Abstract
Ebola virus (EBOV), an enveloped, single-stranded, negative-sense RNA virus, causes severe hemorrhagic fever in humans and nonhuman primates. The EBOV glycoprotein (GP) gene encodes the nonstructural soluble glycoprotein (sGP) but also produces the transmembrane glycoprotein (GP1,2) through transcriptional editing. A third GP gene product, a small soluble glycoprotein (ssGP), has long been postulated to be produced also as a result of transcriptional editing. To identify and characterize the expression of this new EBOV protein, we first analyzed the relative ratio of GP gene-derived transcripts produced during infection in vitro (in Vero E6 cells or Huh7 cells) and in vivo (in mice). The average percentages of transcripts encoding sGP, GP1,2, and ssGP were approximately 70, 25, and 5%, respectively, indicating that ssGP transcripts are indeed produced via transcriptional editing. N-terminal sequence similarity with sGP, the absence of distinguishing antibodies, and the abundance of sGP made it difficult to identify ssGP through conventional methodology. Optimized 2-dimensional (2D) gel electrophoresis analyses finally verified the expression and secretion of ssGP in tissue culture during EBOV infection. Biochemical analysis of recombinant ssGP characterized this protein as a disulfide-linked homodimer that was exclusively N glycosylated. In conclusion, we have identified and characterized a new EBOV nonstructural glycoprotein, which is expressed as a result of transcriptional editing of the GP gene. While ssGP appears to share similar structural properties with sGP, it does not appear to have the same anti-inflammatory function on endothelial cells as sGP.
INTRODUCTION
Zaire ebolavirus (ZEBOV) is the type species of the genus Ebolavirus in the family Filoviridae (11) and a causative agent of a severe hemorrhagic fever in primates, with case fatality rates as high as 90% in humans (12). All Ebola viruses (EBOV) possess a nonsegmented, negative-sense RNA genome with seven linear genes that encode seven structural proteins. The nucleoprotein (NP), virion protein 30 (VP30) and VP35, and RNA-dependent RNA polymerase (L) are components of the nucleocapsid structures, which are the active transcription/replication complexes. VP24, VP40, and the transmembrane glycoprotein (GP1,2) are associated with the viral membrane. GP1,2 is the only surface protein and forms trimeric spikes that facilitate virus entry by receptor binding and fusion with target cells (39).
EBOV undergoes site-specific transcriptional editing of the glycoprotein (GP) gene (see Fig. 1A), comparable to the RNA editing that is commonly observed in the phosphoprotein (P) genes of viruses from the family Paramyxoviridae (16, 24). The primary product of the GP gene is the soluble glycoprotein (sGP), a nonstructural secreted glycoprotein, which is expressed from unedited RNA transcripts (40, 51). GP1,2 is expressed only following transcriptional editing, which occurs at a series of seven uridine residues within the genomic RNA, resulting in an additional adenosine (A) residue in the transcript (40, 51). The subsequent +1 shift results in an extended open reading frame (ORF). The expression of an additional, as yet unidentified nonstructural protein, designated small soluble glycoprotein (ssGP), has long been proposed. This product would occur as a result of transcriptional editing that leads to a +2 shift, resulting in a truncated ORF. Thus, all GP gene products have identical N-terminal primary sequences of 295 amino acids (aa) but differ at their C-terminal portions following the transcriptional editing site (14, 39) (Fig. 1A).
Fig. 1.
Ebola virus glycoprotein gene RNA editing results in multiple gene products. (A) Organization of the Ebola virus glycoprotein gene. (Top) Putative open reading frames (ORFs) for the different GP gene products (sGP, GP1,2, and ssGP). (Bottom) The primary structures of glycoprotein gene products are shown in an alignment of the primary amino acid sequences of sGP, GP1,2, and ssGP. All three proteins share the first 295 aa, including the signal peptide (aa 1 to 32), but differ in their carboxy-terminal portions. (B) Detection of ssGP transcripts in vitro and in vivo. Vero E6 and Huh7 cells were infected with ZEBOV, and mice were infected with MA-ZEBOV. RNA was extracted from infected cell cultures, virus particles, and mouse liver. The editing site region for mRNA and vRNA was amplified (330-bp fragment), cloned, and sequenced as described in Materials and Methods. In total, we analyzed 224 clones from Vero E6 cells, 132 from Huh7 cells, 260 from mouse liver, and 109 from viral particles. (C) Specificity of transcriptional editing. The vast majority of clones from mRNA (97%) contained either 7 (sGP), 8 (GP1,2), or 9 (ssGP) adenosine (A) residues. The remaining 3% of clones analyzed contained multiple adenosine residues in the editing site, which would lead to the incorporation of additional lysine residues in the primary amino acid sequence. With the rare exception of a single adenosine deletion (in clones encoding ssGP), no deletions of adenosine residues were found in the editing site.
Most of the published molecular work on EBOV has used ZEBOV strain Mayinga. Here we identified ssGP transcripts generated during in vitro and in vivo ZEBOV replication and demonstrated that ssGP was expressed and secreted during infection. In addition, we determined that transcriptional editing was the mechanism of expression. Thus, we have identified a new EBOV nonstructural glycoprotein. Biochemical and structural characterization identified ssGP as a secreted homodimer containing N-linked carbohydrates. Therefore, ssGP appears to have biochemical and structural features similar to those of sGP (1, 2, 10); however, ssGP did not rescue barrier function following treatment of endothelial cells with tumor necrosis factor alpha (TNF-α)—a function that has been described previously for sGP (56). This result suggests that these proteins do not share biological functions.
(This work is part of the Ph.D. thesis of M. Mehedi, Department of Medical Microbiology, University of Manitoba, Winnipeg, Canada.)
MATERIALS AND METHODS
Virus, cell culture, and animals.
ZEBOV strain Mayinga (GenBank accession no. AF086833) was used for all in vitro infectious experiments. Vero E6 (a monkey kidney cell line), Huh7 (a human liver cell line), and 293T (a human embryonic kidney cell line) cells were cultured in Dulbecco's minimal essential medium (DMEM) (Sigma-Aldrich, St. Louis, MO) supplemented with 10% fetal bovine serum (FBS) (Wisent, Saint-Jean-Baptiste, Quebec, Canada) under 5% CO2 at 37°C. BALB/c mice (Charles River, MA) were infected with mouse-adapted ZEBOV (MA-ZEBOV) strain Mayinga (GenBank accession no. AF499101) (4).
In vitro ZEBOV infection for transcript analysis.
