Abstract
Passive administration of porcine reproductive and respiratory syndrome virus (PRRSV) neutralizing antibodies (NAbs) can effectively protect pigs against PRRSV infection. However, after PRRSV infection, pigs typically develop a weak and deferred NAb response. One major reason for such a meager NAb response is the phenomenon of glycan shielding involving GP5, a major glycoprotein carrying one major neutralizing epitope. We describe here a type II PRRSV field isolate (PRRSV-01) that is highly susceptible to neutralization and induces an atypically rapid, robust NAb response in vivo. Sequence analysis shows that PRRSV-01 lacks two N-glycosylation sites, normally present in wild-type (wt) PRRSV strains, in two of its envelope glycoproteins, one in GP3 (position 131) and the other in GP5 (position 51). To determine the influence of these missing N-glycosylation sites on the distinct neutralization phenotype of PRRSV-01, a chimeric virus (FL01) was generated by replacing the structural genes of type II PRRSV strain FL12 cDNA infectious clone with those from PRRSV-01. N-glycosylation sites were reintroduced into GP3 and GP5 of FL01, separately or in combination, by site-directed mutagenesis. Reintroduction of the N-glycosylation site in either GP3 or GP5 allowed recovery of in vivo and in vitro glycan shielding capacity, with an additive effect when these sites were reintroduced into both glycoproteins simultaneously. Although the loss of these glycosylation sites has seemingly occurred naturally (presumably by passage through cell cultures), PRRSV-01 virus quickly regains these glycosylation sites through replication in vivo, suggesting that a strong selective pressure is exerted at these sites. Collectively, our data demonstrate the involvement of an N-glycan moiety located in GP3 in glycan shield interference.
INTRODUCTION
Porcine reproductive and respiratory syndrome virus (PRRSV) is the causative agent of an economically important disease in swine characterized by late-term reproductive failure in pregnant sows and respiratory disorder in young pigs (9, 47). PRRSV, together with equine arteritis virus (EAV), lactate dehydrogenase-elevating virus (LDV), and simian hemorrhagic fever virus (SHFV), belongs to the family Arteriviridae in the order Nidovirales (7). The virus is classified into 2 types, type I (European) and type II (North American) based on the genetic and antigenic differences among the PRRSV strains. The viral genome consists of a positive-sense, single-strand RNA molecule of ∼15 kb that contains 9 open reading frames (ORFs) (10, 30, 51). ORF1a and ORF1b encode nonstructural proteins (NSPs) responsible for the replication and transcription of the viral genome (30, 39). ORF2a, ORF2b, and ORF3 to ORF7 encode 7 structural proteins, four of which are glycoproteins (GPs), namely, GP2, GP3, GP4, and GP5 (30, 39, 51). GP5 is considered the major envelope glycoprotein, while GP2, GP3, and GP4 are considered the minor glycoproteins due to the relative abundance of these proteins on the virions (13, 28, 31). GP5 forms a heterodimer with M, the membrane protein (3, 27, 49). The GP5-M heterodimer is of cardinal importance for the formation of viral particles (49). In addition, GP5 of both type I and type II PRRSVs contains a major neutralizing epitope in its ectodomain (34–36, 46, 50). GP2, GP3, and GP4 interact with each other to form a multiprotein complex that is dispensable for viral particle formation yet important for viral infectivity (11, 49). It has been reported recently that GP2 and GP4 of the type II PRRSV strain FL12 interact with CD163, a receptor for PRRSV entry (11). GP4 of the type I PRRSV strain Lelystad contains a neutralizing epitope located at the hypervariable region spanning amino acids 40 to 79 (32, 43). There are several lines of evidence for the existence of neutralizing epitopes in other minor GPs of PRRSV; however, the locations of these epitopes have not been mapped (6, 19, 22).
Pigs exposed to PRRSV develop a prolonged viremia followed by persistent infection in lymphoid tissues for extended periods of time, suggesting that the host immune system does not effectively function at rapidly eliminating the infection (2, 48). The pig's response to PRRSV infection is characterized by a meager induction of innate immunity, the slow appearance of virus-specific gamma interferon (IFN-γ)-producing cells, and the weak and delayed development of neutralizing antibodies (NAbs) (1, 4, 26, 29). Passive transfer of NAbs to pigs prior to challenge with the homologous virulent PRRSV strain results in complete protection of the pigs against infection, demonstrating the important role of NAbs in protective immunity (25, 33). The stimulation of NAbs should therefore be considered an important goal for PRRSV vaccine development.
As previously observed for viruses such as simian immunodeficiency virus (38), human immunodeficiency virus (45), influenza virus (44), hepatitis C virus (24), and Ebola virus (16), PRRSV also relies on glycosylation modification of its envelope proteins to evade the host immune response (3, 15). Our laboratory has previously demonstrated that removal of N-glycosylation sites surrounding the neutralizing epitope located in GP5 of a PRRSV strain (FL12) renders the virus extremely susceptible to antibody neutralization (3). More importantly, the mutant viruses carrying glycosylation deletions in GP5 elicited significantly greater NAb responses than did the wild-type (wt), fully glycosylated virus (3). While the effects of glycosylation of GP5 on NAb development had been demonstrated, the role that glycosylation of other PRRSV proteins may play on glycan shielding immune evasion is not known yet.
We found a type II PRRSV field isolate, herein designated PRRSV-01, which is able to elicit an atypically rapid and robust NAb response in infected pigs. In addition, PRRSV-01 is extremely susceptible to neutralization by several different heterologous antisera. Analysis of structural genes of PRRSV-01 revealed that the virus naturally lacks two N-glycosylation sites in its envelope glycoproteins, one in GP3 at position 131 and the other in GP5 at position 51. The objective of this study was to investigate the influence of the absence of N-glycosylation sites in GP3 and GP5 on the susceptibility of PRRSV-01 to NAbs and its ability to induce NAb production.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
MARC-145 cells (20) were used for viral RNA electroporation and virus propagation and titration. Swine monocyte-derived macrophages were prepared as previously described (40) and used for examination of viremia levels and for isolation of virus from serum samples. The porcine reproductive and respiratory syndrome virus (PRRSV) field isolate, designated PRRSV-01, was isolated at the Iowa State University Veterinary Diagnostic Laboratory (ISU-VDL) in 2001 from a case of PRRSV infection associated with clinical disease. Monoclonal antibody specific for PRRSV nucleocapsid (N) protein (SDOW-17) was purchased from the National Veterinary Services Laboratories (Ames, IA). Anti-mouse, Alexa Fluor 488-conjugated antibody was purchased from Invitrogen (Eugene, OR). Anti-GP5 polyclonal antibody was kindly provided by Carl Gagnon (University of Quebec, Montreal, Canada). Anti-GP3 polyclonal antibody was previously described (14). Swine hyperimmunized antisera against PRRSV strains NSVL 97-7895, VR2332, MN184, and Lelystad were prepared as previously described (33).
Construction of chimeric virus and mutants derived from it.
