Abstract
Monellin is a highly potent sweet-tasting protein but relatively little is known about how it interacts with the sweet taste receptor. We determined X-ray crystal structures of 3 single-chain monellin (MNEI) proteins with alterations at 2 core residues (G16A, V37A, and G16A/V37A) that induce 2- to 10-fold reductions in sweetness relative to the wild-type protein. Surprisingly, no changes were observed in the global protein fold or the positions of surface amino acids important for MNEI sweetness that could explain these differences in protein activity. Differential scanning calorimetry showed that while the thermal stability of each mutant MNEI was reduced, the least sweet mutant, G16A-MNEI, was not the least stable protein. In contrast, solution spectroscopic measurements revealed that changes in protein flexibility and the C-terminal structure correlate directly with protein activity. G16A mutation-induced disorder in the protein core is propagated via changes to hydrophobic interactions that disrupt the formation and/or position of a critical C-terminal poly-(L-proline) II helix. These findings suggest that MNEI interaction with the sweet taste receptor is highly sensitive to the relative positions of key residues across its protein surface and that loss of sweetness in G16A-MNEI may result from an increased entropic cost of binding.
Keywords: spectroscopy, sweet, taste, T1R2, T1R3, X-ray crystallography
Introduction
The protein monellin is a highly potent sweet stimulus to humans. It is many thousands of times sweeter than sucrose on a molar basis (Morris and Cagan 1972) and has significant potential as an alternative to carbohydrate or artificial sweeteners. Monellin is one of a handful of known sweet-tasting proteins that with sugars, artificial sweeteners, and some D-amino acids forms a large and diverse array of sweet-tasting stimuli detected by a single heteromeric G protein–coupled receptor (GPCR) (Vigues et al. 2009; Yarmolinsky et al. 2009). This sweet taste receptor is composed of 2 class C GPCR subunits, T1R2 and T1R3, each with a large (∼55 kDa) extracellular N-terminal domain linked to a C-terminal 7-transmembrane domain by a cysteine-rich region (Vigues et al. 2009).
Strikingly, the heterodimeric T1R2:T1R3 receptor contains at least 4 distinct binding sites, some of which are species specific. The T1R2 and T1R3 N-terminal domains each contain binding sites for natural sugars and the deoxychlorosugar sucralose (Nie et al. 2005, 2006), whereas the dipeptide sweeteners aspartame and neotame appear to only bind the N-terminal domain of human T1R2 (Xu et al. 2004). Two other artificial sweeteners, cyclamate and neohesperidin dihydrochalcone, bind within the transmembrane domain of human T1R3 (Xu et al. 2004; Jiang et al. 2005; Winnig et al. 2005). The binding sites of the sweet proteins monellin, thaumatin, brazzein, and the taste-modifying protein neoculin are also distinct but are currently less well defined (Vigues et al. 2009). Activation of the T1R2:T1R3 receptor by brazzein is dependent upon the cysteine-rich region (Jiang et al. 2004), but modeling and functional mapping studies using chimeric T1Rs and mutagenesis suggest that the major binding sites for monellin, thaumatin, and brazzein reside within the T1R2 N-terminal domain (Temussi 2002; Zhao et al. 2003; Assadi-Porter et al. 2010). More recently, neoculin has also been shown to interact at the amino terminal of T1R3 in a pH-dependent manner (Koizumi et al. 2007; Nakajima et al. 2008).
Natural monellin is composed of 2 chains, A and B, of 44 and 50 amino acids, respectively (Kim et al. 1989). To enhance the thermal stability of monellin, recombinant single-chain monellin proteins were created where the 2 natural chains were either directly connected (single chain monellin [SCM]) (Kim et al. 1989) or joined by a dipeptide linker (MNEI) (Tancredi et al. 1992). Despite extensive mutagenesis and structural characterization, including solution (Spadaccini et al. 2003) and high-resolution X-ray (Hobbs et al. 2007) structures of MNEI itself, little molecular detail is known about the interaction of monellin with T1R2:T1R3. An early concept that sweet proteins might share a common structural motif (Kim et al. 1991), a so-called “sweet finger” that in some way mimics the binding of small molecular weight ligands, has been largely discredited (Tancredi et al. 2004), and more recent modeling studies have suggested that the interactions of sweet proteins with T1R2:T1R3 may be more extensive than for small molecule sweeteners (Temussi 2002). Such a recognition mechanism, with the high affinity it suggests, offers a plausible explanation for the high potency and persistent aftertaste of sweet proteins.
To better understand the molecular basis of sweetness for monellin and other sweet proteins, we determined X-ray crystal structures of 3 MNEI mutants of 2- to 10-fold reduced sweetness (Iijima and Morimoto 1995), G16A-, V37A-, and G16A/V37A-MNEI, for comparison with the previously determined wild-type MNEI structure (Hobbs et al. 2007). Surprisingly, these structures show few conformational changes that could explain the loss of sweetness due to these mutations in the protein core. This finding led us to characterize each protein's stability, structure, and flexibility in solution using differential scanning calorimetry (DSC), circular dichroism (CD) spectroscopy, and Raman optical activity (ROA) spectroscopy, respectively. Changes in protein flexibility centered on a poly-(L-proline) II (PPII) helix structure at the MNEI C-terminus, but not changes in the overall protein thermal stability, correlated with MNEI mutant sweetness.
Materials and methods
MNEI protein expression and purification
Plasmids based on pET22b for expression of wild-type (WT-MNEI) and G16A mutant (G16A-MNEI) proteins were provided by P. Temussi (Niccolai et al. 2001; Spadaccini et al. 2003). The V37A mutation was made in the wild-type sequence by QuikChange site-directed mutagenesis (Stratagene) to produce V37A-MNEI. DNA encoding the double mutant (G16A/V37A-MNEI) was chemically synthesized (GeneArt) and subcloned into the same expression plasmid by swapping the DNA insert. Each protein was expressed and purified as described previously (Hobbs et al. 2007).
