Abstract
Recently, we showed that the Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV) VP80 protein is essential for the formation of both virion types, budded virus (BV) and occlusion-derived virus (ODV). Deletion of the vp80 gene did not affect assembly of nucleocapsids. However, these nucleocapsids were not able to migrate from the virogenic stroma to the nuclear periphery. In the current paper, we constructed a baculovirus recombinant with enhanced-green fluorescent protein (EGFP)-tagged VP80, allowing visualization of the VP80 distribution pattern during infection. In baculovirus-infected cells, the EGFP-VP80 protein is entirely localized in nuclei, adjacent to the virus-triggered F-actin scaffold that forms a highly organized three-dimensional network connecting the virogenic stroma physically with the nuclear envelope. Interaction between VP80 and host actin was confirmed by coimmunoprecipitation. We further showed that VP80 is associated with the nucleocapsid fraction of both BVs and ODVs, typically at one end of the nucleocapsids. In addition, the presence of sequence motifs with homology to invertebrate paramyosin proteins strongly supports a role for VP80 in the polar transport of nucleocapsids to the periphery of the nucleus on their way to the plasma membrane to form BVs and for assembly in the nuclear periphery to form ODVs for embedding in viral occlusion bodies.
INTRODUCTION
During evolution, viral structural proteins are tested continuously for the efficient transfer of viral genetic information from cell to cell in order to spread infection optimally and to ensure virus transmission to a new host. Hence, studying the features and functions of viral structural proteins is crucial to understand in detail all steps of viral infection, including entry, intracellular transport, replication, assembly, and egress.
Baculoviruses constitute a unique group of viruses specific for arthropods, mainly insects. Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV), the model virus of the genus Alphabaculovirus of the Baculoviridae family, is an enveloped virus with a circular, double-stranded DNA genome of ∼130 kbp (4) wrapped in a rod-shaped nucleocapsid. During the infection cycle, two types of virions are formed. Budded virus (BV) is derived from nucleocapsids leaving the cell nucleus and budding through the plasma membrane. On the other hand, occlusion-derived virus (ODV) is formed from nucleocapsids retained in the nucleus, where envelopment occurs prior to embedding of the nucleocapsids in polyhedron-shaped occlusion bodies (see reference 50 for a review). BV mediates the spread of infection from cell to cell, while ODV is responsible for horizontal virus transmission between insects.
Entry of the BV form of AcMNPV into host cells is mediated by clathrin-dependent endocytosis (34), although direct fusion of BVs with the plasma membrane has also been recorded (14). Upon internalization, nucleocapsids are released into the cytoplasm and immediately translocated toward the cell nucleus by actin-mediated movement, which is driven by the viral P78/83 capsid protein (open reading frame [ORF] 1629) and the host Arp2/3 complex (22, 42). Myosin motor functions also appear to be involved in this process (14). Recently, it has been demonstrated that AcMNPV nucleocapsids enter the cell nucleus through nuclear pores (42). In the nucleus, the nucleocapsids are uncoated, upon which early gene transcription immediately starts (42). The nuclear viral replication factory, the so-called virogenic stroma, is a site of viral transcription (36, 45), DNA replication (8, 26), and progeny nucleocapsid assembly (55). Little is known about the mechanisms of assembly, DNA packaging, and egress of progeny nucleocapsids. The participation of several AcMNPV proteins in the assembly and exit of nucleocapsids has been reviewed (12), but their precise roles remain unclear. Furthermore, baculovirus morphogenesis is highly dependent on the host filamentous-actin (F-actin) cytoskeleton (41), which is dramatically rearranged upon infection. The microtubule cytoskeleton has to be taken into account as well (18).
Recently, we have provided evidence for the involvement of the AcMNPV VP80 structural protein in the packaging of nucleocapsids and in their egress from the nucleus toward the cell periphery (39). The vp80 gene is transcribed late in infection as a 2.1-kb transcript with the capacity to encode an 80-kDa protein (35). VP80 is a component of both BV (58) and ODV (6) and was first identified as P87 in Orgyia pseudotsugata MNPV (OpMNPV) (40). Homologues of the vp80 gene are found only in alphabaculoviruses (12). The vp80 homologue in Choristoneura fumiferana MNPV (CfMNPV) is expressed as 72- and 82-kDa protein variants, and only the 82-kDa form is associated with ODV (33). Functional analysis of a Bombyx mori MNPV (BmMNPV) vp80 deletion mutant showed that VP80 is essential for BV production (52), and this was confirmed for AcMNPV (39). More recently, interactions have been identified between VP80 and both the viral core protein 38K (60) and the Helicoverpa armigera NPV (HearNPV) HA100 protein (46).
To better understand the function(s) of VP80 in AcMNPV infection, we constructed an enhanced-green fluorescent protein (EGFP)-tagged VP80 baculovirus, allowing the visualization of VP80, to study its spatiotemporal distribution during infection. We show that VP80 enters the cell nucleus, where it associates with the virus-induced nuclear F-actin cytoskeleton, which forms a highly organized three-dimensional (3D) network penetrating the viral replication factories and connecting them with the nuclear envelope. Moreover, we reveal a paramyosin (PMYO) homology sequence in VP80 and show that VP80 associates with one end of the nucleocapsids. Taken together, our data support the involvement of VP80 in the egress of progeny nucleocapsids from the virogenic stroma toward the nuclear periphery, which is dependent on functional, actin-based myosin motor functions.
MATERIALS AND METHODS
Bioinformatics-based tools.
The VP80 sequence was searched against the protein Conserved Domain Database (CDD) (37, 38) and the SMART version 6 (30, 49), Pfam hidden Markov model (HMM) (20), and SilkDB (15) databases to identify functional protein domains. Protein domain identification is limited by the sensitivity of the search system and the diversity of protein domains in the databases. For this reason, the information in all databases was combined to increase both the sensitivity and the protein diversity. Short sequence motifs, targeting patterns, and posttranslation modification sites were also searched using the ScanProsite (13) and MyHits (43, 44) tools on the EXPASY server (3). Multiple sequence alignment was constructed using the MUSCLE (16) algorithm and manually edited in JalView (59).
Cells and viruses.
Spodoptera frugiperda cell type 9 (Sf9) cells (Invitrogen) were maintained in SF900-II serum-free medium (Invitrogen) at 27°C under standard conditions. All recombinant AcMNPV bacmids and viruses were derived from the commercially available bacmid bMON1427226 (Invitrogen) or its modified version, the Ac-Δvp80 bacmid (39), both propagated in the Escherichia coli strain DH10β. To facilitate VP80 immunodetection, a baculovirus with an N-terminally Flag-tagged VP80 protein (39) was used (Ac-Δvp80-Flag:vp80). To prepare ODVs, AcMNPV strain E2 (51) was used. A recombinant bacmid-derived baculovirus with an egfp reporter gene inserted in the polyhedrin (polh) locus under p10 promoter control and designated the wild-type genotype (Ac-wt) (39) was used as a control.
DNA constructions.
The AcMNPV vp80 ORF was cleaved from pJet1.2-vp80 (39) and subcloned between SacI and XbaI sites in plasmid pIZ (Invitrogen) to create pIZ-vp80. Subsequently, the DNA fragment encoding the vp80 ORF was subcloned between KpnI and XbaI sites of pEGFP-C1 (Clontech) to form pEGFP:vp80. The error-free DNA cassette with the egfp-vp80 fusion gene was then cleaved with NheI and XbaI from pEGFP:vp80 and subcloned into the SpeI site of pIZ (Invitrogen) to generate plasmid pIZ-egfp:vp80.
To construct EGFP-tagged VP80 recombinant baculovirus, donor plasmid pFastBacDUAL (Invitrogen) was modified. A 254-bp fragment of the AcMNPV vp80 promoter sequence was amplified using the forward primer Pvp80-F (5′-GGAACAAAGGCCTGAGCTCAAAGTAAGACCTTTACTGTCC-3′) and the reverse primer Pvp80-R (5′-GGTTAATTGAGCTCATAAGGTTATATCGAATG-3′). The amplified fragment was cloned between Bst1107I and SacI sites of pFastBacDUAL to generate pFB-Pvp80. Then, the egfp-vp80 gene was excised from the pEGFP:vp80 plasmid and subcloned into the SpeI site of pFB-Pvp80 to generate pFB-Pvp80-egfp:vp80. The constructs were verified by DNA sequencing. The developed expression cassette was finally transposed into the polh locus of the Ac-Δvp80 bacmid (39) by following the Bac-to-Bac protocol (Invitrogen). Screening of transposition-positive Ac-Δvp80-egfp:vp80 bacmids was done by a triplex PCR as previously described (39).
