Abstract
During studies of APOBEC3 (A3) anti-human immunodeficiency virus type 1 (anti-HIV-1) mechanisms, we identified a single cysteine at position 320 (C320) that disrupts A3DE activity. This residue is located in the recently identified DNA binding domain in A3G. Replacing C320 with a corresponding tyrosine from A3F (Y307) increased A3DE antiviral activity more than 20-fold. Conversely, replacing A3F Y307 with a cysteine or inserting a similar cysteine into A3B or A3G disrupted the anti-HIV activity of A3. Further investigation uncovered that C320 significantly reduces A3DE catalytic activity.
TEXT
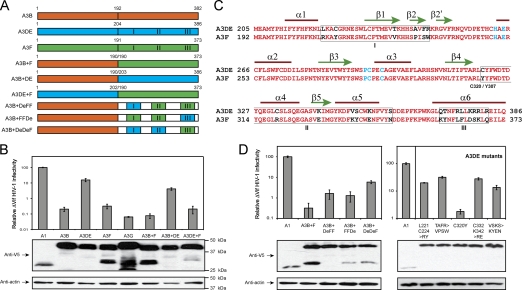
The A3DE gene was originally proposed as two genes, i.e., separated A3D and A3E genes (8). Later, we and others found that they are expressed as a single gene (4, 13), which was renamed A3DE (4). Despite sharing high homology with other A3 proteins that have two cytidine deaminase (CDA) motifs, A3DE exhibits relatively low levels of anti-human immunodeficiency virus type 1 (anti-HIV-1) activity, and Vif still neutralizes this activity (4). To map the domain responsible for its low activity, we used overlap extension PCR to create three chimeras. The A3B+F chimera contains the A3B N-terminal domain (NTD) and A3F C-terminal domain (CTD), and A3B+DE and A3DE+F chimeras contain other combinations of the NTD and CTD from A3B, A3DE, or A3F (Fig. 1A). We then compared the anti-ΔVif HIV-1 activity of the chimeras with those of wild-type proteins. ΔVif HIV-1 was produced from 293T cells ectopically expressing each of these proteins, and viral infectivity was analyzed using GHOST cells. APOBEC1 (A1) was used as a negative control since it does not have any antiretroviral activity. It was found that despite having similar levels of protein expressions, A3B, A3DE, A3F, A3G, A3B+F, A3B+DE, and A3DE+F reduced ΔVif HIV-1 infectivity 485-, 6-, 313-, 1,565-, 1,316-, 24-, or 500-fold, respectively (Fig. 1B). Thus, A3B+F and A3DE+F have anti-HIV-1 activity as potent as that of A3B, A3F, and A3G, whereas A3B+DE has anti-HIV-1 activity as poor as that of A3DE, indicating that the A3DE CTD is responsible for its low activity.
Fig. 1.
Mapping of A3DE residues for low anti-HIV-1 activity. (A) Schematic presentation of chimeras created in this study. A3B, A3DE, and A3F are shown in orange, blue, and green, respectively. Three variable regions I, II, and III are indicated. (C) Alignment of A3DE and A3F CTD amino acid sequences. Conserved residues are shown in red, nonconserved residues are shown in black, and residues in the CDA motif are shown in blue. Three C-terminal variable regions (I, II, III) are boxed, and both A3DE C320 and A3F Y307 residues are indicated. Secondary structure elements used in making the A3DE model in Fig. 3A are shown on top of the sequence. (B and D) Anti-HIV-1 activity of A3 chimeras and A3DE mutants. ΔVif HIV-1 luciferase reporter viruses were produced in 293T cells in the presence of each of these proteins, and their infectivity was determined after infection of GHOST cells. The value of infectivity was calculated from quantitation of luciferase enzyme produced in the GHOST cells, after normalization by levels of viral input (p24Gag). The infectivity of virions produced in the presence of A1 was considered 100%, and others were calculated as relative values according to this standard. The expression of indicated proteins in 293T cells was also determined by Western blotting (lower panels). The standard errors of the means (SEM) were calculated from three independent experiments.
The CTDs of A3DE and A3F are expected to have six α helixes and five β sheets, and they share 88% sequence similarity (Fig. 1C). Nonetheless, the CTDs contain three variable regions (I, II, III), which may account for the different activity levels of A3DE and A3F. We then created three other chimeras that have the A3B NTD but have different CTDs containing three combinations of these variable regions, from A3DE or A3F. Chimera A3B+DeFF contains A3DE I and A3F II and III, A3B+FFDe contains A3F I and II and A3DE III, and A3B+DeDeF contains A3DE I and II and A3F III regions (Fig. 1A). When the anti-ΔVif HIV-1 activity of these chimeras was compared, it was found that A3B+DeDeF had much lower activity than A3B+DeFF and A3B+FFDe, although they were expressed at similar levels (Fig. 1D). Since both A3B+DeFF and A3B+FFDe proteins lack the A3DE region II, it is apparent that this region is responsible for the low activity of A3DE.