X-ray crystal structure determination
Crystallization and structure determination of WT-MNEI was reported previously (Hobbs et al. 2007). Crystallization conditions for mutant MNEI proteins were screened at 18 °C by both hanging and sitting drop vapor diffusion, using drops containing an equal volume of crystallization reagent and protein (6.6–8.8 mg/ml; Table 1).
Table 1.
Wild-type and mutant MNEI crystallization and X-ray crystallographic data collection, refinement, and model statistics
| WT-MNEI | G16A-MNEI | V37A-MNEI | G16A/V37A-MNEI | |
| [Protein] (mg/ml) | 7.6 | 6.6 | 7 | 8.8 |
| Crystallization conditionsa | 200 mM (NH4)2SO4 and 30% w/v PEG4000 | 3.6 M sodium formate and 10% v/v glycerol | 0.2 M Li2SO4 0.1M HEPES and 25% w/v PEG4000 | 200 mM NaCl, 100 mM phosphate-citrate and 20% w/v PEG8000 |
| Space group | P21 | P41212 | P1 | P21 |
| High resolution (Å) | 1.15 | 2.0 | 1.8 | 2.4 |
| Unit-cell parameters | ||||
| a, b, c (Å) | 27.1, 66.3, 27.2 | 48.7, 48.7, 114.8 | 29.9, 39.7, 45.2 | 31.4, 144.1, 45.8 |
| α, β, γ (°) | 90.0, 111.6, 90.0 | 90.0, 90.0, 90.0 | 84.9, 80.2, 83.9 | 90.0, 90.9, 90.0 |
| Redundancyb | 3.8 (3.7) | 13.6 (13.7) | 3.5 (3.5) | 7.3 (5.5) |
| Total observations | 178 910 | 135 715 | 62 318 | 122 444 |
| Unique observations (hkl) | 28 254 | 9968 | 17 417 | 16 708 |
| Completeness (%) | 88.5 (88.5) | 100 (100) | 92.8 (89.5) | 98.4 (85.0) |
| Rmerge (%)c | 4.3 (25.4) | 5.5 (23.7) | 7.8 (13.0) | 10.7 (53.1) |
| I/σI (%) | 8.9 (2.7) | 7.6 (1.4) | 17.9 (6.4) | 16.0 (4.3) |
| Solvent content (%) | 32.9 | 58.6 | 53.0 | 52.6 |
| Non-H protein atoms in refinement | 1074 | 863 | 1839 | 3202 |
| Solvent molecules | 143 | 61 | 228 | 203 |
| Rwork (%)d | 13.2 | 23.1 | 17.7 | 18.4 |
| Rfree (%)d | 16.2 | 27.0 | 21.9 | 24.5 |
| Overall B factor (Å2) | 15.6 | 33.8 | 23.6 | 27.8 |
| Ramachandran analysis (%) | ||||
| Favorable | 92.4 | 87.8 | 91.4 | 96.3 |
| Allowed | 7.6 | 12.2 | 8.6 | 3.7 |
| Others | 0 | 0 | 0 | 0 |
| RMS deviations from ideal geometry | ||||
| Bond lengths (Å) | 0.026 | 0.018 | 0.014 | 0.008 |
| Bond angles (°) | 1.99 | 1.66 | 1.46 | 1.04 |
| PDB codes | 2O9U | 3PYJ | 3PXM | 3Q2P |
HEPES, 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
Crystallization experiments performed as sitting drop (WT-MNEI) or hanging drop (all mutants) vapor diffusion.
Values in parenthesis correspond to the highest resolution shell.
Rmerge = ΣhΣi|I(h)i − <I(h)>|/ΣhΣiI(h)i, where I(h)i is the ith observation of the intensity of reflection h and <I(h)> is the mean value of all I(h)i.
R = Σhkl‖Fobs| − |Fcalc‖/Σ|Fobs|, where |Fobs| and |Fcalc| are the observed and calculated structure factor amplitudes for reflection hkl, applied to the work (Rwork) and test (Rfree) sets, respectively.
Crystallization conditions for G16A-MNEI provided suitable protection for cryocooling in liquid N2 without modification. V37A-MNEI and G16A/V37A-MNEI crystals were dragged through perfluoropolyether (Hampton Research) prior to cryocooling to provide cryoprotection. X-ray diffraction data were collected at 100 K at the European Synchrotron Radiation Facility ID14-2 (G16A-MNEI) or on a Rigaku laboratory X-ray generator with Raxis-IV detector (V37A-MNEI and G16A/V37A-MNEI). Data were processed and scaled with XDS/XSCALE (Kabsch 1993) or MOSFLM/SCALA (Leslie 1992) using additional programs of the CCP4 suite (Bailey 1994), as required. Crystal characteristics and data collection statistics are summarized in Table 1.
All mutant MNEI crystal structures were solved by molecular replacement using MOLREP via the CCP4 suite (Bailey 1994). Initial models were the structure of SCM (PDB code 1IV9) for G16A-MNEI and V37A-MNEI and WT-MNEI (PDB code 2O9U) for G16A/V37A-MNEI. The Rfree reflection set (10%) used for WT-MNEI was flagged and omitted from refinement of the mutant structures. The top molecular replacement solution for G16A-MNEI and V37A-MNEI were refined using cycles of minimization, positional B factor, and simulated annealing protocols (3000 K) in the Crystallography & NMR System (CNS) software package (Brunger et al. 1998). The top G16A/V37A-MNEI solution underwent 3 rounds of refinement in phenix.refine, including bulk solvent, xyz coordinate, and B factor refinements. Each model was manually inspected in Coot (Emsley and Cowtan 2004), and each mutation could be unambiguously identified in the difference density map (e.g., Figure 1A). Following model adjustment and addition of water molecules, final rounds of refinement were performed using REFMAC (Murshudov et al. 1997). Analysis of each final model using PROCHECK (Laskowski et al. 1993) indicated that the structures are of high quality with all amino acids in only the favored or allowed regions of the Ramachandran plot (Table 1). Due to the flexible nature of the engineered linker L23, residues in this region were ill defined for some protein chains. In G16A-MNEI, 3 residues in this region, Asn49, Glu50 and Arg53, were refined with occupancies of 0.5.