Approximately 90% confluent Vero E6 or Huh7 cells were infected with ZEBOV at a multiplicity of infection (MOI) of 1 and were maintained in Opti-MEM (Invitrogen, CA). The supernatant was removed, and total cellular RNA was extracted at 4 days postinfection using Trizol LS reagent (Invitrogen, CA) according to the manufacturer's instructions. ZEBOV GP cDNA was produced from extracted RNA using a GP-specific oligo(dT) primer (ACCGGTTTTTTTTTTTTTTT) with SuperScript III reverse transcriptase (Invitrogen, CA). Subsequently, a fragment covering the editing site of the GP gene was amplified using sequence-specific primers (forward, ACACCACAGTTTCTGCTCCAGC; reverse, TGACTGTGCACTTGAACCATTGC) and high-fidelity Taq DNA polymerase (Roche, Mannheim, Germany). The amplification product (330 bp) was PCR purified using the QIAquick PCR purification kit (Qiagen, CA) and was cloned into the TOPO TA 2.1 PCR cloning vector (Invitrogen, CA) using chemically competent One Shot Top10 Escherichia coli cells and superoptimal broth with catabolite repression (SOC) medium (Invitrogen, CA). Subsequently, bacteria were plated onto LB agar containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal; Invitrogen, CA) for blue-white colony screening, followed by sequencing of bacterial clones to determine the ratios of GP gene-specific transcripts encoding sGP, GP1,2, and ssGP. A similar approach was applied to viral genomic RNA (vRNA) derived from ZEBOV particles harvested from Vero E6 cell supernatants at day 4 postinfection. A vRNA-specific primer (AGAGTAGGGGTCGTCAGGTCC) binding upstream of the GP gene was used to generate cDNA, followed by amplification of the GP gene editing region (330 bp) using the primers described above to determine the ratio of distinct vRNA in viral particles. In addition, a synthetic positive-sense RNA template (AUAGAAUUCUCGGGGAGUGGGCCUUCUGGGAAACUAAAAAAACCUCACUAGAAAAAUUCGCAGUGAAGAGUUGUCGAAUUCCGU) (Thermo Fisher Scientific, MA) was used to generate clones by the same procedure to control for Taq DNA polymerase errors as the source for the introduction of multiple adenosine residues.
In vivo MA-ZEBOV infection for transcript analysis.
BALB/c mice were infected intraperitoneally with 1,000 LD50 (1,000 times the 50% lethal dose [dose leading to death in 50% of the animals]) of MA-ZEBOV. Mice were euthanized 4 days postinfection, and liver samples were collected. RNA was extracted from liver tissue with the RNeasy kit (Qiagen, CA) according to the manufacturer's instructions. To determine the ratio of the GP gene transcripts, we applied the strategy described above for the in vitro procedure.
Expression of recombinant proteins (r.ssGP and r.sGP).
The coding sequence for ZEBOV ssGP lacking the signal peptide (SP) was amplified by reverse transcription-PCR (RT-PCR) from purified vRNA using specific primers (forward, AGCCGGCCAGATCTATCCCACTTGG; reverse, GCCTGCAGTTAACTAGTGAGGTTTTTTTTTAGTTTCCCAGAAGG) (Eurofins MWG Operon, Huntsville, AL). The amplification product was cloned into the eukaryotic expression vector pDisplay (Invitrogen, CA) using BglII and PstI restriction sites. This vector directs the expression of foreign glycoproteins through a vector-specific SP and adds an N-terminal hemagglutinin (HA) tag. We have published previously on a similar pDisplay-based recombinant sGP (r.sGP) expression plasmid (55). Clones that express r.ssGP and r.sGP with the authentic SP and no HA tag were also produced by PCR amplification using specific primers (common forward primer, ATCGGAATTCATGGGCGTTACAGG; reverse primers, ATCTCGAGTTACTAGTGAGGTTTTTTTTTAGTTTCCCAG for r.ssGP and TTCTCGAGTTAGCGCCGGACTCTGAC for r.sGP). The PCR-amplified products were cloned into the eukaryotic expression vector pCAGGS (30) using EcoRI and XhoI restriction sites. All constructs were sequenced at the DNA Core facility at the National Microbiology Laboratory (Winnipeg, Manitoba, Canada). r.ssGP and r.sGP were expressed following transient transfection of pDisplay-HAtag-sGP/ssGP into 293T cells and of pCAGGS-sGP/ssGP into Vero E6 cells using FuGENE 6 (Roche, Mannheim, Germany). Supernatants were collected 72 h posttransfection, and HA-tagged proteins were subsequently purified using an anti-HA affinity column (Roche, Mannheim, Germany) according to the manufacturer's instructions. Proteins were concentrated with Amicon Ultra centrifugal filters (Millipore, MA) with a molecular weight (MW) cutoff of 30,000. The final concentration of proteins was determined by the DC protein assay (Bio-Rad, CA). All proteins were aliquoted and were stored at −20°C. The expression of r.sGP and r.ssGP was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) under reducing (addition of β-mercaptoethanol) and nonreducing conditions and was verified by immunoblotting following transfer to polyvinylidene difluoride (PVDF) membranes. PVDF membranes were blocked overnight with 5% skim milk in phosphate-buffered saline (PBS). For HA-tagged proteins (r.ssGP and r.sGP), membranes were incubated for 1 h at room temperature with a 1/10,000 dilution of a peroxidase-conjugated rat antibody against HA (anti-HA-peroxidase) (Roche, Mannheim, Germany). For untagged proteins (r.ssGP and r.sGP), membranes were incubated with a 1/10,000 dilution of mouse monoclonal antibody (MAb) 42/3.7 (against GP1,2, sGP, and ssGP) (kindly provided by Ayato Takada, Hokkaido University, Sapporo, Japan), followed by incubation with a 1/10,000 dilution of goat anti-mouse IgG(H+L) (Kirkegaard & Perry Laboratories [KPL], Gaithersburg, MD). The ECL Plus Western blotting detection kit (GE Healthcare, Buckinghamshire, United Kingdom) was used to visualize protein bands.
Site-directed mutagenesis of ssGP.
The cysteine (Cys) residue in position 53 of ssGP was mutated into glycine (Gly) in pDisplay-HAtag-ssGP (see above) using a site-directed mutagenesis kit (Stratagene, CA) and the appropriate mutagenesis primers (forward, GTCGACAAACTAGTTGGTCGTGACAAACTGTCATCC; reverse, GGATGACAGTTTGTCACGACCAACTAGTTTGTCGAC). Following sequence confirmation, the mutated r.ssGP was expressed in 293T cells and was analyzed as described above.
Glycosylation analysis of ssGP.
The glycosylation status of r.ssGP and r.sGP was determined using endoglycosidases. All N-linked glycans were removed by digestion with peptide N-glycosidase F (PNGase F) (New England Biolabs, MA), and high-mannose-type N-linked glycans on r.ssGP were removed by digestion with endoglycosidase H (Endo H) (New England Biolabs, MA). To remove O-linked glycans, r.ssGP was treated with O-glycanase (endo-α-N-acetylgalactosaminidase) (18) in combination with different exoglycosidases [sialidase A, β(1-4)-galactosidase, β-N-acetylglucosaminidase, and N-glycanase] (Prozyme, CA). Reactions were performed for 3 h according to the manufacturer's instructions, and results were analyzed by SDS-PAGE and immunoblotting as described above using the anti-HA-peroxidase antibody (Roche) or MAb 42/3.7 at a 1/10,000 dilution.
Detection of ssGP during viral infection.
Vero E6 cells were infected with ZEBOV (MOI, 1). Cell culture supernatants were collected 4 days postinfection and were clarified by centrifugation at 2,000 × g for 10 min at 4°C to remove cell debris, followed by centrifugation at 21,000 × g for 30 min at 4°C to remove viral particles. The clarified supernatant was collected and concentrated with Amicon Ultra centrifugal filters (MW cutoff, 30,000) (Millipore, MA). The concentrated supernatant was treated with SDS (final concentration, 1%) and was heat inactivated at 100°C for 10 min. The inactivated supernatant was treated overnight with PNGase F (New England Biolabs, MA) to remove all the N-linked carbohydrates, and proteins were subjected to 15% SDS-PAGE and were transferred to PVDF membranes. Immunoblotting was performed as described above.
