Abstract
Caveolin 1 (Cav-1), the scaffold protein of a specific membrane lipid raft called caveolae, has been reported to suppress HIV-1 replication. However, the mechanism by which Cav-1 inhibits HIV replication remains unclear. In this study, we investigated the mechanism by which Cav-1 inhibits HIV replication at the level of gene expression. Our results show that Cav-1 represses viral gene expression and that this suppression involves the NF-κB pathway. We used several approaches in different cell types, including primary CD4+ T cells and macrophages, to demonstrate the role of nuclear factor κB (NF-κB) in Cav-1-mediated inhibition of viral expression. A mutational analysis of the cis-acting element shows that the two NF-κB sites in the U3 region of the long terminal repeat (LTR) are critical for Cav-1-mediated inhibition of viral expression. In the presence of Cav-1, phosphorylation of IKKβ, IKKα, IκBα, and NF-κB p65 is dramatically reduced, while viral gene expression is suppressed. In addition, translocation of NF-κB p65 to the nucleus decreases substantially in the presence of Cav-1. Furthermore, significant inhibition of NF-κB activation and binding to target DNA are evident in the presence of Cav-1. These results establish evidence that Cav-1 inhibits HIV replication by transcriptional repression of viral gene expression and contributes to HIV's persistent infection of macrophages.
INTRODUCTION
Caveolae are flask-shaped small invaginations, 50 to 100 nm in diameter, on the plasma membrane that are highly enriched in cholesterol, phospholipids, and sphingolipids and are abundant in various cell types (43, 46, 54, 55, 61). These organelles provide scaffolding for compartmented cellular processes and participate in multiple cellular functions, including endocytosis, transcytosis, cholesterol transport, suppression of cell transformation, and signal transduction (15, 16, 29, 32, 33, 42, 46, 47, 53, 60). The caveolae structure is composed of proteins known as caveolins (Cav), and three types (Cav-1, Cav-2, and Cav-3) have been identified (27, 42). Cav-1 is the major coat protein responsible for caveolae assembly (14, 28) and is highly expressed in quiescent or terminally differentiated cells, including dendritic cells and monocyte/macrophages (17, 19, 36, 43, 46, 51, 54, 55, 61). The Cav-1 protein is generally believed to be absent from lymphocytes. This protein can positively or negatively regulate signaling by interacting directly or indirectly with caveolae-resident proteins (6, 7, 18, 36, 46, 50, 60).
The caveolin scaffolding domain (CSD), residues 82 to 101 in Cav-1, is essential for both caveolin oligomerization and the interaction of caveolins with other proteins (11). Interactions with other proteins through the CSD help provide coordinated and efficient signal transduction (51, 60). The CSD serves as a receptor for binding proteins containing sequence motif ϕXϕXXXXϕ, ϕXXXXϕXXϕ, or ϕXϕXXXXϕXXϕ (ϕ representing any aromatic amino acid and X any other amino acid) (11). By using the CSD domain as a receptor, a conserved Cav-1 binding motif, WNNMTWMQW, is identified in the ectodomain (the C-terminal heptad repeat region) of human immunodeficiency virus type 1 (HIV-1) transmembrane envelope glycoprotein gp41 (21, 23). This motif in the HIV Env has been shown to interact specifically with the CSD domain of Cav-1. Our group revealed the interaction of cellular Cav-1 and HIV Env (via gp41) within the lipid rafts and further demonstrated that Cav-1 modulates Env-induced bystander apoptosis through the interaction with gp41, as well as fusion mediated by HIV Env (58). We have also shown that HIV infection in primary human monocyte-derived macrophages (MDMs) results in a dramatic upregulation of Cav-1 expression mediated by HIV Tat (31). The overexpression of Cav-1 significantly inhibits HIV replication (31, 34), implicating Cav-1 as playing a role in HIV's persistent infection of macrophages. However, the mechanism by which Cav-1 inhibits HIV replication remains to be determined.
HIV-1 virus replication is tightly regulated at the transcriptional level through the long terminal repeat (LTR), with specific interaction of cellular transcription factors binding to a variety of cis-acting DNA sequences within the LTR (12). HIV-1 LTR is comprised of the U3, R, and U5 regions. The U3 region contains an upstream regulatory element, which includes an enhancer with two binding sites for the nuclear factor κB (NF-κB) and nuclear factor of activated T cells (NFAT) proteins and the core promoter, composed of a TATA box and three binding sites for the specificity protein 1 (Sp1) (26). Several mechanisms are proposed for the restriction of HIV replication in monocytes/macrophages. These include either a lack of host transcriptional activators, the presence of host transcriptional repressors, dysfunctional Tat, the modulation of the chromatin environment, or downregulation of HIV transcripts by host microRNAs (5, 30). NF-κB sites located at nucleotide positions −81 to −91 and −95 to −104 relative to the transcriptional start site are one of the main regulators of the HIV-1 LTR in all cell types (2, 37, 41, 52). NF-κB is composed of homo- or heterodimers of Rel family proteins, including p65 (RelA), p50, c-Rel, RelB, and p52 (25). All of the proteins contain an N-terminal Rel homology domain, which functions in DNA binding, dimerization, and interaction with the inhibitory IκB proteins. p65 (RelA), RelB, and c-Rel contain a transactivation domain in the C terminus (25, 37).
