Abstract
Mutations within MHC class I-restricted epitopes have been studied in relation to T cell-mediated immune escape, but their impact on NK cells via interaction with killer Ig-like receptors (KIRs) during early HIV infection is poorly understood. In two patients acutely infected with HIV-1, we observed the appearance of a mutation within the B*57-restricted TW10 epitope (G9E) that did not facilitate strong escape from T cell recognition. The NK cell receptor KIR3DL1, carried by these patients, is known to recognize HLA-B*5703 and is associated with good control of HIV-1. Therefore, we tested whether the G9E mutation influenced the binding of HLA-B*5703 to soluble KIR3DL1 protein by surface plasmon resonance, and while the wild-type sequence and a second (T3N) variant were recognized, the G9E variant abrogated KIR3DL1 binding. We extended the study to determine the peptide sensitivity of KIR3DL1 interaction with epitopes carrying mutations near the C termini of TW10 and a second HLA-B*57-restricted epitope, IW9. Several amino acid changes interfered with KIR3DL1 binding, the most extreme of which included the G9E mutation commonly selected by HLA-B*57. Our results imply that during HIV-1 infection, some early-emerging variants could affect KIR-HLA interaction, with possible implications for immune recognition.
INTRODUCTION
During HIV-1 infection, the complex interactions between the host and pathogen shape the clinical course of disease. The development of sophisticated sequencing technologies, including single-genome amplification (SGA), now enables tracking of the transmitted/founder virus during acute infection, giving insight into the immune responses that shape HIV evolution in vivo (11, 30). CD8+ T cells play a pivotal role in the control of HIV-1, and a large proportion of the mutations selected in acute infection map within, or in proximity to, HLA class I-restricted epitopes, resulting in escape from immune recognition.
As HLA class I molecules also represent ligands for receptors of the innate immune system, it is plausible that certain mutations in HIV-1 infection could impact innate immune responses. This has recently been described in relation to the leukocyte Ig-like receptors (LILRs), a family of HLA class I binding immunomodulatory receptors expressed primarily on myelomonocytic cells (dendritic cells [DCs], macrophages, and monocytes). An escape mutation that affects CD8+ T cell recognition of the HIV-1 HLA-B*27-restricted KK10 epitope also increased binding to the inhibitory LILR receptor ILT4, resulting in impaired DC function (16). However, despite the increased activity of NK cells during acute HIV-1 infection (1, 3), the impact of early-emerging viral variants on NK receptors has not previously been described.
For another family of NK receptors, the killer cell immunoglobulin-like receptors (KIRs), epidemiological evidence has highlighted the importance of specific HLA class I-KIR pairs in determining disease outcomes (21). The KIRs are a polymorphic group of receptors expressed by NK cells and a subset of mature CD8+ T cell lymphocytes and comprise both inhibitory and activating allotypes with either two or three immunoglobulin-like extracellular domains. The inhibitory receptors have long cytoplasmic tails encoding ITIM motifs, while the activating receptors have short cytoplasmic tails and interact with ITAM-bearing adaptor molecules, such as DAP12. The binding of KIRs to their respective HLA class I ligands is determined by polymorphic regions in the HLA class I alpha 1 domain. Thus, a dimorphism at position 80 dictates binding of HLA-C to the two-domain KIRs, whereas the three-domain 3DL1 receptors preferentially bind HLA-A and -B allotypes containing the serological Bw4 motif (positions 77 to 83 in the alpha 1 helix) (18). Both the two- and three-domain KIRs interact with amino acids near the C terminus of the bound peptide (27, 28).
In HIV-1 infection, certain inhibitory KIR3DL1 allotypes, in conjunction with their known HLA class I ligands, associate with different disease progression rates, susceptibility to opportunistic infection, and viral-load set point. The combinations conferring the greatest protection with respect to outcome include Bw4-encoding HLA-B*57 subtypes (including B*5701, B*5703, and the closely related B*5801 subtype) in conjunction with the highly expressed 3DL1 allotypes (8). Although carriage of B*57 alone is associated with a favorable outcome (which is currently attributed to both the nature of the epitopes presented and the functional quality of CD8+ T cells selected [15, 23]), its protective quality is enhanced in combination with specific inhibitory KIR3DL1 allotypes (8, 19). Here, we studied two African-American HLA-B*5703-positive patients, CH77 and CH58, who were KIR3DL1 positive (and KIR3DS1 negative) and in whom early T cell responses against the immunodominant HLA-B*57-restricted gag TSTLQEQIGW peptide (TW10) were generated (11). In both patients, two mutations emerged within TW10 that mapped to separate viral isolates: a glycine-to-glutamate change at position 9 in the epitope (G9E) and a threonine-to-asparagine change at position 3 (T3N). The latter mutation is specifically selected by T cells, and although it is not clear to what extent the T3N mutation impacts immune control, the positioning of T3N within the p24 capsid, its attenuating impact on viral growth in vitro, and the propensity of T3N variants to revert upon transmission to B*57-negative hosts all suggest that selection of this mutation is important (15, 22). In line with previous findings, we show that the T3N change abrogates T cell recognition of the TW10 epitope; however, the G9E mutation, which occurred earlier in infection, had only a modest effect. Therefore, we tested binding of soluble KIR3DL1 protein to HLA-B*5703 protein refolded with both variants of the TW10 epitope and scaled up the study to include the analysis of all possible amino acid substitutions adjacent to the C-terminal anchor residue. We observed that the interaction between KIR3DL1 and HLA-B*5703 was greatly affected by mutations at the penultimate residue of the epitope peptide, most strikingly for the G9E change. It is possible, therefore, that early-emerging mutations that map to T cell epitopes could also affect both KIR-expressing NK and T cells in vivo.
