Abstract
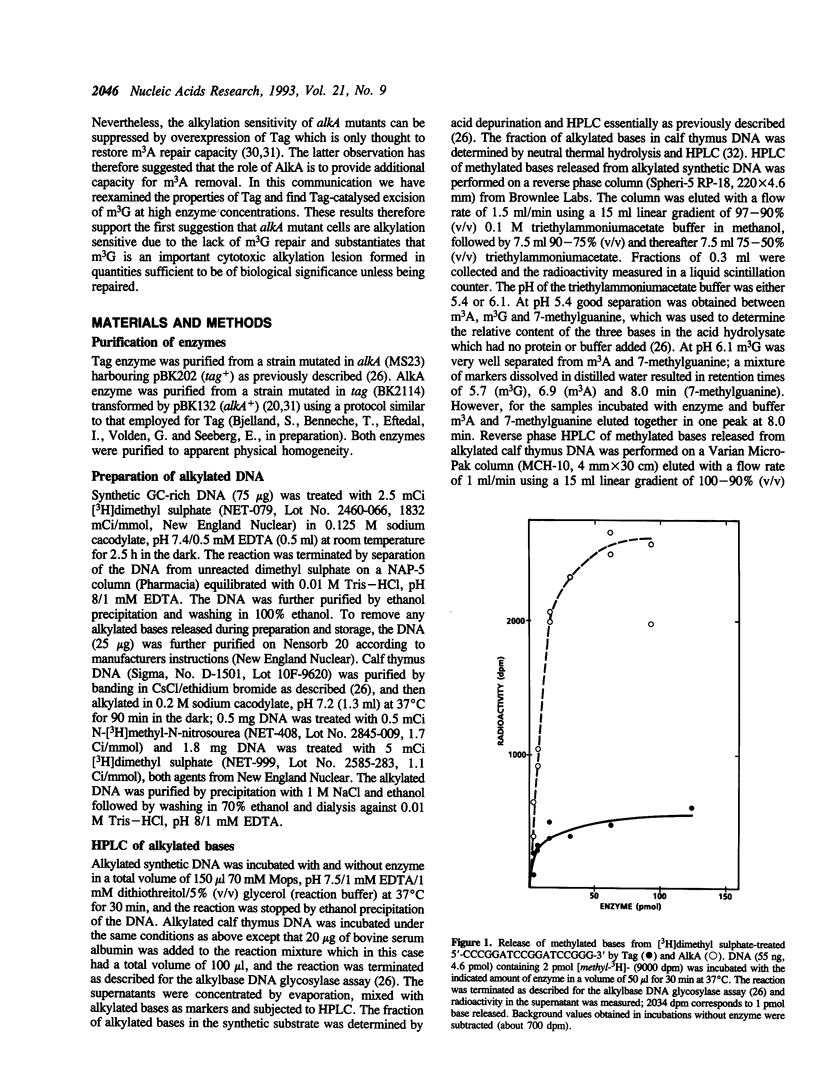
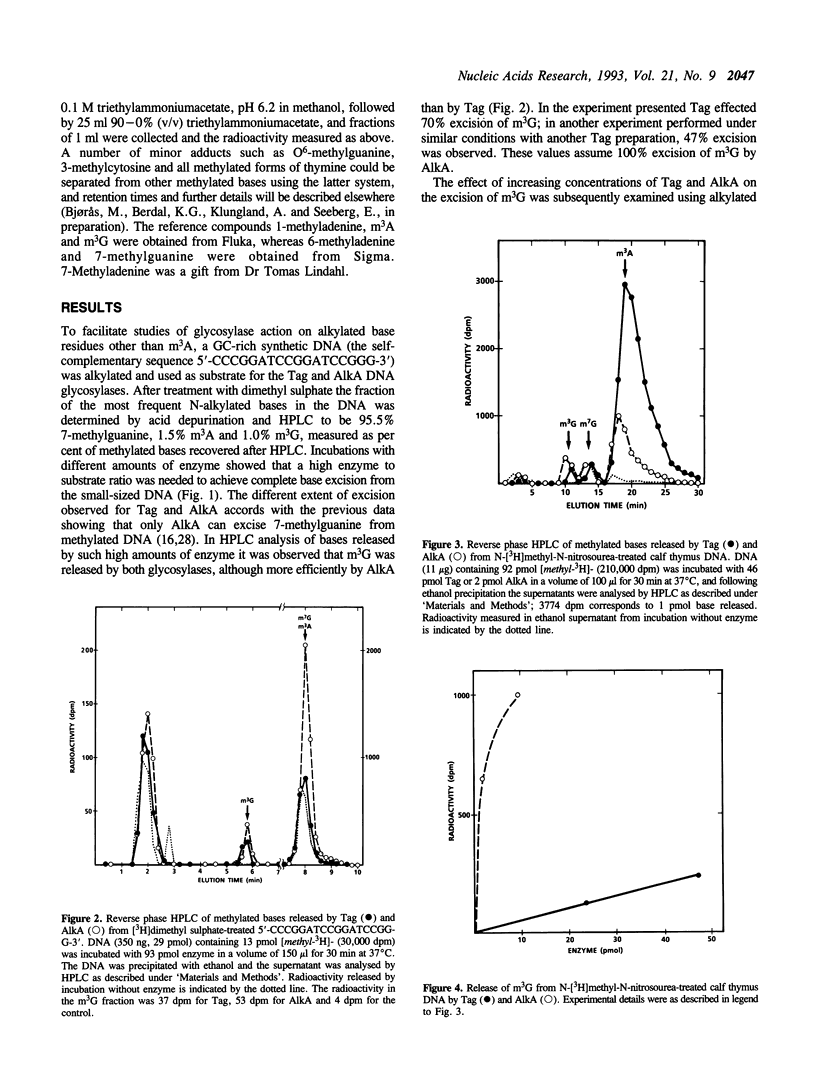
Escherichia coli has two DNA glycosylases for repair of DNA damage caused by simple alkylating agents. The inducible AlkA DNA glycosylase (3-methyladenine [m3A] DNA glycosylase II) removes several different alkylated bases including m3A and 3-methylguanine (m3G) from DNA, whereas the constitutively expressed Tag enzyme (m3A DNA glycosylase I) has appeared to be specific for excision of m3A. In this communication we have reexamined the substrate specificity of Tag by using synthetic DNA rich in GC base pairs to facilitate detection of any possible methyl-G removal. In such DNA alkylated with [3H]dimethyl sulphate, we found that m3G was excised from double-stranded DNA by both glycosylases, although more efficiently by AlkA than by Tag. This was further confirmed using both N-[3H]methyl-N-nitrosourea- and [3H]dimethyl sulphate-treated native DNA, from which Tag excised m3G with an efficiency that was about 70 times lower than for AlkA. These results can explain the previous observation that high levels of Tag expression will suppress the alkylation sensitivity of alkA mutant cells, further implying that m3G is formed in quantity sufficient to represent an important cytotoxic lesion if left unrepaired in cells exposed to alkylating agents.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barrows L. R., Magee P. N. Nonenzymatic methylation of DNA by S-adenosylmethionine in vitro. Carcinogenesis. 1982;3(3):349–351. doi: 10.1093/carcin/3.3.349. [DOI] [PubMed] [Google Scholar]
- Bartsch H., Montesano R. Relevance of nitrosamines to human cancer. Carcinogenesis. 1984 Nov;5(11):1381–1393. doi: 10.1093/carcin/5.11.1381. [DOI] [PubMed] [Google Scholar]
- Beranek D. T. Distribution of methyl and ethyl adducts following alkylation with monofunctional alkylating agents. Mutat Res. 1990 Jul;231(1):11–30. doi: 10.1016/0027-5107(90)90173-2. [DOI] [PubMed] [Google Scholar]
- Beranek D. T., Weis C. C., Swenson D. H. A comprehensive quantitative analysis of methylated and ethylated DNA using high pressure liquid chromatography. Carcinogenesis. 1980 Jul;1(7):595–606. doi: 10.1093/carcin/1.7.595. [DOI] [PubMed] [Google Scholar]
