Abstract
Mutant huntingtin proteolysis mediated by various proteases plays a key role in Huntington's disease (HD) pathogenesis. In this issue of Neuron, Miller et al. have identified 11 proteases, including matrix metalloproteinases (MMPs), that when inhibited reduce huntingtin proteolysis and produce beneficial therapeutic effects. These findings provide new insights into huntingtin proteolysis and its potential as a therapeutic target.
Huntington's disease, the most frequent of a group of nine inherited neurodegenerative polyglutamine disorders, is caused by an expanded CAG triplet repeat in exon 1 of the huntingtin gene that encodes a stretch of polyglutamine (polyQ) residues close to the N-terminus of the huntingtin (Htt) protein. Expansion of the polyglutamine repeat interferes with the interaction of N-terminal 17 amino acid sequence of huntingtin (NT-17) with the nuclear exporter translocated promoter region (Tpr), causing accumulation of mutant huntingtin in the nucleus. Huntingtin aggregates, predominantly composed of N-terminal fragments of polyQ expanded Htt, in the nucleus and cytoplasm of affected neurons are a pathological hallmark of HD (Wheeler et al., 2000; DiFiglia et al., 1997).
Htt is known to be cleaved by various intracellular proteases, including caspases 1, 3, 6, 7, and 8; calpain; and an unidentified aspartic protease. Caspases and calpains are the most widely studied and cleave Htt within the N-terminal region between amino acids 469 and 586 (Gafni et al., 2004; Wellington et al., 2000). Recent work identified Htt cleavage sites closer to the N terminus, between amino acids 105 and 167 (Lunkes et al., 2002; Ratovitski et al., 2007).
Inhibition of mutant Htt cleavage reduces toxicity, indicating an important role for Htt proteolysis in HD pathogenesis (Graham et al., 2006; Gafni et al., 2004). An initial transgenic mouse model expressing an N-terminal fragment with about 150 CAG repeats, the R6/2 mouse, developed a robust behavioral phenotype, striatal atrophy, and reduced survival (Mangiarini et al., 1996). The N-terminal 171 amino acid Htt fragment (N171-82Q mice) is also toxic, whereas the N-terminal 117 amino acid fragment (N117) does not convey the HD phenotype in the “shortstop” mice (Slow et al., 2005). As a general rule, shorter N-terminal Htt fragments produce a more robust phenotype in transgenic mice than does full-length Htt. In vitro studies showed that calpain cleavage of mutant Htt plays a critical role in its pathogenicity (Gafni et al., 2004). Furthermore, a putative aspartyl protease site near the N terminus of mutant Htt has been implicated in the generation of toxic fragments (Lunkes et al., 2002). Studies also showed that a caspase 6 cleavage site plays a critical role in disease pathogenesis, since mutating this site so that it cannot be cleaved abolishes the pathogenicity of mutant Htt in vivo (Graham et al., 2006). More recent studies have demonstrated that, apart from the proteolytic cleavage, N-terminal modifications such as phosphorylation of serines 13 and 16 play a critical role in the toxicity of mutant Htt in vivo (Gu et al., 2009). The studies to date have therefore shown that proteolytic cleavage of mutant Htt, as well as N-terminal modifications, can play a critical role in the disease pathogenesis.
