Abstract
The molecular mechanism for packaging of the adenovirus (Ad) genome into the capsid is likely similar to that of DNA bacteriophages and herpesviruses—the insertion of viral DNA through a portal structure into a preformed prohead driven by an ATP-hydrolyzing molecular machine. It is speculated that the IVa2 protein of adenovirus is the ATPase providing the power stroke of the packaging machinery. Purified IVa2 binds ATP in vitro and, along with a second Ad protein, the L4 22-kilodalton protein (L4-22K), binds specifically to sequences in the Ad genome that are essential for packaging. The efficiency of binding of these proteins in vitro was correlated with the efficiency of packaging in vivo. By utilizing a virus unable to express IVa2, pm8002, it was reported that IVa2 plays a role in assembly of the empty virion. We wanted to address the question of whether the ATP binding, and hence the putative ATPase activity, of IVa2 was required for its role in virus assembly. Our results show that ATPase activity was not required for the assembly of empty virus particles. In addition, we present evidence that particles were assembled in the absence of IVa2 by using two viruses null for IVa2—a deletion mutant virus, ΔIVa2, and the previously described mutant virus, pm8002. Empty virus particles produced by these IVa2 mutant viruses did not contain detectable viral DNA. We conclude that the major role of IVa2 is in viral DNA packaging. A characterization of the empty particles obtained from the IVa2 mutant viruses compared to wild-type empty particles is presented.
INTRODUCTION
The double-stranded DNA genome of adenovirus (Ad) is packaged with at least 12 different viral proteins; 7 of these proteins, hexon, penton, fiber, IIIa, VI, VIII, and IX, associate to form the protein capsid. Proteins V, VII, and mu are associated with the DNA forming the core (38, 44). These proteins are believed to condense the Ad DNA and mediate interactions between the core and capsid. Terminal protein is covalently linked to the 5′ ends of the genome and is the primer for replication; Ad protease is essential for maturation of the assembled particle with the Ad DNA (14, 18). The most abundant protein of the capsid is the trimer of polypeptide II, referred to as hexon. Hexons comprise about 60% of the protein of the virion. The 240 hexons, assembled 12 per facet of the icosahedral structure, have remarkable molecular flexibility, allowing the hexons to occupy four different environments. Hexons are associated with pentons at the 12 vertices; penton bases associate with a trimer of polypeptide IV called fiber. Additional minor components, IIIa, VI, VIII, and IX, contribute to capsid structure and stability (4, 21, 36, 37).
The molecular mechanism for the encapsidation process with Ad is unknown. However, there is evidence to suggest that the process in Ad is likely similar to what is observed for herpesviruses and DNA bacteriophages (reviewed in references 35 and 40). That is, the DNA genome of the virus is inserted into a preformed procapsid through a portal assembly at a unique vertex of the capsid. The movement of the DNA into the capsid is facilitated by an ATPase-driven molecular motor. In the case of Ad, particles devoid of viral DNA, empty particles, can be isolated and separated from mature particles by virtue of their different densities in cesium chloride gradients: 1.29 to 1.3 g/cm3 and 1.34 g/cm3, respectively (19, 22, 41). It has not been determined whether these empty particles are dead end products or assembly intermediates. However, they do contain several Ad capsid proteins in their precursor forms, pVI, pVIII, and IIIa, and maturation of these proteins by Ad protease occurs as a post-DNA-packaging event (6). Light intermediate particles band in cesium chloride gradients at a density of ∼1.30 g/cm3; it is not clear how they differ from empty virus particles. Furthermore, incomplete particles found in a range of cesium chloride densities between that of the empty and mature particles contain Ad DNA of increasing lengths. The sizes of the DNA increase with the increasing density of the particle and are due to extensions from the left end (5, 42). These observations are consistent with polar packaging of the Ad genome, with the initiation of packaging occurring at the left end. In support of this is the observation that sequences at the left end of the Ad type 5 (Ad5) genome, between nucleotides (nt) 200 and 400, are absolutely required for genome packaging (10, 16, 33). These packaging sequences can function at the right end of the Ad genome as well (13). A reverse in the packaging polarity is seen with virus with a functional domain at the right end of the DNA and not at the left end.
Several Ad5 proteins have been shown to bind to the packaging domain. Ad5 proteins IVa2 and L4 22-kilodalton protein (L4-22K) bind specifically to DNA motifs within the packaging domain both in vitro and in vivo (8, 26, 31, 34, 43, 45, 48). It has been shown that there is a direct correlation between packaging efficiency in vivo with the efficiency of IVa2 and L4-22K binding in vitro to the motifs within the packaging domain (31). In addition, the Ad L1-52/55K protein has been shown to specifically associate with the packaging domain in vivo (31, 34). Ad5 mutants in the genes encoding these proteins have defects late in the Ad life cycle consistent with defects in packaging of the virus genome (11, 26, 49). An Ad5 temperature-sensitive (ts) mutant in the L1–52/55K protein, ts369, yields incomplete particles when grown at the nonpermissive temperature (15). Less than genome-length fragments of DNA extending from the left end can be isolated from these particles that accumulate at a density in cesium chloride of approximately 1.3 g/ml (15). A virus that lacks the ability to synthesize the L1-52/55K protein is affected at the late phase of the virus life cycle, and empty particles are produced that lack viral DNA (11). The analysis of these L1-52/55K mutants suggests that this protein plays a role in both the initiation and the completion of encapsidation of Ad DNA. Similarly, a mutant virus unable to synthesize L4-22K is blocked at a late stage, after replication and late gene expression, consistent with a packaging defect (26). The role for IVa2 in the packaging of Ad was presumed to be more complex, involving playing a role in both packaging and assembly of the empty virus particle (49).
The IVa2 protein not only contains domains involved in DNA binding but also contains motifs associated with the binding and hydrolysis of ATP, the Walker A and B boxes (20). These motifs are conserved in IVa2 proteins across all species of Ad so far sequenced. The predicted secondary structure of the IVa2 protein places it in the class of additional strand catalytic E (ASCE) ATPases along with, though distinct from, the packaging ATPases of bacteriophages and other viruses (1). These observations led to the idea that IVa2 may be the ATPase component of the motor that would drive the DNA into the capsid, as seen with bacteriophages (35). Mutations introduced into the IVa2 Walker A or B boxes have been assayed in the context of the Ad genome by transfection. These genomes were unable to produce progeny virus (32). Furthermore, it was demonstrated that purified IVa2 bound ATP in vitro and that mutations in either the Walker A or B box reduced the observed binding (27). However, at present, there has been no report of ATPase activity with the purified form of the IVa2 protein, suggesting that an additional protein may be required for function. These results suggest a role for IVa2 in the recognition of Ad DNA, providing specificity of packaging, and also a role as part of the packaging machine. Consistent with these roles is the observation, by using immunoelectron microscopy, that IVa2 protein is localized at a unique vertex of the mature Ad capsid (3).