- Bjelland S., Seeberg E. Purification and characterization of 3-methyladenine DNA glycosylase I from Escherichia coli. Nucleic Acids Res. 1987 Apr 10;15(7):2787–2801. doi: 10.1093/nar/15.7.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiteux S., Huisman O., Laval J. 3-Methyladenine residues in DNA induce the SOS function sfiA in Escherichia coli. EMBO J. 1984 Nov;3(11):2569–2573. doi: 10.1002/j.1460-2075.1984.tb02175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennand J., Margison G. P. Reduction of the toxicity and mutagenicity of alkylating agents in mammalian cells harboring the Escherichia coli alkyltransferase gene. Proc Natl Acad Sci U S A. 1986 Sep;83(17):6292–6296. doi: 10.1073/pnas.83.17.6292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke N. D., Kvaal M., Seeberg E. Cloning of Escherichia coli genes encoding 3-methyladenine DNA glycosylases I and II. Mol Gen Genet. 1984;197(3):368–372. doi: 10.1007/BF00329931. [DOI] [PubMed] [Google Scholar]
- Dosanjh M. K., Singer B., Essigmann J. M. Comparative mutagenesis of O6-methylguanine and O4-methylthymine in Escherichia coli. Biochemistry. 1991 Jul 16;30(28):7027–7033. doi: 10.1021/bi00242a031. [DOI] [PubMed] [Google Scholar]
- Ellison K. S., Dogliotti E., Connors T. D., Basu A. K., Essigmann J. M. Site-specific mutagenesis by O6-alkylguanines located in the chromosomes of mammalian cells: influence of the mammalian O6-alkylguanine-DNA alkyltransferase. Proc Natl Acad Sci U S A. 1989 Nov;86(22):8620–8624. doi: 10.1073/pnas.86.22.8620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evensen G., Seeberg E. Adaptation to alkylation resistance involves the induction of a DNA glycosylase. Nature. 1982 Apr 22;296(5859):773–775. doi: 10.1038/296773a0. [DOI] [PubMed] [Google Scholar]
- Kaasen I., Evensen G., Seeberg E. Amplified expression of the tag+ and alkA+ genes in Escherichia coli: identification of gene products and effects on alkylation resistance. J Bacteriol. 1986 Nov;168(2):642–647. doi: 10.1128/jb.168.2.642-647.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karran P., Hjelmgren T., Lindahl T. Induction of a DNA glycosylase for N-methylated purines is part of the adaptive response to alkylating agents. Nature. 1982 Apr 22;296(5859):770–773. doi: 10.1038/296770a0. [DOI] [PubMed] [Google Scholar]