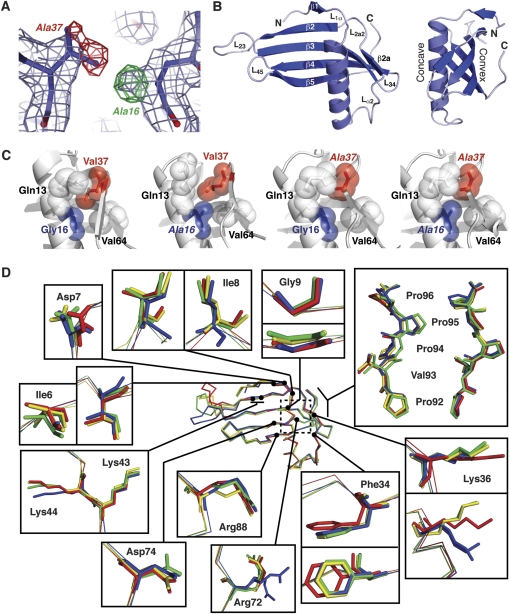
Figure 1.
Crystal structures of wild-type and mutant MNEI proteins. (A) Example of electron density quality at the mutated residues. The wild-type structure (G16/V37) is shown in 2Fo - Fc density (light blue) and Fo - Fc difference density (positive and negative shown in green and red, respectively) generated using the wild-type model and G16A/V37A-MNEI X-ray data. Changes required in the mutant model are indicated next to each peak in the difference map. (B) Cartoon of the WT-MNEI X-ray crystal structure shown in 2 views related by a 90° rotation about the vertical axis. Protein termini, β-strands and loops, including the engineered loop L23, are indicated. Amino acids 16 and 37 are located on the α-helix and strand β2a, respectively. (C) Organization of the MNEI protein core surrounding the mutated positions 16 and 37. Sidechains are shown with van der Waal's radii (semitransparent spheres) with mutated residues indicated with italic font. (D) Backbone alignment of WT- (blue), G16A- (red), V37A- (gold), and G16A/V37A-MNEI (green) with zoomed views of key surface residues for monellin sweetness. The dashed box indicates the protein core containing the mutated residues, shown in panel (C).
The data processing and refinement statistics are summarized in Table 1. All structure factors and coordinates were deposited in the RCSB Protein Data Bank (Bernstein et al. 1977) with accession codes shown in Table 1. Crystallographic figures were generated using PyMol (DeLano, 2002).
Differential scanning calorimetry
Protein melting studies on the wild-type and mutant MNEI proteins were carried out using a VP-DSC microcalorimeter (MicroCal). Samples (0.05 mg/ml) in 50 mM sodium phosphate buffer, pH 6.0, and 150 mM sodium chloride were degassed at room temperature using a ThermoVac (MicroCal) prior to loading into the sample cell. Data were collected over a linear temperature gradient from 50 to 90 °C. Baseline and reference data from a buffer–buffer scan were subtracted and melting curves fit using a nonlinear least-squares method in the MicroCal Origin software to deduce the protein melting temperature (Tm).
ROA spectroscopy
WT-, G16A-, V37A-, and G16A/V37A-MNEI were exhaustively dialyzed against 50 mM sodium phosphate buffer, pH 6.0, with 150 mM sodium chloride at 9, 10, 9, and 8 mg/ml, respectively. Each sample was pipetted into a quartz microfluorescence cell, and ROA spectra were measured using a ChiralRaman Spectrometer (BioTools Inc.) at a wavelength of 532 nm, spectral resolution of 7 cm−1, laser power at the sample of 0.65 W, and total data accumulation times of 6–24 h. Spectra were baseline corrected and mutant protein data normalized to the concentration and collection time used for WT-MNEI to aid visual comparison. The ROA spectrum of G16A/V37A-MNEI was smoothed using a 5-point Fast Fourier Transform function.
CD spectroscopy
CD spectroscopy was conducted on a Jasco J-810 spectropolarimeter using protein samples (0.1 mg/ml) dialyzed against 50 mM sodium phosphate buffer, pH 6.0, and 150 mM sodium chloride. CD spectra from 260 to 200 nm wavelength were collected using 0.1 cm path length quartz cell. Spectra were collected in triplicate as ellipticity (Θ in mdeg), averaged, and converted to standard [Θ]mrw units using the equation:
where c is the protein concentration (in mg/ml), l is the cell path length in centimeters, and Mmrw is the mean residue molecular weight.
Results
We previously reported the crystal structure of WT-MNEI (PDB code 2O9U) (Hobbs et al. 2007) and a preliminary analysis of the G16A mutant (Hobbs et al. 2008); solution nuclear magnetic resonance (NMR) structures of both proteins have also been reported by Temussi and colleagues (Spadaccini et al. 2001, 2003). Like natural monellin, MNEI has a structure that contains a 17-residue α-helix (α1) cradled in the concave face of a 5-stranded antiparallel β-sheet (Figure 1B). In MNEI, the 2 monellin chains are fused through an engineered loop (L23, residues 47–56) between strands β2 and β3 that includes a Gly-Phe insertion (residues 51–52). This loop is conformationally dynamic and is disordered in some MNEI structures. The residues of L23 were therefore excluded from the comparative analyses described below. Sigma A–weighted 2Fo − Fc electron density maps for all other regions of each MNEI mutant showed good continuous density for all residues. Most importantly, each mutation at position 16 and/or 37 was clearly visible in Fo − Fc difference density maps along with small changes in the positions of adjacent residues.
