Abstract
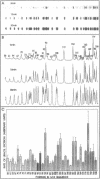
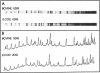
We have purified uracil DNA-glycosylase (UDG) from calf thymus 32,000-fold and studied its biochemical properties, including sequence specificity. The enzyme is apparently closely related to human UDG, since it was recognised by a polyclonal antibody directed towards human UDG. SDS-PAGE and western analysis indicate an apparent M(r) = 27,500. Bovine UDG has a 1.7-fold preference for single stranded over double stranded DNA as a substrate. Sequence specificity for uracil removal from dsDNA was examined for bovine and Escherichia coli UDG, using DNA containing less than one dUMP residue per 100 nucleotides and synthetic oligonucleotides containing one dUMP residue. Comparative studies involving about 40 uracil sites indicated similar specificities for both UDGs. We found more than a 10-fold difference in rates of uracil removal between different sequences. 5'-G/CUT-3' and 5'-G/CUG/C-3' were consensus sequences for poor repair whereas 5'-A/TUAA/T-3' was a consensus for good repair. Sequence specificity was verified in double stranded oligonucleotides, but not in single stranded ones, suggesting that the structure of the double stranded DNA helix has influence on sequence specificity. Rate of uracil removal appeared to be slightly faster from U:A base pairs as compared to U:G mis-matches. The results indicate that sequence specific repair may be a determinant to be considered in mutagenesis.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baer R., Bankier A. T., Biggin M. D., Deininger P. L., Farrell P. J., Gibson T. J., Hatfull G., Hudson G. S., Satchwell S. C., Séguin C. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984 Jul 19;310(5974):207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- Brash D. E., Seetharam S., Kraemer K. H., Seidman M. M., Bredberg A. Photoproduct frequency is not the major determinant of UV base substitution hot spots or cold spots in human cells. Proc Natl Acad Sci U S A. 1987 Jun;84(11):3782–3786. doi: 10.1073/pnas.84.11.3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown T. C., Jiricny J. Repair of base-base mismatches in simian and human cells. Genome. 1989;31(2):578–583. doi: 10.1139/g89-107. [DOI] [PubMed] [Google Scholar]
- Davison A. J., Scott J. E. The complete DNA sequence of varicella-zoster virus. J Gen Virol. 1986 Sep;67(Pt 9):1759–1816. doi: 10.1099/0022-1317-67-9-1759. [DOI] [PubMed] [Google Scholar]
- Delort A. M., Duplaa A. M., Molko D., Teoule R., Leblanc J. P., Laval J. Excision of uracil residues in DNA: mechanism of action of Escherichia coli and Micrococcus luteus uracil-DNA glycosylases. Nucleic Acids Res. 1985 Jan 25;13(2):319–335. doi: 10.1093/nar/13.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dianov G., Price A., Lindahl T. Generation of single-nucleotide repair patches following excision of uracil residues from DNA. Mol Cell Biol. 1992 Apr;12(4):1605–1612. doi: 10.1128/mcb.12.4.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetsch P. W., Henner W. D., Cunningham R. P., Toney J. H., Helland D. E. A highly conserved endonuclease activity present in Escherichia coli, bovine, and human cells recognizes oxidative DNA damage at sites of pyrimidines. Mol Cell Biol. 1987 Jan;7(1):26–32. doi: 10.1128/mcb.7.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domena J. D., Mosbaugh D. W. Purification of nuclear and mitochondrial uracil-DNA glycosylase from rat liver. Identification of two distinct subcellular forms. Biochemistry. 1985 Dec 3;24(25):7320–7328. doi: 10.1021/bi00346a045. [DOI] [PubMed] [Google Scholar]