2D gel electrophoresis.
Supernatants derived from r.ssGP- and r.sGP-expressing Vero E6 cells (transient transfection) and ZEBOV-infected Vero E6 cells were desalted and cleaned using a 2-dimensional (2D) Clean-Up kit (GE Healthcare, Buckinghamshire, United Kingdom) and were resuspended in 2D rehydration buffer {7 M urea, 2 M thiourea, 2% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS)}. Subsequently, 15 μg of each protein was loaded onto a 7-cm, pH 4 to 7 nonlinear (NL) immobilized pH gradient (IPG) strip (GE Healthcare, Buckinghamshire, United Kingdom) using the following treatment profile: 15 h of passive rehydration at 30 V, 0.2-kVh step up to 300 V, 0.3-kVh gradient to 1,000 V, 4.5-kVh gradient to 5,000 V, 3-kVh step up to 5,000 V. The IPG strip was equilibrated for 15 min in equilibration buffer (75 mM Tris-HCl [pH 8.8], 6 M urea, 30% glycerol, 2% SDS, 0.002% bromophenol blue) containing 1% (wt/vol) dithiothreitol (DTT) followed by 15 min in equilibration buffer containing 2.5% iodoacetamide (IAA). The IPG strip was applied to a 12% Bis-Tris gel (Invitrogen, CA). The gel was transferred to a PVDF membrane using the iBlot dry blotting system (Invitrogen, CA), and the immunoblot was probed with MAb 42/3.7 at a 1/10,000 dilution, followed by peroxidase-labeled goat anti-mouse IgG(H+L) (KPL, Gaithersburg, MD) at a 1/10,000 dilution. Detection was performed with the ECL Plus Western blotting detection kit (GE Healthcare).
Endothelial barrier function rescue.
Impedance spectroscopy was used to measure the transendothelial electrical resistance (TER) of a cultured endothelial cell monolayer. The TER predominantly reflects the changes in paracellular permeability (8, 33) with a greater sensitivity and higher time resolution than transwell filter systems. Human umbilical vein endothelial cells (HUVEC) were isolated and cultured as described previously (42). Once HUVEC were confluent, they were equilibrated for 2 h to establish a baseline TER prior to the addition of TNF-α (1 ng/ml) (R&D Systems, Minneapolis, MN) and ssGP or sGP (30 μg/ml) to cells. The TER was determined as described previously (43). Briefly, an alternating voltage was applied, and the impedance magnitude was measured at frequencies between 10 Hz and 1 MHz between the electrode area of the indium tin oxide slide and a counterelectrode. The TER was calculated from the resultant spectra (43). All electrical resistance data are presented as normalized to baseline resistance values (TER/TER0). TER data are shown as means ± standard errors. Data were compared by an unpaired t test. Values were considered to be statistically significant when P was <0.05.
Neutrophil binding assay.
Purified polymorphonuclear leukocytes (PMN) (99%) were isolated from venous blood of healthy donors in accordance with a protocol approved by the Institutional Review Board for Human Subjects, National Institute of Allergy and Infectious Diseases. PMN were resuspended in Roswell Park Memorial Institute (RMPI) medium with 10 mM HEPES. Purified PMN (106 cells) were incubated with 20 μg/ml of either purified HA-tagged sGP or ssGP for 30 min on ice. Cells were spun for 7 min at 456 × g at 4°C and were washed with ice-cold wash buffer (PBS with 2% goat serum). Cells were stained with 100 μl of a phycoerythrin-conjugated anti-HA (anti-HA-PE) (Miltenyi Biotec Inc., CA) antibody (1/10 dilution in wash buffer) for 30 min on ice. A mouse IgG1-PE antibody (Miltenyi Biotec Inc., Auburn, CA) was used as an isotype control. Subsequently, cells were washed twice in 250 μl ice-cold wash buffer and were analyzed by fluorescence-activated cell sorting (FACS).
Statement.
All work with infectious EBOV was performed in the “biosafety level 4” (BSL4) laboratory at the National Microbiology Laboratory of the Public Health Agency of Canada. All animal experiments were performed under an approved-animal-use document and according to the guidelines of the Canadian Council on Animal Care (CCAC).
RESULTS
ssGP RNA transcripts are produced during ZEBOV infection.
The ZEBOV GP gene undergoes site-specific transcriptional editing to produce GP1,2. The primary GP gene product, sGP, is produced from unedited RNA transcripts. The percentages of transcripts that encode sGP and GP1,2 were previously identified as approximately 80 and 20%, respectively (14, 39, 51). However, it is hypothesized that RNA editing produces another nonstructural glycoprotein, designated ssGP, the existence of which has not been verified (Fig. 1A).
As indirect evidence for ssGP expression, we first determined the ratios of different transcripts produced from the EBOV GP gene. For this purpose, we infected Vero E6 cells with ZEBOV, harvested RNA from infected cells, and transcribed the RNA into cDNA. Vero E6 cells were chosen because this is the cell line most widely used for the propagation of EBOV. A fragment of 330 bp covering the GP gene editing site was amplified by PCR, cloned into the TOPO TA vector, and subsequently sequenced. Using this strategy, we obtained 224 clones that differed from each other only by the number of adenosine residues present at the editing site of the GP gene. The percentages of RNA transcripts encoding sGP, GP1,2, and ssGP were determined to be 71, 24, and 5%, respectively (Fig. 1B). A similar percentage of ssGP-specific transcripts (4%) was also determined for ZEBOV-infected Huh7 cells (Fig. 1B). A liver cell line was chosen because the liver is one of the primary target organs for EBOV replication in humans and nonhuman primates. Next, we determined the ratio of transcripts from mice infected with MA-ZEBOV by using the same strategy. Total cellular RNA was extracted from infected mouse livers, and sequence determination of 260 clones revealed the percentages of RNA transcripts to be 67, 31, and 2% for sGP, GP1,2, and ssGP, respectively (Fig. 1B), results similar to those obtained in vitro.
To control for the incorporation of edited genomes into infectious ZEBOV particles, we utilized the same approach. Of 109 clones generated from vRNA derived from ZEBOV particles, the vast majority (94%) showed wild-type sequence at the editing site (7 uridine residues, vRNA sense). Genomes containing 8 uridine residues and thus directly encoding GP1,2 were found in 6% of the clones, a finding that was not unexpected, since such a virus was previously plaque purified and genetically engineered (38, 52). We did not identify vRNA genomes with uridine residues coding directly for ssGP (Fig. 1B), indicating that such viruses might be noninfectious.
Editing was largely a specific event, but in a small proportion of the clones analyzed (3%), we identified multiple additional adenosine residues in the GP-specific transcripts derived from in vitro and in vivo infection. The number of adenosine residues inserted ranged from 1 to 24, but interestingly, adenosine deletions were rare, and only transcripts with at least 6 adenosine residues were detected (Fig. 1C). Higher numbers of adenosine insertions were found particularly in RNA transcripts derived from ZEBOV-infected Vero E6 cells (Fig. 1C). To exclude the possibility that these additional adenosine residues were artificially introduced through polymerase stuttering during PCR amplification, we generated clones from a synthetic RNA using the same strategy and protocol, followed by sequence determination. No addition or deletion of adenosine or other nucleotide residues was observed, confirming that adenosine residues are introduced as a result of editing during viral transcription and not as a result of Taq DNA polymerase amplification (data not shown).
ssGP is secreted as a homodimer.