MATERIALS AND METHODS
Patients and samples.
Patients 700010058 (CH58) and 700010077 (CH77) were enrolled in the acute-infection arm of the CHAVI 001 cohort as described previously (11). Peripheral blood mononuclear cells (PBMCs) were isolated over a Ficoll gradient via centrifugation. Following subsequent wash steps, the PBMCs were cryopreserved at approximately 107cells/vial for downstream analyses.
KIR genotyping.
The KIR genotype and KIR3DL1 subtypes were determined by a multiplex PCR–sequence-specific priming (SSP) methodology as previously described (20, 34).
Gamma interferon (IFN-γ) enzyme-linked immunospot (ELISPOT) assay.
Cryopreserved PBMCs were thawed and rested for 2 h prior to being plated as described previously (11). The cells were plated at 105 cells per well and incubated with peptide over a concentration of 10−5 to 10−11 M at 37°C in 5% CO2 overnight. Responses in excess of 50 spot-forming cells (SFCs) per million PBMC, and greater than 4 times the negative control (i.e., PBMCs plus medium only), were considered positive. For all assays, six negative wells and at least one positive well (10 μg/ml phytohemagglutinin [PHA]) were included, and sample wells were set up in triplicate.
Peptide generation.
Both UV photolabile and conventional peptide epitopes were synthesized based on Fmoc (9-fluorenylmethoxy carbonyl) chemistry. Conventional peptides included variants of the HLA-B*5703-restricted gag epitopes TW10 (TSTLQEQIGW239-249) and IW9 (ISPRTLNAW147-155), incorporating all 20 amino acid substitutions at minus 1 relative to the C-terminal tryptophan anchor residues (C−1), and a subset of peptides with substitutions at minus 2 relative to the C-terminal anchor (C−2). These peptides are referred to here as TW10 and IW9, C−1/C−2 variants. A photolabile version of IW9, comprising a UV-sensitive 3-amino-3-(2-nitrophenyl)-propionic acid residue replacement at position C−1 (9MT4 epitope), was used to generated UV-sensitive HLA-B*5703-β2 M-peptide complexes. The refolding of HLA-B*5703-β2m-9MT4 and subsequent biotinylation, purifications, and UV photocleavage/peptide exchange were performed as outlined previously (29). Following biotinylation, 0.5 μM (approximately 25 μg/ml) of UV-sensitive HLA-B*5703-β2 M-9MT4 monomers was incubated with 50 μM “rescue” peptide in polypropylene 96 (V-shaped)-well plates (Greiner Bio-One), and the final volume was adjusted to 125 μl by adding exchange buffer (20 mM Tris, pH 7, 150 mM NaCl). The samples were then incubated (on ice) in a Camag UV cabinet fitted with a long-wave UV lamp (366 nm). Photoillumination was performed for a total of 60 min, after which the plates were centrifuged to remove aggregated material. The purity of the peptide variants and the integrity of HLA-B*5703 UV-sensitive monomers (pre-UV exchange) were evaluated using a Ultraflex matrix-assisted laser desorption ionization–time of flight (MALDI TOF)/TOF mass spectrometer (Bruker Daltonics) prior to major histocompatibility complex (MHC) class I tetramer generation and surface plasmon resonance (SPR) binding studies.
Construct design for soluble KIR3DL1 proteins.
The extracellular domain (ECD) of KIR3DL1*002 was amplified by PCR (forward primer containing a SalI site [underlined], GGGTCGACCACGTGGGTGGTCAGGAC; reverse primer containing an XhoI site [underlined], GGCTCGAGGTTACCAGATTTGGAGCTTGG), from Image clone 8069046 (Bioscience), and then digested with XhoI and SalI. The presence of an internal XhoI site in the KIR3DL1 ECD necessitated the cloning of the ECD fragment in two steps. The short SalI-XhoI 5′ fragment and the longer XhoI-XhoI 3′ fragment were sequentially inserted into the XhoI site of a modified pMe18s vector containing the leader sequence of the murine SLAM (CD150) gene and human IgG-Fc (a kind gift of L. Lanier, University of California—San Francisco) (7). All constructs were confirmed mutation free by automated sequencing using an ABI3730 (Applied Biosystems).
Expression and purification of soluble KIR3DL1-Fc protein.
HEK-293T cells were cultured in RPMI 1640 medium supplemented with 10% fetal calf serum, 50 units/ml penicillin, 50 μg/ml streptomycin, and 2 mM l-glutamine. Thirty to 70% subconfluent cells in 14-cm-diameter culture plates (grown in Dulbecco's modified Eagle's medium [DMEM], supplemented as described above) were transfected with calcium phosphate precipitates of 10 μg of plasmid DNA, and the medium was changed to CHO-S-SFM II (Invitrogen) supplemented with 50 units/ml penicillin, 50 μg/ml streptomycin, and 2 mM l-glutamine after 8 to 10 h. The cells were grown for a further 3 days before soluble Fc-tagged protein was purified. Typically, 40 to 80 plates were transfected for each construct. Culture supernatants were centrifuged for at 4°C for 20 min (4,000 rpm in a Beckman J2-21 centrifuge), filtered through 0.22-μm PES bottle-top filters (Corning), and then applied under gravity flow to columns containing 2 ml (packed volume) Protein A Sepharose 4B Fast Flow (Sigma-Aldrich) previously equilibrated in DPBS (Sigma-Aldrich). The columns were washed with 50 ml of Dulbecco's phosphate-buffered saline (DPBS) prior to the elution of bound protein with 0.1 M glycine, pH 3.2. Fractions (1 ml) were collected into tubes containing 50 μl of 2 M Tris buffer, and the fractions containing eluted protein were identified spectrophotometrically using a NanoDrop ND1000 (Thermo Scientific). Appropriate fractions were pooled and dialyzed three times (a minimum of 4 h per dialysis) against 2 liters of DPBS and then concentrated using a 10,000-molecular-weight-cutoff Amicon Ultra centrifuge filter (Millipore). Aliquots of the concentrated protein were stored at −80°C.