Utilizing a mutant virus, pm8002, that is unable to express IVa2, Zhang and Imperiale showed that this virus was defective for obtaining mature, infectious virus (49). Furthermore, a characterization of the infection process with this mutant virus showed that there was no accumulation of empty virus particles as well (49). The interpretation of these results altered the role of IVa2 to include a role in empty particle assembly and raised the question of whether assembly and packaging with Ad are linked.
As a first step in unraveling the role of IVa2 in packaging and assembly, we set out to determine if ATP binding, and hence the putative ATPase activity, is required for both packaging and assembly. Toward this end, we generated several Ad virus genomes with mutations in the IVa2 gene that either eliminated the expression of IVa2 or specifically affected the ATP binding/hydrolysis domain. In order to propagate these mutant viruses, we generated a cell line that inducibly expresses the Ad5 IVa2 protein. In contrast to the previously published report (49), we find that Ad5 mutants that do not express the IVa2 protein produce empty virus particles. The ATPase motif mutant virus also produced empty virus particles but was defective for packaging of viral DNA. Empty virus particles produced by these IVa2 mutant viruses did not contain detectable viral DNA. We conclude that the IVa2 protein is not required for empty virus particle assembly and that the IVa2 ATPase domain is required for viral DNA packaging. We present a characterization of the empty particles made in these infections.
MATERIALS AND METHODS
Cells, viruses, and infections.
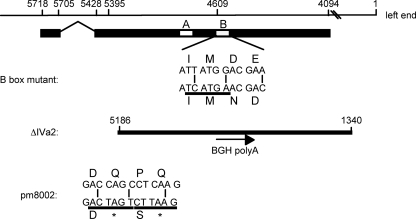
The permissive cell line, N52.E6-Cre, was a gift from G. Schiedner and S. Kochanek (University of Ulm). N52.E6-Cre expresses the adenovirus E1 gene and Cre recombinase (39); Cre recombinase was not important for these experiments. Cells were maintained in Dulbecco's modified Eagle medium supplemented with 10% Fetalclone III serum (HyClone), penicillin, and streptomycin. An N52.E6-Cre subclone, Tet-C4-IVa2, that expresses IVa2 under the control of a Tet-inducible promoter was generated; the new cell line is termed N52.E6-Cre–IVa2. N52.E6-Cre–IVa2 cells were maintained in Dulbecco's modified Eagle medium supplemented with 10% FetalClone III serum, penicillin, streptomycin, 200 μg/ml Geneticin, 200 μg/ml hygromycin B, 10 μg/ml blasticidin S, and 0.1 μg/ml doxycycline. Doxycycline was withdrawn from the culture medium to induce IVa2 expression. ΔIVa2, a IVa2 B box mutant, and pm8002 viruses (27, 49) (Fig. 1) were made by isolating single plaques following transfection of PacI-digested Ad5 infectious clones (2) carrying the respective mutations (pTG3602-ΔIVa2, -Bbox−−, and -pm8002) into the IVa2-inducible cell line. Viruses were amplified by two additional passages, and the titers of final stock lysates were determined by a plaque assay, as described previously (29). The purity of these virus stocks was confirmed by Southern blot analyses of viral extracted DNA from infected cells by the method described by Hirt (17). Southern analyses were done using an Amersham AlkPhos system (GE Healthcare Life Sciences), as described previously (30). The plasmid pTG3602-pm8002 (49) was a gift from M. Imperiale (University of Michigan). The ΔIVa2 and B box mutants were recombined into pTG3602 using the methods described by Evans and Hearing (7). The B box mutant contained two amino acid changes, amino acid 280 D to N and amino acid 281 E to D, and the introduction of a silent mutation in the IVa2 protein to introduce a BspHI restriction enzyme site for screening purposes.
Fig. 1.
Schematic diagram of the Ad5 IVa2 gene and viral mutants. The viral genome is represented by the thin line, shown in a right to left direction with Ad5 nucleotide (nt) numbers following a left-to-right convention. Hash marks indicate discontinuous representation of the Ad5 genome. The coding region of IVa2 is represented by a thick line, and the white boxes represent the Walker A and B boxes (letters A and B). Shown are the changes introduced into the Ad5 genome to generate the B box mutant, ΔIVa2, and pm8002 viruses. For the B box mutant (27), wild-type Ad5 DNA sequences from nt 4609 to 4598 are shown along with the substitutions indicated by vertical lines. The amino acid sequence is indicated by its single-letter code and is shown above and below the DNA sequences for the wild-type and mutant sequences, respectively. The nucleotide sequence underlined indicates the diagnostic BspHI site generated by the nucleotide substitutions. For ΔIVa2, the solid line indicates the deletion that includes Ad5 nt 5186 to nt 1340. BGH polyA refers to the insertion of the bovine growth hormone gene poly(A) signal, and the arrow shows the 5′-to-3′ direction of the insertion. For pm8002 (49), Ad5 nt 5395 to 5384 are shown, with the wild-type sequence above the mutated sequence and substitutions indicated by vertical lines. Amino acid sequences are indicated by single letters for wild-type and mutant proteins above and below the DNA sequences, respectively. Asterisks indicate stop codons. The introduction of diagnostic SpeI (solid line) and AflII (dotted line) sites is shown below the mutant sequence.
N52.E6-Cre cells (2 × 108 to 3 × 108) were infected with wild-type Ad5 and the Ad5 mutant viruses at a multiplicity of infection of 5 PFU/cell, as previously described (29). At 72 h postinfection, cells and media were collected, and the cells were pelleted, resuspended in 7.5 ml TD buffer (25 mM Tris [pH 7.4], 137 mM NaCl, 5 mM KCl, 0.7 mM Na2HPO4), and frozen and thawed at room temperature for 3 cycles. The lysates were cleared by centrifugation at 3,162 × g at 4°C for 30 min. The cleared supernatants were applied to step gradients of 1.25 g/cm3 and 1.40 g/cm3 cesium chloride in TD buffer and spun at 175,117 × g at 16°C for 1 h. Material that concentrated at the interface of the steps was removed and mixed with 1.29 g/cm3 CsCl and spun for 24 h at 174,150 × g at 16°C. Visible bands were collected from the equilibrium gradient, the refractive indices of the fractions were measured using a Carl Zeiss refractometer, and results were converted to densities.