Viral RNA isolated from culture supernatant of field isolate PRRSV-01 was used as the template for reverse transcription-PCR (RT-PCR) to obtain the entire structural genes and the 3′ untranslated region (3′UTR). The PCR product was digested with EcoRV and PacI, and the released fragment containing most of ORF2a, all of ORF3 to ORF7, and the 3′UTR was cloned into the pFL12 plasmid (41) using the same restriction enzyme sites to generate chimeric clone pFL01 (see Fig. 3). Using pFL01 as a backbone, site-directed mutagenesis was carried out by overlap extension using PCR as described previously (18) to reintroduce N-glycosylation sites into GP3 (to produce pFL01/GP3-D131N), GP5 (to produce pFL01/GP5-S51N), or both of them simultaneously (to produce pFL01/DM). The primers used to reintroduce N-glycosylation sites into GP3 and GP5 are shown in Table 1.
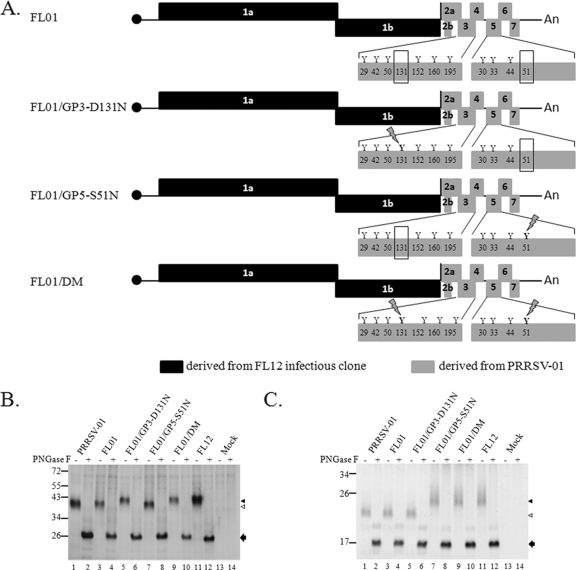
Fig. 3.
(A) Schematic representations of the genomes of chimeric virus FL01 and mutants derived from it. The letter “Y” and the numbers below them represent potential N-glycosylation sites and their relative positions on the corresponding proteins. The absence of N-glycosylation sites at position 131 in GP3 and position 51 in GP5 is highlighted by rectangular boxes. N-glycosylation sites that are artificially reintroduced to the genome are shown with a lightning bolt symbol over the site. (B and C) Electrophoretic mobility of GP3 and GP5, respectively. MARC-145 cells were mock infected or infected with the indicated virus. At 36 h postinfection, proteins were radiolabeled as described in Materials and Methods and immunoprecipitated with anti-GP3 antibodies (B) or anti-GP5 antibodies (C). The immunoprecipitated proteins were left untreated (−) or were treated with PNGase F (+) and analyzed by electrophoresis on SDS-polyacrylamide gel. The approximate molecular masses (in kilodaltons) of the marker proteins are identified to the left of the gel. The positions of fully glycosylated proteins (black triangles) and of the protein species lacking one N-glycosylation site (white triangles) are shown to the right of the gel. Protein bands indentified by black arrows are the products of PNGase F digestion.
Table 1.
Primers used to reintroduce the N-glycosylation sites into GP3 and GP5
| Protein | Primera | Nucleotide sequenceb (5′→3′) |
|---|---|---|
| GP3 | D131N For | GAGATATTTGGGATAGGGAATGTGAGTAAAGTTTATG |
| D131N Rev | CATAAACTTTACTCACATTCCCTATCCCAAATATCTC | |
| GP5 | S51N For | GCTATGCGAGCTGAATGGCACAGATTGGCTG |
| S51N Rev | CAGCCAATCTGTGCCATTCAGCTCGCATAGC |
For, forward; Rev, reverse.
Mutated nucleotides are underlined.
In vitro transcription and electroporation.
In vitro transcription and viral RNA electroporation were done as previously described (3, 41). Briefly, plasmid containing full-length PRRSV genomes was digested with AclI. Purified, linear DNA was used as the template for in vitro transcription reaction using the mMESSAGE mMACHINE Ultra T7 kit per the manufacturer's instructions (Ambion, Austin, TX) to generate the capped, viral RNA transcript. Subconfluent MARC-145 cells were trypsinized and resuspended in fetal bovine serum (FBS)-free Dulbecco modified Eagle medium (DMEM) containing 1.25% (vol/vol) dimethyl sulfoxide (DMSO) at a concentration of 6 × 106 cells/ml. Four hundred microliters of cells was electroporated with approximately 5 μg of in vitro viral RNA transcript together with 5 μg of carrier RNA isolated from naïve MARC-145 cells using Bio-Rad Gene Pulser Xcell at 250 V and 950 μF in a 4.0-mm cuvette. Electroporated cells were diluted in DMEM containing 10% FBS and 1.25% DMSO and placed in a 60-mm tissue culture dish. When a clear cytopathic effect (CPE) was observed, culture supernatant containing rescued virus was collected and passed into naïve MARC-145 cells once or twice to obtain enough virus stock for future studies.
Metabolic labeling and protein analysis.
Metabolic labeling and protein analysis were done as previously described (3, 11). MARC-145 cells in 6-well plate were infected with PRRSV at a multiplicity of infection (MOI) of approximately 0.1 50% tissue culture infectious dose (TCID50) per cell. At 36 h postinfection (p.i.), the cells were washed with phosphate-buffered saline (PBS) (pH 7.4) and starved in methionine- and cysteine-free DMEM for 1 h. The proteins were radiolabeled with 0.6 ml of methionine- and cysteine-free DMEM containing 30 μCi of Expre35S35S protein labeling mix (NEN Life Sciences, MA) for 4 h. After radiolabeling, cell lysates were prepared, and the viral GP3 or GP5 was immunoprecipitated using anti-GP3 or anti-GP5 antibodies, respectively. After that, the precipitated proteins were subjected to peptide N-glycosidase F (PNGase F) digestion (3). The proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and detected by fluorography as described earlier (3, 11).
Viral growth kinetics and plaque morphology.
MARC-145 cells were infected with wt or mutant viruses at an MOI of 5. Aliquots of culture supernatants from infected cells were collected at various time points postinfection, and virus titers were determined and expressed as TCID50 per ml. Plaque morphology was examined in MARC-145 cells as previously described (3).
Animal experiments.
Twenty-four recently weaned piglets (of about 3 weeks of age) purchased from a specific-pathogen-free herd certified not to have PRRSV infection were allocated into six groups and accommodated in isolated biosafety level 2 (BSL-2) rooms. Each group of pigs was separately infected with one of the following viruses: FL12, PRRSV-01, FL01, FL01/GP3-D131N, FL01/GP5-S51N, or FL01/DM. Likewise, three additional pigs were inoculated with PRRSV-01 for initial characterization of the immune response stimulated by this isolate. In all cases, the pigs were injected intramuscularly with 105.0 TCID50 of virus diluted in 2 ml of DMEM. Serum samples were collected periodically from all animals until 49 days postinfection (76 days postinfection for the initial experimental inoculation with PRRSV-01, which is shown in Fig. 1) and stored in small aliquots at −80°C for serological and virological analyses.
Fig. 1.