SPR analyses.
SPR studies were performed using a BIAcore 3000 (GE Healthcare). Immediately following UV photoillumination, C-terminal biotinylated HLA-B*5703-peptide exchange complexes were immobilized via coupling to streptavidin-coated SA sensor chips (GE Healthcare) at levels of 2,000 to 2,500 response units (RU). For each round of screening runs, three of the four flow cells contained individual immobilized HLA-B*5703–peptide complexes, while the fourth flow cell was reserved for a negative control (biotinylated human CD4 protein). A fixed concentration of soluble KIR3DL1-Fc (1 mg/ml) was injected over all flow cells at a rate of 10 μl per minute. Following KIR3DL1-Fc dissociation, the proportion of immobilized B*5703-β2 M peptide complexes comprising correctly folded material was determined by saturation binding, using the MHC class I-reactive and conformation-dependent W6/32 antibody (Cambridge Bioscience Ltd.). Binding responses were determined by subtracting the response measured in the control flow cell, and the final data sets were normalized to correct for differences in the proportion of peptide-MHC complexes immobilized.
RESULTS
Evolution of TW10 variants and CD8+ T cell recognition during acute infection.
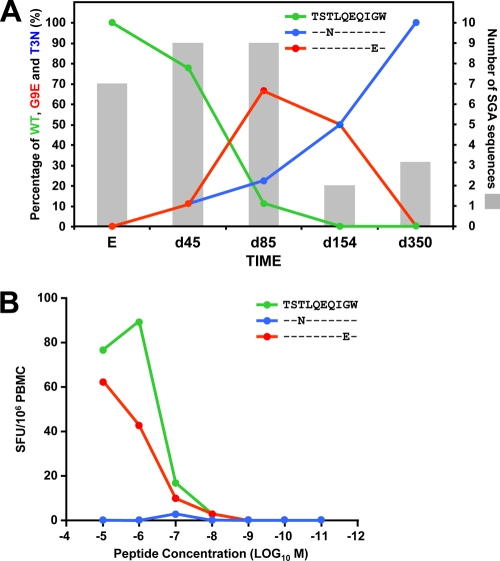
SGA and sequence analysis of samples acquired from two HLA-B*5703-positive individuals during Fiebig stage 2 tracked the emergence of two distinct mutations within the TW10 (gag 240 to 249) epitopes as described previously (11, 30). In donor CH77, a mutation within TW10 at position 9 (G9E) was detected on day 159 after screening (during acute peak viremia) and preceded a separate variant (detected on day 592) carrying mutations at positions 3 (T3N), 8 (V8I), and 9 (G9A). In patient CH58, two distinct viral variants were detected on day 45, one encoding the T3N and the other a G9E mutation, and the latter became the predominant variant for a short period before the final escape to T3N (Fig. 1A). First, we asked how these mutations affected CD8+ T cell recognition and tested PBMCs from donor CH58 for recognition of these variants when presented by autologous cells over a range of concentrations in an IFN-γ ELISPOT assay. Whereas the T3N mutation abrogated T cell-mediated recognition completely, in agreement with published findings (15), the G9E mutation had only a modest impact on T cell recognition (Fig. 1B). Hence, there was a marked difference in the abilities of the early-emerging T3N and G9E variants to escape T cell recognition.
Fig. 1.
Viral evolution and CD8+ T cell recognition of the TW10 G9E variant in patient CH58. (A) Sequence analysis of the TW10 epitope in single genome amplified (SGA) viral RNA for patient CH58, illustrating the gradual decline of the wildtype (WT) TW10 epitope, coincident with the emergence of the G9E and T3N variants. Times are given in days from enrollment (E). G9E was detected as the dominant virus between days 45 and 85, and gradually declined thereafter. The T3N variant subsequently emerged as the dominant variant and was prevalent from day 154 onwards (data are from reference 11). (B) Interfeon-γ ELISPOT assays were used compare recognition of the founder viral TW10 epitope (TSTLQEQIGW) with that of the G9E and T3N variants in donor CH58. PBMCs were incubated overnight in the presence of varying amounts of each peptide (10−11 to 10−5 M). Responses are reported as spot-forming cells (SFC) per million PBMCs.
KIR3DL1-mediated recognition of the wild-type TW10 and the early-emerging variants.
