Abstract
Cross-linking and immunoprecipitation coupled with high-throughput sequencing was used to identify binding sites within 6,304 genes as the brain RNA targets for TDP-43, an RNA binding protein which when mutated causes Amyotrophic Lateral Sclerosis (ALS). Use of massively parallel sequencing and splicing-sensitive junction arrays revealed that levels of 601 mRNAs are changed (including Fus/Tls, progranulin, and other transcripts encoding neurodegenerative disease-associated proteins) and 965 altered splicing events are detected (including in sortilin, the receptor for progranulin), following depletion of TDP-43 from mouse adult brain with antisense oligonucleotides. RNAs whose levels are most depleted by reduction in TDP-43 are derived from genes with very long introns and which encode proteins involved in synaptic activity. Lastly, TDP-43 was found to auto-regulate its synthesis, in part by directly binding and enhancing splicing of an intron within the 3′ untranslated region of its own transcript, thereby triggering nonsense mediated RNA degradation. (147 words)
Introduction
Amyotrophic Lateral Sclerosis (ALS) is an adult-onset disorder in which premature loss of motor neurons leads to fatal paralysis. Most cases of ALS are sporadic with only 10% of patients having a familial history. A breakthrough in understanding ALS pathogenesis was the discovery that TDP-43, which in the normal setting is primarily nuclear, mislocalizes and forms neuronal and glial cytoplasmic aggregates in ALS1, 2, Frontotemporal lobar degeneration (FTLD) and in Alzheimer’s and Parkinson’s disease (reviewed in ref. 3). Dominant mutations in TDP-43 were subsequently identified as causative in sporadic and familial ALS cases and in rare patients with FTLD4–7. At present, it is unresolved as to whether neurodegeneration is due to a loss of TDP-43 function or a gain of toxic property or a combination of the two. However, a striking feature of TDP-43 pathology is TDP-43 nuclear clearance in neurons containing cytoplasmic aggregates, consistent with pathogenesis driven, at least in part, by a loss of TDP-43 nuclear function1, 7.
Several lines of evidence suggested an involvement of TDP-43 in multiple steps of RNA metabolism including transcription, splicing or transport of mRNA3, as well as microRNA metabolism8. Misregulation of RNA processing has been described in a growing number of neurological diseases9. The recognition of TDP-43 as a central player in neurodegeneration, and the recent identification of ALS-causing mutations in FUS/TLS10, 11, another RNA/DNA binding protein, has reinforced a crucial role for RNA processing regulation in neuronal integrity. However, a comprehensive protein-RNA interaction map for TDP-43 and identification of post-transcriptional events that may be crucial for neuronal survival remain to be established.
A common approach for identifying specific RNA-binding protein targets or aberrantly spliced isoforms related to disease has been through selection of candidate genes. However, recent advances in DNA-sequencing technology have provided powerful tools for exploring transcriptomes at remarkable resolution12. Moreover, cross-linking, immunoprecipitation and high-throughput sequencing (CLIP-seq or HITS-CLIP) experiments demonstrated that a single RNA binding protein can have previously unrecognized roles in RNA processing and affect many alternatively spliced transcripts13–15. We have now used these approaches to identify a comprehensive TDP-43 protein-RNA interaction map within the central nervous system. After depletion of TDP-43 in vivo, RNA sequencing and splicing-sensitive microarrays were used to determine that TDP-43 is crucial for maintaining normal levels and splicing patterns of >1,000 mRNAs. The most downregulated of these TDP-43-dependent RNAs have pre-mRNAs with very long introns that contain multiple TDP-43 binding sites and encode proteins related to synaptic activity. The nuclear TDP-43 clearance widely reported in TDP-43 proteinopathies1, 7 will lead to a disruption of this role on long RNAs that are preferentially expressed in brain, thereby contributing to neuronal vulnerability.
Results
Protein-RNA interaction map of TDP-43 in mouse brain
We used CLIP-seq to identify in vivo RNA targets of TDP-43 in adult mouse brain (Fig. S1a). After UV irradiation to stabilize in vivo protein-RNA interactions, we immunoprecipitated TDP-43 with a monoclonal antibody16 that had a higher immunoprecipitation efficiency than any of the commercial antibodies tested (Fig. S1b). Complexes representing the expected molecular weight of a single molecule of TDP-43 bound to its target RNAs were excised (Fig. 1a) and sequenced. We also observed lower mobility protein-RNA complexes whose abundance was reduced by increased nuclease digestion. Immunoblotting of the same immunoprecipitated samples prior to radioactive labeling of the target RNAs demonstrated that TDP-43 protein was a component of both the ~43kD and more slowly migrating complexes (Fig. 1a).
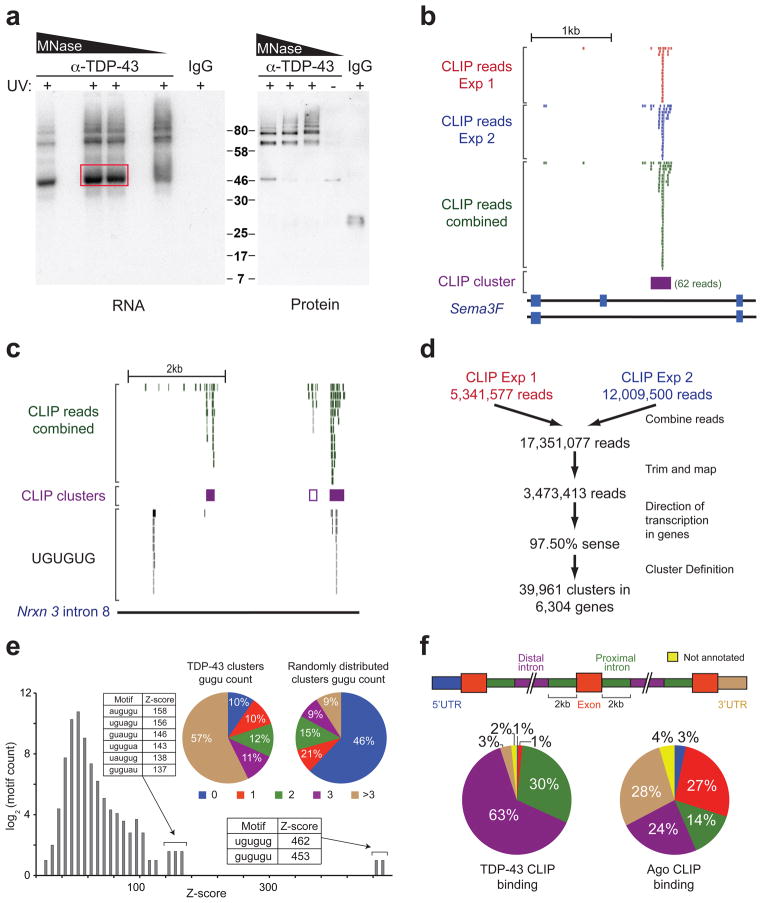
Figure 1. TDP-43 binds distal introns of pre-mRNA transcripts through UG-rich sites in vivo.
(a) Autoradiograph of TDP-43-RNA complexes trimmed by different concentrations of micrococcal nuclease (MNase) (left panel). Complexes within red box were used for library preparation and sequencing. Immunoblot showing TDP-43 in ~46kD and higher molecular weight complexes dependent on UV-crosslinking (UV) (right panel). (b) Example of a TDP-43 binding site (CLIP-cluster) on Semaphorin 3F defined by overlapping reads from 2 independent experiments surpassing a gene-specific threshold. (c) UCSC browser screenshot of neurexin 3 intron 8 (mm8; chr12:89842000-89847000), displaying four examples of TDP-43 binding modes. The right-most CLIP cluster represents a ‘canonical’ binding site coinciding GU-rich sequence motifs while the left-most cluster lacks GU-rich sequences and a region containing multiple GU-repeats shows no evidence of TDP-43 binding. The second CLIP cluster (middle purple-outlined box) with weak binding was found only when relaxing cluster-finding algorithm parameters. (d) Flow-chart illustrating the number of reads analyzed from both CLIP-seq experiments. (e) Histogram of Z-scores indicating the enrichment of GU-rich hexamers in CLIP-seq clusters compared to equally sized clusters, randomly distributed in the same pre-mRNAs. Sequences and Z-scores of the top 8 hexamers are indicated. Pie-charts enumerate clusters containing increasing counts of (GU)2 compared to randomly distributed clusters (p≈0, χ2=21,662). (f) Pre-mRNAs were divided into annotated regions (upper panel). Distribution of TDP-43 (left panel), or previously published Argonaute binding sites21 as a control (right panel), showed preferential binding of TDP-43 in distal introns.
We performed two independent experiments and obtained 5,341,577 and 12,009,500 36 bp sequence reads, respectively, out of which 1,047,642 (20%) and 4,533,626 (38%) mapped uniquely to the repeat-masked mouse genome (Fig. S1c). Mapped reads of both experiments were predominantly within protein-coding genes with ~97% of them oriented in direction of transcription, confirming little DNA contamination. The positions of mapped reads from both experiments were highly consistent, as exemplified by TDP-43 binding on the semaphorin 3F transcript (Fig. 1b).
A cluster-finding algorithm with gene-specific thresholds that accounted for pre-mRNA length and variable expression levels15, 17 was used to identify TDP-43 binding sites from clusters of sequence reads (Fig. 1b), using a conservative threshold (the number of reads mapped to a cluster had to exceed the expected number by chance at a p-value of <0.01). This stringent definition will miss some true binding sites, but was intentionally chosen to identify the strongest bound sites while minimizing false positives. Indeed, additional probable binding sites could be identified by inspection of reads mapped to specific RNAs (see, for example, the reads above the white box in neurexin 3 intron 8 (Fig. 1c) which marks a binding cluster not called by this stringent definition). Moreover, similarly defined clusters from the low mobility complexes (Fig. S1b) showed 92% overlap with those from the monomeric complexes (Fig. 1a), consistent with the reduced mobility complexes comprising multiple TDP-43s (or other RNA binding proteins) bound to a single RNA.
Genome-wide comparison of our replicate experiments revealed (Fig. S1d) that the vast majority (90%) of TDP-43 binding sites in experiment 1 overlapped with those in experiment 2 (compared to an overlap of only 8% (p≈0, Z=570) when clusters were randomly distributed across the length of the pre-mRNAs containing them). Combining the mapped sequences yielded 39,961 clusters, representing binding sites of TDP-43 within 6,304 annotated protein-coding genes, approximately 30% of the murine transcriptome (Fig. 1d). We computationally sampled reads (in 10% intervals) from the CLIP sequences and found a clear logarithmic relationship (Fig. S1e), from which we calculated that our current dataset contains ~84% of all TDP-43 RNA targets in mouse brain. Comparison with the mRNA targets identified from primary rat neuronal cells18 by RNA-immunoprecipitation (RIP) (an approach with the serious caveat that absence of cross-linking allows re-association of RNAs and RNA-binding proteins after cell lysis, as previously documented19) revealed 2,672 of the genes with CLIP-seq clusters in common. As expected from our CLIP-seq analysis in whole brain, we found strong representation of neuronal (see Fig. 3 below) and glial mRNA targets – including Glutamate Transporter 1, Glt1, (Table S6), myelin-associated glycoprotein (Mag), and myelin oligodendrocyte glycoprotein, (Mog).
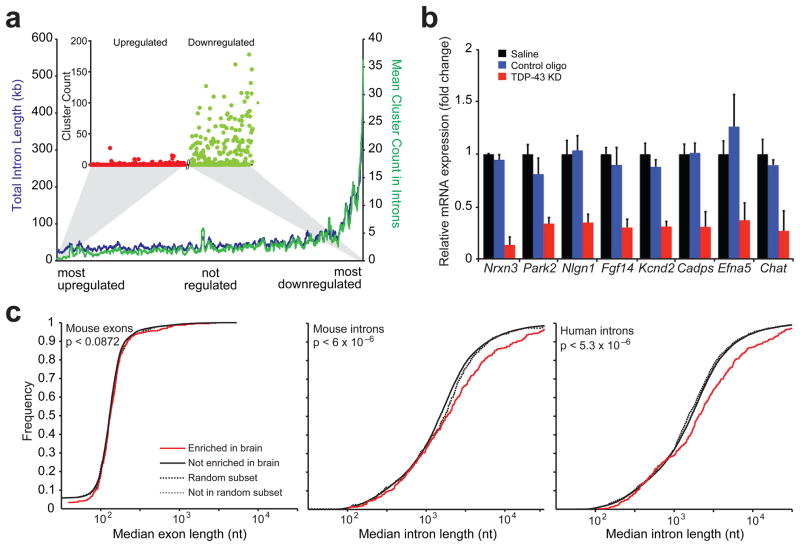
Figure 3. Binding of TDP-43 on long transcripts enriched in brain sustains their normal mRNA levels.
(a) Correlation between RNA-seq and CLIP-seq data. Genes were ranked upon their degree of regulation after TDP-43 depletion (x-axis) and the mean number of intronic CLIP clusters found in the next 100 genes from the ranked list were plotted (y-axis; green line). Similarly, the mean total intron length for the next 100 genes was plotted (y-axis; red line). The cluster count for each up-regulated gene (red dots) and each down-regulated gene (green dots) was plotted using the same ordering (inset). (b) Quantitative RT-PCR for selected down-regulated genes with long introns (except for Chat) revealed significant reduction of all transcripts when compared to controls (p < 8×10−3). Standard deviation was calculated within each group for 3–5 biological replicates. (c) Cumulative distribution plot comparing exon length (left panel) or intron length (middle panel) across mouse brain tissue enriched genes (388 genes) and non-brain tissue enriched genes (15,153 genes). Genes enriched in brain have significantly longer median intron length compared to genes not enriched in brain (solid red line and black lines, p-value < 6.2×10−6 by two-sample Kolmogorov Smirnov goodness-of-fit hypothesis test) while a random subset of 388 genes shows no difference in intron length (dashed lines). Similar analysis across human brain tissue enriched genes (387 genes) and non-brain tissue enriched genes (17,985 genes) also showed significantly longer introns in brain enriched genes (solid red and black lines, p < 5.3×10−6) while a random subset of 387 genes shows no difference in intron length (dashed lines) (right panel).