Neutralization phenotype of PRRSV-01. (A) Susceptibility of PRRSV-01 to cross-neutralization by heterologous antisera. The neutralizing activity of the indicated antiserum was separately measured against PRRSV-01 or against the homologous virus (i.e., the virus used for immunization to prepare each of the reference antisera). (B) Homologous neutralizing antibody (NAb) responses of pigs infected with PRRSV-01. Three recently weaned pigs (pigs 2533, 6180, and 7368) were inoculated with PRRSV-01 as described in Materials and Methods. Homologous neutralization titers of serum samples at the indicated time (in days) postinfection (PI) are shown. Neutralization titers were expressed as the reciprocal of the highest dilution that showed 90% or greater reduction in the number of fluorescent foci presenting in the control wells.
Serum neutralization assay.
Neutralizing antibody titers of serum samples against a specific PRRSV studied were determined using a fluorescent focus neutralization assay described previously (51). Heat-inactivated antisera were diluted 2-fold serially in 50 μl of DMEM supplemented with 5% FBS on a 96-well plate and then incubated with an equal volume (50 μl) containing 100 TCID50 of challenge virus for 1 h at 37°C. The entire mixture of serum and virus (100 μl/well) was transferred to another 96-well plate containing confluent MARC-145 cells that had been seeded 48 h earlier. The plate was further incubated for 36 h at 37°C in a humidified atmosphere containing 5% CO2. The expression of N protein was detected by indirect immunofluorescence assay using anti-N monoclonal antibody SDOW-17, and the presence of PRRSV was observed with a fluorescence microscope. Neutralization titers were expressed as the reciprocal of the highest dilution that showed 90% or greater reduction in the number of fluorescent foci presenting in the control wells.
Statistical analysis.
Before statistical analysis, the neutralization titers were transformed into log base 2. One-way analysis of variance (ANOVA) was used to evaluate the difference among the mean neutralization titers. After that, Tukey's honestly significant difference test was used for multiple comparison.
RESULTS
Identification and primary characterization of the PRRSV field isolate PRRSV-01 that exhibits a unique neutralization phenotype.
During studies involving subtyping of porcine reproductive and respiratory syndrome virus (PRRSV) field isolates by cross-neutralization (unrelated investigations [data not shown here]), we identified a PRRSV field isolate, herein designated PRRSV-01, that possesses a distinct phenotype in relation to this isolate's susceptibility to antibody neutralization and its ability to elicit neutralizing antibodies (NAbs). PRRSV-01 was isolated at the Iowa State University Veterinary Diagnostic Laboratory (ISU-VDL) in 2001 from a case of PRRSV infection associated with clinical disease. After PRRSV-01 was isolated, it was subjected to at least 7 passages in MARC-145 cells at ISU-VDL and then in our laboratory. We have observed that the current PRRSV-01 isolate resulting from those passages in MARC-145 cells was extremely susceptible to neutralization by different heterologous antisera. A complete battery of reference antisera used in our laboratory that are specific for different PRRSV strains, including VR2332 (9), NSVL 97-7895 (41), MN184 (17), and Lelystad (47), was able to neutralize PRRSV-01 at endpoint titers of 1:64 or greater (Fig. 1A). Interestingly, the antisera to VR2332 and MN184 neutralized PRRSV-01 at titers that were 2-fold higher than the titers of each individual antiserum against its respective homologous virus. After 3 naïve pigs were infected with PRRSV-01, we verified that this PRRSV isolate was able to mount an atypically rapid and robust homologous NAb response in the infected pigs (Fig. 1B). NAbs were detected in 2 out of 3 pigs infected with PRRSV-01 as early as 14 days postinfection (p.i.) and in all of the infected pigs at 21 days p.i. In addition, the NAb titers quickly increased and reached titers from 1:512 to 1:2,048 in all of the infected pigs at 42 days p.i. Remarkably, the NAb titer in one of the PRRSV-01-infected pigs reached a peak of 1:4,096 at 76 days p.i. (Fig. 1B). This constitutes a unique NAb response upon experimental inoculation with wt PRRSV. In our experience, neutralization titers above 1:128 in pigs infected with a wt PRRSV strain are rarely seen (3) (see Fig. 5).
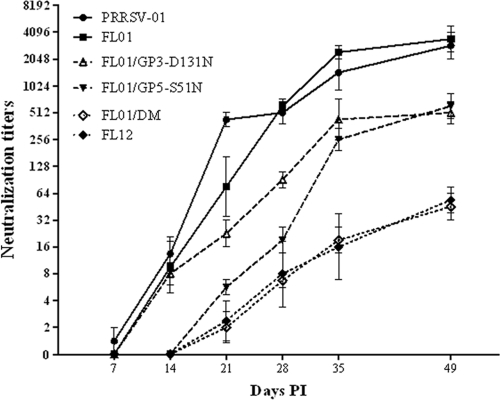
Fig. 5.
Kinetics of NAb response to homologous viruses. Each group of 4 recently weaned pigs was separately inoculated with the indicated virus as described in Materials and Methods. The neutralizing activities of serum samples at the indicated time (in days) postinfection (PI) were measured against homologous virus (i.e., the same virus used for infection). Neutralization titers are expressed as means ± SEMs.
Analysis of PRRSV-01 structural genes indicates that this isolate naturally lacks two N-glycosylation sites in the sequences of its envelope glycoproteins.
The entire structural region of the PRRSV-01 genome was amplified by RT-PCR and sequenced (GenBank accession no. JF422072). Overall, the structural region of the PRRSV-01 genome shares 92% nucleotide identity with strain VR2332, the PRRSV type II prototype and 64% with Lelystad, the type I prototype (data not shown), thus confirming that PRRSV-01 belongs to type II genotype of PRRSV. Analysis of structural genes of PRRSV-01 revealed that the virus possesses alterations in the N-glycosylation patterns of its envelope glycoproteins. GP3 of type II PRRSV typically contains 7 potential N-glycosylation sites, all of which are well conserved in different PRRSV strains (Fig. 2A). Recent studies demonstrate that the site at position 195 of PRRSV strain FL12 is not glycosylated (12). PRRSV-01 GP3 lacks one potential N-glycosylation site compared to other PRRSV strains due to the N131D substitution (Fig. 2A). The number of potential N-glycosylation sites on GP5 of type II PRRSV strains may vary between 3 and 5 (Fig. 2B). While the N-glycosylation sites at N44 and N51 are well conserved, the number and position of N-glycosylation sites upstream of N44 are highly variable. PRRSV-01 GP5 harbors 3 N-glycosylation sites at positions N29, N32, and N44 (Fig. 2B). However, compared to FL12 and other wt PRRSV strains, PRRSV-01 GP5 possesses an N51S substitution that disrupts the N-glycosylation site at this position (Fig. 2B). There are no changes in the N-glycosylation patterns in GP2 and GP4 of PRRSV-01 (data not shown).
Fig. 2.
PRRSV-01 virus naturally lacks two N-glycosylation sites in its envelope glycoproteins. Multiple-sequence alignments of GP3 (A) and GP5 (B) collected from the NCBI database. Only regions that cover the potential N-linked glycosylation sites are shown. Asparagine (N) residues that contain the potential N-glycosylation sites are shown in boldface type. Positions in the PRRSV-01 sequence where the glycosylation sites are absent compared to other porcine reproductive and respiratory syndrome virus (PRRSV) strains are shown on a gray background.