The GP gene products GP1,2 and sGP have identical N-terminal amino acid sequences (aa 1 to 295) but differ in their C-terminal portions and their structures (1, 2, 10, 14, 39). This is also expected for the hypothesized nonstructural ssGP, which theoretically should represent a truncated (266-aa) version of nonstructural sGP (292 aa) (Fig. 1A). All GP gene-specific products carry the same signal peptide (SP), and it is expected that they use the same processing pathway in undergoing similar co- and posttranslational modifications. As a result, nonstructural sGP and the putative nonstructural ssGP are expected to have similar molecular weights and biochemical properties.
For characterization and control purposes, we generated recombinant eukaryotic expression plasmids based on pDisplay and pCAGGS producing both nonstructural glycoproteins (r.ssGP and r.sGP) with or without an N-terminal HA tag. For detection purposes, we mainly used immunoblot analysis with MAb 42/3.7, a monoclonal antibody recognizing an epitope in the N-terminal portion of the proteins common to all GP gene products (47, 48), and we therefore expected to detect ssGP also. In addition, a commercial anti-HA antibody was also used to detect HA-tagged recombinant proteins (r.ssGP and r.sGP). Expression and secretion of r.sGP and r.ssGP were demonstrated after transfection of Vero E6 cells, and the two proteins appeared similar in size (approximately 50 kDa) under reducing SDS-PAGE conditions (Fig. 2A, lanes 3 and 4). Expression was optimal at 72 h posttransfection, a time point that was used for harvesting supernatants from transfected cells for all experiments unless otherwise stated.
Fig. 2.
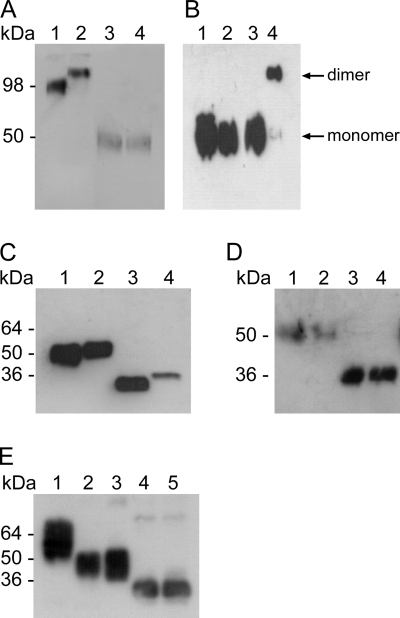
Structure and biochemical properties of ssGP. (A) ssGP is a homodimer. Vero E6 cells were transfected with the appropriate expression plasmids, and supernatants were collected from transfected cells after 72 h. The proteins were separated by reducing or nonreducing SDS-PAGE and were detected by immunoblotting using MAb 42/3.7 (dilution, 1:10,000). Lane 1, r.ssGP (nonreducing); lane 2; r.sGP (nonreducing); lane 3, r.ssGP (reducing); lane 4, r.sGP (reducing). (B) Cysteine at position 53 is responsible for dimerization. Site-directed mutagenesis was performed at amino acid position 53 (cysteine to glycine), and the resulting plasmid was transfected into 293T cells. Proteins were detected by immunoblotting with a peroxidase-conjugated anti-HA antibody (dilution, 1:10,000). Lane 1, r.ssGP Cys53Gly mutant (reducing); lane 2, r.ssGP (reducing); lane 3, r.ssGP Cys53Gly mutant (nonreducing); lane 4, r.ssGP (nonreducing). (C) ssGP contains N-linked carbohydrates. Vero E6 cells were transfected with the appropriate expression plasmids, and supernatants were collected from transfected cells after 72 h. The proteins were treated with PNGase F, separated by reducing SDS-PAGE, and detected by immunoblotting using MAb 42/3.7 (dilution, 1:10,000). Lane 1, untreated r.ssGP; lane 2, untreated r.sGP; lane 3, PNGase F-treated r.ssGP; lane 4, PNGase F-treated r.sGP. (D) ssGP does not contain high-mannose-type N-linked carbohydrates. 293T cells were transfected with the appropriate plasmid expressing HA-tagged r.ssGP, and HA-tagged r.ssGP was purified from the supernatants 72 h posttransfection (see Materials and Methods). HA-tagged r.ssGP was treated with Endo H and/or PNGase F and was analyzed by reducing SDS-PAGE followed by immunoblotting using peroxidase-conjugated anti-HA (dilution, 1:10,000). Lane 1, untreated r.ssGP; lane 2, Endo H-treated r.ssGP; lane 3, r.ssGP treated with Endo H and PNGase F; lane 4, r.ssGP treated with PNGase F. (E) ssGP does not contain O-linked carbohydrates. HA-tagged r.ssGP treated with different exoglycosidases, O-glycanase, N-glycanase, or combinations of these glycosidases was analyzed as described in Materials and Methods. Lane 1, untreated r.ssGP; lane 2, r.ssGP treated with the exoglycosidases [sialidase A, β(1–4)-galactosidase, and β-N-acetylglucosaminidase]; lane 3, r.ssGP treated with the exoglycosidases and O-glycanase; lane 4, r.ssGP treated with the exoglycosidases, O-glycanase, and N-glycanase; lane 5, r.ssGP treated with N-glycanase.
GP1,2 and sGP have been shown to be disulfide-linked homotrimers or homodimers, respectively (1, 2, 10, 41). Therefore, we analyzed the oligomerization of r.ssGP under nonreducing SDS-PAGE conditions, followed by immunoblotting using MAb 42/3.7. Under nonreducing conditions, r.ssGP migrated as an approximately 100 kDa protein, slightly faster than homodimeric r.sGP (Fig. 2A, lanes 1 and 2), suggesting that ssGP also has a dimeric structure. While under reducing conditions sGP and ssGP have near-identical apparent MWs, under nonreducing conditions there is a larger than expected difference in apparent MW, possibly suggesting that their tertiary structures may be different. Cysteine residues at positions 53 (Cys53) and 306 (Cys306) are responsible for intermolecular disulfide bridge formation in sGP (1, 10). Since Cys306 is not present in the ssGP ORF, we replaced Cys53 with a glycine residue by site-directed mutagenesis and analyzed the expressed proteins under reducing and nonreducing SDS-PAGE conditions. The lack of Cys53 prevented the formation of ssGP oligomers, indicating that oligomerization of ssGP is also dependent on an intermolecular disulfide bond between two Cys53 molecules (Fig. 2B).
ssGP is N glycosylated.