Transfection-infection assays.
Spodoptera frugiperda (Sf9) cells (Invitrogen) were maintained in SF900-II serum-free medium (Invitrogen) at 27°C. Five micrograms of bacmid DNA was used to transfect 1 × 106 cells with Cellfectin II (Invitrogen). At 5 days posttransfection (p.t.), the BV-enriched culture supernatant was centrifuged for 5 min at 2,000 × g and used to infect 1.5 × 106 Sf9 cells. Viral propagation was followed by fluorescence microscopy at the time points p.t. or postinfection (p.i.) indicated in the figure. The Ac-wt bacmid/virus carrying the egfp reporter gene under the control of the p10 promoter served as a control. To monitor BV release, 1 million cells were transfected with 5 μg bacmid DNA or infected at a multiplicity of infection (MOI) of 1 50% tissue culture infectious dose (TCID50) unit/cell. Cell culture medium was harvested at various time points and analyzed for the presence of infectious BV by endpoint dilution assays. The averages of the infectious titers from three independent transfections/infections were calculated and plotted into graphs.
Purification and fractionation of BV and ODV virions.
To produce BVs, 3.0 × 107 Sf9 cells were infected with Ac-Δvp80-egfp:vp80, Ac-Δvp80-Flag:vp80, or control Ac-wt virus at an MOI of 1. Six days p.i., BV-enriched medium was collected and centrifuged at 1,500 × g for 10 min. The supernatant was ultracentrifuged at 80,000 × g (Beckman SW28 rotor) for 60 min at 4°C. The BV pellet was resuspended in 350 μl 0.1× TE (10 mM Tris, 1 mM EDTA, pH 7.5) buffer, loaded onto a linear sucrose gradient (25 to 56%, wt/vol), and ultracentrifuged at 80,000 × g (Beckman SW55 rotor) for 90 min at 4°C. The formed BV band was collected and diluted in 12 ml 0.1× TE. The BV preparation was concentrated at 80,000 × g for 60 min at 4°C, and the final virus pellet was resuspended in 150 μl of 0.1× TE.
To produce ODVs, 6.0 × 107 Sf9 cells were coinfected with Ac-Δvp80-Flag:vp80 (MOI = 25 TCID50 units/cell) and AcMNPV strain E2 (51) (MOI = 5 TCID50 units/cell). Five days p.i., the infected cells were harvested, and ODVs were purified from viral occlusion bodies as previously described (7). The final ODV pellet was resuspended in 0.5 ml of 0.1× TE. The purified BV and ODV virions were separated into envelope and nucleocapsid fractions as described previously (7). Final fractions were processed for SDS-PAGE and immunoblotted against either mouse monoclonal anti-Flag antibody (Stratagene), rabbit polyclonal anti-VP39 antiserum (directed against the major capsid protein [kindly provided by Lorena Passarelli, Kansas State University]), rabbit polyclonal anti-GP64 antiserum (directed against the BV envelope protein GP64) (57), or rabbit polyclonal per os infectivity factor 1 (PIF-1) antiserum (directed against PIF-1, an ODV-specific envelope protein) (46).
Immunogold labeling of nucleocapsids for EM.
The BV virions were immunostained against Flag- and EGFP-specific epitopes by the direct immunoelectron microscopy (DIEM) technique. To this aim, BVs were adsorbed on carbon-coated electron microscopy grids, activated with glow discharge. Grids were then incubated on drops of 0.02% Triton X-100 in 0.1× TE (10 mM Tris, 1 mM EDTA, pH 7.5) for 20 s to remove the virion envelope. The grids were twice washed in 1× phosphate-buffered saline (PBS), followed by incubation with primary mouse anti-Flag (Stratagene) or anti-EGFP (Molecular Probes) antibodies, both diluted 1:1,000. Finally, the grids were washed intensively in 1× PBS and exposed to colloidal-gold (5-nm particle)-conjugated secondary goat antibody (Aurion). Grids were washed again in 1× PBS and sterile distilled H2O, contrasted on a drop of 2% phosphotungstic acid (pH 7.0) for 15 s, and dried. Samples were observed in a Philips CM12 electron microscope.
Confocal microscopy.
For confocal imaging, Sf9 cells (1.5 × 106) were seeded in 35-mm glass bottom culture dishes (MatTek) and infected with either Ac-Δvp80-egfp:vp80 or Ac-wt (control) virus at an MOI of 10. After a 45-min adsorption, unbound viruses were gently washed away and fresh SF900-II serum-free medium was added. To avoid rapid temperature changes and cytoskeleton depolymerization at low temperatures, all procedures were performed at 27°C with prewarmed media and solutions. Expression and the subcellular localization of either EGFP-VP80 or EGFP alone were monitored at the time points p.i. indicated in the figures by confocal microscopy using a Leica TCS SP2 AOBS confocal microscope operating with an argon laser (458, 476, 488, 496, and 514 nm) and an HeNe laser (543 and 594 nm). Immediately before observation, cell nuclei were stained with Hoechst 33342 (Invitrogen), and F-actin filaments were stained with rhodamine-phalloidin (Molecular Probes), which does not recognize G-actin, using the manufacturer's protocol. Colocalization measurements expressed as both Pearson's coefficient (PC) and intensity correlation quotient (ICQ) values were performed in JaCoP (5) implemented in ImageJ (1). To build up 3D isosurface volume data from 2D confocal data, 30 scans in Z-axis (400 nm per each depth step) were merged and integrated with the help of ImageSurfer (24). Final adjustment of volume data was done with Chimera (47) software.
For transient-expression assays, Sf9 cells (1.5 × 106) were seeded in 35-mm glass bottom culture dishes (MatTek) and transfected with 5 μg of pIZ-egfp:vp80 or a pIZ-4×EGFP control plasmid. At 24 h p.t., cell nuclei were stained with Hoechst 33342 (Invitrogen), and cells were observed under a confocal microscope as noted above. The pIZ-4×EGFP construct was obtained from Fang Xu (Laboratory of Virology, Wageningen University) and constructed by inserting an HpaI and NheI EGFP cassette from pTetra-EGFP (which was a kind gift from Christian Beetz, Universitätsklinikum Jena, Germany) into pIZ-EGFP-N3 between HindIII and XhoI sites. To this aim, all restriction sites were made blunt ended.
Immunoprecipitation.
To verify VP80 interaction with the host actin, Sf9 cells (3.0 × 107 cells) were infected with Ac-Δvp80-Flag:vp80 (MOI = 20). Two days p.i., the cells were harvested and washed once in 1× PBS. The Flag-tagged protein immunoprecipitation kit (Sigma-Aldrich) was used to precipitate protein complexes according to the manufacturer's protocol. Bound proteins were analyzed on Western blots with mouse anti-Flag (Stratagene) or anti-actin (ImmunO) antibodies. The Sf9 cells infected with Ac-wt virus (MOI = 20) were used as a negative control for immunoprecipitation reactions.
Drug treatments.
To evaluate the effects of 2,3-butanedione monoxime (BDM), a myosin motor inhibitor, and cytochalasin D (CD), an actin-depolymerizing drug, Sf9 cells (1 × 106/well) were seeded in 6-well plates and infected with Ac-wt at an MOI of 10 under standard conditions. At 12 h p.i., BDM (Sigma) or CD (Merck) was added to the culture media at final concentrations of 10 or 30 mM for BDM and 2 or 4 μM for CD. At this time, 100-μl aliquots of culture media were taken to measure residual BV titers (baselines). The cells were further cultivated at 27°C until 48 h p.i., when cell culture supernatants were harvested to measure BV titers by endpoint dilution. Levels of inhibition were calculated by the following formula: percent BV release = 100% × (BV release [with inhibitory drug] − BV baseline [with inhibitory drug])/(BV release [without inhibitory drug] − BV baseline [without inhibitory drug]). In all experiments, each measure point was obtained in triplicate, and means and standard deviations for each experimental condition were determined and plotted into graphs.
RESULTS
Construction of a baculovirus with EGFP-tagged VP80.