Although important caspase and calpain cleavage sites have been identified, it is unknown whether other proteases play an important role in Htt cleavage and pathogenicity. A study by Miller et al. (2010) in this issue of Neuron examined huntingtin proteolytic processing by screening for enzymes that cleave mutant Htt. In an elegant and novel approach using an unbiased RNAi screen consisting of 514 siRNAs targeting the repertoire of all known and predicted human protease genes, they identified proteases that produce toxic N-terminal Htt proteolytic fragments. Htt cleavage sites closer to the N terminus produce smaller, and potentially more toxic, mutant Htt fragments. They used a high-throughput western-blot-based screen to detect the generation of small N-terminal Htt fragments with the 1C2 antibody, which selectively recognizes conformationally altered stretches of increased polyglutamines. In their system, a 55 kDa product was the smallest detectable Htt cleavage product; therefore, they predicted that blocking production of this fragment would prevent Htt-mediated cell death. The screen identified 11 proteases that, when inhibited, reduced huntingtin fragment accumulation detected by the 1C2 anti-polyQ antibody. The proteases identified include members of the calpain family, which are increased in HD transgenic mice and postmortem human brain tissue (CAPN5 and CAPN7); the signal peptide protease-like IMP5 and an amino-terminal signal peptide protease (SPC18); members of the secreted serine-protease kallikrein family (KLK10 and KLK11); the transmembrane-E3-ubiquitin ligase, RNF128; the MMP-2 interacting integrin, ITGA2B; and three members of the matrix metalloproteinase family (MMP-10, -14, -23B). Nine of the 11 proteases are expressed in striatal cells, and their knockdown significantly reduced Htt-mediated striatal cell death in a secondary cellular toxicity screen. They identified several interesting proteases including (1) IMP5, an unusual aspartyl protease that mediates clearance of signal peptides by proteolysis within endoplasmic reticulum and (2) RFN128, a ubiquitin E3 ligase that promotes proteasomal degradation. Further studies are needed to establish the interaction between Htt and these proteases.
Of the nine proteases validated by the secondary screen, Miller et al. found that three are matrix metalloproteinase (MMP) family members (MMP-10, -14, -23B) and one (ITGA2B) interacts with MMP-2, indicating an important role of MMPs in Htt proteolysis and toxicity. MMPs are a family of zinc-containing metalloproteinases that normally remodel the extracellular matrix and are implicated in neurodegeneration. They initially confirmed that MMP-10 and -14 are present in Hdh111Q/111Q striatal cells by western blotting. Then, they found that the knockdown of MMP-10 and -14 using siRNA correlated with a reduction in caspase activity. Then, they showed that a pharmacologic inhibitor, NNGH, which is a non-peptidic, potent inhibitor of MMPs, and two known endogenous inhibitors of MMPs, TIMP1 and TIMP3, blocked Htt-mediated toxicity in the striatal Hdh111Q/111Q cells. The MMPs were processed to their active forms, and the levels of MMP-10 and -14 were increased in Hdh111Q/111Q cells, as compared to HdhQ7/Q7 cells. MMP-10 enzymatic activity was significantly increased in Hdh111Q/111Q cells as compared to HdhQ7/Q7 cells. These findings were recapitulated when tested in the brains of a full-length Htt (YAC128) and N-terminal fragment mouse model (R6/2) of HD.
They examined whether MMPs cleave mutant Htt directly by incubating cell lysates expressing Htt with recombinant MMPs. They found that Htt (wild-type or mutant) expressed in cellular lysates is a substrate for MMP-10, but not for MMP-14 or -2. Using in vitro translation studies, they demonstrated that MMP-10 knockdown suppresses Htt toxicity, through a direct effect on Htt cleavage. They examined the effects of reducing the expression of MMPs on Htt-induced neuronal dysfunction in a HD Drosophila model. In this model system, neuron-specific expression of expanded Htt leads to quantifiable progressive motor deficits. Of the two Drosophila MMP genes, only Dm2-MMP is expressed in postembryonic brain, and heterozygous loss-of-function alleles of Dm2-MMP showed robust and significant effects in improving the motor performance of the flies. Partial loss of function in Drosophila homologs of CAPN5, CAPN7, IMP5, and RNF128 also ameliorated motor deficits in the HD Drosophila model. Photoreceptor degeneration is also reduced in Drosophila expressing NT-Htt[128Q] with decreased levels of CalpA, Sol, and Dm2-MMP.