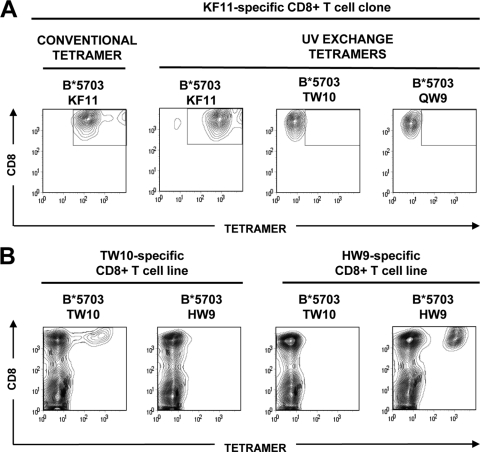
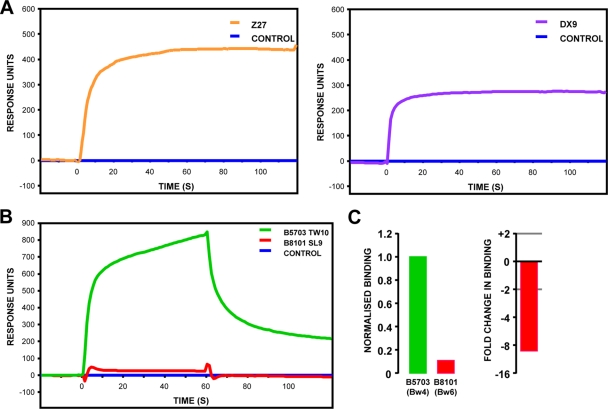
Both CH58 and CH77 carry KIR3DL1 allotypes, which, in combination with certain Bw4 subtypes, including HLA-B*5703, confer enhanced protection against HIV-1 disease progression (8, 21). Therefore, we asked whether the G9E and T3N mutations affected KIR3DL1-mediated recognition. To evaluate the binding of KIR3DL1 to HLA-B*5703-β2m in complex with the wild-type TW10 epitope and the G9E and T3N variants, we expressed and folded both components as recombinant proteins. We generated HLA-B*5703 in complex with the variant TW10 peptides using UV-ligand exchange technology (35) as, in our experience, this method facilitates the production of more stable B*5703-TW10 monomers than with conventional MHC class I folding methods. To confirm the specificity of the UV-peptide exchange, we generated a small panel of UV-exchanged (“conditional”) HLA-B*5703 tetramers. Our ability to produce a variety of HIV-1-specific HLA-B*5703-β2m tetramers, as evidenced by the specific staining of B*57-restricted CD8+ T cell lines and clones, verified the success of UV-peptide exchange (Fig. 2). We also evaluated the integrity and conformation of the KIR3DL1-Fc fusion protein by SPR, where binding of the KIR3DL1-specific antibodies to immobilized KIR3DL1-Fc was evaluated. The proportion of functional protein was estimated at 61% by comparing the ratios of Z27 (which binds a linear epitope) and DX9 (which binds folded protein [17]) (Fig. 3A) binding. We confirmed the specificity of KIR3DL1-Fc by demonstrating its preferential binding to MHC class I molecules bearing a Bw4 motif (Fig. 3B and C).
Fig. 2.
Generation of specific HLA-B*5703 tetramers by UV-ligand exchange. Biotinylated, UV-sensitive HLA-B*5703-9MT4 complexes were photoilluminated and rescued in the presence of HLA-B*5703-restricted KAFSPEVIPMF (KF11), TSTLQEQIGW (TW10), HTQGYFPDW (HW9), and QASQEVKGW (QW9) epitopes prior to tetramer generation via conjugation to an extravidin-phycoerythrin (PE)-labeled fluorochrome. (A) The functionality and specificity of UV-exchanged B*5703-KF11 tetramers was confirmed by staining a B*5703-KF11-restricted CD8+ T cell clone with conventional KF11 tetramer and KF11, TW10, and QW9 tetramers produced by UV exchange. Comparable staining was observed using both KF11 tetramers (open boxes), while the irrelevant UV exchange tetramers failed to bind KF11-specifc T cells. (B) The specificities of B*5703-TW10 and HW9 tetramers were evaluated by staining HW9- and TW10-specific CD8+ T cell lines (day 12) derived from a single donor in vitro.
Fig. 3.
Integrity and specificity of soluble KIR3DL1-Fc. (A) The integrity of KIR3DL1-Fc was assessed by antibody binding, using the KIR3DL1-specific antibodies Z27 (which binds a linear epitope) and DX9 (conformation dependent). After the level of saturation binding of each antibody was corrected for the amount of KIR3DL1 protein immobilized in the SPR flow cell, the proportion of DX9-reactive material (correctly folded) to Z27-reactive material (total protein) was calculated as 61%. (B) The specificity of KIR3DL1 was assessed by comparing its binding to immobilized HLA-B*5703 (Bw4 motif) and B*8101 (Bw6 motif) complexes. (C) The preferential binding of KIR3DL1 to Bw4 serotypes does not reflect differences in the amounts of protein immobilized in the SPR flow cell. The data were corrected for the amount of MHC class I protein immobilized in each flow cell and are shown as both normalized binding (with the level of binding to HLA-B5703 set at 1) and fold change in binding of HLA-B*8101 compared with HLA-B*5703.
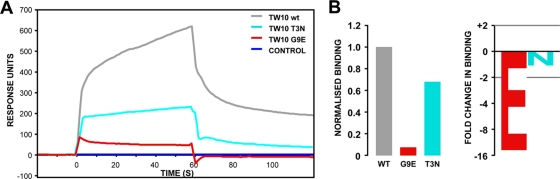
Having established that the KIR3DL1 protein was functional, we analyzed the specific binding of KIR3DL1 to UV-exchanged B*5703 monomers in complex with the consensus TW10 versus the variant epitopes. As demonstrated in Fig. 4, the HIV-1 B clade consensus sequence incorporating a glycine at position 9 (wild-type TW10) facilitated binding to KIR3DL1, and the T3N variant had a modest impact and reduced binding to KIR3DL1 by 1.5-fold relative to the wild-type TW10 peptide. However, the incorporation of glutamate at this position virtually abrogated the interaction and reduced binding by almost 16-fold relative to the glycine variant. Therefore, of the early-emerging TW10 variants, G9E had the most detrimental impact on the binding of KIR3DL1 to HLA-B*5703.