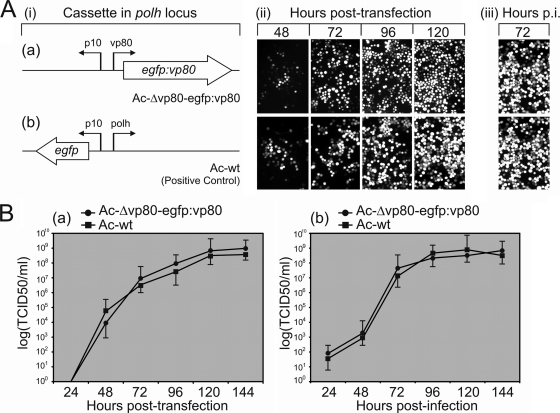
Functional analysis of the AcMNPV Δvp80 genome has shown that the N terminus of VP80, in contrast to its conserved C terminus, is permissive to engineering (39). To construct EGFP-tagged VP80 baculovirus as a tool to study the function of VP80, we constructed an N-terminally egfp-fused vp80 cassette and transposed this into the polh locus of an Ac-Δvp80 replication-deficient bacmid (39) (Fig. 1Ai). Sf9 cells were transfected with the developed Ac-Δvp80-egfp:vp80 bacmid to reconstitute recombinant Ac-Δvp80-egfp:vp80 virus. Correct synthesis of the EGFP-VP80 fusion protein (∼108.83 kDa) was proven by Western blot analysis (data not shown). The Ac-Δvp80-egfp:vp80 propagation capacity was first monitored by fluorescence microscopy (Fig. 1Aii to iii). The results showed widespread egfp-vp80 expression, indicating that the reconstituted EGFP-VP80-producing virus is able to efficiently spread from cell to cell. We did not observe any replication handicap of the EGFP-tagged virus compared with the replication capacity of Ac-wt. Accordingly, determination of growth curves of Ac-Δvp80-egfp:vp80 virus showed replication kinetics similar if not identical to those of Ac-wt propagation (Fig. 1B). The results thus confirmed that the N terminus of VP80 is highly permissive to engineering to display foreign peptide/protein, without any negative effects on viral fitness, at least in cell culture.
Fig. 1.
Construction of EGFP-displaying VP80 baculovirus and its fitness analysis. (A) Comparison of the viral replication capacities of Ac-Δvp80-egfp:vp80 (a) and Ac-wt (b). (i) Diagram of the expression cassettes transposed into the polh locus of a vp80-deleted (a) or an Ac-wt (b) bacmid. (ii) Time course fluorescence microscopy showing the propagation of the infection in Sf9 cells transfected with the resulting bacmid constructs. The progress of viral infection was checked by EGFP detection (converted to white) at the indicated times p.t. At 120 h p.t., the cell culture supernatants were collected to initiate a secondary infection. (iii) Secondary infection assay. (B) One-step growth curves of both the Ac-Δvp80-egfp:vp80 and Ac-wt constructs following transfection (a) or infection (MOI = 1) (b). Cell culture supernatants were harvested at the indicated times p.t. or p.i. and analyzed for the release of infectious BV by endpoint dilution. Infectivity was monitored by EGFP expression. The points indicate the average titer derived from three independent transfections/infections, and the error bars represent the standard deviations (SD).
VP80 is linked to one end of nucleocapsids.
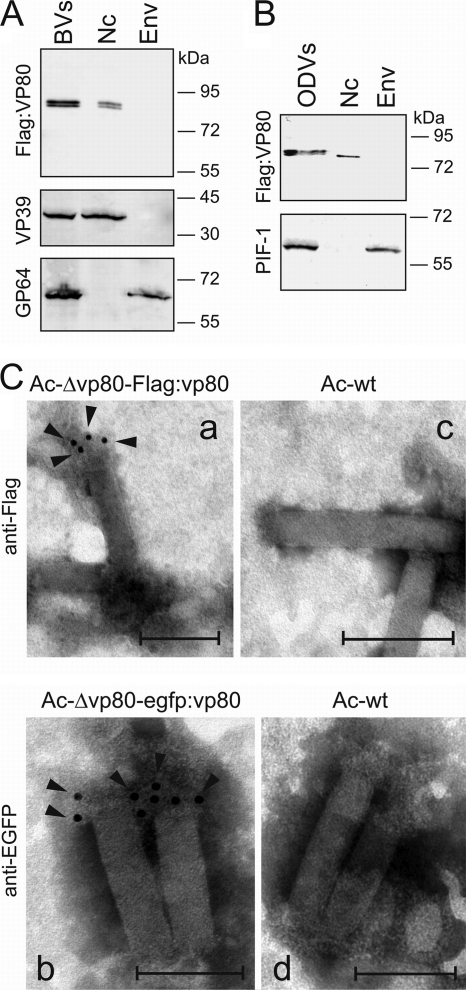
The VP80 protein has been shown to be a component of both BV (58) and ODV (6). To determine whether VP80 is a component of nucleocapsid or envelope, Ac-Δvp80-Flag:vp80 BV was fractionated into nucleocapsid and envelope fractions. Final fractions were subjected to Western blot analysis. Flag-VP80 was detected only in the nucleocapsid fraction as a band of ∼80 kDa (Fig. 2A, upper panel). Correct separation of BV into nucleocapsid and envelope fractions was checked with antibodies against the major capsid protein (VP39) and the BV envelope protein (GP64) (Fig. 2A, middle and lower panels). In order to demonstrate that VP80 is also an ODV nucleocapsid-associated protein, Sf9 cells were coinfected with Ac-Δvp80-Flag:vp80 and wild-type (polyhedron-producing) AcMNPV viruses. Five days p.i., ODV virions were released from produced polyhedra and analogously separated into nucleocapsid and envelope fractions. Western blot analysis proved that VP80 is associated with ODV-derived nucleocapsids and that it migrates as a single band of ∼80 kDa (Fig. 2B, upper panel). Proper fractionation into nucleocapsid and envelope fractions was verified using anti-PIF-1 antiserum, recognizing an ODV membrane-specific protein (Fig. 2B, lower panel).
Fig. 2.
VP80 copurifies with nucleocapsids of both BV and ODV virions and is associated with one end of the nucleocapsid. (A) Ac-Δvp80-Flag:vp80 BV virions were separated into nucleocapsid (Nc) and envelope (Env) fractions. The Flag-VP80 protein was detected on immunoblots with anti-Flag antibody (upper panel). Correct separation into Nc and Env fractions was controlled with anti-VP39 and anti-GP64 antibodies (bottom panels). (B) The ODV virions produced in Sf9 cells coinfected with the Ac-Δvp80-Flag:vp80 (MOI = 25) and AcMNPV strain E2 (MOI = 5) viruses were separated similarly into Nc and Env fractions and immunoblotted with anti-Flag antibody (upper panel). Proper fractionation was controlled using anti-PIF-1 antiserum (bottom panel). Equal amounts of protein (10 μg) were loaded in the lanes. (C) Immunoelectron microscopy of BV nucleocapsids derived from either the Ac-Δvp80-Flag-vp80 (a) or Ac-Δvp80-egfp:vp80 (b) genotype. (c, d) Nucleocapsids purified from the Ac-wt genotype-propagated BVs were used as a negative control. Purified nucleocapsids were adsorbed to EM grids and immunostained with mouse anti-Flag (a, c) or anti-EGFP (b, d) antibody, followed by goat anti-mouse immunoglobulin conjugated with 5-nm gold particles. The black arrowheads point to specific localizations of 5-nm gold particles. Bars represent 200 nm.
To explore VP80 localization in more detail, direct immunoelectron microscopy was carried out to locate VP80 on the nucleocapsid structure. Nucleocapsid rods derived from BVs of either Ac-Δvp80-egfp:vp80 or Ac-Δvp80-Flag:vp80 were prepared to facilitate VP80 immunodetection. EM observations showed that gold particles specifically localized to one end of the nucleocapsids (Fig. 2Ca and b), with use of anti-Flag and anti-EGFP antibody, respectively. The pattern was the same for both tested BVs (Ac-Δvp80-Flag:vp80 and Ac-Δvp80-egfp:vp80). No gold labeling was observed when control Ac-wt BV-derived nucleocapsids were analyzed (Fig. 2Cc and d). The EM results indicate that VP80 is anchored to one end of the nucleocapsid. Due to electron microscope resolution limits, we are not able to provide information about whether VP80 preferentially associates with the basal region or the apical end of the nucleocapsid.
VP80 is present in a highly organized 3D network in cell nuclei.