To map the responsible residues, we exchanged A3DE residues in I and II regions with the corresponding A3F residues and created five A3DE mutants. The L221C224>RY mutant has L221 and C224 residues replaced with R and Y; the TAFR>VPSW mutant has T238, A243, F245, and R246 replaced with V, P, S, and W; the C320Y mutant has C320 replaced with Y; the C332K342>RE mutant has C332 and K342 replaced with R and E; and the VSKS>KYEN mutant has V350, S351, K354, and S359 replaced with K, Y, E, and N, respectively. It was found that despite comparable protein expression levels among the mutants, only the C320Y mutant showed increasing anti-HIV-1 activity, which was at least 20-fold higher than the wild-type protein activity (Fig. 1D). This activity is comparable to A3B and A3F activities, although it is still lower than A3G activity. Thus, a single C320Y mutation recovers A3DE anti-HIV-1 activity, indicating that C320 is responsible for the low activity of A3DE.
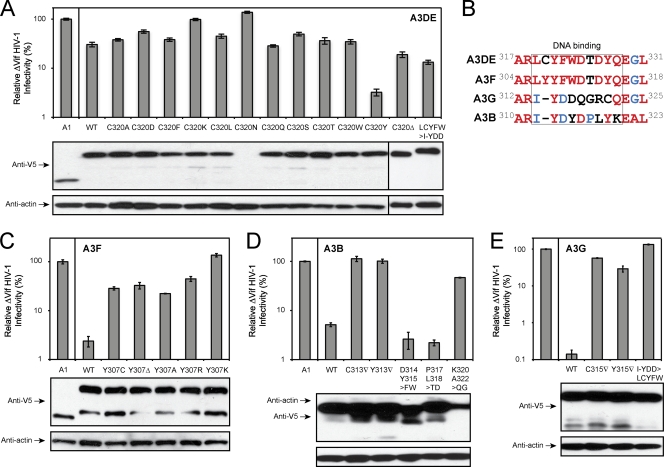
To obtain more insight into this gain-of-function mechanism, we replaced A3DE C320 with other types of amino acids and determined antiviral activity. Initially, we replaced C320 with alanine (A) and leucine (L) that have nonpolar, aliphatic R groups, serine (S), threonine (T), asparagine (N), and glutamine (Q) that have polar, uncharged R groups, lysine (K) that has a positively charged R group, and aspartate (D) that has a negatively charged R group. It was found that although they were well expressed except for the C320N mutant, all C320A, C320L, C320S, C320T, C320Q, C320K, and C320D mutations failed to recover the anti-HIV-1 activity of A3DE (Fig. 2A). Next, we replaced C320 with phenylalanine (F) and tryptophan (W), which have aromatic R groups similar to that of tyrosine. However, both C320F and C320W mutations were still poorly active (Fig. 2A). We also created a C320 deletion mutant, C320Δ, and a LCYFW>I-YDD mutant that has the LCYFW subdomain replaced with a corresponding A3G I-YDD subdomain. Again, both mutations failed to increase the anti-HIV activity of A3DE (Fig. 2A). These results indicate that charge, polarity, and hydrophobicity do not play any role at this position and that only the tyrosine rescues A3DE activity.
Fig. 2.
Characterization of A3 DNA binding domain. The anti-HIV-1 activity of A3DE mutants (A), A3F mutants (C), A3B mutants (D), and A3G mutants (E) were determined as in Fig. 1. Lower panels show protein expression in viral producer cells, which were determined by Western blotting with the indicated antibodies. (B) Amino acid sequence alignment of A3B, A3DE, A3F, and A3G regions, where the A3DE C320 residue is located. Completely conserved, partially conserved, and nonconserved residues are shown in red, blue, or black, respectively. WT, wild type.
The A3DE C320 residue is located in a domain that contains 9 to 10 amino acids, which are heterologous among A3B, A3DE, A3F, and A3G (Fig. 2B). It is replaced with a tyrosine in A3F, and it does not exist in A3G and A3B. We then created multiple A3B, A3F, and A3G mutants to study this domain's function. Initially, we first targeted the A3F Y307 residue and created four substitution mutants, the Y307C, Y307A, Y307R, and Y307K mutants, and one deletion mutant, the Y307Δ mutant. It was found that all these mutations disrupted A3F anti-HIV-1 activity (Fig. 2C). Next, we introduced mutations into A3B and A3G. Five A3B mutants were created: C313▿ that has a cysteine insertion, Y313▿ that has a tyrosine insertion, and D314Y315>FW, P317L318>TD, and K320A322>QG mutants that have the corresponding residues replaced with those from A3DE. Three A3G mutants were created, including two insertion mutants, C315▿ and Y315▿, and one substitution mutant, I-YDD>LCYFW, that has the I-YDD subdomain replaced with the corresponding A3DE LCYFW subdomain. It was found that despite having similar levels of protein expression, only the A3B D314Y315>FW and P317L318>TD mutants were still active and all the others lost activity (Fig. 2D, and 2E). Thus, this domain is also very critical for A3B, A3F, and A3G, although not every residue is equally important.