The G16A and V37A mutations only subtly perturb the MNEI protein core
Mutation of residues G16 or V37 to alanine in MNEI was reported to cause an approximately 10- and 2-fold increase in sweet taste detection threshold, respectively (Iijima and Morimoto 1995). However, the effects of these mutations are not additive as the G16A/V37A double-mutant protein causes only a 4-fold threshold increase (Iijima and Morimoto 1995). To determine if these mutations result in gross changes in protein structure, we superposed each mutant MNEI structure with that of WT-MNEI. The very low root mean square deviation (RMSD) (0.68 − 0.86 Å) for each comparison indicated that all the monellin structures are globally almost identical.
Closer comparison of the G16A- and WT-MNEI structures indicates that only very limited local changes are associated with this mutation (Figure 1C). The G16A mutation is located on the protein core face of the MNEI α-helix, opposite the short β2a strand. The addition of a methyl group in G16A-MNEI subtly perturbs the surrounding residues. The terminal methyl groups on V37, positioned almost directly opposite the site of mutation, are rotated away and displaced by approximately 0.6 Å to avoid a steric clash with A16, which would otherwise be only 2.4 Å away. The position of V64, which lies opposite A16, is also altered with its backbone carbonyl reoriented to face toward the protein core. Q13, situated above A16 on the α-helix, is also displaced in the G16A-MNEI structure (Figure 1C). In contrast, fewer alterations are observed in the core of the V37A mutant, with only a minor adjustment observed in the position of the V64 side chain and the α-helix (Figure 1C). Introducing alanines at both positions 16 and 37 removes the potential steric clash between these positions that is responsible for the alterations in the core of the G16A mutant (Figure 1C). V64 adopts a wild-type position in the double mutant. Thus, the subtle structural perturbations in the protein core that result from the G16A mutation as well as their restoration by a “compensating” V37A mutation each correlate with the relative sweetness of wild-type and mutant MNEI proteins.
As G16 and V37 are situated in buried positions, their mutation is unlikely to directly cause a reduction in sweetness. To quantify potential wider effects of these changes on MNEI structure, we compared each mutant protein volume and surface area to that of WT-MNEI. Only very small differences in volume (less than 1%) were observed for each mutant protein (Table 2). Similarly, the total surface area of the mutant MNEI proteins did not differ from WT-MNEI by more than 1.8% (Table 2) and, most significantly, the degree of change did not correlate with the loss and restoration of sweetness.
Table 2.
Protein-exposed surface area and volume for wild-type and mutant MNEI
| Protein | Surface area (Å2)a | Volume (Å3)a |
| WT-MNEI | 5439 | 9462 |
| G16A-MNEI | 5541 (+1.8%) | 9530 (+0.7%) |
| V37A-MNEI | 5405 (−0.6%) | 9546 (+0.8%) |
| G16A/V37A-MNEI | 5345 (−1.7%) | 9372 (−0.9%) |
Values in parentheses are % change relative to WT-MNEI.
Structural perturbations in the protein core do not extend to the surface
In the absence of any gross structural changes, we next asked whether the subtle effects of core mutations might propagate to induce more significant or extensive changes in critical amino acids on the protein surface. Individual alteration of amino acids on the monellin surface can impact sweetness, as exemplified by mutagenesis (Kohmura et al. 1992a, 1992b; Ariyoshi and Kohmura 1994; Somoza et al. 1995) and surface survey experiments (Niccolai et al. 2001) that identified a group, comprising ∼15–20% of all residues in monellin, as important for this activity. For all these critical residues, the overall average sidechain RMSD value for alignment with WT-MNEI was identical for G16A-MNEI and G16A/V37A-MNEI, whereas V37A-MNEI differed by only 0.1 Å. Inspection of all aligned proteins revealed that only 5 of these residues, Q13, D7, F34, K36, and R72, exhibited modestly altered sidechain positions in some or all the mutant proteins as compared with WT-MNEI (Figures 1C,D). However, these changes did not correlate well with the relative sweetness of each protein, for example, altered from the wild-type conformation in G16A-MNEI but restored in the G16A/V37A-MNEI. For at least one of the residues with a larger reorientation, Q13 (Figure 1C), the sidechain displacement in the G16A-MNEI structure is almost certainly due to a crystal contact with an adjacent molecule as previously observed in one chain of another wild-type single-chain monellin structure (1MOL) (Somoza et al. 1993). Thus, we conclude that the subtle changes in core amino acid positions induced by the mutations at amino acids 16 and 37 do not result in significant or extensive conformational changes to the MNEI surface that could alter interaction with the sweet taste receptor.
Reductions in MNEI stability do not correlate with function
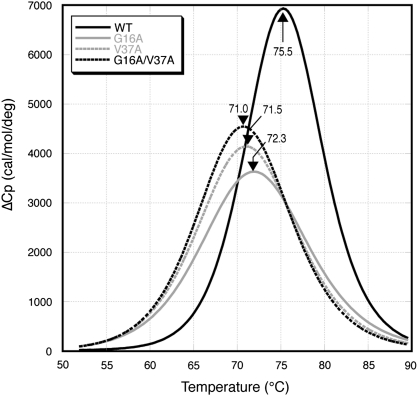
To test whether the reduction in sweetness in G16A-MNEI is associated with a change in protein stability, we measured the melting temperature (Tm) of WT-MNEI and each mutant protein using DSC. The wild-type protein is the most stable with a Tm of 75.2 °C (Figure 2). All 3 mutants exhibit reduced stability (Tm values 71.0–72.3 °C) with an order of stability WT > G16A > V37A > G16A/V37A. The stability of these proteins was previously measured by both chemical denaturation and susceptibility to protease degradation (Iijima and Morimoto 1995). As with DSC, both approaches showed that WT-MNEI was significantly more stable than each mutant. Susceptibility to protease degradation followed the same order as our measurements of protein Tm, whereas the order of stability against chemical denaturation was WT > G16A/V37A > G16A > V37A (Iijima and Morimoto 1995). However, the relative sweetness of the proteins does not correlate with the order of stability produced regardless of the measure of protein stability.
Figure 2.
The stability of MNEI is reduced by the A16 and/or A37 mutations. Normalized DSC analysis of WT-MNEI and each mutant MNEI. Melting temperatures for each protein are shown indicated by an arrow.