Electron microscopy.
Particles from 25-μl droplets of fractions from the CsCl equilibrium gradients were adhered to 300 mesh, Formvar/carbon-supported nickel grids (Electron Microscopy Sciences, Hatfield, PA.) for 2 to 5 min at room temperature. Grids were floated on 400-μl droplets of the following solutions, particle side down, for the indicated times at room temperature: three times for 2 min each on 0.5× TNE buffer (10 mM Tris [pH 7.5], 250 mM NaCl, and 0.5 mM EDTA); two times for 2.5 min each on 5% glutaraldehyde (electron microscopy [EM] grade; Electron Microscopy Sciences, Hatfield, PA)/0.5× TNE buffer; and three times for 3 min each on 0.5× TNE buffer. Excess liquid was removed, and particles were negatively stained with methylamine vanadate (NanoVan; Nanoprobes, Yaphank, NY) by floating grids on 30-μl of stain for 15 s and then removing excess liquid. Samples were viewed with an FEI Tecnai 12 BioTwinG2 transmission electron microscope at 80 kV. Digital images were acquired with an AMT XR-60 charge-coupled-device (CCD) digital camera system and compiled using Adobe Photoshop CS5.
Silver staining and Western blot analyses.
Samples were prepared for gel electrophoresis by removing cesium chloride by precipitation: samples were diluted in two volumes of water and six volumes of ethanol stored at −20°C overnight followed by centrifugation at 16,000 × g at 4°C for 30 min. The samples were washed one time with 75% ethanol and resuspended in 1× sample buffer (60 mM Tris [pH 6.8], 2% SDS, and 20% glycerol). Protein concentrations were determined using the Pierce bicinchoninic acid (BCA) protein assay kit (Thermo Scientific, Rockford, IL). Protein from each empty particle sample (1.8 μg) and protein from the mature wild-type virus particles (1.5 μg) were separated on a 12.5% SDS gel and silver stained utilizing the method described in reference 25. For Western blot analyses, equivalent amounts of protein samples were loaded based on hexon content and were separated on a 7.5% SDS gel, transferred to nitrocellulose overnight at 4°C, and blocked in a solution of 1% casein in Tris-buffered saline for 1 h at room temperature. The membrane was incubated with rabbit polyclonal antibody to Ad5 hexon (GeneTex, Inc., Irvine, CA) diluted in 1% casein/Tris-buffered saline (TBS) with sodium azide for 1 h. The membrane was washed five times for 5 min in TBS with 0.5% Tween 20 and incubated with IRdye800 conjugated goat anti-rabbit immunoglobulin G (Rockland Immunochemicals, Gilbertsville, PA) followed by washes as previously described (26). The same samples were electrophoresed, blotted to membranes, and incubated with rabbit polyclonal antibody directed against IVa2 (28) diluted in 1% casein-TBS-sodium azide overnight at 4°C. A separate membrane was incubated with rabbit anti-L1-52/55K protein antibody (31). The membrane was washed as described above, incubated with a solution of IRDye 680LT conjugated goat anti-rabbit immunoglobulin G (LiCor, Lincoln, NE) diluted in 1% casein-TBS for 1 h and then washed by the procedures described above. Membranes were scanned using an Odyssey system (LiCor, Lincoln, NE). The L4-22K protein was similarly analyzed using a rabbit anti-L4-22K protein antibody (26) but detected using enhanced chemiluminescence (Immobilon) according to the manufacturer's instructions to augment sensitivity.
RESULTS
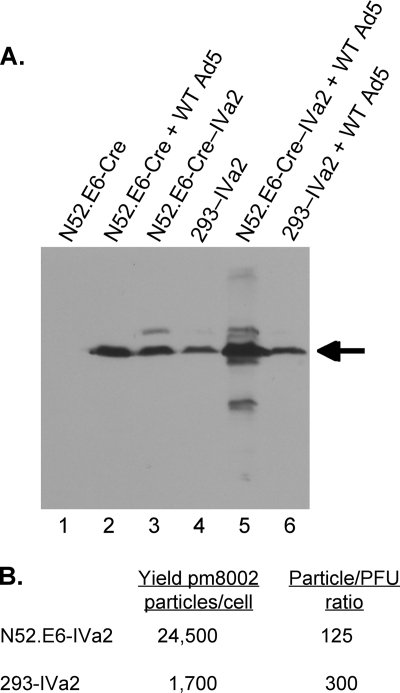
In order to examine the role of ATP binding, and hence the putative ATPase activity of the IVa2 protein in virus assembly, we generated two mutant viruses utilizing the Ad5 infectious genomic clone, pTG3602 (2) (Fig. 1). pTG3602-Bbox− has point mutations in the Walker B box domain that result in the substitutions of aspartic acid with asparagine and the neighboring glutamic acid with an aspartic acid. By homology to known ATPases, these amino acids are implicated in the binding and hydrolysis of ATP (46), and purified IVa2 protein with these mutations was shown to be markedly reduced for ATP binding in vitro (27). The other Ad5 mutant genome was deleted for IVa2 sequences; pTG3602-ΔIVa2 lacks Ad nt 1339 to 5186. This deletion removes a large C-terminal segment of the IVa2 reading frame as well as region E1B and part of region E1A. The poly(A) sequences from the bovine growth hormone gene were inserted in order to ensure expression of the Ad polymerase and terminal protein. This was necessary since the polyadenylation site for the Ad polymerase and terminal protein transcripts overlaps the IVa2 gene region that was deleted. Propagation of the resulting ΔIVa2 virus is dependent upon the expression of the IVa2 and the E1 genes using a complementing cell line. The deletion also removed the coding region of protein IX. However, this did not affect the viability of ΔIVa2 virus as was anticipated from previous results (4, 9). If a stable mRNA was made and spliced, it would encode a protein of 112 amino acids that includes the first 84 amino acids of the IVa2 protein. The advantage of this mutant virus is that it is unable to undergo homologous recombination with the IVa2 gene in the complementing cell to generate a revertant virus that restores IVa2 expression. This is not the case with the IVa2 mutant pm8002, for which mutant virus stocks had to be carefully screened for the presence of revertants. Two stop codons introduced into this mutant genome substitute for the 17th and 19th amino acids of the IVa2 protein (49). ΔIVa2 retains the first 252 nucleotides of the IVa2 gene, including the intron and splice sites. Stocks of the mutant viruses were obtained by transfection of the genomic clones, followed by plaque isolation and amplification using a newly developed IVa2 