The classical NF-κB complex (p50/p65) is sequestered in the cytoplasm by interaction with the inhibitory IκB proteins, including IκBα, IκBβ, IκBε, and IκBγ, of which IκBα is the best characterized. Upon a variety of stimuli and cell activation, IκBα is phosphorylated at the N-terminal residues S32 and S36 by the IκB kinases (IKKs), resulting in ubiquitination and subsequent degradation. This then leads to the release of the p50/p65 complex and allows it to translocate to the nucleus, where it can bind to the promoter and activate gene expression (25). Several studies have shown that NF-κB plays pivotal roles in transcriptional regulation of the HIV LTR, with implications for viral latency, reactivation, and persistent infection (2, 37, 41, 52). One study demonstrated that IκBα in T cells regulated NF-κB shuttling between the nucleus and cytosol, contributing to the lower activation levels of HIV-1 LTR and viral latency (9). NF-κB is involved in the stimulation of HIV-1 replication in monocytes by lipopolysaccharide (LPS), interleukin-6 (IL-6), and tumor necrosis factor alpha (TNF-α) (39, 48, 49). The deletion or introduction of site-specific mutations in the NF-κB sites diminished LTR activity, leading to reduced infectious-virus replication (2). An HIV-infected promonocytic cell line (U937) has an enhanced IκBα degradation due to IKK activation (3). These findings suggest that cellular factors that can negatively affect the NF-κB pathway can result in reduced virus replication and contribute to persistent/chronic macrophage infection. The observations that Cav-1 is upregulated by Tat in HIV-infected cells and that overexpression of Cav-1 inhibits virus production suggest a potential Cav-1-mediated repression of viral gene expression. In this report, we establish for the first time the mechanism of Cav-1-mediated inhibition of HIV replication and provide evidence that Cav-1 inhibits HIV replication through transcriptional repression of viral gene expression by modulating the NF-κB pathway.
MATERIALS AND METHODS
Plasmids and reagents.
The HIV proviral constructs pNL4-3 and pNL4-3.Luc.R−E− were kindly provided by the NIH AIDS Research and Reagent Program (1, 20). An expression plasmid for vesicular stomatitis virus (VSV) envelope G protein (pCI-VSV) was kindly provided by Garry Nolan of Stanford University. A green fluorescent protein (GFP)-expressing plasmid, pCMV-GFP, was constructed by cloning the cytomegalovirus (CMV) promoter into the AgeI and SspI sites of pEGFP.luc (Invitrogen, Carlsbad, CA). Similarly, a plasmid expressing GFP under the control of the HIV LTR, pLTR-GFP, was generated by first performing PCR amplification of HIV-1 LTR sequences from pNL4-3 provirus and then cloning them into the ClaI and AgeI restriction enzyme sites of pEGFP.luc. The plasmid pTatz, expressing HIV-1 Tat, was constructed by inserting the HIV-1 Tat coding sequence downstream from a CMV promoter (45). The Cav-1-expressing plasmid (pCZ-Cav-1) was also generated by placing the Cav-1 coding sequence downstream from a CMV promoter (58). pCZ-vector is a control construct without the Cav-1 coding sequence. The LTR-GFP mutation plasmids with mutations in the NF-κB sites, NF-κBM1 (mutation in only one NF-κB site) and NF-κBM2 (mutation in both NF-κB sites), the NFAT5 site (NFAT5M), and the Sp1 sites (Sp1M) were generated using a site-directed mutagenesis kit according to the manufacturer's protocol (Stratagene). Briefly, the mutations were generated by PCR amplification using pLTR-GFP as the template and the following pairs of primers: for the Sp1 mutation, 5′-CTTTCCAGGGATTTGTGGCCTGTTCGGGACTGGTTAGTGGCGAGCC-3′ and 5′-GGCTCGCCACTAACCAGTCCCGAACAGGCCACAAATCCCTGGAAAG-3′; for the NFAT5 mutation, 5′-TCCGCTGGGGACTTTTTAGGGAGGTGTGGCCTGGG-3′ and 5′-CCCAGGCCACACCTCCCTAAAAAGTCCCCAGCGGA-3′; for NF-κB mutation 1, 5′-GAGCTTTCTACAACTCACTTTCCGCTGGG-3′ and 5′-CCCAGCGGAAAGTGAGTTGTAGAAAGCTC-3′; and for NF-κB mutation 2, NF-κB mutation 1 for the PCR template and primers 5′-CTCACTTTCCGCTGCTCACTTTCCAGGGAGG-3′ and 5′-CCTCCCTGGAAAGTGAGCAGCGGAAAGTGAG-3′. The PCR products were digested with the restriction enzyme DpnI to destroy template plasmids and were then transformed into DH5α competent cells. Introductions of the mutations were confirmed by sequence analysis. The plasmid pGL4.32[luc2P/NF-κB-RE/Hygro], with the luciferase coding sequence under the control of a minimal promoter containing five NF-κB sites, was purchased from Promega. The NF-κB competitive inhibitor peptide SN50 and the mutant control SN50M were purchased from Sigma-Aldrich.