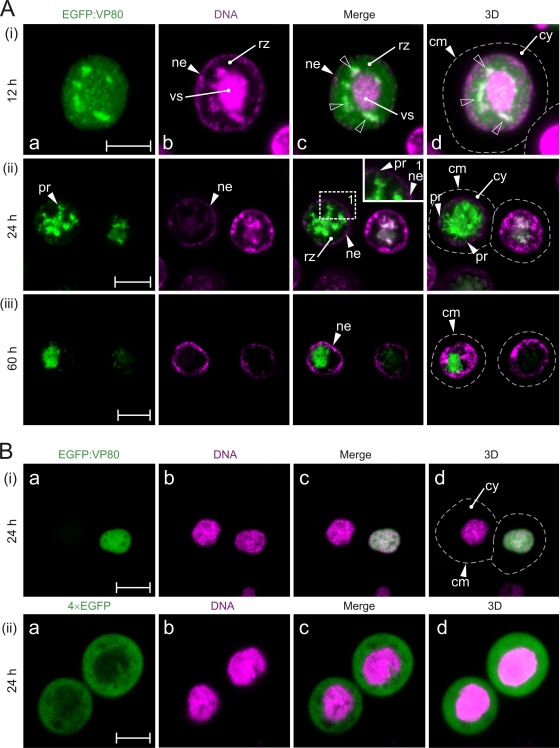
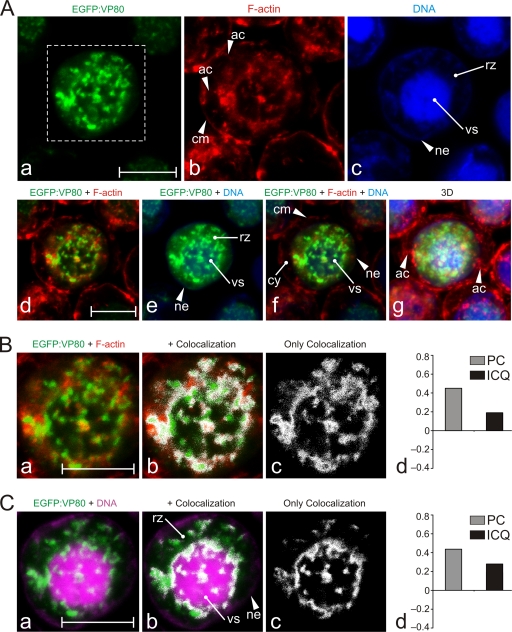
The spatiotemporal distribution of VP80 during viral infection was analyzed by live time course imaging. Sf9 cells were infected with the Ac-Δvp80-egfp:vp80 virus (producing EGFP-fused VP80), followed by confocal imaging (Fig. 3A). Confocal observation showed that EGFP-VP80 strongly accumulates in the nuclei of infected cells, where it forms or associates with a distinct structure(s), which became more apparent later in infection. EGFP-VP80 production was almost undetectable until 12 h p.i., when EGFP-VP80 was mostly homogeneously distributed in cell nuclei but started to accumulate at distinct spots around the DNA-enriched core, the viral replication factory, or “virogenic stroma” (Fig. 3Ai). The EGFP-VP80 spots colocalized with DNA-labeled loci, suggesting a potential function of VP80 in DNA genome processing and packaging into capsids. At this time, in a certain number (∼5%) of infected cells, thick VP80- and DNA-containing filaments directing toward the nuclear periphery were observed (data not shown). In the late phase of infection (24 h p.i.), the strongest EGFP-VP80 signal was localized in the central region of cell nuclei, suggesting the association of EGFP-VP80 with the virogenic stroma (Fig. 3Aii). At this time point, the EGFP-VP80 distribution pattern showed distinct protrusions, routing from the center of the nucleus toward the nuclear envelope (Fig. 3Acii). The intensity of the EGFP-VP80 signal was maximal in the central area of the nucleus, presumably the virogenic stroma region, and gradually decreased toward the ring zone and the contacting nuclear envelope. The term “ring zone” defines a peristromal region enclosing the virogenic stroma inside the baculovirus-infected cell nucleus. Typically, this is the place where ODV virions are embedded into viral occlusion bodies (50).
Fig. 3.
Baculovirus VP80 enters the cell nucleus. (A) Confocal microscopy of Sf9 cells infected with Ac-Δvp80-egfp:vp80 virus (MOI = 10) expressing EGFP-fused VP80 (green) and stained with Hoechst 33342 for DNA (magenta). The cells were observed at 12 h (i), 24 h (ii), and 60 h (iii) p.i. The white open arrowheads point at colocalization sites of EGFP-VP80- and DNA-labeled regions. (B) Transient expression of EGFP-VP80. Confocal microscopy of Sf9 cells transfected with the pIZ-egfp:vp80 (green) (i) or pIZ-4×EGFP (green) (ii) plasmid and stained with Hoechst 33342 for DNA (magenta). The cells were observed at 24 h p.t. Dashed lines represents cell surfaces. All images were collected in green (a) and blue (b) channels and merged (c), and, finally, 3D reconstructions of confocal Z-stack data sets were performed (d). Signals from the blue channel are expressed in magenta for better visualization. Abbreviations: cm, cytoplasmic membrane; cy, cytoplasm; ne, nuclear envelope; nu, nucleus; pr, VP80 protrusion-like structures; rz, ring zone; vs, virogenic stroma. Scale bars represent 10 μm.
With 3D reconstructions of confocal Z-stack data sets, the EGFP-VP80 architecture appeared to form a star-like structure with a highly dense core and protrusions pointing toward the ring zone area and from there toward the nucleus periphery (Fig. 3Adii). Occasionally, crossing of individual beams resembling a highly organized web-like structure could be seen. In the very late phase of infection (60 h p.i.), the EGFP-VP80 signal was still concentrated in the central region of cell nuclei (virogenic stroma localization). Nevertheless, the typical protrusions connecting the virogenic stroma with the nuclear periphery had disappeared (Fig. 3iii). Importantly, no EGFP-VP80 signal was observed by confocal imaging in the cytoplasm at any time point during infection, indicating that VP80 protein is very efficiently targeted into the cell nucleus immediately after synthesis. It is important to mention that a similar VP80 distribution pattern was observed by indirect immunofluorescence using a baculovirus with Flag-tagged VP80, preclusive of artifacts due to EGFP fusion (data not shown).
To verify the specificity of observed phenotypes, Sf9 cells were infected with Ac-wt virus (expressing egfp alone) and also subjected to confocal imaging. In this experiment, we observed a homogenous distribution of the EGFP signal in both the cytoplasmic and nuclear compartments, as has been observed before (e.g., reference 61).
VP80 localization in the absence of viral infection.
When EGFP-VP80 was transiently expressed in insect cells in the absence of any other viral protein, nuclear localization of EGFP-VP80 was seen again (Fig. 3Bi). On the other hand, no nuclear labeling was observed when 4× EGFP (a protein of the same size as the EGFP-VP80 fusion protein) was produced alone (Fig. 3Bii). The results demonstrate that VP80 has an active nuclear localization signal (NLS), which is in accordance with bioinformatic predictions (putative bipartite NLS amino acids [aa] 424 to 439). Notably, transiently expressed EGFP-VP80 did not occupy or form any distinct intranuclear structures in insect cells, as has been seen in baculovirus-infected cells. The results collected so far suggest that intranuclear VP80-specific structures seen during infection are the consequence of complex recruitment and orchestration of viral, and possibly cellular, factors (e.g., the host cytoskeletal proteins) but not of VP80 alone.
Myosin-like sequences in VP80.
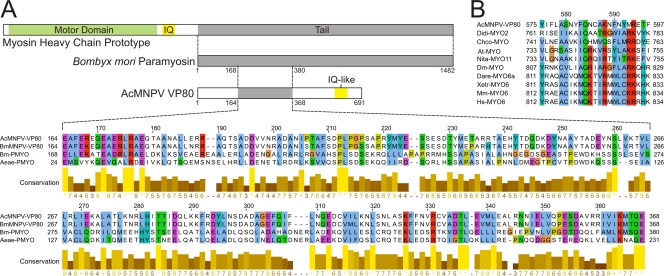
To further elucidate the function of VP80 in the viral replication cycle, the VP80 sequence has been searched against protein databases to identify putative functional domains. No unique conserved domain has been definitively revealed; however, most homology-based tools pointed out a relationship with actin cytoskeleton-interacting proteins, especially with proteins in the myosin protein family. For instance, using the PSI-BLAST (2) approach, a similarity between VP80 and tail domains of myosin proteins was identified, while the SMART server (29) revealed a certain resemblance with myosin motor domains. The MyHits tool on the EXPASY server revealed an IQ-like motif, which is a calmodulin-binding domain present in all myosin motor proteins, in the C-terminal region (aa 575 to 597) of VP80 (Fig. 4B). On the other side, a myosin motor-specific ATP-binding motif (P-loop) responsible for the coordination and hydrolysis of ATP was not identified in VP80.
Fig. 4.