Increased flexibility in the PPII helix of G16A-MNEI
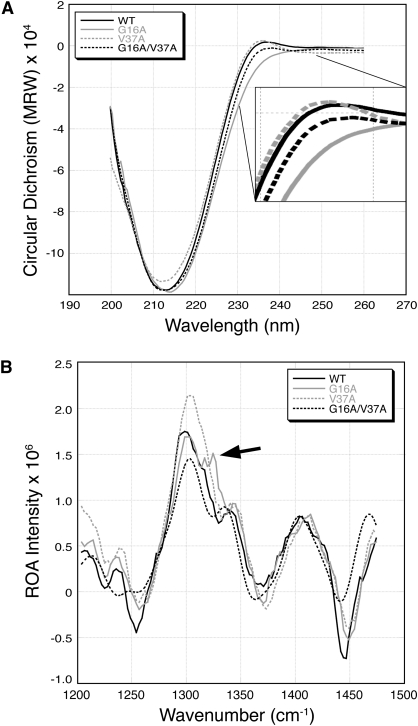
The protein secondary structure of WT-MNEI and each mutant MNEI protein was assessed in solution by CD spectroscopy. We have previously noted that the 3 C-terminal amino acids of WT-MNEI (P94-P95-P96) form a PPII helix and that this structure is visible as a small positive peak in the CD spectrum centered on 230 nm (Hobbs et al. 2007), as expected for a PPII structure (Mandel and Holzwart.G 1973). This peak is present in both WT- and V37A-MNEI spectra but absent in the G16A-MNEI spectrum (Figure 3A). The G16A/V37A-MNEI spectrum contains a peak at 230 nm that is reduced in intensity compared with WT-MNEI. Thus, the loss of the PPII structure with the G16A mutation is partially restored by addition of the V37A mutation, exactly mirroring the effect of these mutations on sweetness threshold (Iijima and Morimoto 1995).
Figure 3.
Monitoring the changes in stability of secondary structures. (A) CD spectrum of each MNEI protein with the region corresponding to PPII structure enlarged (inset). (B) ROA spectrum of each MNEI protein centered on the 1325 cm-1 band visible only for G16A-MNEI (arrow).
Because the PPII structure was present in each of the wild-type and mutant MNEI X-ray crystal structures, we questioned whether the G16A mutation might induce increased flexibility in this region of the protein. ROA spectroscopy measures a small difference in Raman scattering from chiral molecules in circularly polarized light and is a useful probe of order–disorder transitions in proteins (Barron et al. 2002). ROA spectra are characteristic of different aspects of native unfolded protein structures as well as (un)folding transitions in globular proteins (Smyth et al. 2001) with a specific marker band at 1318–1325 cm−1 that is particularly sensitive to PPII helix (Blanch et al. 2000, 2004). We therefore used ROA spectroscopy to further examine the intrinsic flexibility of the 4 MNEI proteins and the structural effect of the core mutations on the surface C-terminal PPII helix.
Each mutant MNEI exhibits clear differences in its ROA spectrum compared with WT-MNEI. The G16A-MNEI ROA spectrum shows intensity decreases at 1255 cm−1 from beta structure and at 1449 cm−1 from aliphatic sidechains, both indicative of sidechain packing destabilization (Figure 3B). This is also true of V37A-MNEI and to a lesser extent, G16A/V37A. Most strikingly, the G16A-MNEI spectrum contains a prominent new band at 1321 cm−1. In contrast, no defined band in this region is observed in the ROA spectra of the WT-, V37A-, or G16A/V37A-MNEI proteins (Figure 3B). In a previous study of human lysozyme and model peptide fragments, an ROA band at ∼1321 cm−1 was assigned to the induced flexible PPII structure (Blanch et al. 2000). Although our X-ray crystal structural analyses indicate an identical PPII helix can form in each MNEI protein, the presence of a 1321 cm−1 band only in the G16A-MNEI ROA spectrum indicates that this mutation specifically perturbs the C-terminal PPII helical structure in solution. By analogy with the prior experiments with lysozyme, we interpret the G16A-MNEI ROA spectra as indicating increased protein flexibility with a specific effect on the PPII helix structure and/or its interaction with the protein core. Critically, this alteration of the PPII helix, observed in both the CD and ROA spectra, correlates directly with the loss and restoration of MNEI sweetness in G16A- and G16A/V37A-MNEI, respectively.
Discussion
Sweet proteins are of potential use as low-calorie sweeteners and could be particularly beneficial to individuals such as diabetics who must control sugar intake. Although the sweet proteins monellin, brazzein, and thaumatin have been extensively characterized by psychophysical, mutagenesis, and structural studies, still relatively little is known about their mechanism of action as chemosensory ligands. Except for one recent study on brazzein (Assadi-Porter et al. 2010), the key determinants of T1R2:T1R3 sweet taste receptor binding and activation remain largely ill defined for this group of proteins. Furthermore, comparisons between the structures of these 3 sweet proteins are not particularly informative as they share no obvious structural similarities (Kant 2005; Temussi 2006). We undertook a biophysical study of wild-type and mutant single-chain monellin (MNEI) proteins that exhibit differing degrees of sweetness due to alterations at 2 amino acids within the protein core. We anticipated that because the mutated residues cannot directly interact with the sweet taste receptor, analysis of these protein structures should reveal more global effects on MNEI structure and thus provide insights complementary to previous studies that have focused on alterations to specific potentially important surface residues (Kohmura et al. 1992a, 1992b; Ariyoshi and Kohmura 1994; Somoza et al. 1995).
A previous NMR study of the G16A-MNEI protein structure suggested that the A16 mutation in the core of the protein results in an extensive rearrangement of the protein surface that might be responsible for reduced interaction with the sweet receptor (Spadaccini et al. 2003). Our crystal structures show that the G16A-MNEI and additional mutants, V37A-MNEI and G16A/V37A-MNEI, are all capable of adopting an essentially wild-type structure. We observed only very minor alterations to amino acid positions within the protein core, each of which served to relieve potential steric clashes. These changes do not appear to propagate and cause extensive changes in the protein volume or surface. Most importantly, outside these minor changes in the protein core, no structural alterations were identified that correlated with the loss and partial restoration of sweetness in G16A- and G16A/V37A-MNEI, respectively.