The predicted molecular mass for ssGP is approximately 30 kDa. It is known that GP1,2 is heavily N and O glycosylated, whereas sGP seems to be only N glycosylated (13, 14, 39, 41, 53, 54). Therefore, glycosylation as one form of posttranslational processing was predicted for ssGP as well. For glycosylation analysis, untagged r.ssGP and r.sGP were treated overnight with PNGase F to remove all the N-linked carbohydrates and were then subjected to SDS-PAGE and immunoblot analysis. Deglycosylated r.ssGP migrated with a lower molecular size (approximately 30 kDa) than deglycosylated r.sGP (approximately 33 kDa), indicating that ssGP, like sGP, contains N-glycans (Fig. 2C). Treatment of r.ssGP with Endo H, which removes all mannose-type N-linked carbohydrates, did not result in a further MW shift, indicating that ssGP N glycosylation is mainly of the complex hybrid type (Fig. 2D).
r.ssGP was further analyzed for O glycosylation. O-Glycanase removes core O-linked carbohydrates by breaking the bond between Galβ(1–3)GalNAc (N-acetylgalactosamine) and serine or threonine. In order for O-glycanase to work properly, additional monosaccharides need to be removed by exoglycosidases from the core structure. Treatment of r.ssGP with different exoglycosidases in combination with O-glycanase did not result in a MW shift during SDS-PAGE, indicating that ssGP does not carry O-linked carbohydrates (Fig. 2E), a finding similar to results reported previously for sGP (14, 39, 53).
ssGP is produced during infection.
In order to identify expression of ssGP during virus infection, we infected Vero E6 cells with ZEBOV and in parallel transiently expressed r.ssGP and r.sGP in Vero E6 cells. The supernatants from infected and transfected cells were subjected to overnight digestion with PNGase F, and the proteins were analyzed by 15% SDS-PAGE, followed by immunoblotting using MAb 42/3.7 for detection. Two specific bands were identified in the supernatant of ZEBOV-infected cells; the more prominent band displayed a molecular mass of approximately 33 kDa, which correlates in size with deglycosylated r.sGP (Fig. 3A, lanes 2 and 3). The weaker, lower-molecular-mass band (approximately 30 kDa) comigrated with deglycosylated r.ssGP and likely represents ssGP that is produced and secreted during ZEBOV infection (Fig. 3A, lanes 1 and 3).
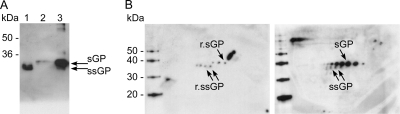
Fig. 3.
Expression of ssGP during Ebola virus infection. Vero E6 cells were infected with ZEBOV at an MOI of 1. Supernatants were collected 4 days postinfection and were treated with PNGase F. Subsequently, proteins were separated using different electrophoresis systems and were detected by immunoblotting using MAb 42/3.7 at a 1/10,000 dilution. For controls, Vero E6 cells were transfected with the appropriate plasmids expressing r.sGP and r.ssGP. Supernatants were collected 72 h posttransfection and were analyzed as described above. (A) Reducing 15% SDS-PAGE. Lane 1, r.ssGP treated with PNGase F; lane 2, r.sGP treated with PNGase F; lane 3, supernatant from ZEBOV-infected Vero E6 cells treated with PNGase F. (B) 2D gel electrophoresis. (Left) r.ssGP and r.sGP treated with PNGase F. (Right) Supernatant from ZEBOV-infected Vero E6 cells treated with PNGase F.
In order to confirm the expression of ssGP during ZEBOV infection, we utilized 2D gel electrophoresis. Vero E6 cells were transfected with plasmids (pCAGGS-ssGP or pCAGGS-sGP), and supernatants were collected at 72 h posttransfection. In parallel, Vero E6 cells were infected with ZEBOV (MOI, 1), and supernatants were harvested at 4 days postinfection. The collected supernatants were subjected to PNGase F treatment overnight, and proteins were subjected to 2D gel electrophoresis using a narrow pH range (pH 4 to 7) in the first dimension and 12% SDS-PAGE in the second dimension. Following the second dimension, immunoblotting was performed using MAb 42/3.7, which detects both proteins. r.sGP and r.ssGP were clearly separated based on their isoelectric points (pIs) and MWs, demonstrating the suitability of the approach for distinguishing between the two similar secreted proteins (Fig. 3B, left). Both proteins appeared in different isoforms, most likely reflecting certain co- or posttranslational modifications besides N glycosylation (Fig. 2C). A very similar pattern was observed when a supernatant derived from ZEBOV-infected cells was applied, confirming the expression of both proteins during ZEBOV infection (Fig. 3B, right). The weaker expression of ssGP correlates with the lower transcript numbers, as determined earlier (Fig. 1B). In addition, we subjected r.ssGP, r.sGP, and a ZEBOV-infected Vero E6 cell supernatant to 2D differential in-gel electrophoresis (2D DIGE) analysis. r.ssGP (labeled with Cy5) and r.sGP (labeled with Cy3) displayed differences in pI and MW similar to those observed by regular 2D gel electrophoresis (Fig. 3B, left). When r.sGP (Cy2 labeled) and r.ssGP (Cy3 labeled) were mixed with Cy5-labeled proteins derived from a ZEBOV-infected supernatant, both proteins were detected at their predicted pIs and MWs (data not shown).
ssGP function.
The structural and biochemical similarities between ssGP and sGP would suggest that ssGP may have a function similar to that of sGP. One of the established functions of sGP is its ability to stabilize the endothelial barrier function (56). To determine whether ssGP could also function as an anti-inflammatory protein, endothelial cells were treated with TNF-α and ssGP. In contrast to the results with sGP, no rescue effect was noted with ssGP; the spectra generated were similar to those obtained with TNF-α alone (Fig. 4A). As previously shown for sGP, ssGP alone did not affect the endothelial barrier function (data not shown). sGP has also been proposed to bind to human neutrophils (20, 59); however, neither ssGP nor sGP was found to bind to neutrophils in this study (Fig. 4B), supporting previous data that could not confirm a specific interaction between sGP and neutrophils (27, 46).
Fig. 4.
Functional analysis of ssGP. (A) Endothelial barrier function rescue assay. Human umbilical vein endothelial cells were equilibrated for 2 h to generate a baseline TER. TNF-α (1 ng/ml) with or without ssGP or sGP (30 μg/ml) was added to the medium as indicated, and the chambers were monitored by impedance spectroscopy. There was a significant difference (P < 0.05) between the TER of cells treated with TNF-α alone and that of cells treated with TNF-α and sGP by 330 min (*), while there was no significant difference between cells treated with TNF-α alone and cells treated with TNF-α and ssGP. (B) Neutrophil binding assay. Purified human neutrophils were incubated with ssGP or sGP (20 μg/ml) and were subsequently stained with anti-HA-PE and analyzed by FACS. The lack of a shift in fluorescence intensity between untreated and treated neutrophils indicates that neither sGP nor ssGP binds to the surfaces of human neutrophils.
DISCUSSION
All known species in the genus Ebolavirus, including the most recently identified, tentative species Bundibugyo ebolavirus (49), possess a highly conserved stretch of seven uridine residues in the genomic RNA sequence (Fig. 1A) and are therefore thought to undergo site-specific transcriptional editing of their GP genes during transcription. This results in the production of different glycoproteins, including the nonstructural glycoprotein sGP and the transmembrane glycoprotein GP1,2 (Fig. 1A) (14, 39); however, transcriptional editing has been experimentally addressed only for ZEBOV (40, 51).