VP80 contains paramyosin-like and myosin-like sequence motifs. (A) Alignment of AcMNPV-VP80 and BmMNPV-VP80 (both with the sequence Glu164-Glu368) with corresponding sequences of paramyosin proteins: Bombyx mori PMYO (Bm-PMYO) (GenBank accession no. XP_637740.1) and Aedes aegypti PMYO (Aeae-PMYO) (XP_001652576.1). (B) AcMNPV VP80 sequence Tyr575-Phe597 aligned with experimentally verified IQ motifs (calmodulin-binding domains) from selected myosin motor proteins: Dictyostelium discoideum MYO2 (Didi-MYO2) (XP_637740.1), Chara corallina MYO (Chco-MYO) (BAB03273.1), Arabidopsis thaliana MYO (At-MYO) (CAA84066.1), Nicotiana tabacum MYO11 (Nita-MYO11) (BAD72949.1), Drosophila melanogaster MYO (Dm-MYO) (CAA47462.1), Xenopus tropicalis MYO6 (Xetr-MYO6) (AAI21230.1), Danio rerio MYO6a (Dare-MYO6a) (AAT92221.1), Mus musculus MYO6 (Mm-MYO6) (NP_001034635.2), and Homo sapiens MYO6 (Hs-MYO6) (NP_004990.3). The alignments were constructed using MUSCLE, and coloring was performed in JalView.
When AcMNPV VP80 or its homologue in Bombyx mori NPV (BmNPV) was searched against the Bombyx mori genome database (15), an obvious similarity to the paramyosin (PMYO) protein was identified. Similarity was also found with the Aedes aegypti PMYO protein (Fig. 4A). For instance, AcMNPV and BmNPV VP80 sequences from Glu164-Glu368 showed 21% and 23% identity, respectively, to the B. mori PMYO sequence Glu168-Glu380. Thus, the in silico searches provided information that baculovirus VP80 is more a putative paramyosin-like or myosin tail-like protein than an active myosin motor protein. Interestingly, cellular paramyosin molecules are essential components of the invertebrate actin-myosin (acto-myosin) complex (17).
Late events in AcMNPV infection are susceptible to BDM and CD.
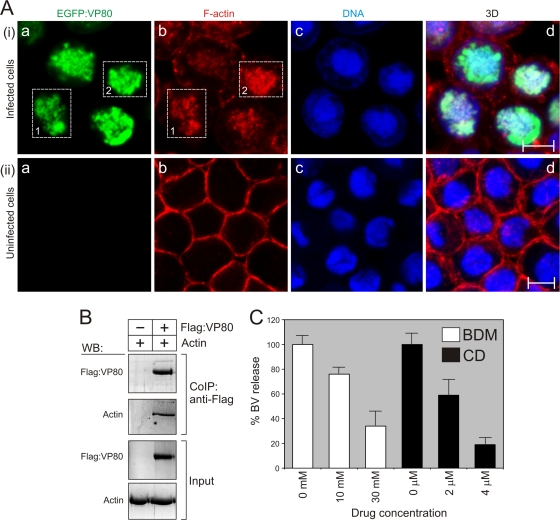
As recently shown (39), VP80 is involved in the transport of progeny nucleocapsids, since deletion of the vp80 gene completely prevents transit of de novo-assembled nucleocapsids toward the nuclear periphery to form viral progeny (BVs and ODVs). These findings, together with the fact that VP80 contains a paramyosin-like domain, led us to the theory that progeny nucleocapsids may be transported toward the nuclear periphery with the help of an acto-myosin complex. To verify this hypothesis, cells in the late phase of AcMNPV infection were treated either with BDM, a myosin motor-specific inhibitor, or CD, an actin-depolymerizing agent. For these purposes, Sf9 cells were infected with Ac-wt and cultured under standard conditions. At 12 h p.i., inhibitors were added at various concentrations (Fig. 5C) to the culture media, and BV production was measured in culture supernatants at 48 h p.i. The presence of each type of inhibitor resulted in a significant inhibition of BV release. The presence of 10 mM BDM reduced BV production to 76% ± 6% and a 30 mM concentration even reduced production to 34% ± 8% of the level in untreated control cells (considered 100%) (Fig. 5C). Similar results were obtained with CD, which reduced BV release to 59% ± 7% (2 μM) and to 19% ± 3% (4 μM) of the control level. The inhibition assays thus support the hypothesis that late events of AcMNPV infection, such as maturation of nucleocapsids and their movement toward the cell periphery and subsequent BV budding, are dependent on actin-based myosin motor functions.
Fig. 5.
VP80 interacts with the virus-triggered, nuclear F-actin cytoskeleton. (A) Confocal microscopy of Sf9 cells infected with Ac-Δvp80-egfp:vp80 virus (MOI = 10) (i) and uninfected Sf9 cells (ii). At 24 h p.i., the cells were fixed and stained with Hoechst 33342 for DNA (blue) and with phalloidin-rhodamine for F-actin (red). All images were collected in green (a), red (b), and blue (c) channels, and, finally, 3D reconstructions of confocal Z-stack data sets were performed (d). The dashed boxes point out overlapping distributions of EGFP-VP80 protein and nuclear F-actin. Scale bar, 10 μm. (B) VP80 associates with host cell actin. Sf9 cells were infected with Ac-Δvp80-Flag:vp80 (MOI = 20), and Flag-VP80 protein was coimmunoprecipitated at 48 h p.i. (CoIP:anti-Flag). Proteins bound to the anti-Flag antibody were analyzed by immunoblotting with anti-Flag and antiactin antibodies. In the experiment, Sf9 cells infected with the Ac-wt virus (MOI = 20) were used as a negative control. Cell lysates are shown as input. WB, Western blot. (C) Sensitivity of AcMNPV infection to BDM and CD treatment. Sf9 cells were infected with Ac-wt and treated with BDM and CD as indicated. BVs were quantified in cell culture supernatants at 48 h p.i. by endpoint dilution. Results of three independent assays, with error bars giving the SD, are shown.
VP80 and F-actin provide traffic connections between the viral replication factory and the nuclear periphery.
To be able to link VP80 functions to actin-based movement, we studied its interactions with the host actin cytoskeleton. To this end, Sf9 cells were infected with the Ac-Δvp80-egfp:vp80 virus and EGFP-VP80 localization was analyzed in relation to the F-actin cytoskeleton by confocal imaging. First, baculovirus infection caused dramatic changes in F-actin network architecture (Fig. 5A), as was shown for the first time by Ohkawa and Volkman (41). Importantly, confocal imaging showed remarkable overlapping distributions of VP80 and F-actin filaments in baculovirus-infected cells (Fig. 5A). Again, the EGFP-VP80 signals were observed only in cell nuclei, where they apparently colocalized with the virus-induced F-actin networks (Fig. 5Ai). Interaction between VP80 and actin was subsequently shown by immunoprecipitation, when host actin was specifically copurified with Flag-tagged VP80 protein (Fig. 5B).
Based on these observations, the question of whether VP80 is involved in the establishment of the virus-induced, nuclear F-actin network was raised. To answer this, Sf9 cells were transfected with either Ac-Δvp80 or Ac-wt bacmid, and F-actin architectures were compared by confocal microscopy. Here, we did not note any dramatic differences in F-actin architectures formed in the absence or presence of VP80 (data not shown). From these data, we concluded that VP80 forms complexes with the host F-actin cytoskeleton but is not necessary for the (re)arrangement of the latter.
Confocal imaging at high magnification revealed that the VP80 protein together with F-actin cables penetrates and encloses the virogenic stroma, the site of virus replication (Fig. 6A). We could observe overt actin-built-up cables permeating the virogenic stroma and going toward its periphery, which were accompanied by the VP80 protein (Fig. 6A). These actin-VP80 tracks apparently connected the virogenic stroma with the nuclear periphery. Furthermore, the F-actin cables appeared to continue toward the cell periphery, while VP80 signal was limited to the nuclear compartment. A colocalization assay provided a spatial map of the overlapping distribution of VP80 with the F-actin cytoskeleton (Fig. 6B). Relatively low values of calculated PC (0.45) and ICQ (0.19) (values of 1 and 0.5, respectively, represent complete colocalization) indicate that VP80 does not directly decorate F-actin filaments but more probably coaligns with or localizes to them at distinct regions. An analogous colocalization assay was done for VP80 and DNA to demonstrate an interaction of VP80 with the virogenic stroma. This analysis provided an encircled colocalization pattern with several highly organized spots inside the virogenic stroma (Fig. 6C).
Fig. 6.