TDP-43 binds GU-rich distal intronic sites
Sequence motifs enriched within TDP-43 binding sites were determined by comparing sequences within clusters to randomly selected regions of similar sizes within the same protein-coding genes. Use of Z-score statistics revealed that the most significantly enriched hexamers consisted of GU-repeats (Z>450) in agreement with published in vitro results20 or a GU-rich motif interrupted by a single adenine (Z=137–158) (Fig. 1e). The majority (57%) of clusters contained at least four GUGU elements compared to only 9% when equally sized clusters were randomly placed in the same pre-mRNAs (Fig. 1e). Furthermore, the number of GUGU tetramers correlated with the “strength” of binding, as estimated by the relative number of reads within each cluster per gene compared to all clusters in other genes (Fig. S2a). Nevertheless, genome-wide analysis revealed that GU-rich repeats were neither necessary nor sufficient to specify a TDP-43 binding site. One example is the left-most binding site in neurexin 3 (Fig. 1c), which does not have a GU motif, while a GU-rich motif 2kb upstream of it is not bound by TDP-43. In fact, only ~3% of all transcribed 300 nucleotide stretches containing more than three GUGU tetramers contained TDP-43 clusters by CLIP-seq, indicating that TDP-43 target genes cannot be identified by simply scanning nucleotide sequences for GU-rich regions.
While the vast majority (93%) of TDP-43 sites lied within introns, a surprising binding preference of TDP-43 was identified with most (63%) intronic clusters being >2kb from the nearest exon-intron boundary (Fig. 1f). This number rises to 82% for clusters >500 bases from the nearest exon-intron boundary (Fig. S2b). Such distal intronic binding is in sharp contrast with published RNA-binding maps for tissue-specific RNA binding proteins involved in alternative splicing, such as Nova or Fox214, 15. The same analysis on published data in mouse brain for the Argonaute proteins17, 21, which are recruited by microRNAs to the 3′ ends of genes in metazoans17, 21, showed a significantly different pattern of binding. Only 24% (or 30%) of Argonaute clusters resided within 2kb (or 500 bases) from the nearest exon-intron boundary, while 28% were within 3′ untranslated regions (3′UTRs) (Figs. 1f and S2b). This prominent concentration of Argonaute binding near 3′ ends is in stark contrast to the uniform distribution of TDP-43 binding sites across the length of pre-mRNAs (Fig. S2c).
RNAs altered after in vivo TDP-43 depletion in mouse brain
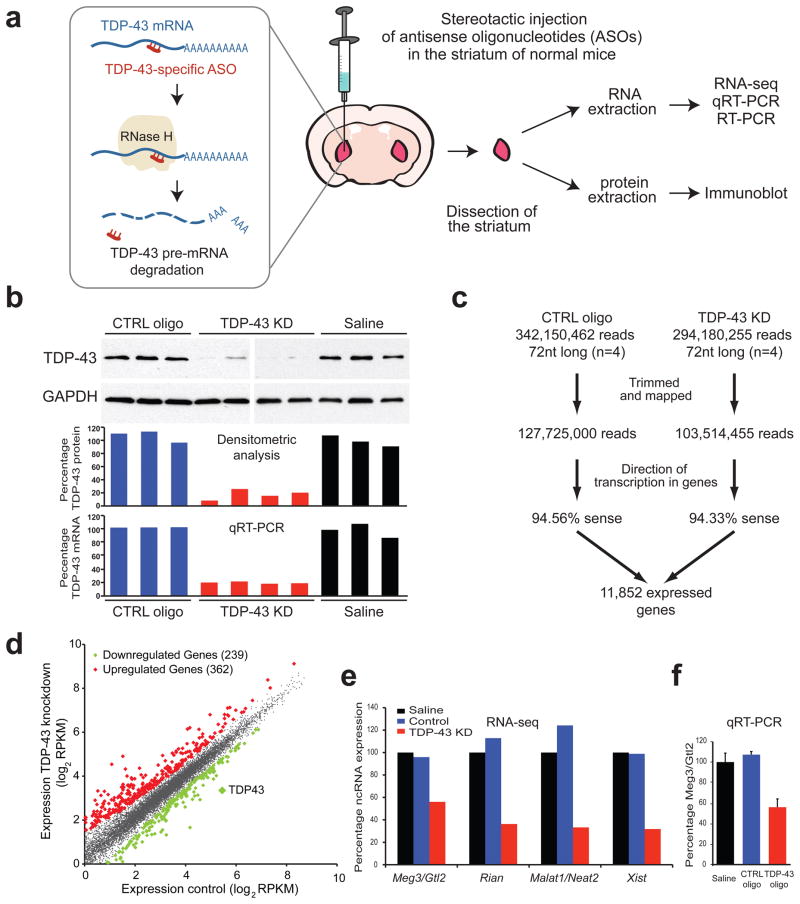
To identify the contribution of TDP-43 in maintaining levels and splicing patterns of RNAs, two antisense oligonucleotides (ASOs) directed against TDP-43 and a control ASO with no target in the mouse genome were injected into the striatum of normal adult mice (Fig. 2a). Striatum is a well-defined structure that is amenable to accurate dissection and isolation, with TDP-43 expression levels comparable to other brain regions. Stereotactic injections of ASOs that target TDP-43, control ASO, or saline were performed in three groups of age and sex matched adult C57BL/6 mice and were tolerated with minimal effects on survival of the animals. Mice were sacrificed after two weeks, and total RNA and protein from striata were isolated (Fig. 2a). Samples treated with TDP-43 ASO showed a significant and reproducible reduction of TDP-43 RNA and protein to approximately 20% of normal levels, when compared to controls (Fig. 2b).
Figure 2. In vivo depletion of TDP-43 in mouse brain with antisense oligonucleotides (ASOs).
(a) Strategy for depletion of TDP-43 in mouse striatum. TDP-43-specific or control ASOs were injected into the striatum of adult mice. TDP-43 mRNA was degraded via endogenous RNase H digestion which specifically recognizes ASO-pre-mRNA (DNA/RNA) hybrids. Mice were sacrificed after 2 weeks and striata were dissected for RNA and protein extraction. (b) Semi-quantitative immunoblot demonstrating significant depletion of TDP-43 protein to 20% of controls in TDP-43 ASO-treated compared to saline and control ASO-treated animals (three upper panels). Quantitative RT-PCR showed similar TDP-43 depletion at the mRNA levels. (c) Flow-chart illustrating the number of reads sequenced and aligned from the RNA-seq experiments. (d) Differentially regulated genes identified by RNA-seq analysis. Scatter-plot revealed the presence of 362 and 239 genes (diamonds) that were significantly up- (red) or down-(green) regulated upon TDP-43 depletion. RNA-seq analysis confirmed that TDP-43 levels were reduced to 20% of control animals. (e) Normalized expression (based on RPKM values from RNA-seq) of four non-coding RNAs that are TDP-43 targets and are downregulated upon TDP-43 depletion. (f) Quantitative RT-PCR validation of non-coding RNA Meg3/Gtl2.
To explore the effects of TDP-43 downregulation on its target RNAs, poly-A enriched RNAs from four biological replicates of TDP-43 or control ASO-treated, as well as three saline-treated animals, were converted to cDNAs and sequenced in a strand-specific manner22, yielding an average of >25 million 72-bp reads per library. The number of mapped reads per kilobase of exon, per million mapped reads (RPKM) for each annotated protein-coding gene was determined to establish a metric of normalized gene expression12. Hierarchical clustering of gene expression values for the independent samples revealed high correlation (R2=0.96) between biological replicates of each condition (TDP-43 and control ASO/saline) (Fig. S3a). Importantly, all control ASO-treated samples were clustered together, as were the samples from TDP-43 ASO-treated animals, consistent with an appreciable impact on gene expression regulation following TDP-43 reduction.
Reads of each treatment group were combined, yielding greater than 100 million uniquely mapped reads per condition (Fig. 2c). Approximately 70% (11,852) of annotated protein-coding genes in mouse satisfied at least 1 RPKM in either condition. Statistical comparison revealed that 362 genes were significantly upregulated and 239 downregulated upon reduction of TDP-43 protein (p < 0.05) (Fig. 2d and Tables S1 and S2). TDP-43 itself was found downregulated by RPKM analysis to 20% of the levels in control treatments, in agreement with quantitative RT-PCR (qRT-PCR) measurements (Fig. 2b). RNAs unique to neurons (including double-cortin, beta-tubulin and choline O-acetyl transferase [Chat]) or glia (including glial fibrillary acidic protein, myelin binding protein, Glt1 and Mag) were highly represented in the RNA-seq data, confirming assessment of RNA levels in multiple cell types, as expected.
Of the set of ~242 literature-curated murine non-coding RNAs23 (ncRNAs), 4 increased and 55 decreased by more than 2-fold upon TDP-43 depletion (p < 10−5, Table S3). Malat1/Neat2, Xist, Rian and Meg3 are ncRNA examples that were both decreased (Fig. 2e, f) and bound by TDP-43, consistent with a direct role of TDP-43 in regulating their levels.
TDP-43 binding to long pre-mRNAs sustains their levels
RNA-seq and CLIP-seq data sets were integrated by first ranking all 11,852 expressed genes by their degree of change upon TDP-43 reduction compared to control treatment. For each group of 100 consecutively ranked genes (starting from the most upregulated gene), the mean number of TDP-43 clusters was determined. No enrichment in TDP-43 clusters within the upregulated genes was identified, indicating that their upregulation was likely an indirect consequence of TDP-43 loss. 49% of RNAs containing TDP-43 clusters were unaffected by TDP-43 depletion, suggesting either that other RNA-binding proteins compensate for TDP-43 loss, or that the remaining 20% of TDP-43 protein suffices to regulate these transcripts. For the 239 RNAs downregulated after TDP-43 depletion, a striking enrichment of multiple TDP-43 binding sites was observed (Fig. 3a). In fact, the 100 most downregulated genes contained an average of ~37 TDP-43 binding sites per pre-mRNA and 12 genes had more than 100 clusters (Fig. 3a and Table S2). We did not observe this bias for multiple TDP-43 binding sites if we randomized the order of genes (Fig. S4a), if we ordered them by their expression levels (RPKM) in either treatment (Fig. S4b, c), or if the Argonaute binding sites were plotted on genes ranked by their expression pattern upon TDP-43 depletion (Fig. S4d). Furthermore, this trend was significant for TDP-43 clusters found within introns (Fig. 3a), but not in exons, 5′or 3′UTRs (Fig. S4e–g).
To address if TDP-43 binding enrichment in the downregulated genes could be attributed to intron size, we performed the same analysis on the ranked list but calculating total (Fig. 3a) or mean (Fig. S4h) intron size instead of cluster counts. This revealed that the most downregulated genes after TDP-43 reduction had exceptionally long introns that were more than 6 times longer (average of 28,707 bp; median length of 11,786 bp) compared to unaffected or upregulated genes (average of 4,532bp; median length of 2,273 bp; p<4×10−18 by t-test). Again, this correlation of downregulation with intron size was not observed for any control condition mentioned above (Fig. S4a–g). Indeed, the enrichment of TDP-43 binding can be largely attributed to intron size differences, as the number of TDP-43 binding sites per kilobase of intron length (cluster density, Fig. S4i) was only slightly increased (p<0.022) for downregulated versus unaffected or upregulated genes (0.072 sites/kb downregulated genes, 0.059 sites/kb other genes). Dividing all mouse protein-coding genes into four groups based on mean intron length (<1kb, 1–10kb, 10–100kb and >100kb) confirmed that the fraction of TDP-43 targets increased (20% to 100%) with intron size (Table 1). Indeed, 83% of genes that contained average intron lengths of 10–100kb, and all 26 genes that contained >100kb long introns were direct targets of TDP-43.
Table 1.
The fraction of downregulated, but not upregulated TDP-43 targets increased with intron size
Number of TDP-43 targets among expressed, downregulated, or upregulated genes, grouped by their mean intron length as indicated. The percentages in parentheses represent the fraction of TDP-43 targets found in each group upon TDP-43 depletion.
| Mean intron length (kb) | Expressed genes | TDP-43 targets | Downregulated genes | Downregulated TDP-43 targets | Upregulated genes | Upregulated TDP-43 targets |
|---|---|---|---|---|---|---|
| 0–1 | 2566 | 510 (20%) | 31 | 8 (26%) | 100 | 7 (7%) |
| 1–10 | 8022 | 4485 (56%) | 80 | 47 (59%) | 252 | 56 (32%) |
| 10–100 | 1238 | 1027 (83%) | 109 | 104 (95%) | 10 | 3 (30%) |
| >100 | 26 | 26 (100%) | 19 | 19 (100%) | 0 | 0 (0%) |
| Total | 11852 | 6048 (51%) | 239 | 178 (74%) | 362 | 66 (18%) |
A highly significant fraction (74%) of all downregulated genes were direct targets of TDP-43 in comparison to genes that were unchanged (52%, p<0.001) or upregulated (18%, p<1×10−17) upon TDP-43 depletion. Remarkably, all 19 down-regulated genes of >100kb long introns were direct TDP-43 targets. In strong contrast, no genes in the same intron length category were upregulated upon TDP-43 depletion, and only 30% of upregulated genes with 10–100kb long introns were TDP-43 targets (Table 1). The crucial role of TDP-43 in maintaining the mRNA abundance of long intron-containing genes was also reflected by the downregulation after depletion of TDP-43 of ~10% of genes with >10kb long introns, the majority (123 of 128, 96%) of which are direct TDP-43 targets.