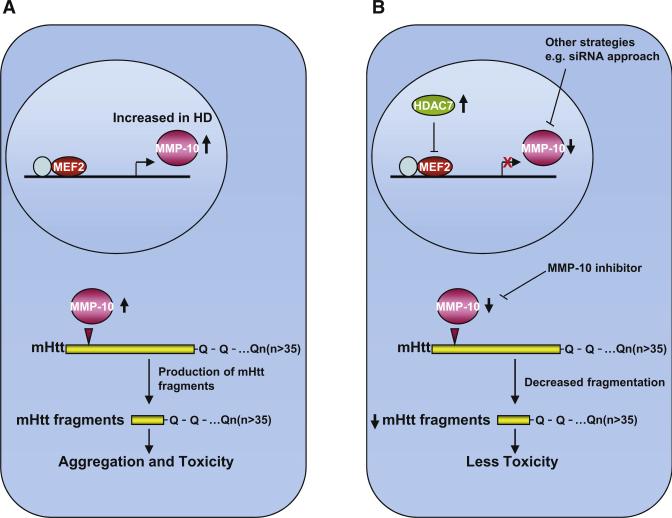
To summarize, Miller et al. identified several novel cleavage enzymes involved in huntingtin proteolytic processing, including MMP-10, a matrix metalloproteinase. They demonstrate increased MMP activity in HD mice, direct cleavage of Htt by MMP-10 (Figure 1A), and prevention of cell death on knockdown of MMP-10 in striatal HdhQ111/Q111 cells and a HD Drosophila model. They localized the MMP-10 cleavage site to a small region (a region closer to the N terminus than the caspase and calpain cleavage products of 55–72 kDa, producing a smaller 48 kDa product) and putative single amino acid residue (amino acid 402). Earlier studies in HD transgenic mice with a regulatable expression of mutant Htt showed that inhibiting expression of mutant Htt in symptomatic mice improved the neurological phenotype and caused the intranuclear inclusions to disappear (Yamamoto et al., 2000; DiFiglia et al., 2007). Mice expressing mutant Htt, resistant to cleavage by caspase-6, maintain normal neuronal function and do not develop striatal neurodegeneration (Graham et al., 2006). Similarly, inhibition of calpains leads to decreased proteolytic processing and aggregation of polyQ expanded Htt, resulting in decreased toxicity in an in vitro cell model (Gafni et al., 2004). These observations indicate that both the behavioral phenotype and the presence of intranuclear inclusions require continuous supplies of the abnormal huntingtin fragments and that the cleavage events are rate-limiting steps in pathogenesis.
Figure 1. MMP-10 in Proteolytic Processing of Huntingtin.
(A) MMP-10 is increased in HD and involved in direct cleavage of Htt, thereby producing small N-terminal toxic fragments.
(B) Pharmacological interventions such as MMP inhibitors or genetic manipulations such as overexpressing HDAC7, which acts as repressor for MMP-10, may provide a therapeutic benefit by reducing the cleavage of mutant Htt.
The importance of the observations of Miller et al. (2010) is that they identify new therapeutic targets for the treatment of HD, and in particular the involvement of MMP-10 in HD pathogenesis. MMP-10 is present in neurons, and it was increased in the striatum of R6/2 and YAC128 transgenic mice. Genetic or pharmacological interventions that block MMP-mediated production of Htt fragments therefore provide a target for therapeutic intervention (Figure 1B). If a selective inhibitor of MMP-10 can be developed, then this could be tested in transgenic mice and eventually in patients. We showed the feasibility of utilizing an MMP inhibitor therapeutically in another neurodegenerative disease, in a transgenic mouse model of ALS (Lorenzl et al., 2006). To investigate this further, it would be of interest to produce a transgenic HD mouse in which the MMP-10 site is mutated, such that it cannot be cleaved, and then to determine whether this abolishes the disease phenotype and pathology, similar to observations with caspase 6. It will also be of interest to cross MMP-10 knockout mice with full-length transgenic mouse models of HD. Activation of MMP-10 is known to occur in response to histone deacetylase 7 (HDAC7) knockdown (Chang et al., 2006); thus, another interesting approach to achieve downregulation of MMP-10 may be to increase expression of HDAC7 in HD cells, or transgenic mice and investigate whether this ameliorates HD pathology.