Fig. 4.
Loss of KIR3DL1 binding to the G9E TW10 variant. (A) Biotinylated B*5703-β2m complexes, irradiated and rescued in the presence of peptides corresponding to the wild-type TW10 epitope and the G9E (red) and T3N (cyan) variants, were immobilized on SA sensor chips, over which a fixed concentration of soluble KIR3DL1-Fc was injected at a constant flow rate. (B) Loss of binding of KIR3DL1 to HLA-B*5703 containing the G9E peptide did not reflect differences in the amount of material immobilized on the surface of the SPR flow cell. The data were corrected for the amount of MHC class I protein immobilized in each flow cell and are shown as both normalized binding (with the level of binding to the wild-type peptide set at 1) and fold change in binding of the G9E and T3N variants compared with the wild type.
Comprehensive analysis of the impact of C−1 substitutions on KIR3DL1 binding.
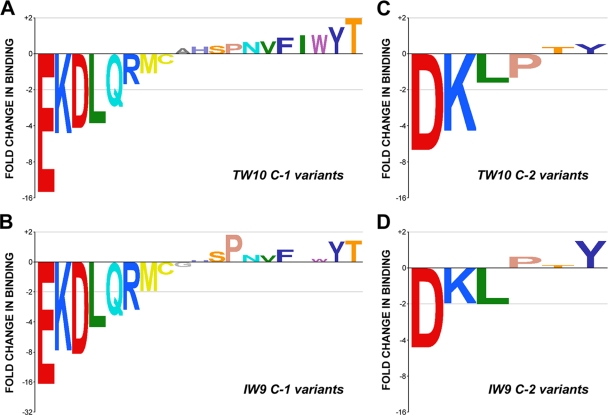
Our initial results confirmed that the interaction between KIR3DL1 and Bw4 HLA class I molecules is sensitive to the amino acid at position −1 relative to the C-terminal anchor residue (C−1). The light-sensitive peptide exchange reaction provided a high-throughput platform that enabled us to evaluate the sensitivity of KIR3DL1 to all amino acid substitutions near the C terminus of the HLA-bound peptide. We produced a panel of B*5703 complexes incorporating all amino acid substitutions at C−1 in both TW10 and the shorter B*57-restricted HIV-1 gag epitope ISPRTLNAW147-155 (IW9). For both sets of epitope variants, we observed similar hierarchies in terms of binding strength, indicating that the KIR3DL1–HLA-B*5703 interaction was highly sensitive to certain amino acid residues adjacent to the C-terminal anchor of the peptide, almost regardless of the sequence context of the rest of the peptide (Fig. 5A and B). Consistent with the results for the TW10 G9E variant, the presence of charged amino acids (both basic and acidic) at C−1 reduced binding to KIR3DL1 relative to the consensus C−1 glycine variant epitope, with the aspartate, lysine, and glutamate substitutions giving rise to the most extreme differences (between 4- and 16-fold reduction) relative to the consensus C−1 glycine peptide. Indeed, for many of the large, branched polar amino acids, C−1 substitution resulted in weaker interactions with KIR3DL1. In contrast, the majority of hydrophobic amino acids facilitated KIR3DL1 binding, particularly the large aromatic amino acids. However, the amino acid variants that enhanced binding relative to the consensus TW10 glycine (and the IW9 alanine) were modest (maximal 2-fold increase) and may reflect experimental “noise” rather than actual binding differences. In contrast, amino acid substitutions that reduced binding to KIR3DL1, most notably aspartate, lysine, glutamate, and leucine incorporations (for both IW9 and TW10), had the most dramatic impact on the interaction between KIR3DL1 and HLA-B*5703.
Fig. 5.
Ligand-specific binding of KIR3DL1 to HLA-B*5703 in complex with C−1 and C−2 TW10 and IW9 variants. The sensitivity of KIR3DL1 binding in the presence of peptide epitope variants incorporating amino acid substitutions is illustrated for the entire panel of TW10 and IW9 C−1 positional variants (A and B) and a subset of C−2 TW10 and IW9 variants (C and D). The data sets were normalized relative to the total amount of MHC class I protein immobilized in the SPR flow cells and are shown as fold change in binding relative to the wild-type amino acid (G for TW10 C−1, I for TW10 C−2, A for IW9 C−1, and N for IW9 C−2) and are ranked in ascending order for the TW10 variants. The IW9 data sets are ranked in the same order as the corresponding TW10 data set. The coloring of the graphs reflects the physical properties of the individual amino acids (see Table S1 in the supplemental material).
Impact of data analysis on the different binding patterns conferred by C−1 mutations.
Given the large impact of C−1 epitope substitutions on KIR3DL1 binding, we asked whether this reflected differences in the recovery of conformationally stable HLA-B*5703/β2M monomers following UV exchange and immobilization. Consequently, for a subset of HLA-B*5703-IW9 C−1 and TW10 C−1 complexes, we evaluated the proportion of conformationally correct (W6/32-reactive) material immobilized. Our findings demonstrated that the large differences in KIR3DL1 binding did not reflect variations related to the quantity of conformationally correct material immobilized (see Fig. S1 in the supplemental material). Accordingly, the KIR3DL1 binding data sets did not vary markedly when adjusted in relation to the total amount of material (RU) or the proportion of W6/32-reactive material immobilized. Consequently, the differences observed with distinct C−1 amino acid substitutions directly reflect disparities in their effects on KIR3DL1-mediated binding to HLA-B*5703 and not differences in the efficiencies of monomer rescue during UV-ligand exchange.