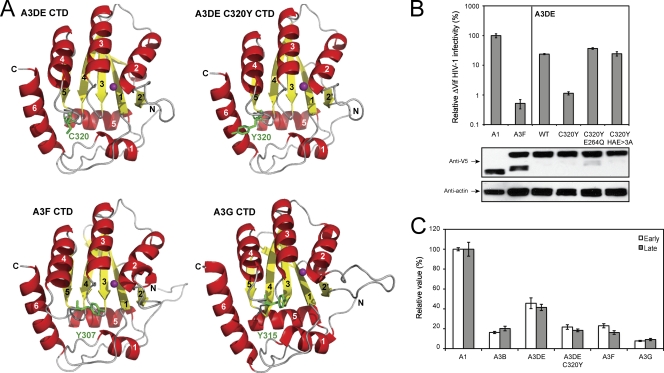
Recently, three groups reported that this domain (Fig. 2B) indeed determines A3G and activation-induced cytidine deaminase (AID) substrate sequence specificity via DNA binding (1, 9, 14). To further understand this domain's function in A3DE, we predicted the A3DE CTD structure based on the known A3G191-384-2K3A crystal structure (12) using YASARA (10) and PYMOL (http://www.pymol.org). After that, we compared A3DE C320, A3F Y307, and A3G Y315 locations (Fig. 3A). All known A3G structures show that the Y315 residue is located in a loop adjacent to the α4 helix, which is accessible to solvent (2, 5–7), and the originally predicted A3G substrate binding surface contains R313, Y315, D316, D317, and R320 in this domain. The importance of R313, Y315, or R320 to A3G cytidine deaminase activity has been experimentally shown (2, 3, 7), and the role of Y315 in base stacking or forming a hydrogen bond with cytosine at position −1 (which is next to the target cytosine at the 5′ side) has been predicted (11). We then decided to study how the C320Y mutation affects A3DE catalytic activity during viral inhibition.
Fig. 3.
(A) Ribbon diagram of A3G and predicted A3DE and A3F CTD three-dimensional structures. A3DE C320, A3DE Y320, A3F Y307, and A3G Y315 residues are shown and labeled in green, and the zinc molecule is shown in magenta. All helices and β strands are numbered, and N and C termini are indicated. (B) Anti-ΔVif HIV-1 activity of the A3DE wild type and C320Y mutants bearing CDA mutations. Protein expression in viral producer cells was detected by Western blotting. (C) Inhibition of HIV-1 reverse transcription by A3 proteins. ΔVif HIV-1 virions carrying A1, A3B, A3DE, A3DE C320Y, A3F, or A3G were used to infect CEM-SS cells. Six hours later, cellular DNAs were extracted from these infected cells, and viral early and late RT products were quantified by real-time PCR.
Initially, we addressed whether the enhancement of A3DE activity by the C320Y mutation is dependent on its catalytic activity. We mutated the 2nd CDA motif, 262HXEX27PCX2C296, by introducing a single E264Q mutation or triple HXE>3A mutations into the C320Y mutant. It was found that these mutations completely disrupted C320Y activity (Fig. 3B), and they did not impair virion packaging capability (data not presented). Thus, the activity of C320Y does require an active CDA. Next, we determined how this mutation affects A3DE inhibition of HIV-1 reverse transcription (RT). Cells were infected with ΔVif HIV-1 virions containing A1, A3B, A3DE, A3DE C320Y, A3F, or A3G, respectively, and viral RT products were quantitated by real-time PCR, as we did before (15). It was found that while A3G and A3DE had the highest or least inhibitory activity, respectively, A3B, A3DE C320Y, and A3F had similar medium levels of activities to reduce early and late RT products (Fig. 3C), indicating that the C320Y mutation increases A3DE inhibition of HIV-1 RT. Finally, we directly sequenced newly synthesized viral cDNAs and compared levels of G-to-A hypermutation, as we did before (4). By sequencing ∼4,000 bp from each sample, it was found that A3DE introduced extremely low levels, whereas the A3DE C320Y mutant introduced very high levels of G-to-A mutations (Fig. 4A). Indeed, the frequency of G-to-A hypermutations caused by A3DE C320Y (1.36%) was similar to that of A3G (1.69%) (Fig. 4B). Such a high mutation rate made it possible to reanalyze A3DE-specific dinucleotide sequence for editing. It was found that while A3G largely preferred 5′-GG sequence (84%) (targeted guanines are underlined), A3DE frequently targeted 5′-GA (51%) and slightly less frequently targeted 5′-GC (37%) (Fig. 4B). Thus, A3DE prefers both 5′-GA and 5′-GC sequences, which confirms our previous observation (4). Taken together, these results demonstrate that the C320Y mutation significantly increases A3DE catalytic activity, which in turn increases its antiviral activity. In addition, since both C320F and C320W mutations did not rescue A3DE activity (Fig. 2A) and the only difference between tyrosine and phenylalanine is a hydroxybenzene versus benzene R group, our results further suggest that instead of aromaticity, an -OH group plays a unique role at position 320 in A3DE substrate recognition.