Gene Ontology (GO) analysis showed that TDP-43 targets whose expression is downregulated upon TDP-43 depletion were highly enriched for synaptic activity and function (Figs. S5, S6 and Table S4). Importantly, several genes with long introns targeted by TDP-43 have crucial roles in synaptic function and have also been implicated in neurological diseases, such as the subunit 2A of the N-methyl-D-aspartate (NMDA) receptor (Grin2a), the ionotropic glutamate receptor 6 (Grik2/GluR6), the calcium-activated potassium channel alpha (Kcnma1), the voltage-dependent calcium channel (Cacna1c), and the synaptic cell-adhesion molecules neurexin 1 and 3 (Nrxn1, Nrxn3) and neuroligin 1 (Nlgn1). We analysed a compendium of expression array data from different mouse organs and human tissues and found that, curiously, genes preferentially expressed in brain have significantly longer introns (p<6×10−6), but not exons (Fig. 3c). The length of these genes is not correlated to the size of the respective proteins and the prevalence of long introns is largely conserved between the corresponding mouse and human genes. Although binding of TDP-43 in long introns can be explained by the increased likelihood to contain UG repeats, the conservation through evolution of this particular gene structure (Fig. S7) suggests that these exceptionally long introns contain important regulatory elements.
To validate the RNA-seq results, we analyzed a selection of brain-enriched TDP-43 targets containing long introns. Genome browser views of neurexin 3 (Nrxn3), Parkin 2 (Park2), neuroligin 1 (Nlgn1), fibroblast growth factor 14 (Fgf14), potassium voltage-gated channel subfamily D member 2 (Kcnd2), calcium-dependent secretion activator (Cadps) and ephrin-A5 (Efna5) revealed a scattered distribution of multiple TDP-43 binding sites across the full length of the pre-mRNA (Fig. S7a), consistent with the results from the global analysis (Fig. 3a). qRT-PCR verified TDP-43-dependent reduction of all these long transcripts tested (Fig. 3b). Chat has a median intron size of <10kb with TDP-43 clusters restricted to a single intronic site (Fig. S7a). Nevertheless, qRT-PCR confirmed the RNA-seq result of a significant reduction in Chat levels after TDP-43 depletion (Fig. 3b).
Only 18% of the upregulated genes were direct targets of TDP-43 (Table 1) and GO analysis revealed an enrichment for genes involved in the inflammatory response (Table 2), suggesting that their differential expression is an indirect consequence of TDP-43 loss. However, of the 66 upregulated RNAs that contained CLIP-seq clusters, 29% harbored TDP-43 binding site(s) within their 3′UTR, a percentage that is 2-fold higher than that of downregulated genes (Fig. S8). This suggests a possible role for TDP-43 to repress gene expression when bound to 3′UTRs.
Table 2.
Main functional categories enriched within the TDP-43-regulated genes
Examples of the most significantly enriched Gene Ontology (GO) terms within the list of up- or downregulated genes upon TDP-43 knockdown, as annotated. Downregulated genes, the majority of which were direct targets of TDP-43, were enriched for synaptic activity and function. In contrast, upregulated genes, most of which were not bound by TDP-43 by CLIP-seq, encoded primarily inflammatory and immune response-related proteins. The p-value indicated was corrected for multiple testing using the Benjamini-Hochberg method.
| Gene Ontology categories | Downregulated genes (239) | Upregulated genes (362) | ||||
|---|---|---|---|---|---|---|
| Gene Ontology Term | Corrected p-value | Gene Ontology Term | Corrected p-value | |||
| Molecular Function | GO:0006811 | ion transport | 2.31E-08 | GO:0006952 | defense response | 2.28E-20 |
| GO:0007268 | synaptic transmission | 1.79E-07 | GO:0006954 | inflammatory response | 4.97E-12 | |
| Cellular Component | GO:0045202 | synapse | 5.33E-08 | GO:0000323 | lytic vacuole | 1.70E-10 |
| GO:0005886 | plasma membrane | 2.35E-13 | GO:0005764 | lysosome | 1.70E-10 | |
| Biological Process | GO:0005216 | ion channel activity | 1.49E-11 | GO:0004197 | cysteine-type endopeptidase activity | 6.00E-04 |
| GO:0022803 | passive transmembrane transporter activity | 7.06E-12 | GO:0003950 | NAD+ ADP- ribosyltransferase activity | 1.11E-02 | |
TDP-43-mediates alternative splicing of its mRNA targets
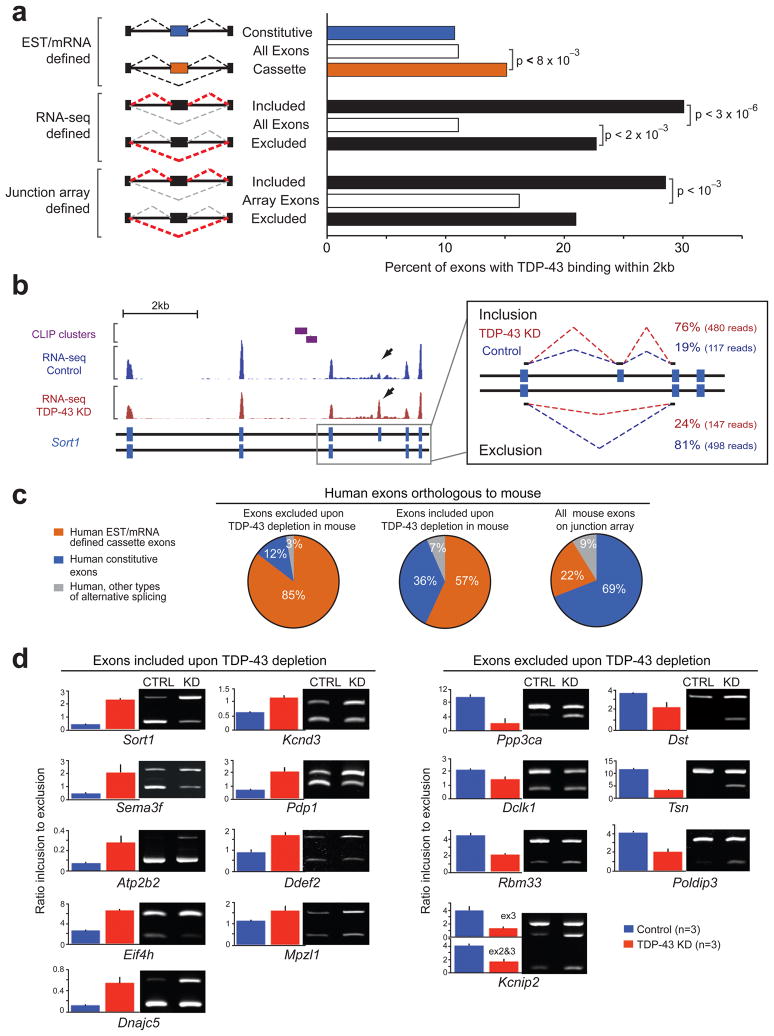
Although TDP-43 binding sites were enriched in distal introns (Fig. 1f), 11% (21,041 out of 190,161) of all mouse exons – including both constitutive and alternative exons – contained TDP-43 binding site(s) within a 2kb window extending from the 5′ and 3′ exon-intron boundaries (Fig. 4a). Compared to all exons, TDP-43 clusters were significantly enriched (p<8×10−3) around exons with transcript evidence for either alternative inclusion or exclusion (i.e., cassette exons). Of the 8,637 known mouse cassette exons, 15.1% contained TDP-43 binding sites in the exon or intron within 2kb of the splice sites. A splice index score for all exons, a measure similar to the “percent spliced in” (or ψ) metric24, was determined by the number of reads that mapped on exons as well as reads that mapped at exon junctions (Fig. 4b). This analysis resulted in identification of 203 cassette exons that were differentially included (93) or excluded (110) (p<0.01) upon TDP-43 depletion. Interestingly, sortilin 1, the gene encoding the receptor for progranulin25, 26, demonstrated the highest splice index score, with exon 18 exclusion requiring TDP-43 (Fig. 4b). Included exons (p<3×10−6) and to a lesser extent, excluded exons (p<2×10−3) identified by RNA-seq, were significantly enriched (~2.7-fold and ~2.0-fold, respectively) for TDP-43 binding when compared to all mouse exons (Fig 4a). Only 33% of RNA-seq-verified TDP-43-regulated cassette exons had previous EST/mRNA evidence for alternative splicing, demonstrating that our approach has identified novel alternative splicing events.
Figure 4. TDP-43-mediates alternative splicing regulation of its RNA targets.
(a) Schematic representation of different exon classes defined by EST and mRNA libraries, RNA-seq or splicing-sensitive microarray data as indicated (left panel). Bar plot displaying the percentage of exons that contain TDP-43 clusters within 2kb upstream and downstream of the exon-intron junctions (right panel). (b) Example of alternative splicing change on exon 18 of sortilin 1 analyzed using RNA-seq reads mapping to the exon body (left panel). Arrows depict increased density of reads in the TDP-43 knockdown samples compared to controls. 76% of the spliced-junction reads in the TDP-43 knockdown samples supported inclusion versus only 19% in the control oligo treated samples (right panel). (c) Comparison of mouse cassette exons detected by splicing-sensitive microarrays to conserved exons in human orthologous genes. 85% and 57% of human exons corresponding to the excluded and included mouse exons, upon TDP-43 depletion, respectively (left and middle pie charts), contained EST and mRNA evidence for alternative splicing. As a control, percentage of human exons orthologous to all mouse exons represented on the splicing-sensitive array, and that have alternative splicing evidence are shown at the right pie chart. (d) Semi-quantitative RT-PCR analyses of selected targets confirmed alternative splicing changes in TDP-43 knockdown samples compared to controls. Representative acrylamide gel pictures of RT-PCR products from control or knockdown adult brain samples (right panel). Quantification of splicing changes from three biological replicates per group (left panels); error bars represent standard deviation.
As an independent method of identifying TDP-43 regulated exons, RNAs from the same ASO-treated animals were analyzed on custom-designed splicing-sensitive Affymetrix microarrays27. Using a conservative statistical cutoff, we detected 779 alternatively spliced events that significantly change upon TDP-43 depletion (Fig. S9). Interestingly, included (p<10−3) but not excluded exons (p<0.3), were significantly enriched for TDP-43 binding (~1.8-fold and ~1.3-fold, respectively), when compared to the unchanged exons on the microarray (Fig. 4a), similar to the trend seen by RNA-seq. The combined RNA-seq and splicing-sensitive microarray data defined a set of 512 alternatively spliced cassette exons whose splicing is affected by loss of TDP-43. The majority of human orthologs of these murine exons (85% of those with excluded and 57% with included exons) have prior EST/mRNA evidence for alternative splicing (Fig. 4c).
Semi-quantitative RT-PCR on selected RNAs validated splicing alterations with more inclusion or exclusion upon TDP-43 reduction (Figs. 4d and S10). Importantly, varying the extent of TDP-43 downregulation (between 40–80%) correlated with the magnitude of splicing changes (Fig. S11). However, the majority of altered splicing events observed upon TDP-43 depletion do not have TDP-43 clusters within 2kb of the splice sites, implicating longer-range interactions or indirect effects of TDP-43 through other splicing factors. Consistent with this latter hypothesis, we have identified TDP-43 binding on pre-mRNAs of RNA-binding proteins including Fus/Tls, Ewsr1, Taf15, Adarb1, Cugbp1, RBFox2 (Rbm9), Tia1, Nova 1 and 2, Mbnl, and neuronal Ptb (or Ptbp2). After TDP-43 depletion, mRNA levels of Fus/Tls (Fig. 6b) and Adarb1 were reduced and exon 5 within the Tia1 transcript was more included, while exon 10 within Ptbp2 was more excluded (Table S5).
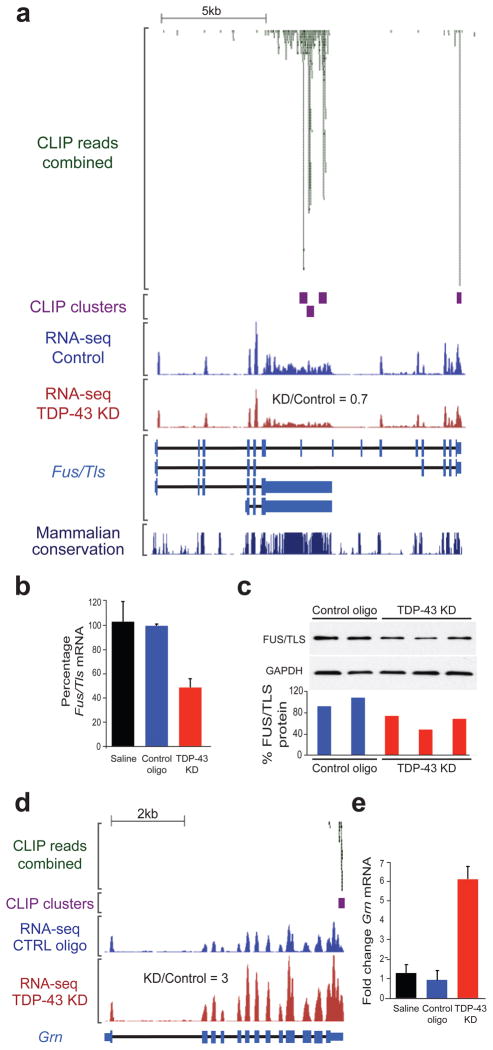
Figure 6. TDP-43 regulates expression of FUS/TLS and Progranulin.