KIR3DL1-mediated recognition of C−2 mutations.
Given the strong influence of C−1 amino acids on the HLA-B*5703-KIR3DL1 interaction, we asked if this dependency also extended to amino acids at C−2. To evaluate this, we generated a select panel of IW9 and TW10 peptides incorporating variant amino acids that either reduced or allowed strong binding to KIR3DL1 at C−1. Our findings depict a clear pattern of KIR3DL1 dependency at position C−2, in agreement with the C−1 changes (Fig. 5C and D). Charged amino acids disrupted binding to KIR3DL1 (albeit less dramatically than the equivalent substitutions at C−1), whereas others, including aromatic and β-branched amino acids, were well tolerated. Collectively, our data suggest KIR3DL1 binding is highly sensitive to the nature of the amino acids at positions in close proximity to the C-terminal anchor residue.
DISCUSSION
During HIV-1 infection, the complex interplay between CD8+ T cell-mediated selection, viral evolution, and fitness cost influences the clinical course of the disease (6, 12, 25). CD8+ T cell-driven escape occurs early during acute infection, with implications for viral control. However, mutations that map to T cell epitopes can also affect recognition of MHC class I-specific receptors of the innate immune system, as previously described for ILT receptors (16). In this study, we followed early-emerging mutations that mapped to the immunodominant gag-derived TW10 epitope in two HLA-B*5703-positive patients. One included the well-described T3N mutation, which, in agreement with previous findings, escaped T cell recognition. In contrast, a second variant incorporating a C-terminal proximal G9E mutation failed to successfully evade TW10-specific T cells. As both patients were heterozygous for inhibitory KIR3DL1 allotypes that comprise receptors for MHC class I Bw4 serotypes, including HLA-B*5703, we tested if these mutations affected recognition of HLA-B*5703 by KIR3DL1. Strikingly, and in contrast to the wild-type TW10 and the T3N mutation, the G9E variant severely reduced interaction between KIR3DL1 and HLA-B*5703.
The side chain of the glutamate in G9E is predicted to point out of the peptide binding groove because of a conserved hydrogen bond between the carbonyl group of the last peptide bond in the epitope and tryptophan 147 in the α2 helix. It also lies close to the Bw4 motif (α1 residues 77 to 81) that is involved in KIR recognition (28). Therefore, we questioned if the relatively fixed conformation of amino acid side chains adjacent to the C terminus anchor affected KIR3DL1-mediated recognition of HLA-B*5703, particularly as diverse C−1 TW10 mutations have been described in HLA-B*57-positive patients. We screened the impacts of all possible C−1 substitutions in two B*5703-restricted epitopes on KIR3DL1-mediated recognition. Collectively, our data illustrated a slight preference for hydrophobic amino acids, in agreement with a recent structural model of a KIR3DL1–HLA-A*24-nef peptide cocomplex highlighting the dominance of hydrophobic interactions at the peptide MHC (pMHC) class I-KIR interface (31). Aromatic amino acids were especially preferred, and presumably, their side chain ring structures conferred a specific role in binding to KIR3DL1. Leucine represented one of the few hydrophobic substitutions to adversely affect binding, while Isoleucine (its isomer) was well tolerated, and this could reflect a precise function conferred by a β-branched amino acid, such as isoleucine, in binding to KIR3DL1. In contrast to hydrophobic C−1 substitutions, many of the large, branched polar amino acid replacements resulted in weaker binding, and for certain polar (neutral) amino acids, steric interference may also play a role. The most striking results, however, were related to the strong dislike of charged amino acids that probably occurred as a consequence of electrostatic effects, of which the G9E variant was the most extreme. Interestingly, the hierarchy of amino acid specificity observed for C−1 substitutions extended to C−2, a secondary position within the epitope that affects KIR recognition (27) and where mutations frequently arise in HIV-1 (as observed in donor CH77) (11). However, the impact of these mutations was less pronounced than for the C−1 substitutions.
Collectively, our data demonstrated that KIR3DL1 exhibits specific sensitivity to the amino acids in proximity to the C-terminal anchor residue, irrespective of the nature of the peptide bound. Our findings support previous studies illustrating differences in the sensitivities of KIR3DL1-expressing NK cell clones to peptide variants presented by HLA-B*2705 molecules (27, 28), particularly in relation to C−1 incorporations that protected target cells from NK cell-mediated lysis (e.g., valine) and those that did not (e.g., glutamate). Our data agree with a second study in which mutagenesis of the HLA-B*2705-restricted Epstein-Barr virus (EBV) EBNA3C258-266 epitope from C−1 glutamate to threonine restored HLA-B27 tetramer binding to KIR3DL1-expressing cells (33). Although HLA-B*5703 and -B*2705 represent Bw4 MHC class I serotypes, their Bw4 motifs and peptide binding specificities are dissimilar, yet the impact of positional substitutions at position C−1 is conserved. Hence, the nature of KIR3DL1-dependent peptide specificity for different MHC class I allotypes is related primarily to (i) the relatively fixed positioning of the peptide's C−1 side chain and (ii) its close proximity to the amino acids comprising the Bw4 motif.