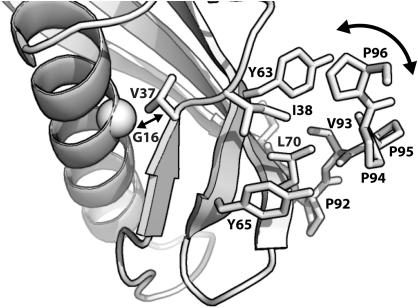
It is well established that protein mutations can result in functional changes even in the absence of obvious structural rearrangements, highlighting the importance of protein dynamics in molecular recognition. For example, energetically favorable mutations rigidify antibodies, thereby increasing their affinity for a particular antigen without causing significant changes in backbone or sidechain positions (Jimenez et al. 2004). Similarly, mutated catabolite activator protein has been shown to bind DNA whilst in an inactive conformation due to favorable changes in the conformational entropy of the protein (Tzeng and Kalodimos 2009). Our DSC analysis of MNEI thermal stability, together with prior analysis of MNEI protein stability through chemical denaturation and protease susceptibility experiments (Iijima and Morimoto 1995), eliminates protein stability as the primary cause of the observed variation in sweetness for these MNEI proteins. However, the CD and ROA spectroscopic approaches used here clearly demonstrate that changes in the structure and flexibility of the MNEI C-terminus directly correlate with MNEI sweetness. Responses to mutational perturbations in protein core residues can be manifest at some distance from the mutated residue, and their effects can be strongly directional (Clarkson et al. 2006). In the case of MNEI, the G16A mutation causes minor alterations to the protein core that lead to wider destabilization of hydrophobic packing interactions and a disruption of the MNEI C-terminal structure (Figure 4). In contrast, although the V37A mutation also decreases protein stability and increases flexibility, it propagates its effect in a different way that results in a more modest (2-fold) reduction in sweetness.
Figure 4.
Model for disruption of the C-terminal PPII helix by G16A mutation. Mutation of G16 to alanine causes a minor reorientation of the adjacent V37 residue (straight double arrow) that is propagated, via changes to the intermediate network of hydrophobic interactions (labeled residues shown as sticks), to increase protein flexibility and disrupt the C-terminal PPII helix structure formed by P94–P96.
The C-terminal residues of MNEI (amino acids 94–96) form a short stretch of PPII helical structure. The PPII helix is a flexible and extended left-handed helix that is typically found on the surface of proteins where it can play an important role in protein–protein interactions and structural integrity. For example, PPII structure is essential in interactions regulating a range of biological processes including gene transcription, cell motility, and the immune response (Bochicchio and Tamburro 2002). The PPII helix of monellin was previously suggested to be important for its sweet taste as deletions in this region (residues 92–96 or 94–96) resulted in a 7- to 20-fold reduction in sweetness (Somoza et al. 1995). However, potential issues with correct protein folding or stability were not addressed. In the mutant MNEI proteins examined here, this C-terminal PPII is present and unaltered in sequence, thus allowing its contribution to MNEI function to be more fully assessed. Involvement of the MNEI PPII helix in the protein's interaction with T1R2:T1R3 is consistent with both the ‘wedge model’ (Temussi 2002) for sweet protein–receptor interaction or as a van der Waals component of a multisite mode of binding, as recently suggested for brazzein (Assadi-Porter et al. 2010). Therefore, although our data cannot resolve the debate over the general model of sweet protein binding to the T1R2:T1R3 receptor, they do identify the PPII structure of monellin as a critical determinant of this interaction.
The structure of WT-MNEI is essentially the same in the crystal (Hobbs et al. 2007) and in solution (Spadaccini et al. 2001). In both cases, the C-terminal PPII helix is positioned against the protein surface where it makes extensive hydrophobic interactions (Figure 4). In contrast, the solution and crystal structures of the G16A-MNEI mutant produce a different picture: although the G16A-MNEI crystal structure, including the PPII helix, is essentially wild type in structure, in solution, the protein appears more globally disordered (Spadaccini et al. 2003). Our solution spectroscopic analysis broadly support a more dynamic structure for all MNEI core mutants but also indicate a specific perturbation of the PPII structure and its interaction with the protein core in G16A-MNEI. Thus, these structural and spectroscopic analyses indicate that the wild-type PPII conformation can be adopted by each of the MNEI mutants but, particularly in the case of G16A-MNEI, only when this more dynamic protein is constrained by protein–protein interactions within a crystal lattice. This observation suggests that this protein conformation is optimal for interaction with the sweet receptor and that the WT-MNEI structure is finely tuned in terms of both stability and rigidity. For the mutant G16A-MNEI to adopt this required conformation brings a significant energetic (entropic) cost that results in a reduced binding affinity and/or residence time on the sweet receptor that is manifest in reduced protein sweetness. Together, these results indicate that the PPII helix of MNEI is a critical factor in protein efficacy as a sweet taste stimulus. Furthermore, alteration of this PPII helix via protein core mutations could potentially be exploited to optimize monellin as a sweetener by fine tuning the MNEI interaction with T1R2:T1R3.
Funding
This work was supported by a grant from the National Institute for Deafness and Other Communication Disorders, National Institutes of Health [DC005786], and a Biotechnology and Biological Sciences Research Council/Cadbury-Schweppes Collaborative Award in Science and Engineering studentship to C.M.T.
Acknowledgments
We thank S.L. Allinson, C.M. Dunham, and beamline staff for assistance with X-ray data collection at the European Synchrotron Radiation Facility (ESRF; ID14.1) and J. Simmons for assistance with CD and DSC experiments.
