligase and PCNA supports conformational switching and efficient ligation of DNA. Mol. Cell. 2006;24:279. doi: 10.1016/j.molcel.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 15.Chen B, et al. ATP ground- and transition states of bacterial enhancer binding AAA+ ATPases support complex formation with their target protein, sigma54. Structure. 2007;15:429. doi: 10.1016/j.str.2007.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Service RF. Structural biology. Protein structure initiative: phase 3 or phase out. Science. 2008;319:1610. doi: 10.1126/science.319.5870.1610. [DOI] [PubMed] [Google Scholar]
- 17.Matthews BW. Protein Structure Initiative: getting into gear. Nat. Struct. Mol. Biol. 2007;14:459. doi: 10.1038/nsmb0607-459. [DOI] [PubMed] [Google Scholar]
- 18.Round AR, et al. Automated sample-changing robot for solution scattering experiments at the EMBL Hamburg SAXS station X33. J. of Appl. Cryst. 2008;41:913. doi: 10.1107/S0021889808021018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Toft KN, et al. High-throughput Small Angle X-ray Scattering from proteins in solution using a microfluidic front-end. Analytical Chemistry. 2008;80:3648. doi: 10.1021/ac800011y. [DOI] [PubMed] [Google Scholar]
- 20.Robin D, et al. Superbend upgrade on the Advanced Light Source. Nuclear Instruments & Methods in Physics Research Section a-Accelerators Spectrometers Detectors and Associated Equipment. 2005;538:65. [Google Scholar]
- 21.Konarev PV, et al. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Cryst. 2003;36:1277. [Google Scholar]
- 22.Petoukhov MV, Konarev PV, Kikhney AG, Svergun DI. ATSAS 2.1 - towards automated and web-supported small-angle scattering data analysis. J. of Appl. Cryst. 2007;40:S223. [Google Scholar]
- 23.Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 1999;76:2879. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mylonas E, Svergun DI. Accuracy of molecular mass determiniation of proteins in solution by small-angle X-ray scattering. J. of Appl. Cryst. 2007;40:s245. [Google Scholar]
- 25.Perez J, et al. Heat-induced unfolding of neocarzinostatin, a small all-beta protein investigated by small-angle X-ray scattering. J. Mol. Biol. 2001;308:721. doi: 10.1006/jmbi.2001.4611. [DOI] [PubMed] [Google Scholar]
- 26.Sokolova AV, Volkov VV, Svergun DI. Prototype of a database for rapid protein classification based on solution scattering data. J. Appl. Cryst. 2003;36:865. [Google Scholar]
- 27.Romanowski MJ, Burley SK. Crystal structure of the Escherichia coli shikimate kinase I (AroK) that confers sensitivity to mecillinam. Proteins. 2002;47:558. doi: 10.1002/prot.10099. [DOI] [PubMed] [Google Scholar]
- 28.Mai X, Adams MW. Purification and characterization of two reversible and ADP-dependent acetyl coenzyme A synthetases from the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 1996;178:5897. doi: 10.1128/jb.178.20.5897-5903.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harrison SC. Comments on the NIGMS PSI. Structure. 2007;15:1344. doi: 10.1016/j.str.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 30.Poole FL, 2nd, et al. Defining genes in the genome of the hyperthermophilic archaeon Pyrococcus furiosus: implications for all microbial genomes. J. Bacteriol. 2005;187:7325. doi: 10.1128/JB.187.21.7325-7332.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Svergun D, Baraberato C, Koch MH. CRYSOL - a Program to evaluate X-ray Solution Scattering of Biological Macromolecules from Atomic Coordinates. J. Appl. Cryst. 1995;28:768. [Google Scholar]
- 32.Petoukhov MV, Svergun DI. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys. J. 2005;89:1237. doi: 10.1529/biophysj.105.064154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Altschul SF, et al. Basic local alignment search tool. J. Mol. Biol. 1990;215:403. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 34.Adams MW, et al. The Southeast Collaboratory for Structural Genomics: a high-throughput gene to structure factory. Acc. Chem. Res. 2003;36:191. doi: 10.1021/ar0101382. [DOI] [PubMed] [Google Scholar]
- 35.Yeh AP, et al. Structures of the superoxide reductase from Pyrococcus furiosus in the oxidized and reduced states. Biochemistry. 2000;39:2499. doi: 10.1021/bi992428k. [DOI] [PubMed] [Google Scholar]
- 36.Bau R, Rees DC, Kurtz DM, Scott RA, Huang H, Adams MWW, Eidsness MK. Crystal Structure of Rubredoxin from Pyrococcus Furiosus at 0.95A Resolution. J. Biol. Inorg. Chem. 1998;3:484. [Google Scholar]
- 37.Nielsen MS, Harris P, Ooi BL, Christensen HE. The 1.5 A resolution crystal structure of [Fe3S4]-ferredoxin from the hyperthermophilic archaeon Pyrococcus furiosus. Biochemistry. 2004;43:5188. doi: 10.1021/bi049942x. [DOI] [PubMed] [Google Scholar]
- 38.Sugar FJ, et al. Comparison of small- and large-scale expression of selected Pyrococcus furiosus genes as an aid to high-throughput protein production. J. Struct. Funct. Genomics. 2005;6:149. doi: 10.1007/s10969-005-3341-3. [DOI] [PubMed] [Google Scholar]
- 39.Jenney FE, Jr, Adams MW. Rubredoxin from Pyrococcus furiosus. Methods Enzymol. 2001;334:45. doi: 10.1016/s0076-6879(01)34457-9. [DOI] [PubMed] [Google Scholar]
- 40.Clay MD, et al. Spectroscopic studies of Pyrococcus furiosus superoxide reductase: implications for active-site structures and the catalytic mechanism. J. Am. Chem. Soc. 2002;124:788. doi: 10.1021/ja016889g. [DOI] [PubMed] [Google Scholar]
- 41.Brereton PS, Verhagen MF, Zhou ZH, Adams MW. Effect of iron-sulfur cluster environment in modulating the thermodynamic properties and biological function of ferredoxin from Pyrococcus furiosus. Biochemistry. 1998;37:7351. doi: 10.1021/bi972864b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goa J. A micro biuret method for protein determination; determination of total protein in cerebrospinal fluid. Scand. J. Clin. Lab Invest. 1953;5:218. doi: 10.3109/00365515309094189. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.