It remains unclear whether the TW10 C−1 mutations that arise during HIV-1 infection represent escape mutations (24). In donors CH77 and CH58, the TW10 position G9E viral variants emerged early and were dominant before T3N, which escapes T cell recognition (22). However, for donor CH58, the G9E variant was well recognized by CD8+ T cells generated in response to the founder viral epitope, consistent with the findings that emerging G9E/D variants fail to fully escape TW10-specific T cell recognition in elite controller groups (24, 26). TW10 position 9 (HXB2 gag248) maps to the side of helix 6 of the p24 capsid at a site where the introduction of a charged residue would disturb the electrostatic charge, with adverse implications for helical packing (22), consistent with the likelihood that G9E viruses are attenuated (24). Given this putative impact on viral fitness, it seems likely that the G9E mutations must have been specifically selected in vivo. So, what drives selection of G9E? It is unlikely that KIR3DL1+ NK cells would directly select G9E, as cells expressing these variants, which fail to bind to the inhibitory receptor, would become more susceptible to elimination by KIR3DL1+ NK cells. Therefore, the most likely explanation is that the slightly impaired T cell recognition of the G9E variant could be enough for it to be selected by TW10-specific T cells, facilitating partial and early immune evasion prior to the emergence of the more successful T3N variants. The rapid appearance of G9E might (i) be related to the ease with which the G9E mutation (underlined) (GGA to GAA) accrues relative to T3N (ACC to AAC) and/or (ii) reflect the cytidine deaminase activity of APOBEC3, resulting in a G-to-A mutation required for the glycine-to-glutamate change. As the G9E variant should incur a fitness cost, it may represent a transitional escape variant prior to the emergence of the less attenuated T3N mutation; this would be consistent with the disappearance of G9E variants in both patients during subsequent screens.
Could the KIR3DL1 receptor have any role in the selection or deselection of the G9E mutant virus? It is possible that, by abrogating the binding of KIR3DL1 to HLA-B*5703, the G9E variant could release NK cells from KIR3DL1-mediated inhibition and that this could facilitate the elimination of G9E variants by KIR3DL1+ NK cells in vivo. It is also possible that the G9E mutation might alter the functional competence of maturing KIR3DL1+ NK cells. Inhibitory KIRs play an important role in NK cell “education” and authorize the extent of their functional capacity during maturation (4, 13–14); this occurs primarily in lymph nodes in the face of vigorous HIV-1 replication (10, 32). Thus, by disrupting the HLA-B*5703–KIR3DL1 interaction, the G9E variant could modulate the education of maturing KIR3DL1+ NK cells. Finally, the G9E mutation might also act at the level of T cells. KIR3DL1 molecules are expressed on mature CD8+ T lymphocyte subsets (during viral persistence and chronic exposure to antigens) that are intrinsically hyporesponsive, partly due to the constitutive recruitment of SHP-1 and -2 phosphatases by KIRs (2, 5, 36). It is possible that KIR3DL1 operates at the T cell surface primarily as a coreceptor to enhance TCR-pMHC class I binding in the synapse. In this model, the inhibitory function of the cytoplasmic domain of KIR3DL1 would be negligible compared to the strong positive signal arising from T cell receptor (TCR) engagement, consistent with the recent demonstration that certain HLA class I peptide combinations can induce KIR clustering in the absence of SHP-1-mediated dephosphorylation of downstream substrates on NK cells (9). By abrogating binding to KIR3DL1, the G9E mutation could decrease the duration of T cell engagement and, hence, the potency of subsequent effector functions.
In summary, our study has demonstrated how the nature and fine specificity of C−1 peptide substitutions affect binding of HLA-B*5703 to KIR3DL1. The early-emerging G9E variant gave rise to the most extreme differences in binding to KIR3DL1. Interestingly, G9E variants are rare, consistent with the mutation making the virus more susceptible to elimination by KIR3DL1-expressing NK cells. In contrast, the G9A mutation, which is common in B clade infections, has little impact on KIR3DL1 binding relative to the consensus G residue. The functional impact of these mutations on KIR3DL1-expressing NK and T cells is the focus of ongoing investigations.
Supplementary Material
ACKNOWLEDGMENTS
We thank Zhanru Yu for peptide synthesis, Alasdair Leslie for providing HLA-B*8101 monomers, and Lewis Lanier for his kind donation of the KIR3DL1*002 construct.
This work was supported by Center for HIV/AIDS Vaccine Immunology grant A1067854 and Dutch Cancer Society grant KWF 2007-3825, in addition to United Kingdom Medical Research Council (MRC) funds.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 23 March 2011.