References
- Ariyoshi Y, Kohmura M. Solid phase synthesis and structure-activity relationships of analogs of the sweet protein monellin. J Synth Org Chem Jpn. 1994;52:359–369. [Google Scholar]
- Assadi-Porter FM, Maillet EL, Radek JT, Quijada J, Markley JL, Max M. Key amino acid residues involved in multi-point binding interactions between brazzein, a sweet protein, and the T1R2-T1R3 human sweet receptor. J Mol Biol. 2010;398:584–599. doi: 10.1016/j.jmb.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey S. The CCP4 suite—programs for protein crystallography. Acta Crystallogr Sect D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Barron LD, Blanch EW, Hecht L. Unfolded proteins studied by Raman optical activity. Adv Protein Chem. 2002;62:51–90. doi: 10.1016/s0065-3233(02)62005-4. [DOI] [PubMed] [Google Scholar]
- Bernstein FC, Koetzle TF, Williams GJB, Meyer EF, Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M. Protein data bank—computer-based archival file for macromolecular structures. J Mol Biol. 1977;112:535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Blanch EW, Gill AC, Rhie AG, Hope J, Hecht L, Nielsen K, Barron LD. Raman optical activity demonstrates poly(L-proline) II helix in the N-terminal region of the ovine prion protein: implications for function and misfunction. J Mol Biol. 2004;343:467–476. doi: 10.1016/j.jmb.2004.08.058. [DOI] [PubMed] [Google Scholar]
- Blanch EW, Morozova-Roche LA, Cochran DA, Doig AJ, Hecht L, Barron LD. Is polyproline II helix the killer conformation? A Raman optical activity study of the amyloidogenic prefibrillar intermediate of human lysozyme. J Mol Biol. 2000;301:553–563. doi: 10.1006/jmbi.2000.3981. [DOI] [PubMed] [Google Scholar]
- Bochicchio B, Tamburro AM. Polyproline II structure in proteins: identification by chiroptical spectroscopies, stability, and functions. Chirality. 2002;14:782–792. doi: 10.1002/chir.10153. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr Sect D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Clarkson MW, Gilmore SA, Edgell MH, Lee AL. Dynamic coupling and allosteric behavior in a nonallosteric protein. Biochemistry. 2006;45:7693–7699. doi: 10.1021/bi060652l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL molecular graphics system. San Carlos (CA): DeLano Scientific LLC; 2002. Available from: http://www.pymol.org. [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr Sect D Biol Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Hobbs JR, Munger SD, Conn GL. Monellin (MNEI) at 1.15 A resolution. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2007;63:162–167. doi: 10.1107/S1744309107005271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs JR, Munger SD, Conn GL. Crystal structures of the sweet protein MNEI: insights into sweet protein-receptor interactions. In: Weerasinghe DK, DuBois GE, editors. Sweetness and sweeteners. American Chemical Society, Washington, DC; 2008. pp. 109–116. [Google Scholar]
- Iijima H, Morimoto K. Controlling susceptibility against protease digestions. Ann N Y Acad Sci. 1995;750:62–65. doi: 10.1111/j.1749-6632.1995.tb19926.x. [DOI] [PubMed] [Google Scholar]
- Jiang P, Cui M, Zhao B, Snyder LA, Benard LMJ, Osman R, Max M, Margolskee RF. Identification of the cyclamate interaction site within the transmembrane domain of the human sweet taste receptor subunit T1R3. J Biol Chem. 2005;280:34296–34305. doi: 10.1074/jbc.M505255200. [DOI] [PubMed] [Google Scholar]
- Jiang PH, Ji QZ, Liu Z, Snyder LA, Benard LMJ, Margolskee RF, Max M. The cysteine-rich region of T1R3 determines responses to intensely sweet proteins. J Biol Chem. 2004;279:45068–45075. doi: 10.1074/jbc.M406779200. [DOI] [PubMed] [Google Scholar]
- Jimenez R, Salazar G, Yin J, Joo T, Romesberg FE. Protein dynamics and the immunological evolution of molecular recognition. Proc Natl Acad Sci U S A. 2004;101:3803–3808. doi: 10.1073/pnas.0305745101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr. 1993;26:795–800. [Google Scholar]
- Kant R. Sweet proteins—potential replacement for artificial low calorie sweeteners. Nutr J. 2005;4:5. doi: 10.1186/1475-2891-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Kang CH, Cho JM. Sweet proteins—biochemical studies and genetic engineering. ACS Symp Ser. 1991;450:28–40. [Google Scholar]
- Kim SH, Kang CH, Kim R, Cho JM, Lee YB, Lee TK. Redesigning a sweet protein - increased stability and renaturability. Protein Eng. 1989;2:571–575. doi: 10.1093/protein/2.8.571. [DOI] [PubMed] [Google Scholar]
- Kohmura M, Nio N, Ariyoshi Y. Highly probable active site of the sweet protein monellin. Biosci Biotechnol Biochem. 1992a;56:1937–1942. doi: 10.1271/bbb.56.1937. [DOI] [PubMed] [Google Scholar]
- Kohmura M, Nio N, Ariyoshi Y. Solid phase synthesis of Asn(A16) monellin, Asn(A22) monellin, Gln(A25) monellin, and Asn(A26) monellin, analogs of the sweet protein monellin. Biosci Biotechnol Biochem. 1992b;56:472–476. doi: 10.1271/bbb.56.472. [DOI] [PubMed] [Google Scholar]
- Koizumi A, Nakajima K, Asakura T, Morita Y, Ito K, Shmizu-Ibuka A, Misaka T, Abe K. Taste-modifying sweet protein, neoculin, is received at human T1R3 amino terminal domain. Biochem Biophys Res Commun. 2007;358:585–589. doi: 10.1016/j.bbrc.2007.04.171. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck—a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- Leslie AGW. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 + ESF-EAMCB Newsl Protein Crystallog. 1992;26 Available from: http://www.mrc-lmb.cam.ac.uk/harry/mosflm/FAQ.html. [Google Scholar]