Transcriptional editing has been described for other negative-stranded RNA viruses, in particular for members of the family Paramyxoviridae, where the phosphoprotein (P) gene undergoes transcriptional editing at a short run of AnGn residues (16, 19). The P proteins from members of the genera Morbillivirus, Respirovirus, Henipavirus, and Avulavirus are expressed from unedited transcripts, whereas the V and W/D (W protein in Nipah virus and D protein in bovine parainfluenza virus 3) proteins require the insertion of one or two guanosine residues into the editing site, respectively (7, 22, 24, 32, 50). Transcriptional editing has also been observed in both the genomic and antigenomic RNA of hepatitis delta virus (HDV) (5, 6, 60). For paramyxoviruses it has been postulated that RNA editing occurs through a mechanism of RNA-dependent RNA polymerase stuttering during template elongation, similar to the polyadenylation of RNA transcripts, indicating that editing sites may have evolved from polyadenylation sites (16). Currently, there are no studies on EBOV addressing the mechanism for RNA editing, but a similarity of the conserved EBOV GP gene editing site (Fig. 1A) with paramyxovirus P gene editing sites is obvious, indicating that a similar mechanism may apply for EBOV.
RNA editing is one of the evolutionary mechanisms allowing a fairly compact RNA virus to optimize its coding capacity. Genes with RNA editing sites encode structural and nonstructural proteins whereby editing is involved in the expression of both, depending on the virus species (9, 16, 22, 24, 29, 39, 40, 55, 57). For example, the P protein of paramyxoviruses is an important component of the nucleocapsid, which is the active transcription/replication complex (3, 24, 29, 58). For HDV, RNA editing of the hepatitis delta antigen (HDAg) p24 gene results in HDAg p27. Both of these proteins are essential: p24 is required for HDV replication, whereas p27 is required for viral RNA packaging (5, 6, 23, 37). For EBOV, the transmembrane glycoprotein GP1,2 plays a major role in viral pathogenesis by mediating receptor binding and fusion (12, 14, 21, 39). In contrast to the well-defined functions for the structural proteins of these viruses, the roles of the nonstructural proteins are not well defined. Nipah virus V and W proteins have been described as interferon (IFN) antagonists (31, 36, 44). No definite role has yet been identified for the nonstructural EBOV product sGP (14, 39). The hypotheses that sGP blocks EBOV neutralizing antibodies (20, 59), to interfere with the innate immune response by binding to CD16b and inhibiting neutrophil activation (59), or counteracts the permeability-increasing effect of proinflammatory mediators such as TNF-α and thus interferes with leukocyte extravasation (10, 55) have been discussed.
This study was designed to identify the expression of a second, previously postulated nonstructural glycoprotein, designated ssGP, through RNA editing during EBOV infection (14, 39). First we demonstrated that ZEBOV infection in vivo and in vitro resulted in the production of three distinct GP gene-specific transcripts. The ratios of transcripts encoding sGP and GP1,2 were similar to those reported previously by others (40, 51); <5% of the transcripts were specific for ssGP (Fig. 1B). The EBOV editing process is not absolutely accurate, as indicated by the insertion of multiple adenosine residues (Fig. 1C). A similar phenomenon has also been seen for paramyxovirus P genes, where as many as 14 nontemplate guanosine residues have been observed to be inserted, and as with EBOV, only rarely were deletions of a nucleotide noted (15, 22, 26, 32). Among paramyxoviruses, the frequency of RNA editing differs, ranging from 31% for Sendai virus to 82% for Nipah virus (22). For EBOV we have no comparisons, since all work on editing has so far focused only on ZEBOV.
All expression products from the GP gene carry the same N-terminal amino acid sequence, including the signal peptide (SP), which directs these proteins to the endoplasmic reticulum (ER) (14, 39). Therefore, all products are potentially exposed to similar co- and posttranslational modifications. The determination of the glycosylation status of ssGP revealed the existence of almost exclusively complex hybrid type N-linked carbohydrates, while no evidence of O glycosylation could be found (Fig. 2C, D, and E). Therefore, the glycosylation of ssGP is similar to that of sGP (2, 10, 35), which also has been shown to carry only N-linked carbohydrates. In this respect, both differ from GP1,2, which is highly N and O glycosylated (13, 17, 35, 41), and the solely O glycosylated delta peptide, the C-terminal proteolytic cleavage fragment of the sGP precursor (54). The ORF encoding ssGP possesses 6 potential attachment sites for N-linked carbohydrates, all of which seem to be used, based on the MW shift after complete deglycosylation following PNGase F treatment (Fig. 2C).
Site-directed mutagenesis determined that mature ssGP forms homodimers via an intermolecular disulfide bond that links two Cys53 residues (Fig. 2B). In the case of sGP, homodimers are stabilized through an additional intermolecular disulfide bond between two Cys306 molecules (1, 10); however, this residue is not present in ssGP. As with sGP, it is expected that the remaining four cysteines (aa 188, 121, 135, and 147) are involved in intramolecular disulfide bond formation and stabilization of the molecule (1, 10). Despite the shared sequence, ssGP did not rescue endothelial barrier function as does sGP (56). This might be explained by the absence in ssGP of Cys306, which functions in stabilizing the sGP homodimer (1, 10) and is essential for the rescue effect on endothelial barrier function (10). Together with the differences in migration patterns under nonreducing conditions (Fig. 2A), these data suggest that ssGP likely has a structure distinct from that of sGP despite the similarities in biochemical properties and primary sequence.
The similar predicted MWs for sGP and ssGP, the expected low abundance of ssGP (<5% of transcripts), and the lack of ssGP-specific antibodies made the detection of ssGP during EBOV infection challenging. Preliminary evidence for ssGP expression was obtained from SDS-PAGE analysis (Fig. 3A). Formic acid or endoproteinase AspN digestion followed by mass spectrometry in order to detect a distinct C-terminal peptide specific for ssGP was unsuccessful despite multiple attempts (data not shown). Therefore, 2D electrophoresis was utilized, since sGP and ssGP differed in their predicted pIs (8.17 and 6.26, respectively). By this method, in combination with deglycosylation, the two different proteins could be separated, providing final proof that ssGP is indeed expressed during EBOV infection (Fig. 3B). Expression was further confirmed through 2D DIGE analysis (data not shown).
To date, all of our attempts have failed to demonstrate any specific binding of sGP and ssGP to peripheral blood mononuclear cells (PBMCs), in particular neutrophils (unpublished data). The previously reported binding of sGP to human neutrophils through an interaction with CD16b (20, 59)—a finding disputed later by others (27, 46)—was also not confirmed here (Fig. 4B). The interaction of a viral surface protein with its receptor is likely mediated through a specific structure of the receptor binding site. The two nonstructural glycoproteins differ from GP1,2 in their tertiary structure. sGP and ssGP are homodimers (1, 10), while GP1,2 is a heterotrimer that consists of the two disulfide-linked proteolytic cleavage fragments GP1 and GP2 (14, 17, 39, 41). Furthermore, both nonstructural products differ from GP1,2 in their glycosylation pattern in that they lack O-linked and high-mannose-type N-linked carbohydrates (Fig. 2). In particular, high-mannose N-glycosylated carbohydrates on surface proteins are known to efficiently enhance interactions with host cell attachment molecules, such as LSECtin, dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin (DC-SIGN), and DC-SIGN-related protein (DC-SIGNR), found on dendritic and endothelial cells, as described for HIV, hepatitis C virus, Marburg virus, and EBOV (25, 28, 34, 45). Therefore, these differences in structure and posttranslational processing may be the reason for the failure to detect binding to PBMCs.