Colocalization of VP80 with F-actin filaments connecting the virogenic stroma with the nuclear envelope. (A) Confocal microscopy of Sf9 cells infected with Ac-Δvp80-egfp:vp80 (MOI = 10) expressing EGFP-fused VP80 (green). At 24 h p.i., the cells were fixed and stained with Hoechst 33342 for DNA (blue) and with phalloidin-rhodamine for F-actin (red). The images were collected in green (a), red (b), and blue (c) channels and differentially merged (d to f), and, finally, 3D reconstructions of confocal Z-stack data sets were performed (g). (B) Colocalization of EGFP-VP80 with nuclear F-actin. (C) Colocalization of EGFP-VP80 with DNA-labeled regions. For panels B and C, the green and red channels from the inset from panel Aa were merged (a). Colocalization regions are depicted in white (b to c). (B and C) The colocalization proportions as represented by PC and ICQ values were plotted into graphs (d). Abbreviations: ac, cytoplasmic F-actin cables; cm, cytoplasmic membrane; cy, cytoplasm; ne, nuclear envelope; rz, ring zone; vs, virogenic stroma. Scale bars represent 10 μm.
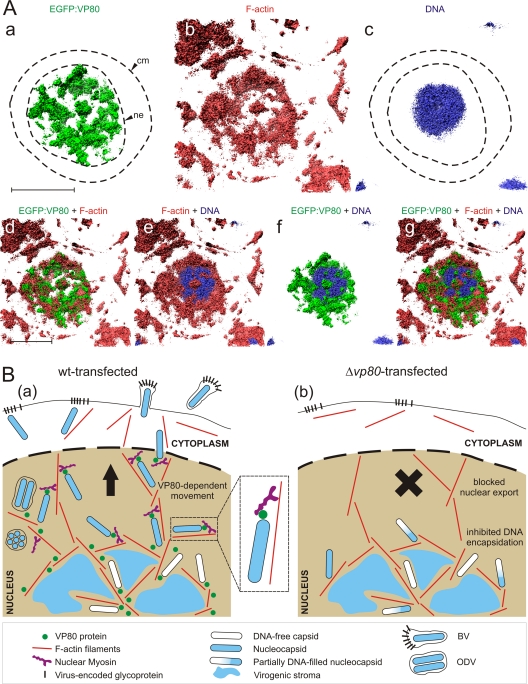
Finally, 3D reconstruction of Z-stack confocal data improved our view on the architecture of baculovirus-infected cells (Fig. 7A). Taken together, our findings show that VP80 protein and F-actin cytoskeleton interact and provide traffic connections between the virogenic stroma and nucleus periphery, presumably to facilitate egress of progeny nucleocapsids.
Fig. 7.
VP80 and the nuclear architecture of baculovirus-infected cells. (A) 3D isosurface rendering based on confocal data. Sf9 cells infected with the Ac-Δvp80-egfp:vp80 virus (green) were fixed and stained with Hoechst 33342 for DNA (blue) and with phalloidin-rhodamine for F-actin (red) and imaged simultaneously by confocal microscopy. The volume data were generated from 30 scans in the Z-axis, using 400 nm per each depth step. The signals are shown separately (a to c) and together (d to g) to demonstrate the intranuclear localization of VP80 to the F-actin cytoskeleton and the DNA-labeled area (virogenic stroma). Abbreviations: cm, cytoplasmic membrane; ne, nuclear envelope. Scale bar, 10 μm. (B) Model for the function of VP80 protein in the baculovirus infection cycle based on the intracellular distribution of VP80, its association with the F-actin cytoskeleton, the Δvp80 phenotype, and the sensitivity of AcMNPV infection to BDM treatment. In Ac-wt-transfected cells (a), a high number of DNA-filled progeny nucleocapsids are transported along F-actin to the cell periphery to form both BV and ODV virions, whereas in Δvp80-transfected cells (b), DNA encapsidation is affected and nucleocapsid release from the virogenic stroma is completely prevented. VP80 is depicted in contact with viral nucleocapsids, nuclear F-actin filaments, and virogenic stroma.
DISCUSSION
In this paper, we studied properties of the AcMNPV VP80 minor capsid protein, its localization on the nucleocapsid, and the kinetics of its subcellular distribution during infection in insect Sf9 cells. We also addressed the question of whether any sequence similarities and, eventually, functional domain topologies between VP80 and proteins deposited in the protein databases can be found.
To address these issues, we engineered the knockout Ac-Δvp80 bacmid to encode “visible” EGFP-tagged VP80 protein. One-step growth kinetics showed that introduction of the egfp-vp80 cassette completely rescued virus replication to the wild-type level (Fig. 1), indicating that fusion of EGFP to VP80 does not negatively affect virus fitness, at least in cell culture. More specifically, we show that the N terminus of VP80 can be targeted to engineer novel virus vectors with designated properties. The constructed Ac-Δvp80-egfp:vp80 virus was then employed as a tool to explore VP80 localization on the nucleocapsid structure, as well as its spatiotemporal distribution during infection.
Immunolabeling of BV-derived nucleocapsids showed that VP80 preferentially associates with one end of the nucleocapsid (Fig. 2), indicating a potential interaction/cooperation with other nucleocapsid end-linked proteins: P78/83 (54) and VLF-1 (53). The polarity of nucleocapsids is in line with earlier observations that ODV nucleocapsids are anchored at one specific end (the cap structure) to envelopes present in the nucleus (21). In infected cells, EGFP-VP80 was observed entirely in the nucleus, and the distribution pattern changed over time (Fig. 3). Initially (12 h p.i.), the EGFP-VP80 signal was homogenously spread throughout the nucleus but subsequently started to colocalize with DNA-containing areas protruding from the virogenic stroma. In the late phase of infection (24 h p.i.), the EGFP-VP80 pattern showed obvious connections between the central area of the nucleus and the nuclear periphery. Interestingly, these EGFP-VP80 tracks that connect the center of the nucleus with the nuclear envelope completely disappeared in the very late phase of infection (60 h p.i.). These spatiotemporal changes in VP80 distribution might apparently be the result of sophisticated baculovirus regulation, functioning as a two-throw switch between the production of two virion types: extracellular BV (late) and intranuclear ODV embedded in viral occlusion bodies (very late).
Confocal imaging showed that EGFP-VP80 accumulates in the nucleus. No EGFP-VP80 signal was observed in the cytoplasm at any time point during infection (Fig. 3 and 5 to 6). This finding is not in agreement with our observations showing the association of EGFP-VP80 with nucleocapsids of both virion types (Fig. 2), theoretically making it possible to detect EGFP-VP80 during nucleocapsid transit through the cytoplasm toward the cell periphery to form BV. The observed inconsistency might be explained by a low abundance of nucleocapsid-bound EGFP-VP80, which is under the detection threshold of confocal microscopy.
Colocalization of VP80 with DNA can indicate that VP80 is a part of a DNA-interacting protein complex or has the ability to bind DNA itself. At the later time (24 h p.i.), EGFP-VP80 was present in a 3D network penetrating and connecting the virogenic stroma with the nuclear envelope. These observations support the hypothesis that VP80 is somehow involved in the packaging of nucleocapsids, which is further based on the relatively high number of only partially DNA-filled capsids (absence of mature capsids) observed in Δvp80-transfected cells (39). In addition, VP80-containing tracks linking the virogenic stroma with the nuclear envelope strongly suggest the involvement of VP80 in progeny virus egress. These findings are in accordance with those of our previous study, where we revealed that deletion of vp80 totally prevents the transport of de novo-assembled nucleocapsids from the virogenic stroma toward the nuclear periphery (39).
In order to identify functional domains, we compared the VP80 sequence with sequences in protein databases. Unexpectedly, a certain similarity was identified between VP80 and the invertebrate paramyosin protein family (Fig. 4A). Strong indications for interactions between viruses and host myosin motors exist (19, 28, 48, 62), but to date, no paramyosin-like proteins have been identified in any viral genome. The paramyosins are a major structural component of muscular thick filaments in a diverse range of invertebrate species, including insects (reviewed in, e.g., reference 23). In general, paramyosins form cofilaments with myosin motor proteins (thick filaments), which interact with actin-based (thin) filaments to convert chemical energy (ATP) into mechanical force (e.g., movement). Importantly, the molar ratio between myosin motors and paramyosin molecules in thick filaments has a crucial impact on the performance of the acto-myosin complex (23). These facts led us to formulate a model scenario (Fig. 7B) where nucleocapsid-linked VP80, a baculovirus paramyosin-like protein, is responsible for interaction with the host acto-myosin complex to facilitate active, directed nucleocapsid transport. In principle, this is nothing unusual, as similar macromolecular assembly movements are frequently executed in cell nuclei, for instance, to export ribosomal subunits (11) or to drive movement of transcription complexes along a DNA template (32, 63). The conserved AcMNPV ODV (6) and BV (58) protein encoded by orf66 (Ac66) has been reported to possess homology to the actin-binding domain of myosin VI (56). Deletion of this gene gives a phenotype similar to that of a vp80 deletion mutant, with impaired nucleocapsid egress from the nucleus as well as a block in ODV synthesis (25). We hypothesize that VP80 and Ac66 may work in concert to transport progeny nucleocapsids to the nuclear periphery.