Cell cultures.
Human embryonic kidney (HEK) 293T cells were obtained from the American Type Culture Collection (Manassas, VA) and cultivated in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, and 100 μg/ml penicillin-streptomycin. Peripheral blood mononuclear cells (PBMCs) were isolated using a Ficoll gradient (Sigma-Aldrich). CD4+ T cells were then isolated from the PBMCs by using a negative cell sorting method (Stem Cell Technologies). The cells were discarded if the purity was below 95% as determined by flow cytometry analysis. CD4+ T cells were maintained in RPMI 1640 containing 10% FBS, 1 M HEPES, 2 mM sodium pyruvate, 0.0075% sodium bicarbonate, and 2 mM l-glutamine. The cells were stimulated with 10 U/ml IL-2 and 2 μg/ml phytohemagglutinin (PHA) (Sigma-Aldrich) for 1 week. Monocytes were isolated by negative selection using a StemSep human monocyte enrichment kit according to the manufacturer's protocol (Stemcell Technologies). The monocyte preparations contained 98% CD14+ cells as determined by flow cytometry. For differentiation of monocytes into macrophages, 1 × 106 monocytes were seeded into Biocoat poly-d-lysine plates (BD Bioscience) and cultured in DMEM supplemented with 10% heat-inactivated human serum, 50 μg/ml gentamicin, 10 μg/ml ciprofloxacin, and 1,000 U/ml macrophage colony-stimulating factor (M-CSF) (Sigma-Aldrich) for 7 days, with half of the culture medium being replaced with fresh medium every 2 days. HeLa-Tat-III/LTR/d1EGFP and HeLa-Tat-III/CMV/d1EGFP cells (44) are derived from the HeLa-TatIII cell line stably expressing GFP under the control of the HIV LTR or CMV promoter. These cell lines were obtained from the NIH AIDS Research and Reference Reagent Program and maintained in DMEM supplemented with 10% FBS, 1 mg/ml G418 (Sigma-Aldrich), and 100 μg/ml penicillin-streptomycin.
Transfection, virus production, and infection.
Transfections of 293T cells were carried out in T75 cell culture flasks by the calcium phosphate method. Seeded cells were transfected with 10 μg of pNL4-3.Luc.R−E− and 10 μg of pCI-VSV. Pseudotyped viral supernatants were harvested 3 days posttransfection and were clarified by centrifugation at 5,000 rpm for 20 min and then by filtering through a 0.45-μm-pore-size filter. Virus production was measured by a p24 enzyme-linked immunosorbent assay (ELISA) method. Adenovirus particles (Ad) for expressing Cav-1 (Ad-Cav-1), LacZ (Ad-LacZ), and GFP (Ad-GFP) were purchased from Vector Biolabs (Philadelphia, PA). Primary CD4+ cells or monocyte-derived macrophages (MDMs) were cultured in 12-well plates at a density of 1 × 106 cells per well and were infected at a multiplicity of infection (MOI) of 50 with Ad-Cav-1 or Ad-GFP for 4 h at 37°C in serum-free medium. The cells were then washed and incubated in serum-containing medium for 36 h and infected with a concentration of 20 ng p24 per 1 × 106 cells of VSV-G Env-pseudotyped HIV-1NL4-3.Luc.R−E− for 4 h at 37°C. These coinfected cells were washed and incubated for an additional 24 h prior to harvest for luciferase assay and immunoblot analysis. HeLa-Tat-III/LTR/d1EGFP and HeLa-Tat-III/CMV/d1EGFP cells were seeded in 6-well culture plates at a concentration of 2 × 105 cells per well and infected with 2 × 107 PFU of Ad-Cav-1 or Ad-LacZ. All other transfections were performed using Fugene 6 according to the protocol described by the manufacturer (Roche, Indianapolis, IN).
Luciferase assay and flow cytometry analysis.