- Domena J. D., Timmer R. T., Dicharry S. A., Mosbaugh D. W. Purification and properties of mitochondrial uracil-DNA glycosylase from rat liver. Biochemistry. 1988 Sep 6;27(18):6742–6751. doi: 10.1021/bi00418a015. [DOI] [PubMed] [Google Scholar]
- Duncan B. K., Miller J. H. Mutagenic deamination of cytosine residues in DNA. Nature. 1980 Oct 9;287(5782):560–561. doi: 10.1038/287560a0. [DOI] [PubMed] [Google Scholar]
- Duncan B. K., Weiss B. Specific mutator effects of ung (uracil-DNA glycosylase) mutations in Escherichia coli. J Bacteriol. 1982 Aug;151(2):750–755. doi: 10.1128/jb.151.2.750-755.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederico L. A., Kunkel T. A., Shaw B. R. A sensitive genetic assay for the detection of cytosine deamination: determination of rate constants and the activation energy. Biochemistry. 1990 Mar 13;29(10):2532–2537. doi: 10.1021/bi00462a015. [DOI] [PubMed] [Google Scholar]
- Georgiadis P., Smith C. A., Swann P. F. Nitrosamine-induced cancer: selective repair and conformational differences between O6-methylguanine residues in different positions in and around codon 12 of rat H-ras. Cancer Res. 1991 Nov 1;51(21):5843–5850. [PubMed] [Google Scholar]
- Glickman B. W. Study of mutational specificity in the lacl gene of Escherichia coli as a window on the mechanisms of mutation. Environ Mol Mutagen. 1990;16(1):48–54. doi: 10.1002/em.2850160109. [DOI] [PubMed] [Google Scholar]
- Haukanes B. I., Helland D. E., Kleppe K. Action of a mammalian AP-endonuclease on DNAs of defined sequences. Nucleic Acids Res. 1989 Feb 25;17(4):1493–1509. doi: 10.1093/nar/17.4.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impellizzeri K. J., Anderson B., Burgers P. M. The spectrum of spontaneous mutations in a Saccharomyces cerevisiae uracil-DNA-glycosylase mutant limits the function of this enzyme to cytosine deamination repair. J Bacteriol. 1991 Nov;173(21):6807–6810. doi: 10.1128/jb.173.21.6807-6810.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M., Wagner R., Radman M. Repair of a mismatch is influenced by the base composition of the surrounding nucleotide sequence. Genetics. 1987 Apr;115(4):605–610. doi: 10.1093/genetics/115.4.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krokan H., Wittwer C. U. Uracil DNa-glycosylase from HeLa cells: general properties, substrate specificity and effect of uracil analogs. Nucleic Acids Res. 1981 Jun 11;9(11):2599–2613. doi: 10.1093/nar/9.11.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T. DNA repair enzymes. Annu Rev Biochem. 1982;51:61–87. doi: 10.1146/annurev.bi.51.070182.000425. [DOI] [PubMed] [Google Scholar]
- McGeoch D. J., Dalrymple M. A., Davison A. J., Dolan A., Frame M. C., McNab D., Perry L. J., Scott J. E., Taylor P. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol. 1988 Jul;69(Pt 7):1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- Muller S. J., Caradonna S. Isolation and characterization of a human cDNA encoding uracil-DNA glycosylase. Biochim Biophys Acta. 1991 Feb 16;1088(2):197–207. doi: 10.1016/0167-4781(91)90055-q. [DOI] [PubMed] [Google Scholar]