(a) CLIP-seq reads and clusters on Fus/Tls transcript showing TDP-43 binding within introns 6 and 7 (also annotated as an alternative 3′UTR) and the canonical 3′UTR. RNA-seq reads from control or TDP-43 knockdown (KD) samples (equal scales) show a slight reduction in Fus/Tls mRNA in the TDP-43 knockdown group and expression values from RNA-seq confirmed that Fus/Tls mRNA was reduced to 70% of control upon TDP-43 reduction. (b) Quantitative RT-PCR confirmed Fus/Tls mRNA downregulation upon TDP-43 depletion. Standard deviation was calculated within each group for 3 biological replicas. (c) Semi-quantitative immunoblot (upper panel) demonstrated a slight but consistent reduction of FUS/TLS protein in ASO-treated mice to ~70% of control levels as quantified by densitometric analysis (lower panel). (d) CLIP-seq reads and clusters on progranulin transcript (Grn) showing a sharp binding within the 3′UTR. RNA-seq reads from control or TDP-43 knockdown (KD) samples (equal scales) show a significant increase in progranulin mRNA in the TDP-43 knockdown group compared to controls. (E) Quantitative RT-PCR confirmed the statistically significant increase (p < 3×10−4) in progranulin mRNA in samples with reduced TDP-43 levels when compared to controls. Standard deviation was calculated within each group for 3 biological replicas.
TDP-43 autoregulation through binding on its 3′UTR
We found TDP-43 binding sites within an alternatively spliced intron in the 3′UTR of the TDP-43 pre-mRNA (Fig. 5a). Interestingly, this binding does not coincide with a long stretch of UG-repeats, suggesting a lower “strength” of binding (Fig. S2a), in agreement with a recent report28. TDP-43 mRNAs spliced at this site (Fig. 5a, isoforms 2 and 3) are predicted to be substrates for nonsense mediated RNA decay (NMD), a process that targets mRNAs for degradation when exon-junction complexes (EJCs) deposited during splicing, located 3′ of the stop codon are not displaced during the pioneer round of translation29. In contrast, TDP-43 mRNAs with an unspliced 3′UTR would not have such a premature termination codon and should escape NMD. This TDP-43 binding implies autoregulatory mechanisms reminiscent to those reported for other RNA-binding proteins30, 31. Indeed, expression in mice of a TDP-43-encoding transgene without the regulatory 3′UTR (ES, S-CL and DWC, unpublished) lead to significant reduction of endogenous TDP-43 mRNA and protein (Fig. 5b, c) within the central nervous system.
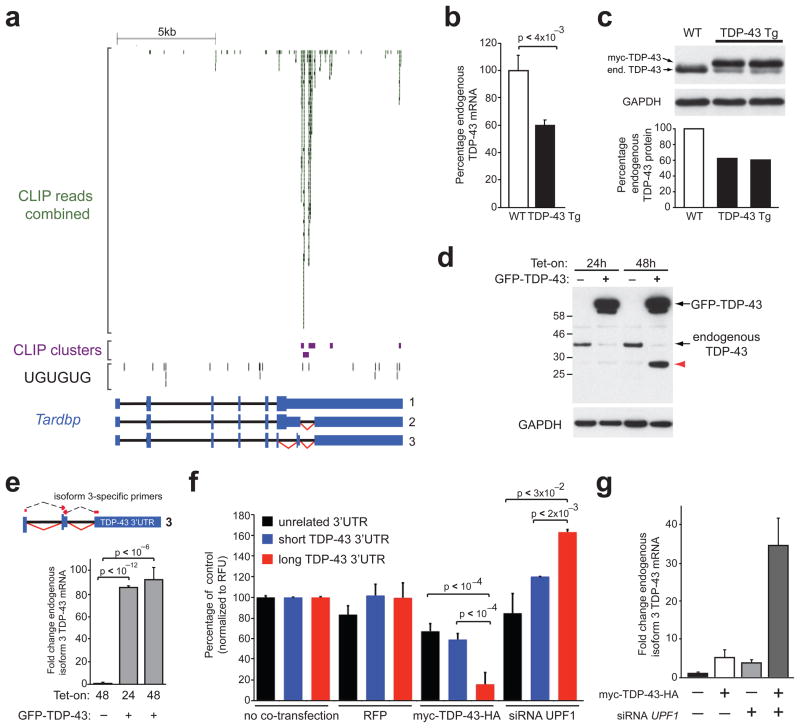
Figure 5. Autoregulation of TDP-43 through binding on the 3′UTR of its own transcript.
(a) CLIP-seq reads and clusters on TDP-43 transcript showing binding mainly within an alternatively spliced part of the 3′ untranslated region (3′UTR), lacking long uninterrupted UG-repeats. (b) Quantitative RT-PCR showing ~50% reduction of endogenous TDP-43 mRNA in transgenic mice overexpressing human myc-TDP-43 not containing introns and 3′UTR. (c) Immunoblots confirming the reduction of endogenous TDP-43 protein (upper panel) to 50% of control levels (by densitometry, lower panel) in response to human myc-TDP-43 overexpression (d) Immunoblot showing reduction of endogenous TDP-43 protein in HeLa cells upon tetracycline induction of a transgene encoding GFP-myc-TDP-43-HA (annotated GFP-TDP-43). The ~30kD product accumulating after 48h (red arrow) was immunoreactive with four TDP-43-specific antibodies. (e) Quantitative RT-PCR using primers spanning the junctions of TDP-43 isoform 3 (upper panel), showed ~100-fold increase in response to tetracycline induction of GFP-TDP-43. TDP-43 isoform 3 was present at very low levels before tetracycline induction. (f) Luciferase assays showing significant reduction of Relative Fluorescence Units (RFUs) in cells expressing renilla luciferase under the control of long TDP-43 3′UTR when co-transfected with myc-TDP-43-HA. Cells treated with siRNA against UPF1 showed a significant increase of RFUs when expressing Luciferase with the long TDP-43 3′UTR, but not in controls. (g) Quantitative RT-PCR scoring levels of endogenous TDP-43 isoform 3 in HeLa cells transiently transfected with myc-TDP-43-HA, UPF1 siRNA or both simultaneously. TDP-43 isoform 3 was increased in response to elevated TDP-43 protein levels, blocking of NMD (by UPF1 siRNA) and there was a synergistic effect in the combined condition.
To identify the molecular basis of this mechanism, we generated HeLa cells in which GFP-myc-TDP-43-HA mRNA lacking introns and 3′UTR was transcribed from a single copy, tetracycline-inducible transgene inserted at a predefined locus by site-directed (Flp) recombinase16. After 24 or 48 hours of GFP-myc-TDP-43-HA induction, a significant reduction of endogenous TDP-43 protein was observed, accompanied by accumulation of a shorter, ~30kD product (Fig. 5d) recognized by four different TDP-43-specific antibodies. While this ~30kD band could be derived from the transgene encoding TDP-43, it was not recognized by anti-myc or anti-HA antibodies and its size is compatible with the endogenous TDP-43 isoform 3. Using qRT-PCR with primers spanning the exon junctions of TDP-43 isoform 3, we found a ~100-fold increase of the spliced isoform 3 upon overexpression of GFP-myc-TDP-43-HA protein (Fig. 5e).
To test if TDP-43 drives splicing of its pre-mRNA through binding to its 3′UTR, we cloned a “long” unspliced version (containing the TDP-43 binding sites) and a “short” spliced version (without TDP-43 binding sites) of the TDP-43 3′UTR downstream of the stop codon of a renilla luciferase gene (Fig. S12a). Both unspliced and spliced 3′UTRs were determined to be present in brain RNAs from mouse and human central nervous systems (Fig. S12a). Both variants, as well as an unaltered luciferase reporter were transfected into HeLa cells along with plasmids driving either increased TDP-43 expression or red fluorescent protein (RFP) (Fig. S12b). Increased levels of TDP-43 protein led to a significant reduction of luciferase produced from the gene carrying the long, intron-containing TDP-43 3′UTR, when compared to the short or unrelated 3′UTR (Fig. 5f). Moreover, co-transfection of the reporters with siRNAs targeting UPF1 (Fig. S12c), an essential component that marks an NMD substrate for degradation32, enhanced luciferase produced by the intron-containing 3′UTR by ~1.5-fold, indicating UPF1-dependent degradation of this mRNA (Fig. 5f). Lastly, the endogenous spliced isoform 3 of TDP-43 was significantly increased, not only upon elevated TDP-43 expression (by transient transfection), but also upon blocking of NMD, with a synergistic effect in the combined conditions (Fig. 5g).
TDP-43 regulates expression of disease-related transcripts
As summarized in Table S6, TDP-43 protein binds and directly regulates a variety of transcripts involved in neurological diseases (Figs. S8 and S13) including Fus/Tls (Fig. 6a) and Grn (Fig. 6d), encoding FUS/TLS and progranulin, mutations in which cause ALS10, 11 or FTLD-U33, 34, respectively. TDP-43 binds to the 3′UTR of Fus/Tls mRNA and in introns 6 and 7, all of which are highly conserved between mammalian species (Fig. 6a). Gene annotation and the presence of RNA-seq reads within these introns are consistent either with an alternative 3′UTR or intron retention. Fus/Tls mRNA and protein were reduced to approximately 40% of their normal levels (Fig. 6b, c). Progranulin mRNA, on the other hand, was markedly increased by ~3–6-fold compared to controls (Fig. 6d, e).
CLIP-seq data also confirmed TDP-43 binding to two RNAs previously reported to be associated with TDP-43: histone deacetylase 6 (Hdac6)35 and low molecular weight neurofilament subunit (Nefl)36. Our RNA-seq data demonstrated that HDAC6, which functions to promote the degradation of polyubiquitinated proteins, was reduced upon TDP-43 depletion (Fig. S14a, b), albeit to a lesser degree in vivo than previously reported in cell culture35. It has been known for many years that Nefl mRNA levels are reduced in degenerating motor neurons from ALS patients37. We identified TDP-43 clusters within the 3′UTR of Nefl (Fig. S14c). Additionally, RNA-seq data confirmed that the mouse Nefl 3′UTR was longer than annotated and Nefl mRNA levels were slightly reduced upon TDP-43 depletion (Fig. S14d). Multiple TDP-43 binding sites were also present in the pre-mRNA from the Mapt gene encoding tau, whose mutation or altered splicing of exon 10 has been implicated in FTD38. However, neither the levels nor the splicing pattern of Mapt RNA were affected by TDP-43 depletion (Table S6). Moreover, we identified multiple TDP-43 intronic binding sites in the Hdh transcript (Fig. S13), encoding huntingtin, the protein whose polyglutamine expansion causes Huntington’s disease in humans39 accompanied by cytoplasmic TDP-43 accumulations40. Moreover, Hdh levels were decreased in mouse brain upon TDP-43 depletion (Table S6). In contrast, we found no evidence for direct binding or TDP-43 regulation of the Sod1 transcript (Table S6), whose aberrant splicing in familial ALS cases41, 42 had raised the possibility that SOD1 missplicing may be involved in the pathogenesis of sporadic ALS.
Discussion
TDP-43 is a central component in the pathogenesis of an ever-increasing list of neurodegenerative conditions. Here we have determined a genome-wide RNA map of >39,000 TDP-43 binding sites in the mouse transcriptome and determined that levels of 601 mRNAs and splicing patterns of 965 mRNAs were altered following TDP-43 reduction in the adult nervous system. Thus, while earlier efforts have implicated TDP-43 as a splicing regulator of a few candidate genes20, 43, 44, our RNA-seq and microarray results established that TDP-43 regulates the largest set (512) of cassette exons thus far reported, demonstrating its broad role in alternative splicing regulation. We also showed that TDP-43 is required for maintenance of 42 non-overlapping, non-coding RNAs.
We have also provided a direct test for how the nuclear loss of TDP-43 widely reported in the remaining motor neurons in ALS autopsy samples1 may contribute to neuronal dysfunction, independent of potential damage from TDP-43 aggregates. Our evidence using in vivo reduction of TDP-43 coupled with RNA sequencing established that TDP-43 is crucial in sustaining levels of 239 mRNAs, including those encoding synaptic proteins, the neurotransmitter acetylcholine, and the disease related Fus/Tls and progranulin. A significant proportion of these pre-mRNAs are directly bound by TDP-43 at multiple sites within exceptionally long introns, a feature that we found most prominently within brain-enriched transcripts (Fig. 3), thereby identifying one component of neuronal vulnerability from TDP-43 loss.
A plausible model for the role of TDP-43 in sustaining the levels of mRNAs derived from long pre-mRNAs is that TDP-43 binding within long introns prevents unproductive splicing events that would introduce premature stop codons and thereby promote RNA degradation. Our results thus identify a novel conserved role for TDP-43 in regulating a subset of these long intron-containing brain-enriched genes. None of our evidence eliminates the possibility that TDP-43 affects RNA levels by additional mechanisms, for example, through transcription regulation or by facilitation of RNA-polymerase elongation, similar to what has been shown for another splicing regulator, SC3545.
FUS/TLS is another RNA-binding protein whose mutation causes ALS10, 11 and in some rare cases FTLD-U. Like TDP-43, FUS/TLS aggregation has also been observed in different neurodegenerative conditions, including Huntington’s disease and spinocerebellar ataxia (reviewed in ref. 3). We have now shown that FUS/TLS mRNA is a direct target of TDP-43 and its level is reduced upon TDP-43 depletion (Fig. 6), thereby identifying a novel FUS/TLS dependency on TDP-43. The latter is also true for two additional disease-relevant proteins, progranulin and its proposed receptor sortilin. Progranulin levels were sharply increased upon TDP-43 reduction and splicing of sortilin was altered. In fact, TDP-43 directly binds and regulates the levels and splicing patterns of transcripts implicated in various neurologic diseases46 (Table S6 and Fig. S13), in agreement with a broad role of TDP-43 in these conditions3.