HIV promoter-driven gene expression was quantified using either a luciferase or a GFP reporter. HEK 293T cells were seeded at a density of 2 × 105 cells per well and transfected with pNL4-3.Luc.R−E− and different concentrations of pCZ-Cav-1 or pCZ-vector. The cells were also transfected with pRL-TK (renilla luciferase expression plasmid; Promega) to monitor transfection efficiency. Luciferase activity was determined from cell lysates with a dual luciferase assay system as described by the manufacturer (Promega, Madison, WI). Primary CD4+ T cells and MDMs were infected with VSV-G-pseudotyped virus, and cell lysates were used to measure luciferase activity. GFP expression was determined by flow cytometry and measuring mean fluorescence intensity. The experiments were performed in triplicates, and the results are reported as normalized means ± standard deviations (SD).
Immunoblot analysis.
Total cellular proteins were extracted in lysis buffer (50 mM Tris, pH 7.5, 100 mM NaCl, 1 mM EDTA, 0.1% [vol/vol] Triton X-100, 10 mM NaF, 1 mM phenylmethylsulfonyl fluoride [PMSF], and 1 mM vanadate), with protein concentration being determined by the Lowry method (Bio-Rad protein assay). Extracted protein was separated by 10 to 15% SDS–PAGE gel electrophoresis and transferred onto a nitrocellulose membrane (Roche). The membrane was blocked with Tris-buffered saline–Tween 20 containing 5% nonfat milk and then incubated with antibodies specific to Cav-1 (Santa Cruz Biotechnology), β-actin (Sigma-Aldrich), Tat (kindly provided by the NIH AIDS Research and Reference Reagent Program), poly(ADP)-ribose polymerase (PARP; Cell Signaling Technology), or NF-κB pathway proteins (total and phosphorylated IKKβ, IKKα, IκBα, and p65; Cell Signaling Technology) overnight at 4°C. The membrane was then incubated with horseradish peroxidase (HRP)-linked anti-mouse or anti-rabbit IgG (Cell Signaling Technology) or anti-human IgG (Abcam, Inc.) for 1 h at room temperature.
Nucleus and cytosol fractionation.
The nucleus and cytosol fractionations were performed as previously described (57). Briefly, cultured cells were washed with cold phosphate-buffered saline (PBS) twice and harvested in 10 mM HEPES-KOH, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM PMSF (pH 7.9) buffer while at 4°C. The cells were further Dounce homogenized, incubated on ice for 15 min, and centrifuged at 14,000 rpm for 10 min at 4°C. The supernatant collected was the cytosolic fraction. The pellet was further lysed in 20 mM HEPES-KOH, 1.5 mM MgCl2, 25% glycerol, 420 mM NaCl, 0.2 mM EDTA, 0.5 mM dithiothreitol (DTT), 0.5 mM PMSF (pH 7.9) buffer for 30 min and centrifuged at 14,000 rpm for 20 min at 4°C. The second supernatant obtained was the nuclear fraction.
NF-κB p65 activity assay.
p65 NF-κB transcriptional activity was analyzed using the ELISA-based kit TransAM according to the manufacturer's protocol (Active Motif North America, Carlsbad, CA), using nuclear protein harvested as described above. Briefly, 10 μg of nuclear extract was diluted with complete lysis buffer provided by the manufacturer in a total volume of 20 μl and mixed with 30 μl of complete binding buffer in a 96-well plate coated with p65 binding-oligonucleotides. The binding assay was performed at room temperature with rocking at 100 rpm for 1 h. After washes, a 1:1,000 dilution of the primary antibody against NF-κB was added and incubated for another hour, which was followed by the addition of 1:1,000-diluted HRP-conjugated secondary antibody.
Statistical analysis.
The differences in the luciferase activities, mean fluorescence intensities, or p65 activities were compared using an unpaired Student's t test. All analyses were performed with SPSS 12.0.1 for Windows and were considered significant at a P value of <0.05.
RESULTS
Cav-1 inhibits gene expression under the control of the HIV LTR.