complementing cell line. This cell line expresses the Ad5 E1 (N52.E6-Cre) (38) and IVa2 gene products (called N52.E6-Cre–IVa2). We characterized the properties of this new cell line in comparison to the 293–IVa2 cell line previously described (49) (Fig. 2). The two cell lines produced similar levels of the Ad5 IVa2 protein as found in infections of noncomplementing cells (N52.E6) with wild-type Ad5 (following doxycycline removal with N52.E6-Cre–IVa2 cells) (Fig. 2A, lanes 2 to 4). The N52.E6-Cre–IVa2 cell line displayed elevated levels of IVa2 protein when infected with Ad5 in comparison to 293–IVa2 cells as well as several minor larger and smaller IVa2 species (Fig. 2A, lanes 5 and 6). The nature of these products is not known, although a smaller IVa2 protein that was initiated at an internal ATG codon has been described (32). We analyzed the total virus yield of the IVa2 mutant virus pm8002 (49) produced by each complementing cell line as well as the particle-to-PFU ratio (P/PFU) of the purified, mature mutant virus particles (Fig. 2B). The N52.E6-Cre–IVa2 cell line produced ∼14-fold more mature pm8002 virus particles per cell than the 293–IVa2 cell line, with a P/PFU ratio of 125 compared to a P/PFU ratio of 300 for 293–IVa2 cells. Thus, the new IVa2 complementing cell line N52.E6-Cre–IVa2 has several useful features for IVa2 mutant virus production.
Fig. 2.
Comparison of IVa2 protein expression in the 293–IVa2 and N52.E6-Cre–IVa2 cell lines. (A) Western blot analysis of IVa2 protein in equivalent amounts of cell extract from uninfected N52.E6 cells (lane 1), N52.E6 cells infected with wild-type Ad5 (lane 2), uninfected N52.E6-Cre–IVa2 and 293–IVa2 cells (lanes 3 and 4), and N52.E6-Cre–IVa2 and 293–IVa2 cells infected with wild-type Ad5 (lanes 5 and 6). For infected cell samples, cell extracts were prepared at 48 h postinfection. Full-length IVa2 protein is indicated by an arrow. (B). Virus yields per cell and the P/PFU ratios for mature pm8002 virus particles produced using each cell line are presented.
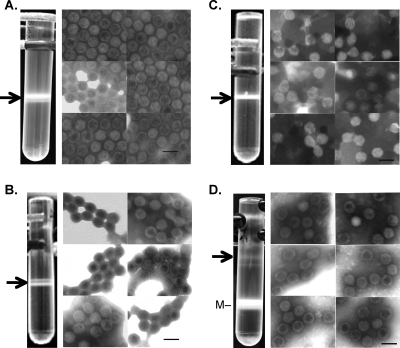
The noncomplementing parental cell line N52.E6-Cre was infected with B box mutant virus, and virus production was analyzed using cesium chloride gradients. Figure 3A shows the results of the CsCl equilibrium gradient with lysates from cells infected with the B box mutant virus. Two bands that migrated too close together for separation were observed; therefore, the bands were pooled for further analysis. The ratio of the two bands varied from experiment to experiment. The material banded at a density of 1.29 to 1.3 g/cm3 and will be referred to as empty particles. Empty particles, devoid of Ad DNA, from infection with wild-type virus have been shown to band at this density (22). No bands were observed lower in the gradient, where mature virus would be expected (1.34 g/cm3). EM images of negatively stained material from the gradient confirmed that virus particles had been assembled. The empty particles from the B box mutant resembled empty particles obtained with wild-type Ad5 (compare Fig. 3A and D). Southern blot analyses of DNA from B box mutant-infected cells showed that the replicated genomes retained the introduced mutations (data not shown). These results showed that ATP binding by the IVa2 protein, and the putative ATPase activity of IVa2, were not required for assembly of an empty virus particle. However, these activities are required to package viral DNA into virions.
Fig. 3.
CsCl equilibrium gradients and electron microscopy of IVa2 mutant viruses. Photographs of centrifuge tubes from CsCl equilibrium gradients from cells infected with B box mutant virus (A), ΔIVa2 mutant virus (B), pm8002 mutant virus (C), and wild-type Ad5 (D). Arrows indicate empty virus particles that were collected. The same number of cells was used for each virus infection. Shown to the right of the CsCl gradients are photographs of six representative images from EM analyses of negatively stained material from the accompanying viruses in panels A to D. (D) M corresponds to mature wild-type Ad5. Dark scale bar in the figure represents 100 nm. Magnification is ×49,000.
A surprising finding, however, was the observation that the IVa2 deletion mutant also produced empty virus particles. Similar to the results with the B box mutant, the mutant virus ΔIVa2 produced empty virus particles that banded in CsCl gradients at a density of 1.29 to 1.3 g/cm3 (Fig. 3B). No other material was seen lower in the gradient at the density of mature virus. EM images of negatively stained material showed that the ΔIVa2 mutant assembled empty virus particles (Fig. 3B). Southern blot analyses of the DNA from ΔIVa2-infected cells showed that the replicated genomes retained the introduced deletion (data not shown). These results raised the possibility that the remaining part of the IVa2 gene in ΔIVa2 (the N-terminal 84 amino acids) could play a role in assembly of empty virus particles. The Walker A and B box domains are located downstream of this region (Fig. 1) and would not be present if this truncated protein was made. A pm8002 virus stock was generated and used to infect N52.E6-Cre cells. As was seen with the B box and ΔIVa2 mutant viruses, material was observed in the equilibrium gradient banding at a density of 1.29 to 1.3 g/cm3, consistent with empty virus particles (Fig. 3C). This result was surprising, since pm8002 previously was reported not to produce empty virus particles (49). Virus particles were present in this material, as determined by EM (Fig. 3C). Southern blot analyses of DNA from the pm8002-infected cells showed that the replicated genomes retained the introduced mutations (data not shown).