Finally, in contrast to a recent study that reported an autoregulatory mechanism for TDP-43 that is independent of pre-mRNA splicing28, our results demonstrate that TDP-43 acts as a splicing regulator to reduce its own expression level by binding to the 3′UTR of its own pre-mRNA. While there may be additional mechanisms beyond NMD28, we found that TDP-43 enhances splicing of an alternative intron in its own 3′UTR thereby autoregulating its levels through a mechanism that involves splicing-dependent RNA degradation by NMD (Fig. 5). TDP-43 autoregulation occurs within the mammalian central nervous system, as shown by significant reduction of endogenous TDP-43 mRNA and protein in response to expression of TDP-43 transgene lacking the regulatory intron, as we have shown here and others have reported47, 48. Both spliced and unspliced TDP-43 RNAs were found in human and mouse brain, consistent with autoregulation at normal TDP-43 levels that substantially attenuates TDP-43 synthesis through production of unstable, spliced RNA.
TDP-43-dependent splicing of its 3′UTR intron as a key component of a TDP-43 autoregulatory loop could participate in a feedforward mechanism enhancing the cytoplasmic TDP-43 aggregates that are hallmarks of familial and sporadic ALS. Following an initiating insult (for example, one that traps some TDP-43 in initial cytoplasmic aggregates), the reduction in nuclear TDP-43 levels would decrease splicing of its 3′UTR, which would in turn produce an elevated pool of stable TDP-43 mRNA. Repeated translation of that TDP-43 mRNA would increase synthesis of new TDP-43 in the cytoplasm whose subsequent co-aggregation into the initial complexes would drive their growth. Disrupted autoregulation – by any event that lowers nuclear TDP-43 – thus provides a mechanistic explanation for what may be a critical, intermediate step in the molecular mechanisms underlying age-dependent degeneration and death of neurons in TDP-43 proteinopathies.
Materials and Methods
CLIP-seq library preparation and sequencing
Brains from 8-week old female C57Bl/6 mice were rapidly and dissociated by forcing through a cell strainer with a pore size of 100μm (BD Falcon) before UV-crosslinking. CLIP-seq libraries were constructed as previously described15, using a custom-made mouse monoclonal anti-TDP-43 antibody16 (40μg of antibody per 400 μL of beads per sample). Libraries were subjected to standard Illumina GA2 sequencing protocol for 36 cycles.
Generation of transgenic mice
All animal procedures were conducted in accordance with the guidelines of the Institutional Animal Care and Use Committee of University of California. cDNAs containing N-terminal myc-tagged full length human TDP-43 were amplified and digested by SalI and cloned into the XhoI-cloning site of the MoPrP.XhoI vector (ATCC #JHU-2). The resultant MoPrP.XhoI-myc-hTDP-43 construct was then digested upstream of the minimal Prnp promoter and downstream of the Prnp exon 3 using BamHI and NotI and cloned into a shuttle vector containing loxP flanking sites. The final construct was then linearized using XhoI, injected into the pro-nuclei of fertilized C56Bl6/C3H hybrid eggs and implanted into pseudopregnant female mice.
Stereotactic injections of antisense oligonucleotides
8–10 week old female C57Bl/6 mice were anesthetized with 3% isofluorane. Using stereotaxic guides, 3 μL of antisense oligonucleotide (ASO) solution – corresponding to a total of 75 μg or 100 μg ASOs – or saline buffer was injected using a Hamilton syringe directly into the striatum. Mice were monitored for any adverse effects for the next two weeks until they were sacrificed. The striatum and adjacent cortex area were dissected, and frozen at −80°C in 1 mL Trizol (Invitrogen). Trizol extraction of RNA and protein was performed according to the manufacturer’s instructions.
RNA quality and RNA-seq library preparation
RNA quality was measured using the Agilent Bioanalyzer system according to the manufacturer’s recommendations. RNA-seq libraries were constructed as described previously22. 8 pM of amplified libraries was used for sequencing on the Illumina GA2 for 72 cycles.
RT and qRT-PCR
cDNA of total RNA extracted from striatum was generated using oligodT and Superscript III reverse transcriptase (Invitrogen) according to the manufacturer’s instructions. To test candidate splicing targets, RT-PCR amplification using between 24 and 27 cycles were performed from at least 3 mice treated with a control ASO and 3 mice with TDP-43 downregulation. Products were separated on 10% polyacrylamide gels followed by staining with SYBR gold (Invitrogen). Quantification of the different isoforms was performed with ImageJ software. Intensity ratio between products with included and excluded exons were averaged for 3 biological replicates per group.
Quantitative RT-PCR for mouse TDP-43 and FUS/TLS were performed using the Express One-Step SuperScript qRT-PCR kits (Invitrogen) and the thermocycler ABI Prism 7700 (Applied Biosystems). cDNA synthesis and amplification were performed according to the manufacturer’s instruction using specific primers and 5′FAM, 3′TAMRA labeled probes. Cyclophilin gene was used to normalize the expression values.
Quantitative RT-PCR for all other genes tested were performed with 3 to 5 mices for each group (treated with saline, control ASO or ASO against TDP-43) and 2 technical replicates using the iQ SYBR green supermix (BioRad) on the IQ5 multicolor real-time PCR detection system (BioRad). The analysis was done using the IQ5 optical system software (BioRad; version 2.1). Expression values were normalized to at least two of the following control genes β-Actin, Actg1 and Rsp9. Expression values were expressed as a percentage of the average expression of the saline treated samples. Inter-group differences were assessed by two-tailed Student’s t-test.
Primers for RT-PCR and qRT-PCR were designed using Primer3 software (http://frodo.wi.mit.edu/primer3/) and sequences are available on request.
Immunoblots
Proteins were separated on custom made 12% SDS page gels and transferred to nitrocellulose membrane (Whatman) following standard protocols. Membranes were blocked overnight in Tris-Buffered Saline Tween-20 (TBST) and 5% non-fat dry milk at 4°C, and then incubated 1 hour at room temperature with primary and then with horseradish peroxidase (HRP)-linked secondary antibodies anti-Rabbit or anti-mouse (GE Healthcare) in TBST with 1% milk. Primary antibodies were: rabbit anti-FUS/TLS (Bethyl Laboratories.Inc; Cat #A300-302A; 1:5000), rabbit anti-TDP-43 (Proteintech; Cat #10782; 1:2,000), rabbit anti-TDP-43 (Aviva system biology; Cat #ARP38942_T100; 1:2,000), custom made mouse anti-TDP-43 (1:1,000)16, custom made rabbit anti-RFP raised against the full length protein (1:7,000), mouse DM1α anti-tubulin (1:10,000), mouse anti-GAPDH (Abcam; Cat #AB8245; 1:10,000).
Cells, cloning and Luciferase assays
HeLa Flp-In cells expressing GFP-myc-TDP-43-HA were generated as previously described16. Isogenic cell lines were grown at 37°C and 5% CO2 in Dulbecco’s modified Eagle medium (DMEM), supplemented with 10% tetracycline-free fetal bovine serum (FBS) and penicillin/streptomycin. Expression of GFP-myc-TDP-43-HA was induced with 1 mg/ml tetracycline for 24–48 hours.
To assess the mechanism of TDP-43 autoregulation, the proximal part of mouse TDP-43 3′ UTR was cloned in the psiCHECK-2 vector (Promega Corporation) that contains a Renilla luciferase and a Firefly luciferase reporter expression cassettes. The following primers were used to amplify 1.7kb of TDP-43 3′ UTR using cDNA from mouse brain: 5′-AAACTCGAGCAGGCTTTTGGTTCTGGAAA-3′ and 5′-AAAGCGGCCGCACCATTTTAGGTGCGGTCAC-3′. We obtained two products of 1.7kb and 1.1kb corresponding respectively to an unspliced and a spliced isoforms of TDP-43 3′ UTR (Fig. S13A). Both products were purified on 1% agarose gel and cloned independently in the psiCHECK-2 vector using NotI and XhoI restriction sites located 3′ to the Renilla luciferase translational stop. Since binding sites for TDP-43 mainly lie in the alternative intron, the spliced isoform was used as a control to assess the effect of TDP-43 protein on its own RNA.
Human myc-TDP-43-HA cDNA (a generous gift from C. Shaw) was cloned into mammalian expression vector, pCl-neo (Promega corporation). RFP (red fluorescence protein) was cloned in the vector pcDNA3 (Invitrogen).
250ng of psiCHECK-2 vector with or without TDP-43 3′ UTR and 250 ng of the vector expressing TDP-43 or RFP were co-transfected in Hela cells using Fugene 6 transfection reagent (Roche) in 12-wells cell culture plates. Luciferase assays were performed 48 hours after transfection using the Dual-Luciferase Reporter 1000 assay system (Promega) according to the manufacturer’s instructions. Five independent experiments were performed and 20ul of lysate were used in duplicate for each condition. Relative fluorescence units (RFU) for Renilla luciferase were normalized to firefly luciferase to control for transfection efficiency. Duplicates were averaged and each condition was expressed as a percentage of the samples without transfection of TDP-43 or RFP cDNA. Inter-group differences were assessed by two-tailed Student’s t-test.
Mouse gene structure annotations
The mouse genome sequence (mm8) and annotations for protein-coding genes were obtained from the University of California, Santa Cruz (UCSC) Genome Browser. Known mouse genes (knownGene containing 31,863 entries) and known isoforms (knownIsoforms containing 31,014 entries in 19,199 unique isoform clusters) with annotated exon alignments to the mouse genomic sequence were processed as follows. Known genes that were mapped to different isoform clusters were discarded. All mRNAs aligned to mm8 that were greater than 300 nt were clustered together with the known isoforms. For the purpose of inferring alternative splicing, genes containing fewer than three exons were removed from further consideration. A total of 1.9 million spliced ESTs were mapped onto the 16,953 high-quality gene clusters to identify alternative splicing events. Final annotated gene regions were clustered together so that any overlapping portion of these databases was defined by a single genomic position. Promoter regions were arbitrarily defined as 1.5 kb upstream of the transcriptional start site of the gene and intergenic regions as unannotated regions in the genome. To identify 5′ and 3′ untranslated regions we relied on the coding annotation in UCSC known genes that we extended 1.5kb downstream or upstream the start and stop codons, respectively.
Comparison to human alternatively spliced exons
To find evidence for conserved alternative splicing patterns in humans, we used the UCSC LiftOver tool to obtain the corresponding human coordinates of the mouse exons that had evidence for differential splicing either from RNA-seq or splicing-sensitive microarray data. These human genome coordinates were then compared with human gene structure annotations constructed analogously to mouse annotations as described above to determine if the exon in the human ortholog was alternatively or constitutively spliced based on transcript evidence in human EST or mRNA databases49.
Computational identification of CLIP-seq clusters
CLIP-seq reads were trimmed to remove adaptor sequences and homopolymeric runs, and mapped to the repeat-masked mouse genome (mm8) using the bowtie short-read alignment software (version 0.12.2) with parameters -q -p 4 -e 100 -y -a -m 10 --best --strata, incorporating the base-calling quality of the reads. To eliminate redundancies created by PCR amplification, all reads with identical sequence were considered a single read. Significant clusters were calculated by first determining read number cutoffs using the Poisson distribution, where λ was the frequency of reads mapped over an interval of nucleotide sequence, k was the number of reads being analyzed for significance, and f (k;λ) returned the probability that exactly k reads would be found. For any desired p-value, p-cutoff, a read number cutoff was calculated by summing the probabilities for finding k or more tags, and determining the minimum value of k that satisfies i=k such that f(k;λ)>p-cutoff. The frequency λ was calculated by dividing the total number of mapped reads by the number of non-overlapping intervals present in the transcriptome. The interval size was chosen based on the average size of the CLIP product, which includes only the selected RNA fragments but not any ligated adapters (150bp). A global and local cutoff was determined using the whole transcriptome frequency or gene-specific frequency, respectively. The gene-specific frequency was the number of reads overlapping that gene divided by the pre-mRNA length. A sliding window of 150bp was used to determine where the read numbers exceed both the global and local cutoffs. At each significant interval, we attempted to extend the region by adding in the next read, ensuring continued significance at the same p-value cutoff.
Log curve analysis to determine target-saturation of CLIP-derived clusters
We used a curve-fitting approach with various sampling rates to estimate the number of CLIP-seq reads required in order to discover additional CLIP clusters. The target rate was calculated by determining the new set of CLIP-derived gene targets found at each step-wise increase in the number of sequenced reads. A scatter plot of targets found versus sampling rate was fitted with a log curve, which was then used to extrapolate the number of targets expected to be found by increasing read counts.
Splicing-sensitive microarray data analysis
Microarray data analysis was performed as previously described, selecting events significant, high-quality events using a q-value > 0.05 and an absolute separation score (sepscore) >0.527. The equation for sepscore = log2[TDP-43 depleted (Skip/Include)/Control treated (Skip/Include)]. For each replicate set, the log2 ratio of skipping to inclusion was estimated using robust least-squares analysis. Previously published work using similar cutoffs have validated about 85% of splicing events by RT-PCR27.
Events on the array were defined in the mm9 genome annotation, and for proper comparison to RNA-seq and CLIP-seq data, cassette events were converted to the mm8 genome annotation using the UCSC LiftOver tool. If an event did not exactly overlap an mm8 annotated exon, it was left out of further analyses.