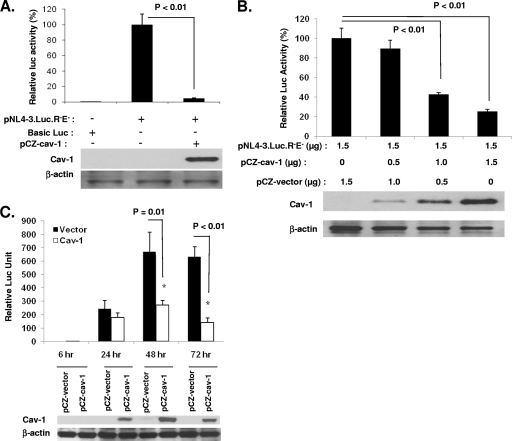
We and others have previously reported that Cav-1 inhibits HIV replication (31, 34). In order to determine the mechanisms of Cav-1-mediated reduction of HIV replication, we evaluated the expression of luciferase under the control of the HIV LTR using a provirus that is defective for Env and has the coding sequence of luciferase placed in the Nef region (pNL4-3.Luc.R−E−). HEK 293T cells were cotransfected with pNL4-3.Luc.R−E− and the Cav-1 expression plasmid (pCZ-Cav-1) or the vector devoid of Cav-1 (pCZ-vector). The level of protein production driven by the HIV LTR was then measured by the luciferase activity. There is no endogenous Cav-1 expression in the 293T cell line. In the presence of Cav-1 expression, the luciferase activity was reduced significantly (about 20-fold with cotransfection of 1.5 μg pCZ-Cav-1 and 1.0 μg pNL4-3.Luc.R−E−) compared to the luciferase activity in cells that received pCZ-vector (Fig. 1A). The transfection of different concentrations of pCZ-Cav-1 along with a constant amount of pNL4-3.Luc.R−E− resulted in reductions of luciferase activity in a dose-dependent manner, reaching 4-fold at a ratio of 1:1 of pCZ-Cav-1 to pNL4-3.Luc.R−E− (Fig. 1B). Time course experiments were performed by harvesting samples at 24, 48, and 72 h posttransfection. Inhibition occurred 48 h after transfection and peaked at 72 h (Fig. 1C).
Fig. 1.
Cav-1 modulates HIV-1 gene expression. (A) Inhibition of luciferase expression encoded in NL4-3.Luc.R−E− by Cav-1. HEK 293T cells were cotransfected with pRL-TK (10 ng), pNL4-3.Luc.R−E− (1.0 μg), or basic luciferase plasmid (1.0 μg) with no promoter and pCZ-Cav-1 (1.5 μg) or pCZ vector (1.5 μg). The cells were harvested 48 h posttransfection and subjected to a dual luciferase assay. The levels of luciferase are expressed relative to the levels in cells transfected with pCZ vector (100%). (B) Cav-1 dose-dependent inhibition of viral gene expression. A constant amount of pNL4-3.Luc.R−E− was cotransfected with various concentrations of pCZ-Cav-1 into HEK 293T cells. Luciferase assay was performed 48 h posttransfection. (C) Time-dependent inhibition of gene expression by Cav-1. HEK 293T cells were cotransfected with pNL4-3.Luc.R−E− and pCZ-Cav-1 or pCZ vector. Cells were harvested at 6, 24, 48, and 72 h posttransfection and subjected to luciferase assay. Results are shown as means ± SD of three determinations. The cells were also harvested and subjected to Western blot analysis for levels of Cav-1 and β-actin.
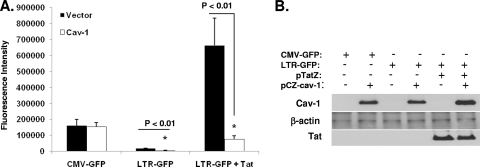
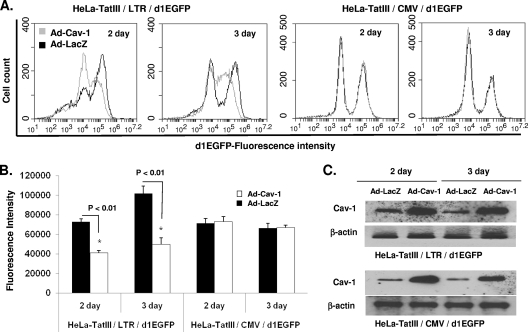
In an effort to quell the argument that other viral factors could be involved in the reduction of the luciferase activity, we directly evaluated the effect of Cav-1 expression on the HIV-1 LTR promoter activity. A GFP construct under the control of the HIV LTR (pLTR-GFP) or CMV promoter (pCMV-GFP) was cotransfected with pCZ-Cav-1 into 293T cells in the presence or absence of Tat, and GFP expression was measured by mean fluorescence intensity using flow cytometry. As shown in Fig. 2, Cav-1 specifically inhibited LTR-driven GFP expression, whereas Cav-1 had no effect on GFP expression under the control of the CMV promoter. Interestingly, the significant inhibition of LTR-driven GFP expression by Cav-1 occurs in both the presence and absence of Tat, indicating a target region for Cav-1-mediated transcriptional repression in the U3 region of the HIV LTR. To further confirm our findings, we tested the influence of Cav-1 on the HIV LTR in HeLa cells stably expressing GFP under the control of the HIV LTR (HeLa-Tat-III/LTR/d1EGFP) or CMV promoter (HeLa-Tat-III/CMV/d1EGFP). These HeLa cell lines also stably express Tat. Cav-1 was introduced into these cells by infection with adenovirus vector containing the Cav-1 expression cassette (Ad-Cav-1). As shown in Fig. 3, the introduction of Cav-1 via an adenovirus vector infection triggered the suppression of LTR-driven GFP expression, as detected by the shift of the fluorescence emission peak at days 2 and 3. GFP expression under the control of the CMV promoter remained the same whether cells were infected with Ad-Cav-1 or not. Infection of HeLa-Tat-III/LTR/d1EGFP or HeLa-Tat-III/CMV/d1EGFP with Ad-LacZ had no effect on GFP expression. These results establish with no ambiguity that Cav-1 represses LTR-driven gene expression, and the finding that gene expression is inhibited with or without Tat suggests the presence of a target region for Cav-1-mediated transcriptional repression in the U3 region of the HIV LTR.