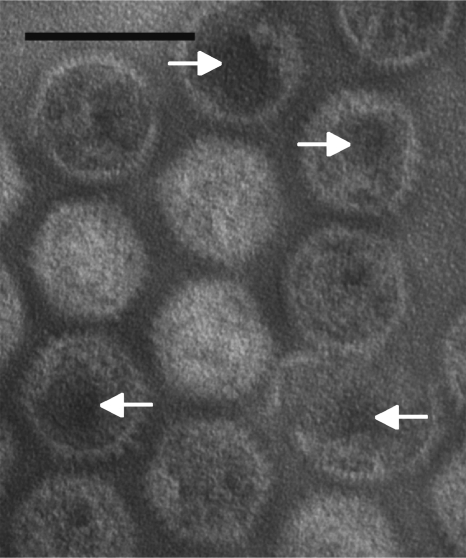
In all cases in the EM images, some of the empty particles appeared to be degraded as exemplified by the results with the B box mutant particles shown in Fig. 4. It is unclear whether the particles were degraded before EM or had degraded during the process of fixing and staining the particles. These results suggest that the empty virus particles are more fragile than mature particles containing DNA, since images from negative staining and EM of mature particles appeared more uniform (data not shown).
Fig. 4.
EM analysis of B box mutant empty virus particles. Photograph of an EM analysis of negatively stained B box mutant empty virus particles at ×180,000 magnification. Arrows point to areas of particles showing disintegration. Dark scale bar in figure represents 100 nm.
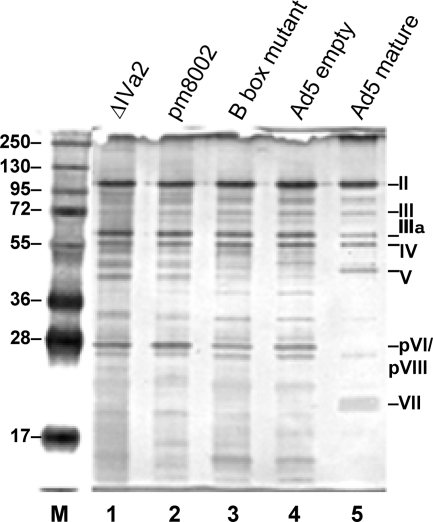
The virus particles obtained from CsCl equilibrium gradients were further characterized by SDS-polyacrylamide gel electrophoresis followed by silver staining and Western blot analysis and for viral DNA content. The results are shown in Fig. 5 and 6. Protein profiles of the empty particles from the three mutant viruses were compared to the profiles from Ad5 mature and empty particles. Figure 5, lane 5, shows the silver stain profile of mature Ad5 virus particles isolated on CsCl gradients. The identities of viral proteins based on molecular weight are indicated, including the major structural proteins: polypeptide II (hexon), III (penton), IV (fiber), and IIIa. Also identifiable are the core proteins, V and VII, the latter being the processed form of pVII. Figure 5, lane 4, shows the profile of the empty particles from wild-type Ad5 isolated from CsCl gradients. The major polypeptides are visible and, as expected, the empty particles lack proteins V and VII and its precursor pVII (6). The empty particles from ΔIVa2, pm8002, and the B box mutant (Fig. 4, lanes 1, 2, and 3, respectively) had profiles very similar to that of the empty particles of wild-type Ad5 with respect to the major polypeptides. In addition, the precursor proteins, pVI and pVIII, were observed with all of the empty particles. The presence of the precursor proteins is consistent with the particles being immature, and the density of the particles is consistent with their being empty virus particles. Subtle differences of unknown origin were evident with the different empty virus particles.
Fig. 5.
Analysis of proteins in empty particles of IVa2 mutant viruses. Proteins from empty particles obtained from CsCl equilibrium gradients were analyzed by SDS-PAGE and silver staining. Lane 1, ΔIVa2 empty particles; lane 2, pm8002 empty particles; lane 3, B box mutant empty particles; lane 4, wild-type Ad5 empty particles; lane 5, wild-type Ad5 mature particles. Numbers on the right indicate polypeptides of virus particles and their designations. M, molecular mass marker proteins; molecular masses (in kilodaltons) are indicated on the left.
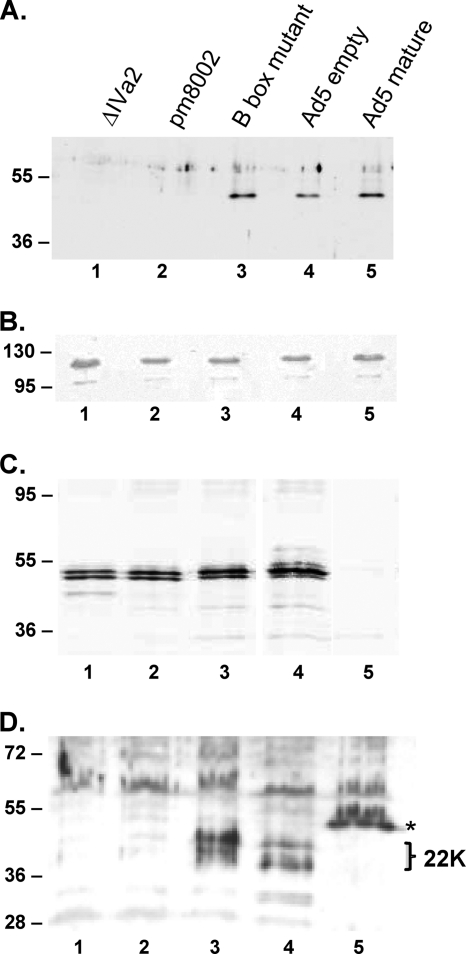
Virus particles were examined for the presence of the IVa2, L1-52/55K, and L4-22K packaging proteins. It had been shown previously that IVa2 is found in mature virus particles (49) but that the L1-52/55K protein is not (11, 15). The proteins from wild-type and mutant infections were separated on SDS-polyacrylamide gels and analyzed by Western blotting. Results in Fig. 6A show that wild-type empty particles contained IVa2 protein (lane 4) as did the mature virus particles (lane 5). In addition, the B box mutant particles also contained the IVa2 protein (lane 3). These results demonstrate that the binding of ATP by IVa2 is not a requirement for its interaction with the virus particle. Both ΔIVa2 and pm8002 empty particles lacked the IVa2 protein, as expected, further confirming that IVa2 is not required for empty virus particle assembly. As a control, virus particles were also Western blotted for hexon protein, and the results in Fig. 6B show similar signals for the different virus particles. Therefore, the relative ratio of IVa2 to hexon in B box mutant empty virus particles appears similar to that of the wild-type virus. Empty virus particles produced by each of the IVa2 mutant viruses contained the L1-52/55K protein (Fig. 6C). In contrast, empty virus particles produced by the two IVa2 null viruses did not contain the L4-22K protein, whereas the IVa2 B box mutant empty particles and wild-type Ad5 empty particles did contain the L4-22K protein (Fig. 6D). Further, like the L1-52/55K protein, the L4-22K protein was absent from wild-type Ad5 mature virus particles.