To determine whether PRRSV-01 GP3 and GP5 actually lack N-glycosylation sites, we examined the electrophoretic mobility of the proteins on SDS-polyacrylamide gels. FL12 GP3 and GP5 were used for comparison. PRRSV-01 GP3 migrates to a position of ∼40 kDa, which is about 2.5 kDa more than for FL12 GP3 (Fig. 3B, compare lanes 1 and 11). Likewise, PRRSV-01 GP5 migrates to a position of ∼22 kDa, which is also ∼2.5 kDa less than for FL12 GP5 (Fig. 3C, compare lanes 1 and 11), although PRRSV-01 GP5 contains the same number of potential N-glycosylation sites as FL12 GP5 does (Fig. 2B). Since each N-glycan moiety contributes ∼2.5 kDa to the protein mass (23), the results indicate that for both GP3 and GP5 of PRRSV-01, each contains one N-glycan moiety less than the corresponding proteins of strain FL12. The results also suggest that 1 of the potential N-glycosylation sites in GP5 of PRRSV-01 is not used for incorporation of the N-glycan moiety.
Reverse genetic system for studying the influence of the absence of N-glycan moiety on the neutralization phenotype of PRRSV-01.
It has been well documented that modification of viral envelope proteins by glycosylation is one of the common strategies viruses employ to evade the host immune system (8, 16, 24, 38, 44). We were therefore interested in studying the possible influence of the altered glycosylation patterns in GP3 and GP5 of PRRSV-01 on the susceptibility of the virus to antibody neutralization in vitro and on the ability of the virus to mount a NAb response in vivo. To facilitate our study, a chimeric cDNA infectious clone (FL01) was generated by replacing most of ORF2a, the entire ORFs for ORF3 to ORF7, and the 3′UTR of FL12 cDNA infectious clone (41) with the counterparts of PRRSV-01 (Fig. 3A). Using FL01 as a backbone, N-glycosylation sites were separately introduced into the glycan-missing positions in GP3 and GP5 by mutating D131 in GP3 and S51 in GP5 to N, respectively. A double mutant was also generated by mutating D131 in GP3 and S51 in GP5 to N simultaneously. Viable viruses were rescued from FL01 and mutants derived from it. The structural genes of the rescued viruses were amplified by RT-PCR and sequenced to confirm the presence of desired mutations and the absence of any other unplanned mutations (data not shown).
To ascertain that the N-glycosylation sites artificially introduced into GP3 and GP5 were used for incorporation of glycan moieties, we examined the electrophoretic mobility of the proteins on SDS-polyacrylamide gels. As shown in Fig. 3B, GP3 of FL01/GP3-N131D and FL01/DM migrated to a position of ∼42 kDa, which is the same molecular mass as FL12 GP3. Similarly, GP5 of FL01/GP5-S51N and FL01/DM migrated to the same position as FL12 GP5 did (Fig. 3C). The results indicated that additional molecular mass was added to GP3 and GP5 when artificial N-glycosylation sites were introduced into these proteins. To confirm that the additional molecular weight was indeed due to the addition of glycan moieties, proteins after precipitation with anti-GP3 and anti-GP5 antibodies were treated with peptide N-glycosidase F (PNGase F), an enzyme that cleaves off all forms of N-linked oligosaccharides from glycoproteins. As shown in Fig. 3B and C, after treatment with PNGase F, all of the proteins migrated to the same position. These results indicated that the additional molecular mass was indeed due to the addition of N-glycan moieties into the proteins. Collectively, our results indicated that the artificial N-glycosylation sites were used for incorporation of glycan moieties.
Influence of the reintroduced N-glycosylation sites on viral replication in vitro.
We had previously reported that deletions of certain N-glycosylation sites in GP3 and GP5 of PRRSV strain FL12 severely affected the replication of the mutated virus in cell culture (3, 12). In the present study, both PRRSV-01 and FL01 grew very efficiently in MARC-145 cells in spite of the absence of N-glycosylation sites in their GP3 and GP5 (Fig. 4). We asked whether introduction of N-glycosylation sites into the glycan-missing positions of FL01 would have any effects on the viral growth properties. To this end, growth kinetics was examined in MARC-145 cells. Overall, no significant difference in growth kinetics was observed among the viruses (Fig. 4A). All of the viruses reached their peak viral titers at times ranging from 24 to 48 h p.i., and the titers gradually declined thereafter. The similar growth kinetics for FL01 and mutants derived from it suggested that reintroducing N-glycosylation sites into the FL01 genome did not affect the efficiency of viral replication in vitro. Plaque morphology was also examined in MARC-145 cells. As shown in Fig. 4B, all of the viruses were able to generate clear and distinct plaques. Plaques generated by FL01 and FL01/GP3-D131N were almost similar in size, and the sizes of their plaques were intermediate between the plaques generated by PRRSV-01 and FL12. Plaques generated by FL01/GP5-S51N and FL01/DM were slightly smaller than those generated by FL01.
Fig. 4.
Effects of the absence of N-glycosylation sites in GP3 and GP5 on the in vitro growth properties. (A) Single-step growth curves of the indicated viruses upon infection of MARC-145 cells at an MOI of 5. Viral titers are expressed as means ± standard errors of the means (SEMs) (error bars) of data obtained from three independent experiments. (B) Plaque morphology in MARC-145 cells.
Influence of the reintroduced N-glycosylation sites on the susceptibility to antibody neutralization in vitro.
We sought to study the influence of the modification of N-glycosylation sites on the susceptibility of FL01 to antibody neutralization. For reference, we used serum samples collected from 4 different pigs that had been infected with FL01. Neutralizing activities of these antisera were separately measured against viruses with different N-glycosylation patterns. As shown in Table 2, the FL01-specific antisera could barely neutralize FL12 in spite of the fact that the genomes of FL01 and FL12 share the same nonstructural region. Conversely, the neutralization titers of the FL01-specific sera measured against PRRSV-01 were not significantly different from the titers measured against FL01, the homologous virus, although the two viral genomes differ in their nonstructural portions. Neutralization titers of the sera were significantly reduced when measured against FL01/GP5-S51N, the virus that contains the reintroduced N-glycosylation site in its GP5. This result corroborates our previous finding which indicated the importance of N-glycan at N51 in GP5 of PRRSV on the susceptibility of the virus to antibody neutralization (3). Strikingly, neutralization titers were also significantly reduced when measured against FL01/GP3-D131N. Neutralization titers were further reduced when measured against FL01/DM, suggesting an additive effect of N-glycans in GP3 and GP5. Collectively, our results indicate that the absence of N-glycans in GP3 and GP5 of PRRSV-01 is responsible for the enhanced susceptibility of this viral isolate to antibody neutralization in vitro.
Table 2.
Susceptibility of different PRRSV strains to antibody neutralization in vitro
| Serum samplea | Neutralization titerb measured against the following virus: |
|||||
|---|---|---|---|---|---|---|
| FL01 | PRRSV-01 | FL12 | FL01/GP3-D131N | FL01/GP5-S51N | FL01/DM | |
| 1 | 2,048 | 2,048 | 8 | 256 | 128 | 64 |
| 2 | 8,192 | 4,096 | 8 | 1,024 | 256 | 64 |
| 3 | 1,024 | 2,048 | ≤4 | 256 | 256 | 64 |
| 4 | 1,024 | 2,048 | 8 | 128 | 256 | 32 |
| Geometric mean | 2,048 A | 2,435.5 A | 4.8 B | 304.4 C | 215.3 CD | 53.8 D |
Serum samples were obtained from pigs that had been infected with FL01 at 49 days postinfection (p.i.).