Fig. 2.
Cav-1 modulates gene expression under the control of the HIV-1 LTR in the presence and absence of Tat. (A) GFP under the control of CMV promoter (pCMV-GFP) or the HIV LTR (pLTR-GFP) was cotransfected with pCZ-Cav-1 or pCZ-vector in the presence or absence of Tat (pTatZ) into HEK 293T cells. The cells were washed with PBS twice and subjected to flow cytometry to detect GFP fluorescence intensity. Results are shown as means ± SD of three determinations. (B) The cells were also harvested and subjected to Western blot analysis to determine the expression levels of Cav-1, Tat, and β-actin.
Fig. 3.
Activity of Cav-1 on gene expression in a stable cell line expressing GFP under the control of the HIV LTR. (A and B) Stable HeLa cell lines expressing GFP under the control of the HIV LTR (HeLa-TatIII/LTR/d1EGFP) or CMV promoter (HeLa-TatIII/CMV/d1EGFP) were infected with adenovirus vector containing Cav-1 (Ad-Cav-1) or LacZ (Ad-LacZ). Infected cells were harvested at days 2 and 3 postinfection and subjected to flow cytometry analysis for GFP fluorescence intensity. Experiments were performed independently three times, and results are expressed as means ± SD. (C) The level of adenovirus vector-delivered Cav-1 expression was determined by Western blot analysis. EGFP, enhanced green fluorescent protein.
NF-κB sites are necessary for the inhibitory effect of Cav-1 on the HIV-1 LTR.
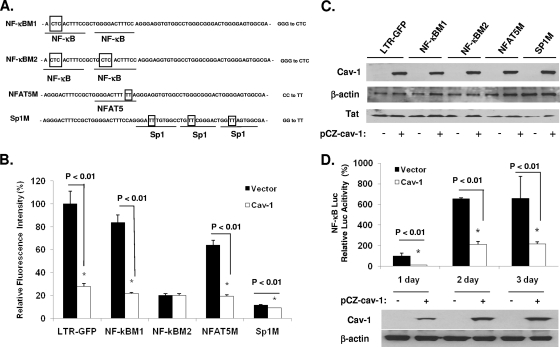
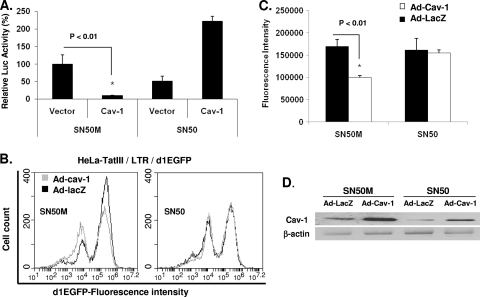
In order to map the target(s) for the Cav-1 suppression of HIV LTR-driven gene expression, we constructed LTR promoter cassettes with site-specific mutations in the NF-κB, Sp1, and NFAT5 cis-acting elements located within the U3 region of the LTR (Fig. 4A). Each of these altered LTRs was placed upstream from the coding sequence of GFP to monitor gene expression. Plasmids containing the mutant forms of the LTR, as well as the wild-type LTR, were cotransfected with a Tat-expressing construct (pTatZ) and pCZ-Cav-1 or pCZ-vector into 293T cells. The level of gene expression was measured by the mean fluorescence intensity and compared to that of the wild-type LTR. In cells that received pCZ-vector, there was only minimal decrease in GFP expression with constructs containing a mutation in one of the NF-κB sites (NF-κBM1), indicating that one NF-κB is sufficient for efficient gene expression (Fig. 4B). Constructs with mutations in NFAT5, the two NF-κBs, or the Sp1 sites showed reductions in GFP expression by 5.2-, 1.6-, or 8.7-fold, respectively, compared to that of the wild-type LTR (Fig. 4B). In the presence of Cav-1, the level of GFP expression was inhibited by 75%, 70%, or 27% with the LTR mutations in the single NF-κB, NFAT5, and the Sp1 sites, respectively, compared to the expression in cells receiving pCZ-vector (Fig. 4B). The ability of Cav-1 to suppress LTR-driven GFP expression, however, was blocked when both NF-κB sites were mutated (Fig. 4B), indicating that Cav-1 repression of LTR-controlled gene expression is mediated by the NF-κB cis-acting elements. In addition, we tested the activity of Cav-1 on a heterologous minimal promoter containing 5 NF-κB sites (pGL4.32[luc2P/NF-κB-RE/Hygro]) by cotransfection along with pCZ-Cav-1 or pCZ-vector and measurement of luciferase activity. As shown in Fig. 4D, luciferase expression was suppressed by 75%, 69%, and 66% at days 1, 2, and 3, respectively, in the presence of Cav-1 compared to the expression in cells transfected with pCZ-vector. To further confirm the NF-κB-mediated Cav-1 suppression of gene expression, we tested gene expression under the control of the HIV LTR in the presence of a peptide (SN50) which blocks the nuclear importation of the p50/p65 complex. The addition of SN50 blocked the activity of Cav-1 in suppressing LTR-driven expression both in transiently expressing cells, as demonstrated by measuring luciferase activity (Fig. 5A), and in the stable GFP-expressing HeLa-Tat-III/LTR/d1EGFP cell line (Fig. 5B, C, and D). The control peptide, SN50M, did not affect the ability of Cav-1 to inhibit LTR-dependent gene expression. These results taken together clearly establish NF-κB's requirement for Cav-1's ability to suppress HIV gene expression.