Fig. 6.
Analysis of viral proteins present in empty particles of IVa2 mutant viruses. Western blot analyses with anti-IVa2 antibody (A), anti-hexon antibody (B), anti-L1-52/55K antibody (C), and anti-L4-22K antibody (D) of proteins in particles isolated by CsCl equilibrium gradient centrifugation. Lane 1, ΔIVa2 empty particles; lane 2, pm8002 empty particles; lane 3, B box mutant empty particles; lane 4, wild-type Ad5 empty particles; lane 5, wild-type Ad5 mature particles. Molecular masses (in kilodaltons) are indicated on the left. (D) The L4-22K protein (22K) appears as a doublet. The asterisk indicates a viral protein in mature particles that cross-reacts with the antibody and corresponds in migration to core protein V (note its absence in empty virus particles).
Finally, we determined by Southern blotting whether empty virus particles produced by these IVa2 mutant viruses contain viral DNA. As a positive control, we isolated empty virus particles from cells infected at the restrictive temperature with the temperature-sensitive mutant virus ts369. ts369 produces a temperature-sensitive L1-52/55K protein, and empty virus particles isolated from cells infected at the restrictive temperature with this mutant contain Ad5 left-end DNA sequences that correspond to the packaging domain and adjacent sequences (15). Small viral DNA was readily apparent in ts369 empty virus particles; in contrast, no detectable viral DNA was evident in empty virus particles produced by each of the IVa2 mutant viruses (data not shown). These results indicate that the particle heterogeneity observed with the IVa2 mutant viruses on CsCl gradients (Fig. 3) does not represent the presence or absence of left-end viral DNA sequences.
DISCUSSION
The Ad5 IVa2 protein is essential for the packaging of the Ad genome into its capsid. We have demonstrated that the IVa2 protein is not required for assembly of an empty viral capsid. This result is opposite to what was previously published (49). In our experiments, empty particles were obtained from infections with mutant viruses containing either a deletion of a significant portion of the IVa2 coding region or containing stop codons that disrupted the expression of the full-length protein (Fig. 3). The replicated genomes that accumulated during these infections were pure mutant genomes as determined by Southern blot analyses; therefore, the accumulation of empty particles was not due to reversion of the mutations. A question arises: why is there a difference between our results and those in the previously published report? The appearance of empty particles is not due to the difference in cell lines used in the present report compared with those used in the previous report. N52.E6-Cre cells were used for the experiments presented here, but infections of 293 cells, the cell type previously used (49) yielded the same results (293 cells maintained in the Hearing laboratory as well as the Imperiale laboratory were tested; data not shown). We believe that the most likely explanation for these differences is the way the virus particles were processed. The Imperiale laboratory lysed infected cells by three cycles of freezing and thawing as well as by sonication (M. Imperiale, personal communication). In our studies, no sonication step was employed. As suggested by the images in Fig. 3 and 4, we believe that the empty virus particles produced by IVa2 mutant viruses are fragile, perhaps disrupted by sonication. We also examined virus production at a later time point (72 h) than in the previously published report (48 h) (49). Finally, we note that the Imperiale laboratory was able to detect empty particles in pm8002 infections when our IVa2 complementing cell line was used to produce the mutant virus stock (M. Imperiale, personal communication). Perhaps this reflects the improved virus yield and P/PFU ratio using IVa2 mutant viruses produced using the N52.E6-Cre–IVa2 cell line.
Utilizing the B box mutant virus, we have determined that preventing ATP binding by the IVa2 protein, and likely inhibiting the putative IVa2 ATPase activity, had no detrimental effect on the assembly of empty Ad virus particles (Fig. 3). Although we did not quantify the amount of empty particles/cell obtained in these infections, the results from the CsCl equilibrium gradients showed that a significant amount of empty particles accumulated. Relatively small amounts of empty particles compared to those of mature particles accumulated during the course of a wild-type Ad5 infection. Previous data implied a precursor/product relationship between the empty and mature capsids (41). If the empty particles we observed are dead-end products rather than procapsids in packaging, this would not negate the fact that assembly occurred in the absence of IVa2 protein. The major effect of preventing ATP binding by the IVa2 protein was the inability of the mutant virus to package DNA. Therefore, it seems likely that IVa2 is an ATPase and that this activity is essential for the packaging of the Ad genome into the capsid. This is consistent with previously published results that localize the IVa2 protein at a unique capsid vertex (3), a hallmark of bacteriophage packaging motors (35).
It is interesting that the IVa2 protein is found in association with the empty particles isolated from infections with wild-type Ad5 and with the B box mutant virus (Fig. 6). This association may be mediated by the L1-52/55K protein which is found in empty Ad particles, since it was previously shown that these proteins interact (12) and L1-52/55K plays a important role in packaging (11, 15). There is no significant difference in the relative amounts of IVa2 protein found in mature Ad5 virus particles and the amounts in the empty particles from the B box mutant virus. At this level of analysis, we could not rule out small changes that might be significant. However, if indeed the empty particles observed are procapsids, then either there are different populations of IVa2, one on the particle and one on Ad DNA-packaging sequences, or the specific selection of Ad DNA by IVa2 occurs at the entry site for the DNA on the procapsid. That empty particles assemble in the absence of IVa2 suggests the possibility that IVa2 associates with capsids and viral DNA late in the assembly process. The empty virus particles lacking IVa2 contained the L1-52/55K protein but not the L4-22K protein (Fig. 6). The IVa2 protein is required for the L4-22K protein to bind to packaging sequences in vitro (8, 26), and the current results suggest that an interaction between these two proteins also is required for the L4-22K protein to enter empty capsids. In contrast, IVa2 B box mutant empty capsids did contain the L4-22K protein, suggesting that ATP binding by IVa2 is not required for such an interaction to occur. Interestingly, wild-type Ad5 mature virus particles lacked the L4-22K protein as previously described for the L1-52/55K protein (11, 15). Ad5 mature virus particles contained the IVa2 protein, as previously reported (49). That the L1-52/55K and L4-22K proteins appear to exit the particle coordinately during capsid maturation may reflect their direct interaction. The L1-52/55K protein associates with the packaging domain in vivo as measured by chromatin immunoprecipitation assays (31, 34), but the IVa2 protein is not required for this interaction (34). Perhaps the L1-52/55K protein is tethered to the packaging domain via interaction with the L4-22K protein.