Neutralization titers were expressed as the reciprocal of the highest dilution that showed 90% or greater reduction in the number of fluorescent foci presenting in the control wells. Means with different letters (A to D) are significantly different (P < 0.05).
Influence of the reintroduced N-glycosylation sites on the ability to elicit NAb response in vivo.
We inoculated six different groups of recently weaned pigs with viruses containing different glycosylation patterns to study the influence of the reintroduced N-glycosylation sites on the ability to elicit neutralizing antibody response in vivo (Fig. 5). First, we measured neutralizing activities of all serum samples taken at different time points, ranging from 7 to 49 days p.i. against the corresponding homologous viruses (the term homologous refers to the virus used for infection in each case). Pigs infected with FL01 and PRRSV-01 mounted very early and robust NAb responses. Homologous NAbs were detected in pigs infected with these 2 viruses at times as early as 14 days p.i., and the antibody titers quickly increased and reached the mean titers of between 1:2,048 and 1:4,096 at 49 days p.i. There were no significant differences in the kinetics of homologous NAb responses between pigs infected with FL01 and pigs infected with PRRSV-01. In sharp contrast, homologous NAbs were not detected in pigs infected with FL01/DM and FL12 until 28 days p.i. At 49 days p.i., the mean neutralization titer of FL01/DM-infected pigs was only 1:45, which was ∼76-fold lower than the homologous titers of pigs infected with FL01 at the same time p.i. Homologous NAb induction in pigs infected with FL01/GP3-D131N and FL01/GP5-S51N was intermediate between FL01/DM and FL01. These results demonstrate that the N-glycan moieties in both GP3 and GP5 of PRRSV-01 affect the ability of the virus to elicit a homologous NAb response.
Next, we wanted to study the effects of N-glycan moieties on the NAb response against FL01/DM, the fully glycosylated virus. Antisera from pigs infected with hypoglycosylated viruses possessed greater neutralizing activities against FL01/DM than did the antisera from FL01/DM-infected pigs (Table 3). However, the differences in neutralizing activities in the different groups of pigs were not statistically significant.
Table 3.
Neutralizing activities of serum samples measured against FL01/DMa
| Virus | Neutralization titerb |
|---|---|
| PRRSV-01 | 98.7 |
| FL01 | 128 |
| FL01/GP3-D131N | 58.7 |
| FL01/GP5-S51N | 64 |
| FL01/DM | 45.3 |
The serum samples were from pigs infected with different viruses, and the samples were collected at 49 days p.i.
The neutralization titers were expressed as the geometric means of endpoint titers (n = 4 pigs per group). The differences observed among the means of titers were not statistically significant (P > 0.05 by one-way ANOVA).
Simultaneous occurrence of viremia and NAbs.
One of the hallmarks of PRRSV infection is the prolonged viremic phase. Our laboratory had previously demonstrated that passive transfer of NAbs to adult sows completely blocked the establishment of viremia to the point of reaching sterilizing conditions (33) and that young pigs with a level of circulating NAbs sufficient to reach a titer of 1:8 in serum can protect the pigs from viremia after challenge with a highly virulent PRRSV strain (25). Since PRRSV-01 and FL01 mounted an early and robust homologous NAb response, we were interested in examining the effects of these early and robust NAb responses on the levels of viremia, as this would have significant implications for the potential use of this strain in an immunization approach. To our surprise, viremia was still detected in all pigs infected with PRRSV-01 as well as in those infected with FL01 at 14 day p.i. in spite of the presence of NAbs at a titer greater than 1:8 (Table 4).
Table 4.
Viremia measured in swine monocyte-derived macrophages
| Group and pig IDa no. | Viral titerb at the following day p.i.: |
||
|---|---|---|---|
| 7 | 14 | 21 | |
| PRRSV-01-infected group | |||
| 327 | 2.25 | 2.25 | 2.75 |
| 343 | 2.50 | 3.25 | ND |
| 527 | ND | 3.00 | 3.25 |
| 972 | 1.75 | 4.75 | 3.50 |
| Mean | 2.17 | 3.31 | 3.17 |
| FL01-infected group | |||
| 310 | 3.00 | 3.50 | 2.25 |
| 318 | 2.00 | 5.00 | 2.00 |
| 337 | 2.50 | 4.50 | 3.00 |
| 355 | 2.25 | 4.75 | 2.75 |
| Mean | 2.44 | 4.44 | 2.50 |
| FL01/DM-infected group | |||
| 311 | 5.50 | 1.75 | 2.50 |
| 317 | 4.25 | 4.75 | ND |
| 322 | 5.50 | 4.50 | ND |
| 325 | 4.75 | 1.75 | 1.75 |
| Mean | 5.00 | 3.19 | 2.13 |
ID, identification.
Viral titers at the indicated times (days) p.i. are expressed as the log10 TCID50/ml. ND, not detected.
Emergence of a NAb escape mutant.
On the basis of previous results reported for lactate dehydrogenase-elevating virus (LDV), a related arterivirus (8), we reasoned that during the process of replication in the host, the PRRSV-01 and FL01 viruses may have acquired mutations that helped them escape the effect of NAbs. To address this possibility, we isolated virus from a serum sample collected from one pig that had been infected with FL01 at 14 days p.i. The virus was designated FL01-14dpi, and the susceptibility of the virus to antibody neutralization was examined. As shown in Fig. 6A, there was a great difference in the susceptibility of FL01-14dpi and FL01 to neutralization by FL01-specific antisera. For instance, all serum samples at 14 days p.i. were able to neutralize FL01 with titers equal to or greater than 1:8, while none of the serum samples were able to neutralize FL01-14dpi. At 49 days p.i., when the mean neutralization titer against FL01 was 1:3,444, the titer against FL01-14dpi was only 1:90. These results indicate that the virus at 14 days p.i. (i.e., FL01-14dpi) is much more resistant to antibody neutralization than the input virus (FL01).
Fig. 6.
Emergence of antibody escape mutant. (A) FL01-14dpi was isolated from a serum sample from a pig that had been infected with FL01 virus at 14 days postinfection (dpi). The neutralizing activities of serum samples from all four FL01-infected pigs at the indicated days postinfection (PI) were measured against FL01-14dpi. Neutralization titers are expressed as means ± SEMs. Kinetics of FL01 homologous antibody response was redrawn from Fig. 5 for comparison. (B) Mutations in the structural genes of FL01-14dpi compared to the corresponding genes of FL01 before infection. Amino acid changes that result in reappearance of N-glycosylation sites are shown in boldface type.