REFERENCES
- Chang S, Young BD, Li S, Qi X, Richardson JA, Olson EN. Cell. 2006;126:321–334. doi: 10.1016/j.cell.2006.05.040. [DOI] [PubMed] [Google Scholar]
- DiFiglia M, Sapp E, Chase KO, Davies SW, Bates GP, Vonsattel JP, Aronin N. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- DiFiglia M, Sena-Esteves M, Chase K, Sapp E, Pfister E, Sass M, Yoder J, Reeves P, Pandey RK, Rajeev KG, et al. Proc. Natl. Acad. Sci. USA. 2007;104:17204–17209. doi: 10.1073/pnas.0708285104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gafni J, Hermel E, Young JE, Wellington CL, Hayden MR, Ellerby LM. J. Biol. Chem. 2004;279:20211–20220. doi: 10.1074/jbc.M401267200. [DOI] [PubMed] [Google Scholar]
- Graham RK, Deng Y, Slow EJ, Haigh B, Bissada N, Lu G, Pearson J, Shehadeh J, Bertram L, Murphy Z, et al. Cell. 2006;125:1179–1191. doi: 10.1016/j.cell.2006.04.026. [DOI] [PubMed] [Google Scholar]
- Gu X, Greiner ER, Mishra R, Kodali R, Osmand A, Finkbeiner S, Steffan JS, Thompson LM, Wetzel R, Yang XW. Neuron. 2009;64:828–840. doi: 10.1016/j.neuron.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzl S, Narr S, Angele B, Krell HW, Gregorio J, Kiaei M, Pfister HW, Beal MF. Exp. Neurol. 2006;200:166–171. doi: 10.1016/j.expneurol.2006.01.026. [DOI] [PubMed] [Google Scholar]
- Lunkes A, Lindenberg KS, Ben-Haiem L, Weber C, Devys D, Landwehrmeyer GB, Mandel JL, Trottier Y. Mol. Cell. 2002;10:259–269. doi: 10.1016/s1097-2765(02)00602-0. [DOI] [PubMed] [Google Scholar]
- Mangiarini L, Sathasivam K, Seller M, Cozens B, Harper A, Hetherington C, Lawton M, Trottier Y, Lehrach H, Davies SW, Bates GP. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- Miller JP, Holcomb J, Al-Ramahi I, de Haro M, Gafni J, Zhang N, Kim E, Sanhueza M, Torcassi C, Kwak S, et al. Neuron. 2010;67(this issue):199–212. doi: 10.1016/j.neuron.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratovitski T, Nakamura M, D'Ambola J, Chighladze E, Liang Y, Wang W, Graham R, Hayden MR, Borchelt DR, Hirschhorn RR, Ross CA. Cell Cycle. 2007;6:2970–2981. doi: 10.4161/cc.6.23.4992. [DOI] [PubMed] [Google Scholar]
- Slow EJ, Graham RK, Osmand AP, Devon RS, Lu G, Deng Y, Pearson J, Vaid K, Bissada N, Wetzel R, et al. Proc. Natl. Acad. Sci. USA. 2005;102:11402–11407. doi: 10.1073/pnas.0503634102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellington CL, Leavitt BR, Hayden MR. J. Neural Transm. 2000;(Suppl.):1–17. doi: 10.1007/978-3-7091-6284-2_1. [DOI] [PubMed] [Google Scholar]
- Wheeler VC, White JK, Gutekunst CA, Vrbanac V, Weaver M, Li XJ, Li SH, Yi H, Vonsattel JP, Gusella JF, et al. Hum. Mol. Genet. 2000;9:503–513. doi: 10.1093/hmg/9.4.503. [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Lucas JJ, Hen R. Cell. 2000;101:57–66. doi: 10.1016/S0092-8674(00)80623-6. [DOI] [PubMed] [Google Scholar]