- Karran P., Lindahl T., Ofsteng I., Evensen G. B., Seeberg E. Escherichia coli mutants deficient in 3-methyladenine-DNA glycosylase. J Mol Biol. 1980 Jun 15;140(1):101–127. doi: 10.1016/0022-2836(80)90358-7. [DOI] [PubMed] [Google Scholar]
- Larson K., Sahm J., Shenkar R., Strauss B. Methylation-induced blocks to in vitro DNA replication. Mutat Res. 1985 Jun-Jul;150(1-2):77–84. doi: 10.1016/0027-5107(85)90103-4. [DOI] [PubMed] [Google Scholar]
- Lindahl T., Sedgwick B., Sekiguchi M., Nakabeppu Y. Regulation and expression of the adaptive response to alkylating agents. Annu Rev Biochem. 1988;57:133–157. doi: 10.1146/annurev.bi.57.070188.001025. [DOI] [PubMed] [Google Scholar]
- McCarthy T. V., Karran P., Lindahl T. Inducible repair of O-alkylated DNA pyrimidines in Escherichia coli. EMBO J. 1984 Mar;3(3):545–550. doi: 10.1002/j.1460-2075.1984.tb01844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabeppu Y., Kondo H., Sekiguchi M. Cloning and characterization of the alkA gene of Escherichia coli that encodes 3-methyladenine DNA glycosylase II. J Biol Chem. 1984 Nov 25;259(22):13723–13729. [PubMed] [Google Scholar]
- Nakabeppu Y., Miyata T., Kondo H., Iwanaga S., Sekiguchi M. Structure and expression of the alkA gene of Escherichia coli involved in adaptive response to alkylating agents. J Biol Chem. 1984 Nov 25;259(22):13730–13736. [PubMed] [Google Scholar]
- Rebeck G. W., Samson L. Increased spontaneous mutation and alkylation sensitivity of Escherichia coli strains lacking the ogt O6-methylguanine DNA repair methyltransferase. J Bacteriol. 1991 Mar;173(6):2068–2076. doi: 10.1128/jb.173.6.2068-2076.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riazuddin S., Lindahl T. Properties of 3-methyladenine-DNA glycosylase from Escherichia coli. Biochemistry. 1978 May 30;17(11):2110–2118. doi: 10.1021/bi00604a014. [DOI] [PubMed] [Google Scholar]