Genomic analysis of CLIP clusters
Two biologically independent TDP-43 CLIP-seq libraries were generated and sequenced on the Illumina GA2. Subjecting reads from each library to our cluster-identification pipeline described above defined 15,344 and 30,744 clusters for CLIP experiment 1 and 2 respectively. A gene was considered to be a TDP-43 target that overlapped in both experiments if it contained an overlapping cluster. We generated 10 randomly distributed cluster sets and compared each to the original clusters. To compute the significance of the overlap, we calculated the standard or Z-score as follows: (percent overlap in the two experiments – mean percent overlap in 10 randomly distributed cluster sets)/standard deviation of percent overlap in the randomly distributed cluster sets. A p-value was computed from the standard normal distribution and assigned significance if it was lower than p<0.01. To generate the final set of TDP-43 CLIP-seq clusters, unique reads from both experiments were combined and subjected to our cluster-identification pipeline. Overall, clusters were at most 300 bases in length, with the median of 142 bases. As a comparison to a published HITS-CLIP/CLIP-seq dataset of a RNA binding protein in mouse brain, we downloaded Ago-HITS-CLIP reads (Brain[A-E]_130_50_fastq.txt) from http://ago.rockefeller.edu/rawdata.php and subjected the combined 1,651,104 reads to our cluster-identification and generated 33,390 clusters in 7,745 genes21. To identify enriched motifs within cluster regions, Z-scores were computed for hexamers as previously published15.
Functional annotation analysis
We used the Database for Annotation, Visualization and Integrated Discovery (DAVID Bioinformatic Resources 6.7; http://david.abcc.ncifcrf.gov/). For all genes down-regulated or up-regulated upon TDP-43 depletion a background corresponding to all genes expressed in brain was used.
Transcriptome and splicing analysis
Strand-specific RNA-seq reads each from control oligonucleotide, saline and TDP43 oligonucleotide treated animals were generated and ~50% mapped uniquely to our annotated gene structure database, using the bowtie short-read alignment software (version 0.12.2, with parameters -m 5 -k 5 --best --un --max -f) incorporating the base-calling quality of the reads. To eliminate redundancies created by PCR amplification, all reads with identical sequence were considered single reads. The expression value of each gene was computed by the number of sense reads that mapped uniquely to the exons per kilobase of exon sequence and normalized by the total number of million mapped sense reads to the genes (RPKM). Each RNA-seq sample was summarized by a vector of RPKM values for every gene and pairwise correlation coefficients were calculated for all replicates using a linear least squares regression against the log RPKM vectors. Hierarchical clustering revealed that the three treated conditions clustered into similar groups. The reads within each condition were combined to identify genes that were significantly up- and downregulated upon TDP-43 depletion. Local mean and standard deviations were calculated for the nearest 1000 genes, as determined by log average RPKM values between knockdown and control and a local Z-score was defined. The resulting Z scores were used to assign significantly changed genes (Z>2 up-regulated, Z<-2 down-regulated), as well as ranking the entire gene list for relative expression changes. Specific parameters, such as intron length or TDP-43 binding sites, were plotted for the next 100 genes.
Exons with canonical splice signals (GT-AG, AT-AC, GC-AG) were retained, resulting in a total of 190,161 exons. For each protein-coding gene, the 50 bases at the 3′ end of each exon were concatenated with the 50 bases at the 5′ end of the downstream exon producing 1,827,013 splice junctions. An equal number of “impossible” junctions was generated by joining the 50-base exon junction sequences in reverse order. To identify differentially regulated alternative cassette exons, we employed a modification of a published method24. In short, the read count supporting inclusion of the exon (overlapping the cassette exon and splice junctions including the exon) are compared to the read count supporting exclusion of the exon (overlapping the splice junction of the upstream and downstream exon. For a splice junction read to be enumerated, we required that at least 18 nucleotides of the read aligned and 5 bases of the read extended over the splice junction with no mismatches 5 bases around the splice junction. For the TDP-43 and control ASOs comparison, we constructed a 2×2 contingency table using of the counts of the reads supporting the inclusion and exclusion of the exon, in 2 conditions. Every cell in the 2×2 table had to contain at least 5 counts for a χ2 statistic to be computed. At a p<0.01, 110 excluded and 93 included single cassette exons were detected to be differentially regulated by TDP-43. As an estimate of false discovery, we observed that ~20 single cassette exons were detected by utilizing the “impossible” junction database.
Pre-mRNA features and tissue-specificity analysis
Affymetrix microarray data representing 61 mouse tissues were downloaded from the Gene Expression Omibus repository (www.ncbi.nih.gov/geo) under accession number GSE113350. Probes on the microarrays were cross-referenced to 15,541 genes in our database using files downloaded from the UCSC Genome Browser (knownToGnf1m and knownToGnfAtlas2). The expression value for each gene was represented by the average value of the two replicate microarray experiments for each tissue. To identify genes enriched in brain, we grouped 13 tissues as “brain” (substantia nigra, hypothalamus, preoptic, frontal cortex, cerebral cortex, amygdala, dorsal striatum, hippocampus, olfactory bulb, cerebellum, trigeminal, dorsal root ganglia, pituitary), and the remaining 44 tissues as “non-brain”, excluding the 5 embryonic tissues (embryo day 10.5, embryo day 9.5, embryo day 8.5, embryo day 7.5, embryo day 6.5). For each gene, the t-statistic was computed as , where μbrain (σ brain) and μnon–brain (σ non–brain) were the average (standard deviation) of the gene expression values in brain and non-brain tissues, respectively. At a t-statistic value cutoff of ≥1.5, 388 genes were categorized as brain-enriched. Concurrently, at a cutoff of <1.5, 15,153 genes were categorized as non-brain enriched. Random sets of 388 genes were selected from the 15,541 genes as controls for the brain-enriched set. To determine if pre-mRNA features were significantly different in the set of brain-enriched genes (or randomly chosen genes) compared to non-brain enriched genes, we performed the two-sample Kolmogorov-Smirnov goodness-of-fit hypothesis test which determines if the distribution of features were drawn from the same underlying continuous population. Cumulative distribution plots of pre-mRNA features were generated to illustrate the differences.
Affymetrix microarray data representing 79 human tissues were downloaded from the Gene Expression Omibus repository under the same accession number GSE113350. Probes on the human microarrays were cross-referenced to 18,372 genes in our database using files downloaded from the UCSC Genome Browser (knownToGnfAtlas2 – hg18). The same analysis as done for the mouse array data was repeated for these human array data. We grouped 17 tissues as “brain” (temporal lobe, globus pallidus, cerebellum peduncles, cerebellum, caudate nucleus, whole brain, parietal lobe, medulla oblongata, amygdala, prefrontal cortex, occipital lobe, hypothalamus, thalamus, subthalamic nucleus, cingulated cortex, pons, fetal brain), and the remaining 62 tissues as “non-brain.” At the same t-statistic cutoffs, 387 and 17,985 genes were categorized as brain-enriched and non-brain enriched, respectively. Random sets of 387 genes were selected from the 18,372 genes as controls for the brain-enriched set.
Supplementary Material
Supplementary Figure 1 TDP-43 CLIP-seq method, reproducibility and saturation of binding sites
(a) Schematic representation of cross-linking, immunoprecipitation and high-throughput sequencing protocol (CLIP-seq). (b) Autoradiographs of CLIP-seq experiments performed in parallel using two different antibodies, a commercially available rabbit polyclonal TDP-43 antibody (Aviva Systems Biology, Cat. No. ARP38942_T100) left and an in-house monoclonal antibody recognizing an epitope within amino acids 251–414 of human TDP-4316. Under identical conditions and exposure times, our monoclonal antibody showed a much higher potency of immunoprecipitating TDP-43-RNA complexes. The orange box depicts the excised low mobility complexes used for the cDNA libraries for comparison to the monomeric TDP-43 complexes. (c) Flow-chart illustrating the number of reads sequenced and aligned from the two independent CLIP-seq experiments. CLIP-seq libraries were subjected to 36-bp sequencing on the Illumina Genome Analyzer 2. In total 1 and 4.5 million reads were mapped uniquely to annotated genes for experiment 1 and 2 respectively. (d) Reproducibility of CLIP-seq experiments on a genome-wide scale. Clusters identified in independent CLIP-seq experiments were considered to overlap if 1 base of a cluster in one experiment extended to a cluster in the other experiment (left panel). A gene containing an overlapping cluster was considered to overlap (right panel). Venn diagrams revealed statistically significant overlap between clusters and genes in replicate CLIP-seq libraries, with randomly distributed clusters shown as a comparison (lower panel). (e) Gene target saturation chart plotting gene targets found by cluster-finding on increasing sampling rates (5% intervals). Data follows a logarithmic expansion with an R2 value of 0.98.
Supplementary Figure 2 Cluster strength and gene-location biases of TDP-43 binding
(a) Box plots of cluster strength (by χ2 analysis) versus GUGU-cluster bins (counts of non-overlapping GUGU tetramer found within the cluster). By t-test comparison to all clusters, bins of 11–15 and 16+ GUGUs were significantly enriched (p < 0.01 and p < 10−37, respectively). On each box, the central mark is the median, the edges of the box are the 25th and 75th percentiles, the whiskers extend to the most extreme data points not considered outliers, and outliers are plotted individually. (b) Pre-mRNAs were divided into annotated 5′ and 3′ untranslated regions, exons, proximal introns (<0.5kb from a splice junction), distal introns (>0.5kb from a splice junction) and unannotated regions. Pie-charts revealed that the majority of TDP-43 binding sites were located in distal intronic regions of pre-mRNA (left panel). For comparison, previously published Argonaute (Ago) binding sites in mouse brain21 are displayed as analogous pie-charts (right panel). (c) CLIP binding sites of TDP-43 in the two replicates, Ago, and random TDP-43 clusters across genes. The fraction of CLIP clusters was plotted depending on the relative gene position from the 5′ to the 3′ ends of each target gene.
Supplementary Figure 3 RNA-seq biological replicas
Correlation between RNA-seq results obtained from mice subjected to different ASO treatments. Heatmap generated from Cluster3 and Matlab, using linear least-squares regression correlation coefficients of the pair-wise comparison between RPKM values of all genes from each experiment. “K” is TDP-43 knockdown samples, “C” is control oligo-, “S” is saline-treated samples, and numbers represent biological replicates.
Supplementary Figure 4 Correlation between RNA-seq and CLIP-seq data using various parameters
(a) Genes were ranked randomly instead of upon their degree of regulation after TDP-43 depletion and the average TDP-43 CLIP clusters found in introns of the next 100 genes in the ranked list were plotted (green line). Similarly, the median of total intron length for the next 100 genes was plotted (blue line). (b) Genes were ranked by ascending expression (RPKM) values in the samples treated with control ASO. (c) Genes were ranked by ascending expression (RPKM) values in the samples treated with TDP-43 ASO. (d) Genes were ranked upon their degree of regulation after TDP-43 depletion and average Ago clusters found in introns of the next 100 genes (green line) or the total intron length for the next 100 genes (blue line) were plotted. (e) Genes were ranked upon their degree of regulation after TDP-43 depletion and average clusters found in 5′UTR for the next 100 genes (green line) or the total 5′UTR length for the next 100 genes (blue line) were plotted. (f) Genes were ranked upon their degree of regulation after TDP-43 depletion and average clusters found in 3′UTR for the next 100 genes (green line) or the total 3′UTR length for the next 100 genes (blue line) were plotted. (g) Genes were ranked upon their degree of regulation after TDP-43 depletion and average clusters found in exons for the next 100 genes (green line) or the total exon length for the next 100 genes (blue line) were plotted. No correlation between CLIP and RNA-seq data was observed (a–f). (h) Genes were ranked upon their degree of regulation after TDP-43 depletion and average clusters found in introns for the next 100 genes (green line) or the average intron length for the next 100 genes (blue line) were plotted. (i) Genes were ranked upon their degree of regulation after TDP-43 depletion and average cluster density (cluster count/intron length) for the next 100 genes was plotted.
Supplementary Figure 5 The calcium signaling pathway is affected by TDP-43 depletion
(a) The calcium-signaling pathway annotated by KEGG (Kyoto Encyclopedia of Genes and Genomes) was found to be significant using the functional annotation tool DAVID. Red-bordered gene groups contain TDP-43 targets that are downregulated upon TDP-43 depletion. (b) List of downregulated TDP-43 targets contributing to calcium-signaling pathway.
Supplementary Figure 6 The long-term potentiation pathway is affected by TDP-43 depletion
(a) The long-term potentiation pathway annotated by KEGG was found to be significant using the functional annotation tool DAVID. Red-bordered gene groups contain TDP-43 targets that are downregulated upon TDP-43 depletion. (b) List of downregulated TDP-43 targets contributing to long-term potentiation pathway.
Supplementary Figure 7 Representative examples of genes with long introns and multiple TDP-43 binding sites
(a) Most transcripts in this group (mean intron length >10kb) displayed multiple intronic TDP-43 binding sites as determined by CLIP-seq (purple bars above each gene). In contrast, Chat is an example of a downregulated gene with short intron length (mean intron length <10kb) that shows a single TDP-43 binding site. The number of clusters per gene (n) is in purple on the left. Note that each cluster represents a binding cluster defined by a collection of overlapping reads as shown for Park2 (inset). (b) Examples of conservation of gene structures in human neurexin 3 and parkin 2 showing similar genic architectures as their mouse orthologues.
Supplementary Figure 8 TDP-43 binding on the 3′UTR of RNAs represses gene expression
(a) Normalized expression (based on RPKM values from RNA-seq) of selected up-regulated genes upon TDP-43 depletion that also contain TDP-43 binding regions in their 3′UTR. (b) Enrichment for binding within the 3′UTR among upregulated TDP-43 targets, compared to unchanged genes. *Upregulated with intronic binding versus unchanged with intronic binding (p<1.8×10−6), **Upregulated with 3′UTR binding versus unchanged with 3′UTR binding (p<8.2×10−3), ***Upregulated with 3′UTR binding versus downregulated with 3′UTR binding (p < 1.4 × 10−2). P-values calculated by chi-square.