In conclusion, we have identified a new protein expressed during EBOV infection that represents a second nonstructural glycoprotein. We have also determined its expression strategy, namely, RNA editing of the GP gene. The protein is processed through the ER pathway, which includes homodimerization through an intermolecular disulfide bond between two Cys53 molecules, mainly complex type N glycosylation and secretion from expressing cells. No function has been assigned to the new protein yet; however, it does not serve the same anti-inflammatory function as sGP. New strategies are needed to address the important question of the functions of both nonstructural proteins produced during EBOV infection.
ACKNOWLEDGMENTS
We thank Anita Mora (Visual Medical Arts, Division of Intramural Research [DIR], NIAID, NIH) for help with graphical design and Addie Whitney (Laboratory of Human Bacterial Pathogenesis, DIR, NIAID, NIH) for help with the neutrophil binding assay.
This study was supported by the National Microbiology Laboratory (NML) of the Public Health Agency of Canada (PHAC) and the Intramural Research Program of NIAID, NIH.
Footnotes
Published ahead of print on 16 March 2011.
REFERENCES
- 1. Barrientos L. G., Martin A. M., Rollin P. E., Sanchez A. 2004. Disulfide bond assignment of the Ebola virus secreted glycoprotein SGP. Biochem. Biophys. Res. Commun. 323:696–702 [DOI] [PubMed] [Google Scholar]
- 2. Barrientos L. G., Martin A. M., Wohlhueter R. M., Rollin P. E. 2007. Secreted glycoprotein from live Zaire ebolavirus-infected cultures: preparation, structural and biophysical characterization, and thermodynamic stability. J. Infect. Dis. 196(Suppl. 2):S220–S231 [DOI] [PubMed] [Google Scholar]
- 3. Bourhis J. M., Canard B., Longhi S. 2006. Structural disorder within the replicative complex of measles virus: functional implications. Virology 344:94–110 [DOI] [PubMed] [Google Scholar]
- 4. Bray M., Davis K., Geisbert T., Schmaljohn C., Huggins J. 1998. A mouse model for evaluation of prophylaxis and therapy of Ebola hemorrhagic fever. J. Infect. Dis. 178:651–661 [DOI] [PubMed] [Google Scholar]
- 5. Casey J. L., Bergmann K. F., Brown T. L., Gerin J. L. 1992. Structural requirements for RNA editing in hepatitis delta virus: evidence for a uridine-to-cytidine editing mechanism. Proc. Natl. Acad. Sci. U. S. A. 89:7149–7153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Casey J. L., Gerin J. L. 1995. Hepatitis D virus RNA editing: specific modification of adenosine in the antigenomic RNA. J. Virol. 69:7593–7600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cattaneo R., Kaelin K., Baczko K., Billeter M. A. 1989. Measles virus editing provides an additional cysteine-rich protein. Cell 56:759–764 [DOI] [PubMed] [Google Scholar]
- 8. Claude P. 1978. Morphological factors influencing transepithelial permeability: a model for the resistance of the zonula occludens. J. Membr. Biol. 39:219–232 [DOI] [PubMed] [Google Scholar]
- 9. Curran J. A., Kolakofsky D. 1987. Identification of an additional Sendai virus non-structural protein encoded by the P/C mRNA. J. Gen. Virol. 68(Pt 9):2515–2519 [DOI] [PubMed] [Google Scholar]
- 10. Falzarano D., et al. 2006. Structure-function analysis of the soluble glycoprotein, sGP, of Ebola virus. Chembiochem 7:1605–1611 [DOI] [PubMed] [Google Scholar]
- 11. Feldmann H., et al. 2005. Filoviridae, p. 645–653 In Fauquet C. M., Mayo M. A., Maniloff J., Desselberger U., Ball L. A. (ed.), Virus taxonomy. Eighth Report of the International Committee on Taxonomy of Viruses Elsevier Academic Press, London, United Kingdom [Google Scholar]
- 12. Feldmann H., Jones S., Klenk H. D., Schnittler H. J. 2003. Ebola virus: from discovery to vaccine. Nat. Rev. Immunol. 3:677–685 [DOI] [PubMed] [Google Scholar]
- 13. Feldmann H., Nichol S. T., Klenk H. D., Peters C. J., Sanchez A. 1994. Characterization of filoviruses based on differences in structure and antigenicity of the virion glycoprotein. Virology 199:469–473 [DOI] [PubMed] [Google Scholar]
- 14. Feldmann H., Volchkov V. E., Volchkova V. A., Stroher U., Klenk H. D. 2001. Biosynthesis and role of filoviral glycoproteins. J. Gen. Virol. 82:2839–2848 [DOI] [PubMed] [Google Scholar]
- 15. Galinski M. S., Troy R. M., Banerjee A. K. 1992. RNA editing in the phosphoprotein gene of the human parainfluenza virus type 3. Virology 186:543–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hausmann S., Garcin D., Delenda C., Kolakofsky D. 1999. The versatility of paramyxovirus RNA polymerase stuttering. J. Virol. 73:5568–5576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ito H., Watanabe S., Sanchez A., Whitt M. A., Kawaoka Y. 1999. Mutational analysis of the putative fusion domain of Ebola virus glycoprotein. J. Virol. 73:8907–8912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Iwase H., Hotta K. 1993. Release of O-linked glycoprotein glycans by endo-α-N-acetylgalactosaminidase. Methods Mol. Biol. 14:151–159 [DOI] [PubMed] [Google Scholar]
- 19. Jacques J. P., Hausmann S., Kolakofsky D. 1994. Paramyxovirus mRNA editing leads to G deletions as well as insertions. EMBO J. 13:5496–5503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kindzelskii A. L., Yang Z., Nabel G. J., Todd R. F., III, Petty H. R. 2000. Ebola virus secretory glycoprotein (sGP) diminishes FcγRIIIB-to-CR3 proximity on neutrophils. J. Immunol. 164:953–958 [DOI] [PubMed] [Google Scholar]
- 21. Kuhn J. H., et al. 2006. Conserved receptor-binding domains of Lake Victoria marburgvirus and Zaire ebolavirus bind a common receptor. J. Biol. Chem. 281:15951–15958 [DOI] [PubMed] [Google Scholar]
- 22. Kulkarni S., et al. 2009. Nipah virus edits its P gene at high frequency to express the V and W proteins. J. Virol. 83:3982–3987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kuo M. Y., Chao M., Taylor J. 1989. Initiation of replication of the human hepatitis delta virus genome from cloned DNA: role of delta antigen. J. Virol. 63:1945–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lamb R. A., Parks G. D. 2007. Paramyxoviridae: the viruses and their replication, p. 1449–1496 In Knipe D. M., Howley P. M., Griffin D. E., Lamb R. A., Martin M. A., Roizman B., Straus S. E. (ed.), Fields virology, 5th ed., vol. 1 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 25. Lin G., et al. 2003. Differential N-linked glycosylation of human immunodeficiency virus and Ebola virus envelope glycoproteins modulates interactions with DC-SIGN and DC-SIGNR. J. Virol. 77:1337–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lo M. K., et al. 2009. Determination of the henipavirus phosphoprotein gene mRNA editing frequencies and detection of the C, V and W proteins of Nipah virus in virus-infected cells. J. Gen. Virol. 90:398–404 [DOI] [PubMed] [Google Scholar]
- 27. Maruyama T., Buchmeier M. J., Parren P. W., Burton D. R. 1998. Ebola virus, neutrophils, and antibody specificity. Science 282:843 [Google Scholar]