To determine the mutations responsible for the resistance of FL01-14dpi to antibody neutralization, the entire structural region of the viral genome was sequenced. Overall, we detected a total of 4 amino acid changes (Fig. 6B). Surprisingly, two of the amino acid changes resulted in the appearance of N-glycosylation sites at the glycan-missing positions both in GP3 and GP5. The results suggested that the reappearance of N-glycosylation sites is most likely responsible for the resistance of FL01-14dpi to antibody neutralization, although the contribution of other mutations may not be ruled out. To determine whether the restoration of N-glycosylation sites also happened in other pigs, seven more viruses were further isolated from serum samples at 14 days p.i., which included 3 viruses from the remaining FL01-infected pigs and 4 viruses from PRRSV-01-infected pigs. After the viruses were isolated from serum samples, ORF3 and ORF5 of these viruses were sequenced. Similar to FL01-14dpi, all of the 7 newly isolated viruses exhibited N-glycosylation sites at the glycan-missing positions (data not shown). In addition, we also observed an S204P mutation in GP3 of 1 virus and a C112F mutation in GP5 of another virus (data not shown). The rapid reappearance of N-glycosylation sites in GP3 and GP5 suggests that there is a strong selective pressure at those sites.
DISCUSSION
Many viruses rely on glycosylation modification of their envelope glycoproteins to evade the host immune system (16, 24, 38, 44, 45). It has been demonstrated in the case of PRRSV that N-glycans surrounding the major neutralizing epitope located in the ectodomain of GP5 protect the virus from antibody neutralization and impair the immunogenicity of the epitope (3, 15). Although the PRRSV virion carries 4 different glycoproteins, namely, GP2, GP3, GP4, and GP5, so far it is not known whether the N-glycans on glycoproteins other than GP5 were involved in immune evasion. In the present study, we demonstrate that the N-glycan in GP3 of a type II PRRSV isolate is as important as its counterpart in GP5 regarding its role in protection of the virus from antibody neutralization.
GP5 of type II PRRSV contains 3 to 5 potential N-glycosylation sites, of which the N-glycosylation sites at N44 and N51 are strongly conserved, while the N-glycosylation sites upstream of N44 are variable (Fig. 2B). Our laboratory had demonstrated that FL12 mutants carrying disrupted N-glycosylation sites at N34 and N51 of GP5 were more susceptible to antibody neutralization and elicited greater NAb responses than did the wt FL12 strain (3). Likewise, Faaberg et al. (15) reported that the PRRSV field isolate HV-1 naturally lacking the N-glycosylation site(s) upstream of N44 mounted a significantly early and strong NAb response which was coincident with the increased antibody binding to the nearby neutralizing epitope. Surprisingly, the authors observed that another PRRSV field isolate named N44 lacking N-glycosylation site at position N44 did not display any enhancement in eliciting NAbs even though this glycosylation site is located inside the neutralizing epitope (15). In the present study, we demonstrate that the absence of N-glycosylation site at N51 in GP5 of PRRSV-01 also increases the susceptibility of the virus to antibody neutralization as well as enhances the ability of the virus to mount a homologous NAb response. Collectively, the available information suggests that N-glycans located upstream of N44 and N51 are important for the virus to escape antibody neutralization, while the N-glycan at N44 might not be as central to that purpose.
A remarkable observation in our study is the involvement of an N-glycan moiety in GP3 of a type II PRRSV isolate in interference with antibody neutralization. GP3 of PRRSV harbors 7 potential N-glycosylation sites, the highest number of glycosylation sites compared to all the other PRRSV envelope glycoproteins (12). Moreover, all the glycosylation sites are strongly conserved in different type II PRRSV strains (Fig. 2A). To examine the role of N-glycosylation of minor glycoproteins in eliciting NAbs, we previously generated a series of mutant viruses, each of which carry one glycosylation deletion or multiple glycosylation deletions. We found that disruption of N-glycosylation sites at N29, N152, and N160 in GP3 of PRRSV strain FL12 did not affect virus susceptibility to antibody neutralization as well as the ability to mount a NAb response (12). We report herein that the absence of N-glycosylation site at N131 in GP3 of PRRSV-01 results in enhanced neutralization susceptibility and increased NAb elicitation. The results suggest that the relative position of the glycosylation site in the glycoprotein might be important for its ability to modulate NAb induction.
We do not know at this time how the N-glycan at N131 in GP3 of PRRSV-01 protects the virus from neutralization by NAbs. There is evidence that GP3 of type II PRRSV might contain one neutralizing epitope, but the location of this epitope has not been mapped (6). It is therefore tempting to speculate that the glycan moiety at N131 in GP3 may prevent the accessibility of NAbs to an epitope located within GP3. On the other hand, GP2, GP3, and GP4 of PRRSV interact each other to form a multiprotein complex (11). Thus, it is also possible that the glycan moiety in GP3 might mask the epitope located in GP2 or GP4.
The neutralizing epitope in GP5 that we initially described has been considered the major neutralizing epitope (34). In this study, the susceptibility to antibody neutralization of FL01/GP3-D131N is not significantly different from that of FL01/GP5-S51N (Table 2). Likewise, both viruses yield similar homologous NAb titers at 49 days p.i., although there seems to be a slight difference in the kinetics of NAb response (Fig. 5). Together, the results indicate that the N-glycan in GP3 is as important as the one in GP5 regarding its role in modulating the NAb response. Therefore, this notion should be taken into account in future PRRSV vaccine design.
The presence of glycan moieties in the viral envelope proteins not only renders the virus resistant to antibody neutralization but also impairs the ability of the virus to elicit NAb responses. Therefore, removal of glycosylation sites increases the susceptibility of the virus to antibody neutralization as well as enhances the ability of the virus to elicit NAbs in vivo (3, 38). In the present study, although all the hypoglycosylated viruses were able to elicit homologous NAb responses greater than the FL01/DM virus, none of them were able to elicit significantly greater neutralization titers measured against FL01/DM (Table 3). We reason that the failure of hypoglycosylated viruses to induce greater NAb titers against FL01/DM was due to the rapid reappearance of glycosylation sites at the glycan-missing positions. It is noteworthy that although the glycosylation sites reappeared within 14 days p.i., FL01 was still able to show a tendency to induce a greater NAb titer against FL01/DM. It is therefore plausible to hypothesize that if the hypoglycosylated form is stable for longer periods of time after infection, then the pigs infected with hypoglycosylated viruses may produce greater NAb titers to the FL01/DM than those infected with FL01/DM.
We had previously reported that removal of the N-glycosylation sites in GP3 or GP5 of the PRRSV strain FL12 severely affected viral replication in MARC-145 cells (3, 12). In this study, we observe the enhanced replication efficiency of PRRSV-01 and FL01 in MARC-145 cells in spite of the absence of the N-glycosylation sites in their GP3 and GP5 (Fig. 4). The discrepancy in the effect of N-glycosylation sites on viral replication might be due to the difference in the ways the N-glycosylation sites are deleted. In our previous studies, the N-glycosylation sites in GP3 and GP5 of FL12 were artificially removed from the genome of the virus, and presumably no other mutations occurred (3, 12). In the current study, the N-glycosylation sites were naturally deleted. We assume that concomitant with the mutations leading to the removal of N-glycosylation sites from GP3 and GP5 of PRRSV-01, other compensational mutations also occurred which together enhance the replication of the virus in cell culture.