Fig. 4.
NF-κB cis-acting elements are essential for the modulation of Cav-1 on HIV-1 LTR activation. (A) Sequence illustrating each mutation introduced in the NF-κB, NFAT5, and Sp1 sites of the HIV LTR. (B) Wild-type LTR-GFP or each of the LTR mutant constructs placed upstream from the GFP-coding sequence were transfected into HEK 293T cells with pCZ-Cav-1 or pCZ-vector along with pTatZ. GFP expression was monitored by using flow cytometry and measuring fluorescence intensity. The levels of fluorescence intensity with mutations in the LTR are expressed relative to the levels in the cells transfected with the wild-type LTR-GFP (100%). (C) The cells were also harvested and subjected to Western blot analysis to determine the expression levels of Cav-1 and Tat. (D) Activity of Cav-1 on gene expression under the control of a heterologous promoter (pGL4.32[luc2P/NF-κB-RE/Hygro]) containing five NF-κB cis-acting elements. pGL4.32[luc2P/NF-κB-RE/Hygro] was cotransfected with pCZ-Cav-1 or pCZ vector into HEK 293T cells. Cells were harvested at days 1, 2, and 3 and assayed for luciferase activity. All results are expressed as means ± SD of three determinations.
Fig. 5.
Cav-1 modulates HIV-1 LTR-driven transcription through NF-κB. (A) HEK 293T cells were cotransfected with pRL-TK, pNL4-3.Luc.R−E−, and pCZ-Cav-1 or pCZ vector in the presence of SN50M or SN50 (50 ng/ml). The cells were harvested 48 h posttransfection and subjected to a dual luciferase assay. The levels of luciferase activity are expressed relative to the levels in cells transfected with pCZ-vector in the presence of SN50M. (B and C) HeLa-TatIII/LTR/d1EGFP cells were infected with Ad-Cav-1 or Ad-LacZ in the presence of SN50 or SN50M (50 ng/ml). The cells were harvested 2 days later and subjected to flow cytometry analysis for d1EGFP fluorescence intensity. (D) The expression levels of Cav-1 delivered using adenovirus vector infection were determined by Western blot analysis. All results are expressed as means ± SD of three independent experiments, and P values are shown.
Cav-1 modulates the NF-κB signal pathways.