- 28. Marzi A., et al. 2004. DC-SIGN and DC-SIGNR interact with the glycoprotein of Marburg virus and the S protein of severe acute respiratory syndrome coronavirus. J. Virol. 78:12090–12095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Matsumura H., Ikemura N., Ito Y., Kuribayashi K. 1999. RNA editing-like phenomenon in paramyxovirus V gene mRNA observed in insect cells infected with a recombinant baculovirus. J. Gen. Virol. 80(Pt 1):117–123 [DOI] [PubMed] [Google Scholar]
- 30. Niwa H., Yamamura K., Miyazaki J. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193–199 [DOI] [PubMed] [Google Scholar]
- 31. Park M. S., et al. 2003. Newcastle disease virus (NDV)-based assay demonstrates interferon-antagonist activity for the NDV V protein and the Nipah virus V, W, and C proteins. J. Virol. 77:1501–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pelet T., Curran J., Kolakofsky D. 1991. The P gene of bovine parainfluenza virus 3 expresses all three reading frames from a single mRNA editing site. EMBO J. 10:443–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Powell D. W. 1981. Barrier function of epithelia. Am. J. Physiol. 241:G275–G288 [DOI] [PubMed] [Google Scholar]
- 34. Powlesland A. S., et al. 2008. A novel mechanism for LSECtin binding to Ebola virus surface glycoprotein through truncated glycans. J. Biol. Chem. 283:593–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ritchie G., et al. 2010. Identification of N-glycans from Ebola virus glycoproteins by matrix-assisted laser desorption/ionisation time-of-flight and negative ion electrospray tandem mass spectrometry. Rapid Commun. Mass Spectrom. 24:571–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rodriguez J. J., Parisien J. P., Horvath C. M. 2002. Nipah virus V protein evades alpha and gamma interferons by preventing STAT1 and STAT2 activation and nuclear accumulation. J. Virol. 76:11476–11483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ryu W. S., Bayer M., Taylor J. 1992. Assembly of hepatitis delta virus particles. J. Virol. 66:2310–2315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sanchez A., Kiley M. P., Holloway B. P., Auperin D. D. 1993. Sequence analysis of the Ebola virus genome: organization, genetic elements, and comparison with the genome of Marburg virus. Virus Res. 29:215–240 [DOI] [PubMed] [Google Scholar]
- 39. Sanchez A., Giesbert T. W., Feldmann H. 2007. Filoviridae: Marburg and Ebola viruses, p. 1409–1448 In Knipe D. M., Howley P. M., Griffin D. E., Lamb R. A., Martin M. A., Roizman B., Straus S. E. (ed.), Fields virology, 5th ed., vol. 1 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 40. Sanchez A., Trappier S. G., Mahy B. W., Peters C. J., Nichol S. T. 1996. The virion glycoproteins of Ebola viruses are encoded in two reading frames and are expressed through transcriptional editing. Proc. Natl. Acad. Sci. U. S. A. 93:3602–3607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sanchez A., et al. 1998. Biochemical analysis of the secreted and virion glycoproteins of Ebola virus. J. Virol. 72:6442–6447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schnittler H. J., Wilke A., Gress T., Suttorp N., Drenckhahn D. 1990. Role of actin and myosin in the control of paracellular permeability in pig, rat and human vascular endothelium. J. Physiol. 431:379–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Seebach J., et al. 2000. Endothelial barrier function under laminar fluid shear stress. Lab. Invest. 80:1819–1831 [DOI] [PubMed] [Google Scholar]
- 44. Shaw M. L., Garcia-Sastre A., Palese P., Basler C. F. 2004. Nipah virus V and W proteins have a common STAT1-binding domain yet inhibit STAT1 activation from the cytoplasmic and nuclear compartments, respectively. J. Virol. 78:5633–5641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Simmons G., et al. 2003. DC-SIGN and DC-SIGNR bind Ebola glycoproteins and enhance infection of macrophages and endothelial cells. Virology 305:115–123 [DOI] [PubMed] [Google Scholar]
- 46. Sui J., Marasco W. A. 2002. Evidence against Ebola virus sGP binding to human neutrophils by a specific receptor. Virology 303:9–14 [DOI] [PubMed] [Google Scholar]
- 47. Takada A., Ebihara H., Feldmann H., Geisbert T. W., Kawaoka Y. 2007. Epitopes required for antibody-dependent enhancement of Ebola virus infection. J. Infect. Dis. 196(Suppl. 2):S347–S356 [DOI] [PubMed] [Google Scholar]
- 48. Takada A., Ebihara H., Jones S., Feldmann H., Kawaoka Y. 2007. Protective efficacy of neutralizing antibodies against Ebola virus infection. Vaccine 25:993–999 [DOI] [PubMed] [Google Scholar]
- 49. Towner J. S., et al. 2008. Newly discovered Ebola virus associated with hemorrhagic fever outbreak in Uganda. PLoS Pathog. 4:e1000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vidal S., Curran J., Kolakofsky D. 1990. Editing of the Sendai virus P/C mRNA by G insertion occurs during mRNA synthesis via a virus-encoded activity. J. Virol. 64:239–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Volchkov V. E., et al. 1995. GP mRNA of Ebola virus is edited by the Ebola virus polymerase and by T7 and vaccinia virus polymerases. Virology 214:421–430 [DOI] [PubMed] [Google Scholar]
- 52. Volchkov V. E., et al. 2001. Recovery of infectious Ebola virus from complementary DNA: RNA editing of the GP gene and viral cytotoxicity. Science 291:1965–1969 [DOI] [PubMed] [Google Scholar]
- 53. Volchkova V. A., Feldmann H., Klenk H. D., Volchkov V. E. 1998. The nonstructural small glycoprotein sGP of Ebola virus is secreted as an antiparallel-orientated homodimer. Virology 250:408–414 [DOI] [PubMed] [Google Scholar]
- 54. Volchkova V. A., Klenk H. D., Volchkov V. E. 1999. Delta-peptide is the carboxy-terminal cleavage fragment of the nonstructural small glycoprotein sGP of Ebola virus. Virology 265:164–171 [DOI] [PubMed] [Google Scholar]
- 55. Wahl-Jensen V., et al. 2005. Role of Ebola virus secreted glycoproteins and virus-like particles in activation of human macrophages. J. Virol. 79:2413–2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wahl-Jensen V. M., et al. 2005. Effects of Ebola virus glycoproteins on endothelial cell activation and barrier function. J. Virol. 79:10442–10450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang L. F., et al. 1998. A novel P/V/C gene in a new member of the Paramyxoviridae family, which causes lethal infection in humans, horses, and other animals. J. Virol. 72:1482–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wiegand M. A., Bossow S., Schlecht S., Neubert W. J. 2007. De novo synthesis of N and P proteins as a key step in Sendai virus gene expression. J. Virol. 81:13835–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yang Z., et al. 1998. Distinct cellular interactions of secreted and transmembrane Ebola virus glycoproteins. Science 279:1034–1037 [DOI] [PubMed] [Google Scholar]
- 60. Zheng H., Fu T. B., Lazinski D., Taylor J. 1992. Editing on the genomic RNA of human hepatitis delta virus. J. Virol. 66:4693–4697 [DOI] [PMC free article] [PubMed] [Google Scholar]