Fig. 4.
Sequencing of newly synthesized viral cDNAs. (A) Nucleotide sequence alignment of HIV-1 cDNA from cells infected with HIV-1 carrying A3DE, A3DE C320Y, or A3G. The pNL4-3 sequence from nucleotides 5699 to 6076 is shown on the top. Only the mutated nucleotides are shown. Dots indicate nucleotides identical to pNL4-3 sequences. (B) Comparison of dinucleotide consensus sequences for A3DE C320Y and A3G editing. Higher frequencies are in bold. Targeted guanines are underlined.
Acknowledgments
We thank the NIH AIDS Research and Reference Reagent Program for reagents.
Y.-H.Z. is supported by grants AI063944 and AI080225 from the National Institutes of Health. H.M. is supported by grant AI073167.
Footnotes
Published ahead of print on 23 March 2011.
REFERENCES
- 1. Carpenter M. A., Rajagurubandara E., Wijesinghe P., Bhagwat A. S. 2010. Determinants of sequence-specificity within human AID and APOBEC3G. DNA Repair (Amsterdam) 9:579–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chen K. M., et al. 2008. Structure of the DNA deaminase domain of the HIV-1 restriction factor APOBEC3G. Nature 452:116–119 [DOI] [PubMed] [Google Scholar]
- 3. Chen K. M., et al. 2007. Extensive mutagenesis experiments corroborate a structural model for the DNA deaminase domain of APOBEC3G. FEBS Lett. 581:4761–4766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dang Y., Wang X., Esselman W. J., Zheng Y. H. 2006. Identification of APOBEC3DE as another antiretroviral factor from the human APOBEC family. J. Virol. 80:10522–10533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Furukawa A., et al. 2009. Structure, interaction and real-time monitoring of the enzymatic reaction of wild-type APOBEC3G. EMBO J. 28:440–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Harjes E., et al. 2009. An extended structure of the APOBEC3G catalytic domain suggests a unique holoenzyme model. J. Mol. Biol. 389:819–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Holden L. G., et al. 2008. Crystal structure of the anti-viral APOBEC3G catalytic domain and functional implications. Nature 456:121–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jarmuz A., et al. 2002. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics 79:285–296 [DOI] [PubMed] [Google Scholar]
- 9. Kohli R. M., et al. 2009. A portable hot spot recognition loop transfers sequence preferences from APOBEC family members to activation-induced cytidine deaminase. J. Biol. Chem. 284:22898–22904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Krieger E., Koraimann G., Vriend G. 2002. Increasing the precision of comparative models with YASARA NOVA—a self-parameterizing force field. Proteins 47:393–402 [DOI] [PubMed] [Google Scholar]
- 11. Rausch J. W., Chelico L., Goodman M. F., Le Grice S. F. 2009. Dissecting APOBEC3G substrate specificity by nucleoside analog interference. J. Biol. Chem. 284:7047–7058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shandilya S. M., et al. 2010. Crystal structure of the APOBEC3G catalytic domain reveals potential oligomerization interfaces. Structure 18:28–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stenglein M. D., Harris R. S. 2006. APOBEC3B and APOBEC3F inhibit L1 retrotransposition by a DNA deamination-independent mechanism. J. Biol. Chem. 281:16837–16841 [DOI] [PubMed] [Google Scholar]
- 14. Wang M., Rada C., Neuberger M. S. 2010. Altering the spectrum of immunoglobulin V gene somatic hypermutation by modifying the active site of AID. J. Exp. Med. 207:141–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang X., et al. 2010. Moloney leukemia virus 10 (MOV10) protein inhibits retrovirus replication. J. Biol. Chem. 285:14346–14355 [DOI] [PMC free article] [PubMed] [Google Scholar]