To test our hypothesis that the acto-myosin complex is important for progeny nucleocapsid transport, we treated AcMNPV-infected cells (at 12 h p.i., the initial time of progeny egress) with either BDM or CD and checked inhibition effects on progeny BV production. We could see strong inhibitory effects on BV release using each drug (Fig. 5C), demonstrating a significant contribution of the host acto-myosin complex in progeny virus egress and budding. It has previously been shown that transport of parental AcMNPV nucleocapsids to and/or into the nucleus is facilitated by the host acto-myosin complex (14, 27, 42). Herein, we provide evidence for participation of the actin-based myosin motor functions also in late events (such as egress and budding) of AcMNPV infection. Nevertheless, it has to be taken into account that the BDM-triggered decrease in BV production might also be the result of side effects of the drug (e.g., Ca2+ release from the endoplasmic reticulum, etc).
To extend our theory, we looked at a potential in vivo interaction between VP80 and the host actin cytoskeleton. Upon AcMNPV infection, we could see a typical rearrangement of the F-actin filaments (Fig. 5A). More interestingly, the nuclear EGFP-VP80 pattern evidently accompanied the virus-triggered F-actin network, confirming our assumptions. Detailed views into the nucleus revealed that VP80 and F-actin filaments together penetrate and link the virogenic stroma with the nuclear periphery (Fig. 6). VP80 localization was limited to the nucleus, while F-actin filaments were seen going out of the nucleus and continuing toward the cell periphery. Additionally, immunoprecipitation assays proved that VP80 forms complexes with host actin (Fig. 5B). It remains unclear whether VP80 directly interacts with or is part of a protein complex interacting with the actin cytoskeleton. However, our colocalization measurement implies that VP80 does not tightly decorate actin filaments but more probably coaligns with them at certain distances. Taken together, we provide evidence that AcMNPV VP80 is a newly identified baculovirus actin-interacting capsid protein, next to major capsid protein VP39 (9, 10), Wiscott-Aldrich Syndrome Protein (WASP) homology phosphoprotein P78/83 (27), and BV/ODV C42 (31).
Baculoviruses replicate and package their viral DNA genomes inside the nuclei of infected cells; hence, transport to and away from the nucleus is a pivotal task of their evolutionary success. A previous study by Ohkawa and coworkers (42) clearly demonstrated that AcMNPV parental nucleocapsids are capable of undergoing undirected, actin-based motility (by means of so-called actin comets) in the cytoplasm to reach the cell nucleus. We do not suppose that this kind of transport is used by progeny nucleocapsids to egress the cell nucleus because of the highly organized structure of the nucleus. Especially in baculovirus-infected cells, where the nuclei are markedly enlarged and the highly dense virogenic stroma represents a labyrinth for progeny nucleocapsids, undirected actin comet-based motility does not appear to be effective. Our data collected so far imply that the escape of progeny nucleocapsids from the virogenic stroma toward the nuclear periphery, and maybe even further toward the cell periphery for BVs, is mediated by a more directed transit, in which both F-actin and myosin motor functions are included. In addition, there is also recent evidence for involvement of the microtubule cytoskeleton in baculovirus progeny egress (18). Hence, it seems that microfilaments and microtubules mutually contribute to the efficient transport of progeny nucleocapsids from the nucleus toward the cell periphery.
Therefore, future research should address the following tasks. Which host myosin motor proteins interact with baculovirus capsid proteins, e.g., VP80? How do these interactions mediate the movement and budding of progeny nucleocapsids? How is transport of viral nucleocapsids timed along F-actin filaments (BV versus ODV phenotype)? Another intriguing question is how baculoviruses in other genera (Betabaculovirus, Gammabaculovirus, and Deltabaculovirus) that do not encode VP80 manage to transport their nucleocapsids to the nuclear periphery.
ACKNOWLEDGMENTS
We are grateful to Daniel Stockholm and Perinne Borel (both are at Généthon, France) for kind assistance in confocal microscopy. We thank Ke Peng for the help with ODV preparation and Feana Francis Devaraj for the immunogold labeling of baculovirus nucleocapsids (both are at Wageningen University, Netherlands).
Martin Marek, Lionel Galibert, and Otto-Wilhelm Merten were financed by the project BACULOGENES of the European Union (contract no. FP6-037541). Monique M. van Oers was financed in part by the Program Strategic Alliances of the Royal Dutch Academy of Sciences.
Footnotes
Published ahead of print on 30 March 2011.
REFERENCES
- 1. Abramoff M. D., Magelhaes P. J., Ram S. J. 2004. Image processing with ImageJ. Biophotonics Int. 11:36–42 [Google Scholar]
- 2. Altschul S., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Appel R. D., Bairoch A., Hochstrasser D. F. 1994. A new generation of information retrieval tools for biologists: the example of the ExPASy WWW server. Trends Biochem. Sci. 19:258. [DOI] [PubMed] [Google Scholar]
- 4. Ayres M. D., Howard S. C., Kuzio J., Lopez-Ferber M., Possee R. D. 1994. The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology 202:586–605 [DOI] [PubMed] [Google Scholar]
- 5. Bolte S., Cordelires F. P. 2006. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224:213–232 [DOI] [PubMed] [Google Scholar]
- 6. Braunagel S. C., Russell W. K., Rosas-Acosta G., Russell D. H., Summers M. D. 2003. Determination of the protein composition of the occlusion-derived virus of Autographa californica nucleopolyhedrovirus. Proc. Natl. Acad. Sci. U. S. A. 100:9797–9802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Braunagel S. C., Summers M. D. 1994. Autographa californica nuclear polyhedrosis virus, PDV, and ECV viral envelopes and nucleocapsids: structural proteins, antigens, lipid and fatty acid profiles. Virology 202:315–328 [DOI] [PubMed] [Google Scholar]
- 8. Carstens E. B. 2009. AcMNPV as a model for baculovirus DNA replication. Virol. Sin. 24:243–267 [Google Scholar]
- 9. Charlton C. A., Volkman L. E. 1993. Penetration of Autographa californica nuclear polyhedrosis virus nucleocapsids into IPLB Sf21 cells induces actin cable formation. Virology 197:245–254 [DOI] [PubMed] [Google Scholar]
- 10. Charlton C. A., Volkman L. E. 1991. Sequential rearrangement and nuclear polymerization of actin in baculovirus-infected Spodoptera frugiperda cells. J. Virol. 65:1219–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cisterna B., Necchi D., Prosperi E., Biggiogera M. 2006. Small ribosomal subunits associate with nuclear myosin and actin in transit to the nuclear pores. FASEB J. 20:1901–1903 [DOI] [PubMed] [Google Scholar]
- 12. Cohen D. P. A., Marek M., Davies B. G., Vlak J. M., van Oers M. M. 2009. Encyclopedia of Autographa californica nucleopolyhedrovirus genes. Virol. Sin. 24:359–414 [Google Scholar]
- 13. de Castro E., et al. 2006. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 34:W362–W365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dong S., et al. 2010. Autographa californica multicapsid nucleopolyhedrovirus efficiently infects Sf9 cells and transduces mammalian cells via direct fusion with the plasma membrane at low pH. J. Virol. 84:5351–5359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Duan J., et al. 2010. SilkDB v2.0: a platform for silkworm (Bombyx mori) genome biology. Nucleic Acids Res. 38:D453–D456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Edgar R. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Epstein H. F., Aronow B. J., Harris H. E. 1976. Myosin-paramyosin cofilaments: enzymatic interactions with F-actin. Proc. Natl. Acad. Sci. U. S. A. 73:3015–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fang M., Nie Y., Theilmann D. A. 2009. AcMNPV EXON0 (AC141) which is required for the efficient egress of budded virus nucleocapsids interacts with β-tubulin. Virology 385:496–504 [DOI] [PubMed] [Google Scholar]
- 19. Feierbach B., Piccinotti S., Bisher M., Denk W., Enquist L. W. 2006. Alpha-herpesvirus infection induces the formation of nuclear actin filaments. PLoS Pathog. 2:0763–0776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Finn R. D., et al. 2008. The Pfam protein families database. Nucleic Acids Res. 36:D281–D288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fraser M. J. 1986. Ultrastructural observations of virion maturation in Autographa californica nuclear polyhedrosis virus infected Spodoptera frugiperda cell cultures. J. Ultrastruct. Mol. Struct. Res. 95:189–195 [Google Scholar]