- Rydberg B., Lindahl T. Nonenzymatic methylation of DNA by the intracellular methyl group donor S-adenosyl-L-methionine is a potentially mutagenic reaction. EMBO J. 1982;1(2):211–216. doi: 10.1002/j.1460-2075.1982.tb01149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakumi K., Nakabeppu Y., Yamamoto Y., Kawabata S., Iwanaga S., Sekiguchi M. Purification and structure of 3-methyladenine-DNA glycosylase I of Escherichia coli. J Biol Chem. 1986 Nov 25;261(33):15761–15766. [PubMed] [Google Scholar]
- Sakumi K., Sekiguchi M. Structures and functions of DNA glycosylases. Mutat Res. 1990 Sep-Nov;236(2-3):161–172. doi: 10.1016/0921-8777(90)90003-n. [DOI] [PubMed] [Google Scholar]
- Sancar A., Sancar G. B. DNA repair enzymes. Annu Rev Biochem. 1988;57:29–67. doi: 10.1146/annurev.bi.57.070188.000333. [DOI] [PubMed] [Google Scholar]
- Sassanfar M., Dosanjh M. K., Essigmann J. M., Samson L. Relative efficiencies of the bacterial, yeast, and human DNA methyltransferases for the repair of O6-methylguanine and O4-methylthymine. Suggestive evidence for O4-methylthymine repair by eukaryotic methyltransferases. J Biol Chem. 1991 Feb 15;266(5):2767–2771. [PubMed] [Google Scholar]
- Singer B. O-alkyl pyrimidines in mutagenesis and carcinogenesis: occurrence and significance. Cancer Res. 1986 Oct;46(10):4879–4885. [PubMed] [Google Scholar]
- Steinum A. L., Seeberg E. Nucleotide sequence of the tag gene from Escherichia coli. Nucleic Acids Res. 1986 May 12;14(9):3763–3772. doi: 10.1093/nar/14.9.3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas L., Yang C. H., Goldthwait D. A. Two DNA glycosylases in Escherichia coli which release primarily 3-methyladenine. Biochemistry. 1982 Mar 16;21(6):1162–1169. doi: 10.1021/bi00535a009. [DOI] [PubMed] [Google Scholar]
- Umbenhauer D., Wild C. P., Montesano R., Saffhill R., Boyle J. M., Huh N., Kirstein U., Thomale J., Rajewsky M. F., Lu S. H. O(6)-methyldeoxyguanosine in oesophageal DNA among individuals at high risk of oesophageal cancer. Int J Cancer. 1985 Dec 15;36(6):661–665. doi: 10.1002/ijc.2910360607. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y., Katsuki M., Sekiguchi M., Otsuji N. Escherichia coli gene that controls sensitivity to alkylating agents. J Bacteriol. 1978 Jul;135(1):144–152. doi: 10.1128/jb.135.1.144-152.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]