- Mandel R, Holzwart G. Ultraviolet circular-dichroism of polyproline and oriented collagen. Biopolymers. 1973;12:655–674. [Google Scholar]
- Morris JA, Cagan RH. Purification of monellin, sweet principle of Dioscoreophyllum cumminsii. Biochim Biophys Acta. 1972;261:114–122. doi: 10.1016/0304-4165(72)90320-0. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Nakajima K, Morita Y, Koizumi A, Asakura T, Terada T, Ito K, Shimizu-Ibuka A, Maruyama J, Kitamoto K, Misaka T, et al. Acid-induced sweetness of neoculin is ascribed to its pH-dependent agonistic-antagonistic interaction with human sweet taste receptor. FASEB J. 2008;22:2323–2330. doi: 10.1096/fj.07-100289. [DOI] [PubMed] [Google Scholar]
- Niccolai N, Spadaccini R, Scarselli M, Bernini A, Crescenzi O, Spiga O, Ciutti A, Di Maro D, Bracci L, Dalvit C, et al. Probing the surface of a sweet protein: NMR study of MNEI with a paramagnetic probe. Protein Sci. 2001;10:1498–1507. doi: 10.1110/ps.30101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Y, Vigues S, Hobbs JR, Conn GL, Munger SD. Distinct contributions of T1R2 and T1R3 taste receptor subunits to the detection of sweet stimuli. Curr Biol. 2005;15:1948–1952. doi: 10.1016/j.cub.2005.09.037. [DOI] [PubMed] [Google Scholar]
- Nie YL, Hobbs JR, Vigues S, Olson WJ, Conn GL, Munger SD. Expression and purification of functional ligand-binding domains of T1R3 taste receptors. Chem Senses. 2006;31:505–513. doi: 10.1093/chemse/bjj053. [DOI] [PubMed] [Google Scholar]
- Smyth E, Syme CD, Blanch EW, Hecht L, Vasak M, Barron LD. Solution structure of native proteins with irregular folds from Raman optical activity. Biopolymers. 2001;58:138–151. doi: 10.1002/1097-0282(200102)58:2<138::AID-BIP30>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Somoza JR, Cho JM, Kim SH. The taste active regions of monellin, a potently sweet protein. Chem Senses. 1995;20:61–68. doi: 10.1093/chemse/20.1.61. [DOI] [PubMed] [Google Scholar]
- Somoza JR, Jiang F, Tong L, Kang CH, Cho JM, Kim SH. Two crystal structures of a potently sweet protein—Natural monellin at 2.75 Å resolution and single-chain monellin at 1.7 Å resolution. J Mol Biol. 1993;234:390–404. doi: 10.1006/jmbi.1993.1594. [DOI] [PubMed] [Google Scholar]
- Spadaccini R, Crescenzi O, Tancredi T, De Casamassimi N, Saviano G, Scognamiglio R, Di Donato A, Temussi PA. Solution structure of a sweet protein: NMR study of MNEI, a single chain monellin. J Mol Biol. 2001;305:505–514. doi: 10.1006/jmbi.2000.4304. [DOI] [PubMed] [Google Scholar]
- Spadaccini R, Trabucco F, Saviano G, Picone D, Crescenzi O, Tancredi T, Temussi PA. The mechanism of interaction of sweet proteins with the T1R2- T1R3 receptor: evidence from the solution structure of G16A-MNEI. J Mol Biol. 2003;328:683–692. doi: 10.1016/s0022-2836(03)00346-2. [DOI] [PubMed] [Google Scholar]
- Tancredi T, Iijima H, Saviano G, Amodeo P, Temussi PA. Structural determination of the active site of a sweet protein—a 1H NMR investigation of pMNEI. FEBS Lett. 1992;310:27–30. doi: 10.1016/0014-5793(92)81138-c. [DOI] [PubMed] [Google Scholar]
- Tancredi T, Pastore A, Salvadori S, Esposito V, Temussi PA. Interaction of sweet proteins with their receptor—a conformational study of peptides corresponding to loops of brazzein, monellin and thaumatin. Eur J Biochem. 2004;271:2231–2240. doi: 10.1111/j.1432-1033.2004.04154.x. [DOI] [PubMed] [Google Scholar]
- Temussi PA. Why are sweet proteins sweet? Interaction of brazzein, monellin and thaumatin with the T1R2-T1R3 receptor. FEBS Lett. 2002;526:1–4. doi: 10.1016/s0014-5793(02)03155-1. [DOI] [PubMed] [Google Scholar]
- Temussi PA. Natural sweet macromolecules: how sweet proteins work. Cell Mol Life Sci. 2006;63:1876–1888. doi: 10.1007/s00018-006-6077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng SR, Kalodimos CG. Dynamic activation of an allosteric regulatory protein. Nature. 2009;462:368–372. doi: 10.1038/nature08560. [DOI] [PubMed] [Google Scholar]
- Vigues S, Dotson CD, Munger SD. The receptor basis of sweet taste in mammals. Results Probl Cell Differ. 2009;47:187–202. doi: 10.1007/400_2008_2. [DOI] [PubMed] [Google Scholar]
- Winnig M, Bufe B, Meyerhof W. Valine 738 and lysine 735 in the fifth transmembrane domain of rTas1r3 mediate insensitivity towards lactisole of the rat sweet taste receptor. BMC Neurosci. 2005;6:22. doi: 10.1186/1471-2202-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Staszewski L, Tang HX, Adler E, Zoller M, Li XD. Different functional roles of T1R subunits in the heteromeric taste receptors. Proc Natl Acad Sci U S A. 2004;101:14258–14263. doi: 10.1073/pnas.0404384101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarmolinsky DA, Zuker CS, Ryba NJ. Common sense about taste: from mammals to insects. Cell. 2009;139:234–244. doi: 10.1016/j.cell.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao GQ, Zhang Y, Hoon MA, Chandrashekar J, Erlenbach I, Ryba NJ, Zuker CS. The receptors for mammalian sweet and umami taste. Cell. 2003;115:255–266. doi: 10.1016/s0092-8674(03)00844-4. [DOI] [PubMed] [Google Scholar]