PRRSV-01 was isolated from a field sample submitted to the Veterinary Diagnostic Laboratory of Iowa State University. Immediately after isolation, the ORF5 of PRRSV-01 was sequenced as a routine surveillance (GenBank accession no. EU556202.1). After that, the virus was propagated in MARC-145 cells for multiple passages. Retrospective analysis of the ORF5 sequence obtained at the time of isolation of the virus revealed that the N-glycosylation site at position N51 was indeed present in the gene (data not shown). Since the ORF3 sequence of the virus right after isolation is not available, we do not know the N-glycosylation pattern of this protein at the time of isolation. We hypothesize that both glycosylation sites at N131 in GP3 and N51 in GP5 of PRRSV-01 were lost during replication of the virus in MARC-145 cells.
The rapid reappearance of N-glycosylation sites in both GP3 and GP5 of PRRSV-01 and FL01 after inoculation into pigs suggests a strong selective pressure at these sites. It is most likely that the host immune response is one of the major driving forces for the restoration of the N-glycosylation sites. In addition, the PRRSV-01 virus used in this study had adapted to MARC-145 cells, which differ from porcine macrophage cells (i.e., the natural host cells of PRRSV) in terms of their expression of receptors for PRRSV entry (5, 21, 42). Thus, the different cell tropism might also contribute to the reappearance of the N-glycosylation sites in GP3 and GP5 of PRRSV-01 during its replication in vivo.
The natural absence of N-glycosylation sites had been observed in other viruses such as lactate dehydrogenase-elevating virus (LDV) (8). The primary envelope glycoprotein VP-3P of LDV strains LDV-C and LDV-v naturally lack two N-glycosylation sites compared to strains LDV-P and LDV-vx (8). Consequently, the LDV-C and LDV-v strains are much more susceptible to antibody neutralization and elicit much greater NAb responses than the LDV-P and LDV-vx strains (8). More importantly, the LDV-C and LDV-v strains also regained N-glycosylation sites after 2 or 3 passages in mice and became resistant to antibody neutralization.
In summary, our results herein confirm the previous findings that the N-glycan moieties in GP5 of type II PRRSV are important for the virus to escape the effect of NAbs (3, 15). In addition, we demonstrate for the first time that the N-glycan in GP3 of type II PRRSV is also important in protecting the virus from antibody neutralization. Likewise, the overall body of results herein presented firmly confirms the important notion that GP3 may be involved in inducing neutralizing antibodies, a concept that has already been suggested by previous investigators (6, 19, 22, 37).
ACKNOWLEDGMENTS
This research has been supported by a grant from the PRRS CAP, USDA NIFA award 2008-55620-19132.
We thank Carl Gagnon (University of Quebec, Montreal, Canada) for kindly providing anti-GP5 antibody and Lalit Beura for comments on the manuscript.
The animals used in this research were housed and handled following the protocols approved by the Institutional Animal Care Committee of the University of Nebraska—Lincoln under protocols 07-09-041D and 07-10-048C.
Footnotes
Published ahead of print on 16 March 2011.
REFERENCES
- 1. Albina E., Carrat C., Charley B. 1998. Interferon-alpha response to swine arterivirus (PoAV), the porcine reproductive and respiratory syndrome virus. J. Interferon Cytokine Res. 18:485–490 [DOI] [PubMed] [Google Scholar]
- 2. Allende R., et al. 2000. Porcine reproductive and respiratory syndrome virus: description of persistence in individual pigs upon experimental infection. J. Virol. 74:10834–10837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ansari I. H., Kwon B., Osorio F. A., Pattnaik A. K. 2006. Influence of N-linked glycosylation of porcine reproductive and respiratory syndrome virus GP5 on virus infectivity, antigenicity, and ability to induce neutralizing antibodies. J. Virol. 80:3994–4004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Beura L. K., et al. 2010. Porcine reproductive and respiratory syndrome virus nonstructural protein 1beta modulates host innate immune response by antagonizing IRF3 activation. J. Virol. 84:1574–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Calvert J. G., et al. 2007. CD163 expression confers susceptibility to porcine reproductive and respiratory syndrome viruses. J. Virol. 81:7371–7379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cancel-Tirado S. M., Evans R. B., Yoon K. J. 2004. Monoclonal antibody analysis of porcine reproductive and respiratory syndrome virus epitopes associated with antibody-dependent enhancement and neutralization of virus infection. Vet. Immunol. Immunopathol. 102:249–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cavanagh D. 1997. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 142:629–633 [PubMed] [Google Scholar]
- 8. Chen Z., Li K., Plagemann P. G. 2000. Neuropathogenicity and sensitivity to antibody neutralization of lactate dehydrogenase-elevating virus are determined by polylactosaminoglycan chains on the primary envelope glycoprotein. Virology 266:88–98 [DOI] [PubMed] [Google Scholar]
- 9. Collins J. E., et al. 1992. Isolation of swine infertility and respiratory syndrome virus (isolate ATCC VR-2332) in North America and experimental reproduction of the disease in gnotobiotic pigs. J. Vet. Diagn. Invest. 4:117–126 [DOI] [PubMed] [Google Scholar]
- 10. Conzelmann K. K., Visser N., Van Woensel P., Thiel H. J. 1993. Molecular characterization of porcine reproductive and respiratory syndrome virus, a member of the arterivirus group. Virology 193:329–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Das P. B., et al. 2010. The minor envelope glycoproteins GP2a and GP4 of porcine reproductive and respiratory syndrome virus interact with the receptor CD163. J. Virol. 84:1731–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Das P. B., et al. 2011. Glycosylation of minor envelope glycoproteins of porcine reproductive and respiratory syndrome virus in infectious virus recovery, receptor interaction, and immune response. Virology 410:385–394 [DOI] [PubMed] [Google Scholar]
- 13. Dea S., Gagnon C. A., Mardassi H., Pirzadeh B., Rogan D. 2000. Current knowledge on the structural proteins of porcine reproductive and respiratory syndrome (PRRS) virus: comparison of the North American and European isolates. Arch. Virol. 145:659–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. de Lima M., et al. 2009. GP3 is a structural component of the PRRSV type II (US) virion. Virology 390:31–36 [DOI] [PubMed] [Google Scholar]
- 15. Faaberg K. S., et al. 2006. Neutralizing antibody responses of pigs infected with natural GP5 N-glycan mutants of porcine reproductive and respiratory syndrome virus. Viral Immunol. 19:294–304 [DOI] [PubMed] [Google Scholar]
- 16. Francica J. R., et al. 2010. Steric shielding of surface epitopes and impaired immune recognition induced by the Ebola virus glycoprotein. PLoS Pathog. 6:e1001098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Han J., Wang Y., Faaberg K. S. 2006. Complete genome analysis of RFLP 184 isolates of porcine reproductive and respiratory syndrome virus. Virus Res. 122:175–182 [DOI] [PubMed] [Google Scholar]
- 18. Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51–59 [DOI] [PubMed] [Google Scholar]