- Olsen L. C., Aasland R., Krokan H. E., Helland D. E. Human uracil-DNA glycosylase complements E. coli ung mutants. Nucleic Acids Res. 1991 Aug 25;19(16):4473–4478. doi: 10.1093/nar/19.16.4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen L. C., Aasland R., Wittwer C. U., Krokan H. E., Helland D. E. Molecular cloning of human uracil-DNA glycosylase, a highly conserved DNA repair enzyme. EMBO J. 1989 Oct;8(10):3121–3125. doi: 10.1002/j.1460-2075.1989.tb08464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percival K. J., Klein M. B., Burgers P. M. Molecular cloning and primary structure of the uracil-DNA-glycosylase gene from Saccharomyces cerevisiae. J Biol Chem. 1989 Feb 15;264(5):2593–2598. [PubMed] [Google Scholar]
- Shlomai J., Kornberg A. Deoxyuridine triphosphatase of Escherichia coli. Purification, properties, and use as a reagent to reduce uracil incorporation into DNA. J Biol Chem. 1978 May 10;253(9):3305–3312. [PubMed] [Google Scholar]
- Slupphaug G., Olsen L. C., Helland D., Aasland R., Krokan H. E. Cell cycle regulation and in vitro hybrid arrest analysis of the major human uracil-DNA glycosylase. Nucleic Acids Res. 1991 Oct 11;19(19):5131–5137. doi: 10.1093/nar/19.19.5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talpaert-Borlé M., Campagnari F., Creissen D. M. Properties of purified uracil-DNA glycosylase from calf thymus. An in vitro study using synthetic DNA-like substrates. J Biol Chem. 1982 Feb 10;257(3):1208–1214. [PubMed] [Google Scholar]
- Talpaert-Borlé M., Clerici L., Campagnari F. Isolation and characterization of a uracil-DNA glycosylase from calf thymus. J Biol Chem. 1979 Jul 25;254(14):6387–6391. [PubMed] [Google Scholar]
- Terleth C., van de Putte P., Brouwer J. New insights in DNA repair: preferential repair of transcriptionally active DNA. Mutagenesis. 1991 Mar;6(2):103–111. doi: 10.1093/mutage/6.2.103. [DOI] [PubMed] [Google Scholar]
- Tomilin N. V., Aprelikova O. N. Uracil-DNA glycosylases and DNA uracil repair. Int Rev Cytol. 1989;114:125–179. doi: 10.1016/s0074-7696(08)60860-8. [DOI] [PubMed] [Google Scholar]
- Varshney U., Hutcheon T., van de Sande J. H. Sequence analysis, expression, and conservation of Escherichia coli uracil DNA glycosylase and its gene (ung). J Biol Chem. 1988 Jun 5;263(16):7776–7784. [PubMed] [Google Scholar]
- Varshney U., van de Sande J. H. Specificities and kinetics of uracil excision from uracil-containing DNA oligomers by Escherichia coli uracil DNA glycosylase. Biochemistry. 1991 Apr 23;30(16):4055–4061. doi: 10.1021/bi00230a033. [DOI] [PubMed] [Google Scholar]
- Vollberg T. M., Siegler K. M., Cool B. L., Sirover M. A. Isolation and characterization of the human uracil DNA glycosylase gene. Proc Natl Acad Sci U S A. 1989 Nov;86(22):8693–8697. doi: 10.1073/pnas.86.22.8693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner H. R., Duncan B. K., Garrett C., Neuhard J. Synthesis and metabolism of uracil-containing deoxyribonucleic acid in Escherichia coli. J Bacteriol. 1981 Feb;145(2):687–695. doi: 10.1128/jb.145.2.687-695.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittwer C. U., Bauw G., Krokan H. E. Purification and determination of the NH2-terminal amino acid sequence of uracil-DNA glycosylase from human placenta. Biochemistry. 1989 Jan 24;28(2):780–784. doi: 10.1021/bi00428a055. [DOI] [PubMed] [Google Scholar]
- Wittwer C. U., Krokan H. Uracil-DNA glycosylase in HeLa S3 cells: interconvertibility of 50 and 20 kDa forms and similarity of the nuclear and mitochondrial form of the enzyme. Biochim Biophys Acta. 1985 Dec 20;832(3):308–318. doi: 10.1016/0167-4838(85)90264-x. [DOI] [PubMed] [Google Scholar]
- Worrad D. M., Caradonna S. Identification of the coding sequence for herpes simplex virus uracil-DNA glycosylase. J Virol. 1988 Dec;62(12):4774–4777. doi: 10.1128/jvi.62.12.4774-4777.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]