Supplementary Figure 9 Splicing changes upon TDP-43 depletion identified by exon-junction arrays
Distribution of alternative splicing events upon TDP-43 depletion detected using splicing-sensitive microarrays. Of the 326 cassette events defined by Affymetrix on mm9 annotation, 287 were annotated by our stringent gene structure annotations in mm8, and were used for comparisons to RNA-seq and CLIP-seq data.
Supplementary Figure 10 Examples of alternative splicing regulation by TDP-43
RNA targets with exons included (a) or excluded (b) upon TDP-43 knockdown (same as in Fig. 4d). Semi-quantitative RT-PCR of selected targets showing splice changes in samples with TDP-43 knockdown compared to controls. Schematic representation of assessed exons with the changed exon(s) colored orange (left). The exon number of the gene is indicated below each box. The red arrows depict the position of the primers used for RT-PCR. Purple boxes indicate TDP-43 binding sites as defined by CLIP-seq. Representative acrylamide gel pictures of RT-PCR products from control or knockdown adult brain samples, as indicated (right panel). Quantification of splicing changes from three biological replicas per group is shown in the middle panel.
Supplementary Figure 11 Splicing changes correlate with the level of TDP-43 knockdown
(a) Acrylamide gels with semi-quantitative RT-PCR samples showing Sortilin 1 splicing changes in mice with various levels of TDP-43 reduction (shown as percentages above each bar). (b) Significant correlation of TDP-43 levels to Sortilin 1 splicing changes (correlation value = −0.905, n=27). (C) Acrylamide gels with semi-quantitative RT-PCR samples showing Kcnip splicing changes in mice with various levels of TDP-43 reduction (shown as percentages above each bar on the lowest panel). (d–e) Significant correlation of TDP-43 levels to Kcnip splicing changes (correlation value = 0.91 for exon 3 and 0.83 for exon 2 and 3, n=12).
Supplementary Figure 12 TDP-43 3′UTRs used in luciferase assays and control immunoblots
(a) Two alternative 3′UTRs of TDP-43 were amplified from mouse or human cDNA using primers whose approximate locations are shown by the small red arrows below the schemes on the right. The first 1.7kb or 1.1kb of each long (unspliced) or short (spliced) 3′UTR respectively were then cloned into a renilla luciferase reporter vector. The latter also contains the firefly luciferase gene that is expressed independently from renilla luciferase and serves as a transfection normalization control. (b) Representative immunoblots of HeLa cell lysates used for luciferase assays in Fig. 5f showing increased expression of TDP-43 in samples transfected with a myc-hTDP-43-HA expressing vector. For control, cells were transfected with a vector expressing an unrelated protein, namely red fluorescence protein (RFP), whose levels were confirmed by immunoblot (second panel). (c) Representative immunoblots of HeLa cell lysates used for Fig. 5f, g showing decreased levels of UPF1 in samples transfected with UPF1 siRNA.
Supplementary Figure 13 TDP-43 binding and regulation of Hdac6 and Nefl transcripts
(a) CLIP-seq reads and clusters on Hdac6 transcript showing intronic TDP-43 binding. RNA-seq reads from control or TDP-43 knockdown (KD) samples (equal scales) show a slight decrease in Hdac6 mRNA in the TDP-43 knockdown group. (b) Quantitative RT-PCR confirmed the slight decrease in Hdac6 mRNA in samples with reduced TDP-43 levels when compared to controls. Standard deviation was calculated within each group for 3 biological replicas. (c) CLIP-seq reads and clusters on Nefl transcript showing two distinct TDP-43 binding sites within the 3′UTR. Our RNA-seq reads indicate that the 3′UTR of mouse Nefl is longer than reported in UCSC database (light blue bar indicates the extention). RNA-seq reads from control or TDP-43 knockdown (KD) samples (equal scales) show a slight decrease in Nefl mRNA in the TDP-43 knockdown group, in accordance with previous reports.
Supplementary Figure 15 TDP-43 binding patterns on transcripts encoding for selecteddisease-related proteins
The number of clusters per gene (n) is in purple on the left. Note that each cluster represents a binding site defined by a collection of overlapping reads as shown for ataxin 1 (inset).
Supplementary Table 1 Genes upregulated upon TDP-43 depletion in adult mouse brain
The number of CLIP-seq clusters, localized in the 3′UTR, the 5′UTR, exonic or intronic regions of regulated each gene, are shown in the respective columns.
Supplementary Table 2 Genes downregulated upon TDP-43 depletion in adult mouse brain
The number of CLIP-seq clusters, localized in the 3′UTR, the 5′UTR, exonic or intronic regions of regulated each gene, are shown in the respective columns.
Supplementary Table 3 ncRNAs with changes in expression levels upon TDP-43 depletion in adult mouse brain
Supplementary Table 4 Enriched Gene Ontology terms in TDP-43 target RNAs that are downregulated upon TDP-43 depletion in adult mouse brain
The p-value indicated is corrected for multiple testing using the Benjamini-Hochberg method.
Supplementary Table 5 TDP-43 binding and regulation of transcripts encoding for proteins involved in RNA metabolism
Supplementary Table 6 TDP-43 binding and regulation of transcripts associated with disease
Z-scores between −1.96 and 1.96 represent p-values greater than 0.05.
Acknowledgments
The authors would like to thank members of Dr. Bing Ren’s lab, especially Zhen Ye, Samantha Kuan and Lee Edsall for technical help with the Illumina sequencing and Dr. Ulrich Wagner for helpful discussions, Kevin Clutario and Jihane Boubaker for technical help, as well as all members of the Yeo and Cleveland laboratories, Dr. Manuel Ares, Jr for generous support, and the neuro-team of ISIS Pharmaceuticals for critical comments and suggestions on this project. MP is the recipient of a Human Frontier Science Program Long Term Fellowship. CLT is the recipient of the Milton-Safenowitz postdoctoral fellowship from the Amyotrophic Lateral Sclerosis Association. DWC receives salary support from the Ludwig Institute for Cancer Research. SCH is funded by an NSF Graduate Research Fellowship. This work was been supported by grants from the NIH (R37 NS27036 and an ARRA Challenge grant) to D.W.C and partially by grants from the US National Institutes of Health (HG004659 and GM084317 to GWY), and the Stem Cell Program at the University of California, San Diego (GWY).
Footnotes
AUTHOR CONTRIBUTIONS
MP, CL-T, JM and TYL performed the experiments. KRH, SCH and TYL conducted the bioinformatics analysis. S-CL developed the monoclonal TDP-43-specific antibody used for CLIP-seq and the tetracycline-inducible GFP-TDP-43-expressing HeLa cells. S-CL and ES generated the transgenic myc-TDP-43 mice. JPD and LS conducted the preliminary splice-junction microarray analyses. MP, CLT, EW, CM, YS, CFB and HK conducted the antisense oligonucleotide experiments. MP, CL-T, KRH, GWY and DWC designed the experiments. MP, CL-T, KRH, SCH, GWY and DWC wrote the paper.
COMPETING INTERESTS STATEMENT
The authors declare no competing financial interests.
Microarray CEL files and sequenced reads have been deposited at the Gene Expression Omnibus database repository and the NCBI Short Read Archive, respectively. All our data (microarray, RNAseq, and CLIPseq) are combined as a “SuperSeries entry” under one accession number XXXXXX.
References
- 1.Neumann M, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 2.Arai T, et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem Biophys Res Commun. 2006;351:602–611. doi: 10.1016/j.bbrc.2006.10.093. [DOI] [PubMed] [Google Scholar]
- 3.Lagier-Tourenne C, Polymenidou M, Cleveland DW. TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum Mol Genet. 2010;19:R46–64. doi: 10.1093/hmg/ddq137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gitcho MA, et al. TDP-43 A315T mutation in familial motor neuron disease. Annals of neurology. 2008;63:535–538. doi: 10.1002/ana.21344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kabashi E, et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat Genet. 2008;40:572–574. doi: 10.1038/ng.132. [DOI] [PubMed] [Google Scholar]
- 6.Sreedharan J, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Deerlin VM, et al. TARDBP mutations in amyotrophic lateral sclerosis with TDP-43 neuropathology: a genetic and histopathological analysis. Lancet Neurol. 2008;7:409–416. doi: 10.1016/S1474-4422(08)70071-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buratti E, et al. Nuclear factor TDP-43 can affect selected microRNA levels. FEBS J. 2010;277:2268–2281. doi: 10.1111/j.1742-4658.2010.07643.x. [DOI] [PubMed] [Google Scholar]
- 9.Cooper T, Wan L, Dreyfuss G. RNA and Disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwiatkowski TJ, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009 doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 11.Vance C, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009 doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 13.Ule J, et al. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- 14.Licatalosi DD, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yeo GW, et al. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat Struct Mol Biol. 2009;16:130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ling SC, et al. ALS-associated mutations in TDP-43 increase its stability and promote TDP-43 complexes with FUS/TLS. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:13318–13323. doi: 10.1073/pnas.1008227107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zisoulis DG, et al. Comprehensive discovery of endogenous Argonaute binding sites in Caenorhabditis elegans. Nat Struct Mol Biol. 2010;17:173–179. doi: 10.1038/nsmb.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sephton CF, et al. Identification of neuronal RNA targets of TDP-43-containing Ribonucleoprotein complexes. J Biol Chem. 2011 doi: 10.1074/jbc.M110.190884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mili S, Steitz JA. Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. Rna. 2004;10:1692–1694. doi: 10.1261/rna.7151404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buratti E, et al. Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. Embo J. 2001;20:1774–1784. doi: 10.1093/emboj/20.7.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parkhomchuk D, et al. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res. 2009;37:e123. doi: 10.1093/nar/gkp596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pang KC, et al. RNAdb--a comprehensive mammalian noncoding RNA database. Nucleic Acids Res. 2005;33:D125–130. doi: 10.1093/nar/gki089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang ET, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu F, et al. Sortilin-mediated endocytosis determines levels of the frontotemporal dementia protein, progranulin. Neuron. 2010;68:654–667. doi: 10.1016/j.neuron.2010.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carrasquillo MM, et al. Genome-wide Screen Identifies rs646776 near Sortilin as a Regulator of Progranulin Levels in Human Plasma. Am J Hum Genet. 2010;87:890–897. doi: 10.1016/j.ajhg.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Du H, et al. Aberrant alternative splicing and extracellular matrix gene expression in mouse models of myotonic dystrophy. Nat Struct Mol Biol. 2010;17:187–193. doi: 10.1038/nsmb.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ayala YM, et al. TDP-43 regulates its mRNA levels through a negative feedback loop. EMBO J. 2011 doi: 10.1038/emboj.2010.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Le Hir H, Moore MJ, Maquat LE. Pre-mRNA splicing alters mRNP composition: evidence for stable association of proteins at exon-exon junctions. Genes & development. 2000;14:1098–1108. [PMC free article] [PubMed] [Google Scholar]
- 30.Wollerton MC, Gooding C, Wagner EJ, Garcia-Blanco MA, Smith CW. Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Mol Cell. 2004;13:91–100. doi: 10.1016/s1097-2765(03)00502-1. [DOI] [PubMed] [Google Scholar]
- 31.Sureau A, Gattoni R, Dooghe Y, Stevenin J, Soret J. SC35 autoregulates its expression by promoting splicing events that destabilize its mRNAs. EMBO J. 2001;20:1785–1796. doi: 10.1093/emboj/20.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perlick HA, Medghalchi SM, Spencer FA, Kendzior RJ, Jr, Dietz HC. Mammalian orthologues of a yeast regulator of nonsense transcript stability. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:10928–10932. doi: 10.1073/pnas.93.20.10928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baker M, et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature. 2006;442:916–919. doi: 10.1038/nature05016. [DOI] [PubMed] [Google Scholar]
- 34.Cruts M, et al. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature. 2006;442:920–924. doi: 10.1038/nature05017. [DOI] [PubMed] [Google Scholar]
- 35.Fiesel FC, et al. Knockdown of transactive response DNA-binding protein (TDP-43) downregulates histone deacetylase 6. Embo J. 2010;29:209–221. doi: 10.1038/emboj.2009.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strong MJ, et al. TDP43 is a human low molecular weight neurofilament (hNFL) mRNA-binding protein. Mol Cell Neurosci. 2007;35:320–327. doi: 10.1016/j.mcn.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 37.Bergeron C, et al. Neurofilament light and polyadenylated mRNA levels are decreased in amyotrophic lateral sclerosis motor neurons. J Neuropathol Exp Neurol. 1994;53:221–230. doi: 10.1097/00005072-199405000-00002. [DOI] [PubMed] [Google Scholar]
- 38.Hutton M, et al. Association of missense and 5’-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 1998;393:702–705. doi: 10.1038/31508. [DOI] [PubMed] [Google Scholar]
- 39.A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. The Huntington’s Disease Collaborative Research Group. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 40.Schwab C, Arai T, Hasegawa M, Yu S, McGeer PL. Colocalization of transactivation-responsive DNA-binding protein 43 and huntingtin in inclusions of Huntington disease. J Neuropathol Exp Neurol. 2008;67:1159–1165. doi: 10.1097/NEN.0b013e31818e8951. [DOI] [PubMed] [Google Scholar]
- 41.Valdmanis PN, et al. A mutation that creates a pseudoexon in SOD1 causes familial ALS. Ann Hum Genet. 2009;73:652–657. doi: 10.1111/j.1469-1809.2009.00546.x. [DOI] [PubMed] [Google Scholar]
- 42.Birve A, et al. A novel SOD1 splice site mutation associated with familial ALS revealed by SOD activity analysis. Hum Mol Genet. 2010;19:4201–4206. doi: 10.1093/hmg/ddq338. [DOI] [PubMed] [Google Scholar]
- 43.Mercado PA, Ayala YM, Romano M, Buratti E, Baralle FE. Depletion of TDP 43 overrides the need for exonic and intronic splicing enhancers in the human apoA-II gene. Nucleic Acids Res. 2005;33:6000–6010. doi: 10.1093/nar/gki897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dreumont N, et al. Antagonistic factors control the unproductive splicing of SC35 terminal intron. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin S, Coutinho-Mansfield G, Wang D, Pandit S, Fu XD. The splicing factor SC35 has an active role in transcriptional elongation. Nat Struct Mol Biol. 2008;15:819–826. doi: 10.1038/nsmb.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tollervey JR, et al. Characterising the RNA targets and position-dependent splicing regulation by TDP-43; implications for neurodegenerative diseases. Nature neuroscience. 2011 doi: 10.1038/nn.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu YF, et al. Wild-type human TDP-43 expression causes TDP-43 phosphorylation, mitochondrial aggregation, motor deficits, and early mortality in transgenic mice. J Neurosci. 2010;30:10851–10859. doi: 10.1523/JNEUROSCI.1630-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Igaz LM, et al. Dysregulation of the ALS-associated gene TDP-43 leads to neuronal death and degeneration in mice. The Journal of clinical investigation. 2011 doi: 10.1172/JCI44867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yeo GW, Van Nostrand EL, Liang TY. Discovery and analysis of evolutionarily conserved intronic splicing regulatory elements. PLoS Genet. 2007;3:e85. doi: 10.1371/journal.pgen.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Su AI, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1 TDP-43 CLIP-seq method, reproducibility and saturation of binding sites
(a) Schematic representation of cross-linking, immunoprecipitation and high-throughput sequencing protocol (CLIP-seq). (b) Autoradiographs of CLIP-seq experiments performed in parallel using two different antibodies, a commercially available rabbit polyclonal TDP-43 antibody (Aviva Systems Biology, Cat. No. ARP38942_T100) left and an in-house monoclonal antibody recognizing an epitope within amino acids 251–414 of human TDP-4316. Under identical conditions and exposure times, our monoclonal antibody showed a much higher potency of immunoprecipitating TDP-43-RNA complexes. The orange box depicts the excised low mobility complexes used for the cDNA libraries for comparison to the monomeric TDP-43 complexes. (c) Flow-chart illustrating the number of reads sequenced and aligned from the two independent CLIP-seq experiments. CLIP-seq libraries were subjected to 36-bp sequencing on the Illumina Genome Analyzer 2. In total 1 and 4.5 million reads were mapped uniquely to annotated genes for experiment 1 and 2 respectively. (d) Reproducibility of CLIP-seq experiments on a genome-wide scale. Clusters identified in independent CLIP-seq experiments were considered to overlap if 1 base of a cluster in one experiment extended to a cluster in the other experiment (left panel). A gene containing an overlapping cluster was considered to overlap (right panel). Venn diagrams revealed statistically significant overlap between clusters and genes in replicate CLIP-seq libraries, with randomly distributed clusters shown as a comparison (lower panel). (e) Gene target saturation chart plotting gene targets found by cluster-finding on increasing sampling rates (5% intervals). Data follows a logarithmic expansion with an R2 value of 0.98.