- 22. Goley E. D., et al. 2006. Dynamic nuclear actin assembly by Arp2/3 complex and a baculovirus WASP-like protein. Science 314:464–467 [DOI] [PubMed] [Google Scholar]
- 23. Hooper S. L., Hobbs K. H., Thuma J. B. 2008. Invertebrate muscles: thin and thick filament structure; molecular basis of contraction and its regulation, catch and asynchronous muscle. Prog. Neurobiol. 86:72–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jen D., et al. 2004. ImageSurfer: a tool for visualizing correlations between two volume scalar fields. In Proceedings of the Conference on Visualization (2004) http://wwwx.cs.unc.edu/∼taylorr/Comp715/papers/imagesurfer.pdf [Google Scholar]
- 25. Ke J., Wang J., Deng R., Wang Z. 2008. Autographa californica multiple nucleopolyhedrovirus ac66 is required for the efficient egress of nucleocapsids from the nucleus, general synthesis of preoccluded virions and occlusion body formation. Virology 374:421–431 [DOI] [PubMed] [Google Scholar]
- 26. Kool M., Ahrens C. H., Vlak J. M., Rohrmann G. F. 1995. Replication of baculovirus DNA. J. Gen. Virol. 76:2103–2118 [DOI] [PubMed] [Google Scholar]
- 27. Lanier L. M., Volkman L. E. 1998. Actin binding and nucleation by Autographa californica M nucleopolyhedrovirus. Virology 243:167–177 [DOI] [PubMed] [Google Scholar]
- 28. Lehmann M. J., Sherer N. M., Marks C. B., Pypaert M., Mothes W. 2005. Actin- and myosin-driven movement of viruses along filopodia precedes their entry into cells. J. Cell Biol. 170:317–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Letunic I., et al. 2006. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 34:D257–D260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Letunic I., Doerks T., Bork P. 2009. SMART 6: recent updates and new developments. Nucleic Acids Res. 37:D229–D232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li K., et al. 2010. The putative pocket protein binding site of Autographa californica nucleopolyhedrovirus BV/ODV-C42 is required for virus-induced nuclear actin polymerization. J. Virol. 84:7857–7868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li Q., Sarna S. K. 2009. Nuclear myosin II regulates the assembly of preinitiation complex for ICAM-1 gene transcription. Gastroenterology 137:1051–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li X., Pang A., Lauzon H. A. M., Sohi S. S., Arif B. M. 1997. The gene encoding the capsid protein P82 of the Choristoneura fumiferana multicapsid nucleopolyhedrovirus: sequencing, transcription and characterization by immunoblot analysis. J. Gen. Virol. 78:2665–2673 [DOI] [PubMed] [Google Scholar]
- 34. Long G., Pan X., Kormelink R., Vlak J. M. 2006. Functional entry of baculovirus into insect and mammalian cells is dependent on clathrin-mediated endocytosis. J. Virol. 80:8830–8833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lu A., Carstens E. B. 1992. Nucleotide sequence and transcriptional analysis of the p80 gene of Autographa californica nuclear polyhedrosis virus: a homologue of the Orgyia pseudotsugata nuclear polyhedrosis virus capsid-associated gene. Virology 190:201–209 [DOI] [PubMed] [Google Scholar]
- 36. Lu A., Miller L. 1995. The roles of eighteen baculovirus late expression factor genes in transcription and DNA replication. J. Virol. 69:975–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Marchler-Bauer A., et al. 2005. CDD: a conserved domain database for protein classification. Nucleic Acids Res. 33:D192–D196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Marchler-Bauer A., et al. 2003. CDD: a curated Entrez database of conserved domain alignments. Nucleic Acids Res. 31:383–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Marek M., van Oers M. M., Deveraj F. F., Vlak J. M., Merten O.-W. 2011. Engineering of baculovirus vectors for the manufacture of virion-free biopharmaceuticals. Biotechnol. Bioeng. 108:1056–1067 [DOI] [PubMed] [Google Scholar]
- 40. Muller R., Pearson M. N., Russell R. L. Q., Rohrmann G. F. 1990. A capsid-associated protein of the multicapsid nuclear polyhedrosis virus of Orgyia pseudotsugata: genetic location, sequence, transcriptional mapping, and immunocytochemical characterization. Virology 176:133–144 [DOI] [PubMed] [Google Scholar]
- 41. Ohkawa T., Volkman L. E. 1999. Nuclear F-actin is required for AcMNPV nucleocapsid morphogenesis. Virology 264:1–4 [DOI] [PubMed] [Google Scholar]
- 42. Ohkawa T., Volkman L. E., Welch M. D. 2010. Actin-based motility drives baculovirus transit to the nucleus and cell surface. J. Cell Biol. 190:187–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pagni M., et al. 2004. MyHits: a new interactive resource for protein annotation and domain identification. Nucleic Acids Res. 32:W332–W335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pagni M., et al. 2007. MyHits: improvements to an interactive resource for analyzing protein sequences. Nucleic Acids Res. 35:W433–W437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Passarelli A. L., Guarino L. A. 2007. Baculovirus late and very late gene regulation. Curr. Drug Targets 8:1103–1115 [DOI] [PubMed] [Google Scholar]
- 46. Peng K., et al. 2009. Identification of protein-protein interactions of the occlusion-derived virus-associated proteins of Helicoverpa armigera nucleopolyhedrovirus. J. Gen. Virol. 91:659–670 [DOI] [PubMed] [Google Scholar]
- 47. Pettersen E. F., et al. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612 [DOI] [PubMed] [Google Scholar]
- 48. Roberts K. L., Baines J. D. 2010. Myosin Va enhances secretion of herpes simplex virus 1 virions and cell surface expression of viral glycoproteins. J. Virol. 84:9889–9896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schultz J., Milpetz F., Bork P., Ponting C. P. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. U. S. A. 95:5857–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Slack J., Arif B. M. 2007. The baculoviruses occlusion-derived virus: virion structure and function. Adv. Virus Res. 69:99–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Summers M. D., Volkman L. E. 1976. Comparison of biophysical and morphological properties of occluded and extracellular nonoccluded baculovirus from in vivo and in vitro host systems. J. Virol. 17:962–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tang X.-D., et al. 2008. Characterization of a Bombyx mori nucleopolyhedrovirus with Bmvp80 disruption. Virus Res. 138:81–88 [DOI] [PubMed] [Google Scholar]
- 53. Vanarsdall A. L., Okano K., Rohrmann G. F. 2006. Characterization of the role of very late expression factor 1 in baculovirus capsid structure and DNA processing. J. Virol. 80:1724–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Vialard J. E., Richardson C. D. 1993. The 1,629-nucleotide open reading frame located downstream of the Autographa californica nuclear polyhedrosis virus polyhedrin gene encodes a nucleocapsid-associated phosphoprotein. J. Virol. 67:5859–5866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Volkman L. E. 1988. Autographa californica MNPV nucleocapsid assembly: inhibition by cytochalasin D. Virology 163:547–553 [DOI] [PubMed] [Google Scholar]
- 56. Volkman L. E. 2007. Baculovirus infectivity and the actin cytoskeleton. Curr. Drug Targets 8:1075–1083 [DOI] [PubMed] [Google Scholar]
- 57. Wang M., et al. 2010. Partial functional rescue of Helicoverpa armigera single nucleocapsid nucleopolyhedrovirus infectivity by replacement of F protein with GP64 from Autographa californica multicapsid nucleopolyhedrovirus. J. Virol. 84:11505–11514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wang R., et al. 2010. Proteomics of the Autographa californica nucleopolyhedrovirus budded virions. J. Virol. 84:7233–7242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Waterhouse A. M., Procter J. B., Martin D. M. A., Clamp M., Barton G. J. 2009. Jalview version 2. A multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wu W., et al. 2008. Autographa californica multiple nucleopolyhedrovirus 38K is a novel nucleocapsid protein that interacts with VP1054, VP39, VP80, and itself. J. Virol. 82:12356–122354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Xu F., Vlak J. M., Eker A. P. M., van Oers M. M. 2010. DNA photolyases of Chrysodeixis chalcites nucleopolyhedrovirus target to the nucleus and interact with chromosomes and mitotic spindle structures. J. Gen. Virol. 91:907–914 [DOI] [PubMed] [Google Scholar]
- 62. Xu X., et al. 2011. VP15R from infectious spleen and kidney necrosis virus is a non-muscle myosin-II-binding protein. Arch. Virol. 156:53–61 [DOI] [PubMed] [Google Scholar]
- 63. Ye J., Zhao J., Hoffmann-Rohrer U., Grummt I. 2008. Nuclear myosin I acts in concert with polymeric actin to drive RNA polymerase I transcription. Genes Dev. 22:322–330 [DOI] [PMC free article] [PubMed] [Google Scholar]