REFERENCES
- 1. Alter G., Altfeld M. 2009. NK cells in HIV-1 infection: evidence for their role in the control of HIV-1 infection. J. Intern. Med. 265:29–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alter G., et al. 2008. Ligand-independent exhaustion of killer immunoglobulin-like receptor-positive CD8+ T cells in human immunodeficiency virus type 1 infection. J. Virol. 82:9668–9677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alter G., et al. 2007. Evolution of innate and adaptive effector cell functions during acute HIV-1 infection. J. Infect. Dis. 195:1452–1460 [DOI] [PubMed] [Google Scholar]
- 4. Anfossi N., et al. 2006. Human NK cell education by inhibitory receptors for MHC class I. Immunity 25:331–342 [DOI] [PubMed] [Google Scholar]
- 5. Anfossi N., et al. 2004. Coordinated expression of Ig-like inhibitory MHC class I receptors and acquisition of cytotoxic function in human CD8+ T cells. J. Immunol. 173:7223–7229 [DOI] [PubMed] [Google Scholar]
- 6. Bhattacharya T., et al. 2007. Founder effects in the assessment of HIV polymorphisms and HLA allele associations. Science 315:1583–1586 [DOI] [PubMed] [Google Scholar]
- 7. Carr W. H., et al. 2007. Cutting edge: KIR3DS1, a gene implicated in resistance to progression to AIDS, encodes a DAP12-associated receptor expressed on NK cells that triggers NK cell activation. J. Immunol. 178:647–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carrington M., Martin M. P., van Bergen J. 2008. KIR-HLA intercourse in HIV disease. Trends Microbiol. 16:620–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fadda L., et al. 2010. Peptide antagonism as a mechanism for NK cell activation. Proc. Natl. Acad. Sci. U. S. A. 107:10160–10165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Folkvord J. M., Armon C., Connick E. 2005. Lymphoid follicles are sites of heightened human immunodeficiency virus type 1 (HIV-1) replication and reduced antiretroviral effector mechanisms. AIDS Res. Hum. Retroviruses 21:363–370 [DOI] [PubMed] [Google Scholar]
- 11. Goonetilleke N., et al. 2009. The first T cell response to transmitted/founder virus contributes to the control of acute viremia in HIV-1 infection. J. Exp. Med. 206:1253–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kawashima Y., et al. 2009. Adaptation of HIV-1 to human leukocyte antigen class I. Nature 458:641–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kim S., et al. 2005. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature 436:709–713 [DOI] [PubMed] [Google Scholar]
- 14. Kim S., et al. 2008. HLA alleles determine differences in human natural killer cell responsiveness and potency. Proc. Natl. Acad. Sci. U. S. A. 105:3053–3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Leslie A. J., et al. 2004. HIV evolution: CTL escape mutation and reversion after transmission. Nat. Med. 10:282–289 [DOI] [PubMed] [Google Scholar]
- 16. Lichterfeld M., et al. 2007. A viral CTL escape mutation leading to immunoglobulin-like transcript 4-mediated functional inhibition of myelomonocytic cells. J. Exp. Med. 204:2813–2824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Litwin V., Gumperz J., Parham P., Phillips J. H., Lanier L. L. 1994. NKB1: a natural killer cell receptor involved in the recognition of polymorphic HLA-B molecules. J. Exp. Med. 180:537–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Long E. O., et al. 1997. Killer cell inhibitory receptors: diversity, specificity, and function. Immunol. Rev. 155:135–144 [DOI] [PubMed] [Google Scholar]
- 19. López-Vázquez A., et al. 2005. Interaction between KIR3DL1 and HLA-B*57 supertype alleles influences the progression of HIV-1 infection in a Zambian population. Hum. Immunol. 66:285–289 [DOI] [PubMed] [Google Scholar]
- 20. Martin M. P., Carrington M. 2008. KIR locus polymorphisms: genotyping and disease association analysis. Methods Mol. Biol. 415:49–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Martin M. P., et al. 2007. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat. Genet. 39:733–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Martinez-Picado J., et al. 2006. Fitness cost of escape mutations in p24 Gag in association with control of human immunodeficiency virus type 1. J. Virol. 80:3617–3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Migueles S. A., et al. 2008. Lytic granule loading of CD8+ T cells is required for HIV-infected cell elimination associated with immune control. Immunity 29:1009–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Miura T., et al. 2009. HLA-B57/B*5801 human immunodeficiency virus type 1 elite controllers select for rare gag variants associated with reduced viral replication capacity and strong cytotoxic T-lymphocyte recognition. J. Virol. 83:2743–2755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Moore C. B., et al. 2002. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science 296:1439–1443 [DOI] [PubMed] [Google Scholar]
- 26. O'Connell K. A., et al. 2010. Control of HIV-1 in elite suppressors despite ongoing replication and evolution in plasma virus. J. Virol. 84:7018–7028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Peruzzi M., Parker K. C., Long E. O., Malnati M. S. 1996. Peptide sequence requirements for the recognition of HLA-B*2705 by specific natural killer cells. J. Immunol. 157:3350–3356 [PubMed] [Google Scholar]
- 28. Peruzzi M., Wagtmann N., Long E. O. 1996. A p70 killer cell inhibitory receptor specific for several HLA-B allotypes discriminates among peptides bound to HLA-B*2705. J. Exp. Med. 184:1585–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rodenko B., et al. 2006. Generation of peptide-MHC class I complexes through UV-mediated ligand exchange. Nat. Protoc. 1:1120–1132 [DOI] [PubMed] [Google Scholar]
- 30. Salazar-Gonzalez J. F., et al. 2009. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J. Exp. Med. 206:1273–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sharma D., et al. 2009. Dimorphic motifs in D0 and D1+D2 domains of killer cell Ig-like receptor 3DL1 combine to form receptors with high, moderate, and no avidity for the complex of a peptide derived from HIV and HLA-A*2402. J. Immunol. 183:4569–4582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stellbrink H. J., et al. 1997. Asymptomatic HIV infection is characterized by rapid turnover of HIV RNA in plasma and lymph nodes but not of latently infected lymph-node CD4+ T cells. AIDS 11:1103–1110 [DOI] [PubMed] [Google Scholar]
- 33. Stewart-Jones G. B., et al. 2005. Crystal structures and KIR3DL1 recognition of three immunodominant viral peptides complexed to HLA-B*2705. Eur. J. Immunol. 35:341–351 [DOI] [PubMed] [Google Scholar]
- 34. Thananchai H., et al. 2007. Cutting edge: allele-specific and peptide-dependent interactions between KIR3DL1 and HLA-A and HLA-B. J. Immunol. 178:33–37 [DOI] [PubMed] [Google Scholar]
- 35. Toebes M., et al. 2006. Design and use of conditional MHC class I ligands. Nat. Med. 12:246–251 [DOI] [PubMed] [Google Scholar]
- 36. van der Veken L. T., et al. 2009. Functional analysis of killer Ig-like receptor-expressing cytomegalovirus-specific CD8+ T cells. J. Immunol. 182:92–101 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.