Detailed analyses of the motor function in the packaging process in bacteriophage phi29 has shown that coordinated, sequential hydrolyzes of four ATPs by four adjacent ATPases leads to the movement of 10 bp of DNA into the procapsid (35). Addition of nonhydrolyzable ATP stalls the motor for the duration of the substrate binding, and stalling occurs even if only one ATPase subunit was inhibited for hydrolysis (24, 47). In contrast to these results, the protease ClpX is a hexameric ATPase that is part of a protease and does not appear to function by a sequential mechanism (23). It was suggested that ATP hydrolysis by ClpX hexamer is probabilistic and that this flexibility may be related to its interaction with a substrate that is not uniform in structure. The fact that we were able to propagate the Ad5 B box mutant in an IVa2 complementing cell line shows that the mutant protein did not act as a dominant negative effector. If the IVa2 protein is an ATPase, then we suggest that the packaging motor may not function in a sequential manner. Interestingly, the molecular mechanism of packaging of the Ad genome may be different from that of the bacteriophage, since Ad DNA is found associated with the core proteins pVII, V, and mu. It is still unknown whether the appearance of these core proteins in an Ad particle occurs concomitant with or after the packaging of viral DNA. In either case, the packaging ATPase for Ad may require more flexibility in providing a power stroke for moving substrates that are not just a double helix of DNA but also have bound protein. It is worth noting that the IVa2 protein has been classified as a member of the p loop, ASCE ATPases by sequence comparisons, but within this large group, it is distinct from the bacteriophage packaging motors and more similar to the ATP-binding cassette (ABC) transporter ATPases (1). Mechanistic questions regarding the role of IVa2 in Ad packaging can be approached using the complementation system that we have developed.
ACKNOWLEDGMENTS
We thank Gudrin G. Schiedner and S. Kochanek (University of Ulm) for the N52.E6-Cre cell line and Kasey Karen for contributions to the development of the Tet-inducible derivative of this cell line. We thank Michael Imperiale and Nicolas Nassar and members of our laboratory for informed discussions. We thank Ilana Shoshani for excellent technical help. We thank Susan Van Horn and the Central Microscopy Imaging Center at Stony Brook University for performing the electron microscopy studies.
This work was supported by NIH grant AI041636.
Footnotes
Published ahead of print on 30 March 2011.
REFERENCES
- 1. Burroughs A. M., Iyer L. M., Aravind L. 2007. Comparative genomics and evolutionary trajectories of viral ATP dependent DNA-packaging systems. Genome Dyn. 3:48–65 [DOI] [PubMed] [Google Scholar]
- 2. Chartier C., et al. 1996. Efficient generation of recombinant adenovirus vectors by homologous recombination in Escherichia coli. J. Virol. 70:4805–4810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Christensen J. B., et al. 2008. Presence of the adenovirus IVa2 protein at a single vertex of the mature virion. J. Virol. 82:9086–9093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Colby W. W., Shenk T. 1981. Adenovirus type 5 virions can be assembled in vivo in the absence of detectable polypeptide IX. J. Virol. 39:977–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Daniell E., Mullenbach T. 1978. Synthesis of defective viral DNA in HeLa cells infected with adenovirus type 3. J. Virol. 26:61–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Edvardsson B., Everitt E., Jornvall H., Prage L., Philipson L. 1976. Intermediates in adenovirus assembly. J. Virol. 19:533–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Evans J. D., Hearing P. 2003. Distinct roles of the adenovirus E4 ORF3 protein in viral DNA replication and inhibition of genome concatenation. J. Virol. 77:5295–5304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ewing S. G., Byrd S. A., Christensen J. B., Tyler R. E., Imperiale M. J. 2007. Ternary complex formation on the adenovirus packaging sequence by the IVa2 and L4 22-kilodalton proteins. J. Virol. 81:12450–12457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ghosh-Choudhury G., Haj-Ahmad Y., Graham F. L. 1987. Protein IX, a minor component of the human adenovirus capsid, is essential for the packaging of full length genomes. EMBO J. 6:1733–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Grable M., Hearing P. 1990. Adenovirus type 5 packaging domain is composed of a repeated element that is functionally redundant. J. Virol. 64:2047–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gustin K. E., Imperiale M. J. 1998. Encapsidation of viral DNA requires the adenovirus L1 52/55-kilodalton protein. J. Virol. 72:7860–7870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gustin K. E., Lutz P., Imperiale M. J. 1996. Interaction of the adenovirus L1 52/55-kilodalton protein with the IVa2 gene product during infection. J. Virol. 70:6463–6467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hammarskjold M. L., Winberg G. 1980. Encapsidation of adenovirus 16 DNA is directed by a small DNA sequence at the left end of the genome. Cell 20:787–795 [DOI] [PubMed] [Google Scholar]
- 14. Hannan C., Raptis L. H., Dery C. V., Weber J. 1983. Biological and structural studies with an adenovirus type 2 temperature-sensitive mutant defective for uncoating. Intervirology 19:213–223 [DOI] [PubMed] [Google Scholar]
- 15. Hasson T. B., Soloway P. D., Ornelles D. A., Doerfler W., Shenk T. 1989. Adenovirus L1 52- and 55-kilodalton proteins are required for assembly of virions. J. Virol. 63:3612–3621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hearing P., Samulski R. J., Wishart W. L., Shenk T. 1987. Identification of a repeated sequence element required for efficient encapsidation of the adenovirus type 5 chromosome. J. Virol. 61:2555–2558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hirt B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365–369 [DOI] [PubMed] [Google Scholar]