- 19. Jiang W., Jiang P., Wang X., Li Y., Du Y. 2008. Enhanced immune responses of mice inoculated recombinant adenoviruses expressing GP5 by fusion with GP3 and/or GP4 of PRRS virus. Virus Res. 136:50–57 [DOI] [PubMed] [Google Scholar]
- 20. Kim H. S., Kwang J., Yoon I. J., Joo H. S., Frey M. L. 1993. Enhanced replication of porcine reproductive and respiratory syndrome (PRRS) virus in a homogeneous subpopulation of MA-104 cell line. Arch. Virol. 133:477–483 [DOI] [PubMed] [Google Scholar]
- 21. Kim J. K., Fahad A. M., Shanmukhappa K., Kapil S. 2006. Defining the cellular target(s) of porcine reproductive and respiratory syndrome virus blocking monoclonal antibody 7G10. J. Virol. 80:689–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim W. I., Yoon K. J. 2008. Molecular assessment of the role of envelope-associated structural proteins in cross neutralization among different PRRS viruses. Virus Genes 37:380–391 [DOI] [PubMed] [Google Scholar]
- 23. Kornfeld R., Kornfeld S. 1985. Assembly of asparagine-linked oligosaccharides. Annu. Rev. Biochem. 54:631–664 [DOI] [PubMed] [Google Scholar]
- 24. Liu M., et al. 2007. Deletion of N-glycosylation sites of hepatitis C virus envelope protein E1 enhances specific cellular and humoral immune responses. Vaccine 25:6572–6580 [DOI] [PubMed] [Google Scholar]
- 25. Lopez O. J., et al. 2007. Protection against porcine reproductive and respiratory syndrome virus (PRRSV) infection through passive transfer of PRRSV-neutralizing antibodies is dose dependent. Clin. Vaccine Immunol. 14:269–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lopez O. J., Osorio F. A. 2004. Role of neutralizing antibodies in PRRSV protective immunity. Vet. Immunol. Immunopathol. 102:155–163 [DOI] [PubMed] [Google Scholar]
- 27. Mardassi H., Massie B., Dea S. 1996. Intracellular synthesis, processing, and transport of proteins encoded by ORFs 5 to 7 of porcine reproductive and respiratory syndrome virus. Virology 221:98–112 [DOI] [PubMed] [Google Scholar]
- 28. Mardassi H., Mounir S., Dea S. 1995. Molecular analysis of the ORFs 3 to 7 of porcine reproductive and respiratory syndrome virus, Quebec reference strain. Arch. Virol. 140:1405–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Meier W. A., et al. 2003. Gradual development of the interferon-gamma response of swine to porcine reproductive and respiratory syndrome virus infection or vaccination. Virology 309:18–31 [DOI] [PubMed] [Google Scholar]
- 30. Meulenberg J. J. 2000. PRRSV, the virus. Vet. Res. 31:11–21 [DOI] [PubMed] [Google Scholar]
- 31. Meulenberg J. J., et al. 1995. Characterization of proteins encoded by ORFs 2 to 7 of Lelystad virus. Virology 206:155–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Meulenberg J. J., van Nieuwstadt A. P., van Essen-Zandbergen A., Langeveld J. P. 1997. Posttranslational processing and identification of a neutralization domain of the GP4 protein encoded by ORF4 of Lelystad virus. J. Virol. 71:6061–6067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Osorio F. A., et al. 2002. Passive transfer of virus-specific antibodies confers protection against reproductive failure induced by a virulent strain of porcine reproductive and respiratory syndrome virus and establishes sterilizing immunity. Virology 302:9–20 [DOI] [PubMed] [Google Scholar]
- 34. Ostrowski M., et al. 2002. Identification of neutralizing and nonneutralizing epitopes in the porcine reproductive and respiratory syndrome virus GP5 ectodomain. J. Virol. 76:4241–4250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pirzadeh B., Dea S. 1997. Monoclonal antibodies to the ORF5 product of porcine reproductive and respiratory syndrome virus define linear neutralizing determinants. J. Gen. Virol. 78(Pt 8):1867–1873 [DOI] [PubMed] [Google Scholar]
- 36. Plagemann P. G., Rowland R. R., Faaberg K. S. 2002. The primary neutralization epitope of porcine respiratory and reproductive syndrome virus strain VR-2332 is located in the middle of the GP5 ectodomain. Arch. Virol. 147:2327–2347 [DOI] [PubMed] [Google Scholar]
- 37. Plana Duran J., et al. 1997. Baculovirus expression of proteins of porcine reproductive and respiratory syndrome virus strain Olot/91. Involvement of ORF3 and ORF5 proteins in protection. Virus Genes 14:19–29 [DOI] [PubMed] [Google Scholar]
- 38. Reitter J. N., Means R. E., Desrosiers R. C. 1998. A role for carbohydrates in immune evasion in AIDS. Nat. Med. 4:679–684 [DOI] [PubMed] [Google Scholar]
- 39. Snijder E. J., Meulenberg J. J. 1998. The molecular biology of arteriviruses. J. Gen. Virol. 79(Pt 5):961–979 [DOI] [PubMed] [Google Scholar]
- 40. Subramaniam S., et al. 2010. Porcine reproductive and respiratory syndrome virus non-structural protein 1 suppresses tumor necrosis factor-alpha promoter activation by inhibiting NF-kappaB and Sp1. Virology 406:270–279 [DOI] [PubMed] [Google Scholar]
- 41. Truong H. M., et al. 2004. A highly pathogenic porcine reproductive and respiratory syndrome virus generated from an infectious cDNA clone retains the in vivo virulence and transmissibility properties of the parental virus. Virology 325:308–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Van Breedam W., et al. 2010. The M/GP(5) glycoprotein complex of porcine reproductive and respiratory syndrome virus binds the sialoadhesin receptor in a sialic acid-dependent manner. PLoS Pathog. 6:e1000730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vanhee M., et al. 2010. A variable region in GP4 of European-type porcine reproductive and respiratory syndrome virus induces neutralizing antibodies against homologous but not heterologous virus strains. Viral Immunol. 23:403–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang C. C., et al. 2009. Glycans on influenza hemagglutinin affect receptor binding and immune response. Proc. Natl. Acad. Sci. U. S. A. 106:18137–18142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wei X., et al. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307–312 [DOI] [PubMed] [Google Scholar]
- 46. Weiland E., Wieczorek-Krohmer M., Kohl D., Conzelmann K. K., Weiland F. 1999. Monoclonal antibodies to the GP5 of porcine reproductive and respiratory syndrome virus are more effective in virus neutralization than monoclonal antibodies to the GP4. Vet. Microbiol. 66:171–186 [DOI] [PubMed] [Google Scholar]
- 47. Wensvoort G., et al. 1991. Mystery swine disease in The Netherlands: the isolation of Lelystad virus. Vet. Q. 13:121–130 [DOI] [PubMed] [Google Scholar]
- 48. Wills R. W., Doster A. R., Galeota J. A., Sur J. H., Osorio F. A. 2003. Duration of infection and proportion of pigs persistently infected with porcine reproductive and respiratory syndrome virus. J. Clin. Microbiol. 41:58–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wissink E. H., et al. 2005. Envelope protein requirements for the assembly of infectious virions of porcine reproductive and respiratory syndrome virus. J. Virol. 79:12495–12506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wissink E. H., et al. 2003. The major envelope protein, GP5, of a European porcine reproductive and respiratory syndrome virus contains a neutralization epitope in its N-terminal ectodomain. J. Gen. Virol. 84:1535–1543 [DOI] [PubMed] [Google Scholar]
- 51. Wu W. H., et al. 2001. A 10-kDa structural protein of porcine reproductive and respiratory syndrome virus encoded by ORF2b. Virology 287:183–191 [DOI] [PubMed] [Google Scholar]