Supplementary Figure 2 Cluster strength and gene-location biases of TDP-43 binding
(a) Box plots of cluster strength (by χ2 analysis) versus GUGU-cluster bins (counts of non-overlapping GUGU tetramer found within the cluster). By t-test comparison to all clusters, bins of 11–15 and 16+ GUGUs were significantly enriched (p < 0.01 and p < 10−37, respectively). On each box, the central mark is the median, the edges of the box are the 25th and 75th percentiles, the whiskers extend to the most extreme data points not considered outliers, and outliers are plotted individually. (b) Pre-mRNAs were divided into annotated 5′ and 3′ untranslated regions, exons, proximal introns (<0.5kb from a splice junction), distal introns (>0.5kb from a splice junction) and unannotated regions. Pie-charts revealed that the majority of TDP-43 binding sites were located in distal intronic regions of pre-mRNA (left panel). For comparison, previously published Argonaute (Ago) binding sites in mouse brain21 are displayed as analogous pie-charts (right panel). (c) CLIP binding sites of TDP-43 in the two replicates, Ago, and random TDP-43 clusters across genes. The fraction of CLIP clusters was plotted depending on the relative gene position from the 5′ to the 3′ ends of each target gene.
Supplementary Figure 3 RNA-seq biological replicas
Correlation between RNA-seq results obtained from mice subjected to different ASO treatments. Heatmap generated from Cluster3 and Matlab, using linear least-squares regression correlation coefficients of the pair-wise comparison between RPKM values of all genes from each experiment. “K” is TDP-43 knockdown samples, “C” is control oligo-, “S” is saline-treated samples, and numbers represent biological replicates.
Supplementary Figure 4 Correlation between RNA-seq and CLIP-seq data using various parameters
(a) Genes were ranked randomly instead of upon their degree of regulation after TDP-43 depletion and the average TDP-43 CLIP clusters found in introns of the next 100 genes in the ranked list were plotted (green line). Similarly, the median of total intron length for the next 100 genes was plotted (blue line). (b) Genes were ranked by ascending expression (RPKM) values in the samples treated with control ASO. (c) Genes were ranked by ascending expression (RPKM) values in the samples treated with TDP-43 ASO. (d) Genes were ranked upon their degree of regulation after TDP-43 depletion and average Ago clusters found in introns of the next 100 genes (green line) or the total intron length for the next 100 genes (blue line) were plotted. (e) Genes were ranked upon their degree of regulation after TDP-43 depletion and average clusters found in 5′UTR for the next 100 genes (green line) or the total 5′UTR length for the next 100 genes (blue line) were plotted. (f) Genes were ranked upon their degree of regulation after TDP-43 depletion and average clusters found in 3′UTR for the next 100 genes (green line) or the total 3′UTR length for the next 100 genes (blue line) were plotted. (g) Genes were ranked upon their degree of regulation after TDP-43 depletion and average clusters found in exons for the next 100 genes (green line) or the total exon length for the next 100 genes (blue line) were plotted. No correlation between CLIP and RNA-seq data was observed (a–f). (h) Genes were ranked upon their degree of regulation after TDP-43 depletion and average clusters found in introns for the next 100 genes (green line) or the average intron length for the next 100 genes (blue line) were plotted. (i) Genes were ranked upon their degree of regulation after TDP-43 depletion and average cluster density (cluster count/intron length) for the next 100 genes was plotted.
Supplementary Figure 5 The calcium signaling pathway is affected by TDP-43 depletion
(a) The calcium-signaling pathway annotated by KEGG (Kyoto Encyclopedia of Genes and Genomes) was found to be significant using the functional annotation tool DAVID. Red-bordered gene groups contain TDP-43 targets that are downregulated upon TDP-43 depletion. (b) List of downregulated TDP-43 targets contributing to calcium-signaling pathway.
Supplementary Figure 6 The long-term potentiation pathway is affected by TDP-43 depletion
(a) The long-term potentiation pathway annotated by KEGG was found to be significant using the functional annotation tool DAVID. Red-bordered gene groups contain TDP-43 targets that are downregulated upon TDP-43 depletion. (b) List of downregulated TDP-43 targets contributing to long-term potentiation pathway.
Supplementary Figure 7 Representative examples of genes with long introns and multiple TDP-43 binding sites
(a) Most transcripts in this group (mean intron length >10kb) displayed multiple intronic TDP-43 binding sites as determined by CLIP-seq (purple bars above each gene). In contrast, Chat is an example of a downregulated gene with short intron length (mean intron length <10kb) that shows a single TDP-43 binding site. The number of clusters per gene (n) is in purple on the left. Note that each cluster represents a binding cluster defined by a collection of overlapping reads as shown for Park2 (inset). (b) Examples of conservation of gene structures in human neurexin 3 and parkin 2 showing similar genic architectures as their mouse orthologues.
Supplementary Figure 8 TDP-43 binding on the 3′UTR of RNAs represses gene expression
(a) Normalized expression (based on RPKM values from RNA-seq) of selected up-regulated genes upon TDP-43 depletion that also contain TDP-43 binding regions in their 3′UTR. (b) Enrichment for binding within the 3′UTR among upregulated TDP-43 targets, compared to unchanged genes. *Upregulated with intronic binding versus unchanged with intronic binding (p<1.8×10−6), **Upregulated with 3′UTR binding versus unchanged with 3′UTR binding (p<8.2×10−3), ***Upregulated with 3′UTR binding versus downregulated with 3′UTR binding (p < 1.4 × 10−2). P-values calculated by chi-square.
Supplementary Figure 9 Splicing changes upon TDP-43 depletion identified by exon-junction arrays
Distribution of alternative splicing events upon TDP-43 depletion detected using splicing-sensitive microarrays. Of the 326 cassette events defined by Affymetrix on mm9 annotation, 287 were annotated by our stringent gene structure annotations in mm8, and were used for comparisons to RNA-seq and CLIP-seq data.
Supplementary Figure 10 Examples of alternative splicing regulation by TDP-43
RNA targets with exons included (a) or excluded (b) upon TDP-43 knockdown (same as in Fig. 4d). Semi-quantitative RT-PCR of selected targets showing splice changes in samples with TDP-43 knockdown compared to controls. Schematic representation of assessed exons with the changed exon(s) colored orange (left). The exon number of the gene is indicated below each box. The red arrows depict the position of the primers used for RT-PCR. Purple boxes indicate TDP-43 binding sites as defined by CLIP-seq. Representative acrylamide gel pictures of RT-PCR products from control or knockdown adult brain samples, as indicated (right panel). Quantification of splicing changes from three biological replicas per group is shown in the middle panel.
Supplementary Figure 11 Splicing changes correlate with the level of TDP-43 knockdown
(a) Acrylamide gels with semi-quantitative RT-PCR samples showing Sortilin 1 splicing changes in mice with various levels of TDP-43 reduction (shown as percentages above each bar). (b) Significant correlation of TDP-43 levels to Sortilin 1 splicing changes (correlation value = −0.905, n=27). (C) Acrylamide gels with semi-quantitative RT-PCR samples showing Kcnip splicing changes in mice with various levels of TDP-43 reduction (shown as percentages above each bar on the lowest panel). (d–e) Significant correlation of TDP-43 levels to Kcnip splicing changes (correlation value = 0.91 for exon 3 and 0.83 for exon 2 and 3, n=12).
Supplementary Figure 12 TDP-43 3′UTRs used in luciferase assays and control immunoblots
(a) Two alternative 3′UTRs of TDP-43 were amplified from mouse or human cDNA using primers whose approximate locations are shown by the small red arrows below the schemes on the right. The first 1.7kb or 1.1kb of each long (unspliced) or short (spliced) 3′UTR respectively were then cloned into a renilla luciferase reporter vector. The latter also contains the firefly luciferase gene that is expressed independently from renilla luciferase and serves as a transfection normalization control. (b) Representative immunoblots of HeLa cell lysates used for luciferase assays in Fig. 5f showing increased expression of TDP-43 in samples transfected with a myc-hTDP-43-HA expressing vector. For control, cells were transfected with a vector expressing an unrelated protein, namely red fluorescence protein (RFP), whose levels were confirmed by immunoblot (second panel). (c) Representative immunoblots of HeLa cell lysates used for Fig. 5f, g showing decreased levels of UPF1 in samples transfected with UPF1 siRNA.
Supplementary Figure 13 TDP-43 binding and regulation of Hdac6 and Nefl transcripts
(a) CLIP-seq reads and clusters on Hdac6 transcript showing intronic TDP-43 binding. RNA-seq reads from control or TDP-43 knockdown (KD) samples (equal scales) show a slight decrease in Hdac6 mRNA in the TDP-43 knockdown group. (b) Quantitative RT-PCR confirmed the slight decrease in Hdac6 mRNA in samples with reduced TDP-43 levels when compared to controls. Standard deviation was calculated within each group for 3 biological replicas. (c) CLIP-seq reads and clusters on Nefl transcript showing two distinct TDP-43 binding sites within the 3′UTR. Our RNA-seq reads indicate that the 3′UTR of mouse Nefl is longer than reported in UCSC database (light blue bar indicates the extention). RNA-seq reads from control or TDP-43 knockdown (KD) samples (equal scales) show a slight decrease in Nefl mRNA in the TDP-43 knockdown group, in accordance with previous reports.
Supplementary Figure 15 TDP-43 binding patterns on transcripts encoding for selecteddisease-related proteins
The number of clusters per gene (n) is in purple on the left. Note that each cluster represents a binding site defined by a collection of overlapping reads as shown for ataxin 1 (inset).
Supplementary Table 1 Genes upregulated upon TDP-43 depletion in adult mouse brain
The number of CLIP-seq clusters, localized in the 3′UTR, the 5′UTR, exonic or intronic regions of regulated each gene, are shown in the respective columns.
Supplementary Table 2 Genes downregulated upon TDP-43 depletion in adult mouse brain
The number of CLIP-seq clusters, localized in the 3′UTR, the 5′UTR, exonic or intronic regions of regulated each gene, are shown in the respective columns.
Supplementary Table 3 ncRNAs with changes in expression levels upon TDP-43 depletion in adult mouse brain
Supplementary Table 4 Enriched Gene Ontology terms in TDP-43 target RNAs that are downregulated upon TDP-43 depletion in adult mouse brain
The p-value indicated is corrected for multiple testing using the Benjamini-Hochberg method.
Supplementary Table 5 TDP-43 binding and regulation of transcripts encoding for proteins involved in RNA metabolism
Supplementary Table 6 TDP-43 binding and regulation of transcripts associated with disease
Z-scores between −1.96 and 1.96 represent p-values greater than 0.05.