- 18. Imelli N., Ruzsics Z., Puntener D., Gastaldelli M., Greber U. F. 2009. Genetic reconstitution of the human adenovirus type 2 temperature-sensitive 1 mutant defective in endosomal escape. Virol. J. 6:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ishibashi M., Maizel J. V., Jr 1974. The polypeptides of adenovirus. V. Young virions, structural intermediate between top components and aged virions. Virology 57:409–424 [DOI] [PubMed] [Google Scholar]
- 20. Koonin E. V., Senkevich T. G., Chernos V. I. 1993. Gene A32 product of vaccinia virus may be an ATPase involved in viral DNA packaging as indicated by sequence comparisons with other putative viral ATPases. Virus Genes 7:89–94 [DOI] [PubMed] [Google Scholar]
- 21. Liu H., et al. 2010. Atomic structure of human adenovirus by cryo-EM reveals interactions among protein networks. Science 329:1038–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Maizel J. V., Jr., White D. O., Scharff M. D. 1968. The polypeptides of adenovirus. II. Soluble proteins, cores, top components and the structure of the virion. Virol 36:126–136 [DOI] [PubMed] [Google Scholar]
- 23. Martin A., Baker T. A., Sauer R. T. 2005. Rebuilt AAA + motors reveal operating principles for ATP-fuelled machines. Nature 437:1115–1120 [DOI] [PubMed] [Google Scholar]
- 24. Moffitt J. R., et al. 2009. Intersubunit coordination in a homomeric ring ATPase. Nature 457:446–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Morrissey J. H. 1981. Silver stain for proteins in polyacrylamide gels: a modified procedure with enhanced uniform sensitivity. Anal. Biochem. 117:307–310 [DOI] [PubMed] [Google Scholar]
- 26. Ostapchuk P., Anderson M. E., Chandrasekhar S., Hearing P. 2006. The L4 22-kilodalton protein plays a role in packaging of the adenovirus genome. J. Virol. 80:6973–6981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ostapchuk P., Hearing P. 2008. Adenovirus IVa2 protein binds ATP. J. Virol. 82:10290–10294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ostapchuk P., Hearing P. 2005. Control of adenovirus packaging. J. Cell. Biochem. 96:25–35 [DOI] [PubMed] [Google Scholar]
- 29. Ostapchuk P., Hearing P. 2003. Minimal cis-acting elements required for adenovirus genome packaging. J. Virol. 77:5127–5135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ostapchuk P., Hearing P. 2001. Pseudopackaging of adenovirus type 5 genomes into capsids containing the hexon proteins of adenovirus serotypes B, D, or E. J. Virol. 75:45–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ostapchuk P., Yang J., Auffarth E., Hearing P. 2005. Functional interaction of the adenovirus IVa2 protein with adenovirus type 5 packaging sequences. J. Virol. 79:2831–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pardo-Mateos A., Young C. S. 2004. A 40 kDa isoform of the type 5 adenovirus IVa2 protein is sufficient for virus viability. Virology 324:151–164 [DOI] [PubMed] [Google Scholar]
- 33. Parks R. J., et al. 1996. A helper-dependent adenovirus vector system: removal of helper virus by Cre-mediated excision of the viral packaging signal. Proc. Nat. Acad. Sci. U. S. A. 93:13565–13570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Perez-Romero P., Tyler R. E., Abend J. R., Dus M., Imperiale M. J. 2005. Analysis of the interaction of the adenovirus L1 52/55-kilodalton and IVa2 proteins with the packaging sequence in vivo and in vitro. J. Virol. 79:2366–2374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rao V. B., Feiss M. 2008. The bacteriophage DNA packaging motor. Annu. Rev. Genet. 42:647–681 [DOI] [PubMed] [Google Scholar]
- 36. Reddy V. S., Natchiar S. K., Stewart P. L., Nemerow G. R. 2010. Crystal structure of human adenovirus at 3.5 Å resolution. Science 329:1071–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. San Martin C., et al. 2008. Localization of the N-terminus of minor coat protein IIIa in the adenovirus capsid. J. Mol. Biol. 383:923–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sato K., Hosokawa K. 1984. Analysis of the interaction between DNA and major core protein in adenovirus chromatin by circular dichroism and ultraviolet light induced cross-linking. J. Biochem. 95:1031–1039 [DOI] [PubMed] [Google Scholar]
- 39. Schiedner G., Hertel S., Kochanek S. 2000. Efficient transformation of primary human amniocytes by E1 functions of Ad5: generation of new cell lines for adenoviral vector production. Hum. Gene Ther. 11:2105–2116 [DOI] [PubMed] [Google Scholar]
- 40. Sun S., Rao V. B., Rossmann M. G. 2010. Genome packaging in viruses. Curr. Opin. Struct. Biol. 20:114–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sundquist B., Everitt E., Philipson L., Hoglund S. 1973. Assembly of adenoviruses. J. Virol. 11:449–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tibbetts C. 1977. Viral DNA sequences from incomplete particles of human adenovirus type 7. Cell 12:243–249 [DOI] [PubMed] [Google Scholar]
- 43. Tyler R. E., Ewing S. G., Imperiale M. J. 2007. Formation of a multiple protein complex on the adenovirus packaging sequence by the IVa2 protein. J. Virol. 81:3447–3454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vayda M. E., Rogers A. E., Flint S. J. 1983. The structure of nucleoprotein cores released from adenovirions. Nucleic Acids Res. 11:441–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yang T. C., Yang Q., Maluf N. K. 2009. Interaction of the adenoviral IVa2 protein with a truncated viral DNA packaging sequence. Biophys. Chem. 140:78–90 [DOI] [PubMed] [Google Scholar]
- 46. Yoshida M., Amano T. 1995. A common topology of proteins catalyzing ATP-triggered reactions. FEBS Lett. 359:1–5 [DOI] [PubMed] [Google Scholar]
- 47. Yu J., Moffitt J., Hetherington C. L., Bustamante C., Oster G. 2010. Mechanochemistry of a viral DNA packaging motor. J. Mol. Biol. 400:186–203 [DOI] [PubMed] [Google Scholar]
- 48. Zhang W., Imperiale M. J. 2000. Interaction of the adenovirus IVa2 protein with viral packaging sequences. J. Virol. 74:2687–2693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang W., Imperiale M. J. 2003. Requirement of the adenovirus IVa2 protein for virus assembly. J. Virol. 77:3586–3594 [DOI] [PMC free article] [PubMed] [Google Scholar]