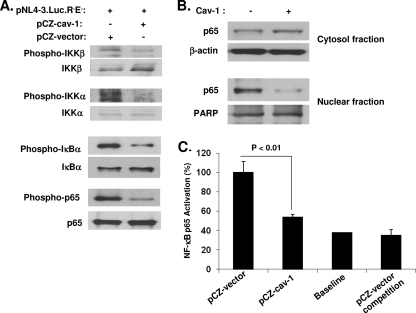
The phosphorylation of IKKs (IKKβ and IKKα), IκBα, and p65 and the translocation of p65 into the nucleus are important for NF-κB-mediated transcriptional activation (25, 37). Therefore, to demonstrate whether Cav-1 modulates the NF-κB pathway in order to transcriptionally inhibit HIV replication, we examined the level of phosphorylation of proteins (IKKβ, IKKα, IκBα, and p65) involved in the NF-κB pathway. HEK 293T cells were transfected with pNL4-3.Luc.R−E− along with pCZ-Cav-1 or pCZ-vector, and samples were analyzed by Western blotting using specific antibodies. As shown in Fig. 6A, phosphorylation of IKKβ and IKKα was significantly reduced in cells transfected with pCZ-Cav-1 compared to that in cells treated with pCZ-vector, suggesting that Cav-1 inhibited IKKα and IKKβ activation. The decrease in IKKα and IKKβ phosphorylation means a reduction in the activation/phosphorylation of IκBα, which thus inhibits IKK-mediated phosphorylation-induced proteosomal degradation of the IκB. This subsequently affects the translocation of the active NF-κB transcription factor subunits to the nucleus and the following induction of target gene expression. To confirm such a notion, we examined whether Cav-1 had an effect on the phosphorylation of IκBα and p65. As we predicted, Cav-1 significantly reduced the phosphorylation of both IκBα and p65 (Fig. 6A). Furthermore, the translocation of p65 from the cytoplasm to the nucleus was significantly decreased in the presence of Cav-1 (Fig. 6B). This reduction in p65 translocation would suggest that with less p65 in the nucleus, there should, therefore, be less activated p65 complex for binding to target DNA, thus lowering gene expression. To investigate whether there is a decreased amount of p65 binding to target DNA, an ELISA-based NF-κB activation assay was performed. A marked reduction of NF-κB p65 DNA binding activity was observed in cells receiving Cav-1 compared to the activity in cells receiving vector only (Fig. 6C). The specificity of p65 binding activity was confirmed by a competition experiment with excess amounts of oligonucleotides containing the wild-type NF-κB binding site (Fig. 6C). These results, therefore, establish that Cav-1 modulates the NF-κB pathway to inhibit HIV replication at the viral gene transcription stage.
Fig. 6.
Cav-1 modulates the NF-κB signal pathway. (A) Phosphorylation proteins of the NF-κB pathway inhibited by Cav-1. HEK 293T cells were cotransfected with pNL4-3.Luc.R−E− and pCZ-Cav-1 or pCZ vector. The cells were harvested at 16 h posttransfection and subjected to Western blot analysis to determine the levels of phosphorylated and total IKKβ, IKKα, IκBα, and p65. (B) Translocation of p65 from the cytoplasm to the nucleus decreased in the presence of Cav-1. Cells treated as described for panel A were harvested at 16 h posttransfection and subjected to cytosol and nucleus fractionation. Western blot analysis was used to determine the levels of p65 in cytosolic and nuclear fractions. β-Actin and poly(ADP)-ribose polymerase (PARP) were used as loading controls for cytosolic and nuclear fractions, respectively. (C) NF-κB p65 binding to specific target DNA inhibited by the presence of Cav-1. Nuclear protein was harvested from the cells cotransfected with pNL4-3.Luc.R−E− and pCZ-Cav-1 or pCZ-vector and subjected to NF-κB p65 DNA binding activity assay using an ELISA-based kit. The levels of p65 binding to target DNA in the presence of Cav-1 are expressed relative to the levels of pCZ-vector. Results shown are means ± SD of three independent experiments.
NF-κB-mediated transcriptional inhibition of HIV gene expression in human primary monocyte-derived macrophages and CD4+ T cells.
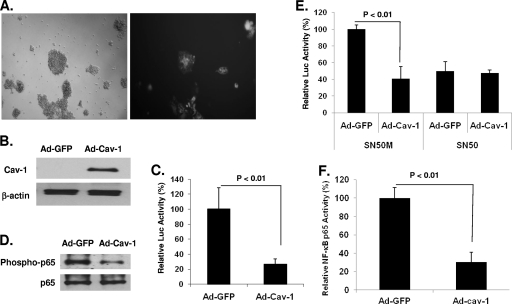
To demonstrate the physiological relevance of our results, we used primary MDMs and CD4+ T cells to show inhibition of HIV gene expression through the NF-κB pathway. Monitoring the level of viral gene expression in primary CD4+ T cells and MDMs was performed by infecting the cells with NL4-3.Luc.R−E− pseudotyped with VSV-G Env and expressing the luciferase gene in one round of replication. Cav-1 was delivered in these cells by infection with an adenovirus vector containing a Cav-1 expression cassette (Ad-Cav-1). Western blot analysis shows no Cav-1 expression in primary CD4+ T cells infected with adenovirus vector containing GFP (Ad-GFP) (Fig. 7A and B), confirming previous findings that human T cells do not express Cav-1. T cells transduced with Ad-Cav-1 and infected with pseudotyped NL4-3.Luc.R−E− significantly inhibited luciferase expression, by 74% compared to expression in T cells transduced with Ad-GFP (Fig. 7C). Phosphorylation of the NF-κB p65 was markedly decreased in CD4+ T cells transduced with Ad-Cav-1 compared to that in cells receiving Ad-GFP (Fig. 7D). Similar to the results in 293T cells, treatment of the primary CD4+ T cells with SN50 resulted in blocking of the inhibition of HIV gene expression by Cav-1 due to SN50's hindrance of the nuclear localization of NF-κB, whereas SN50M had no influence on Cav-1's ability to inhibit luciferase expression (Fig. 7E). An assay for NF-κB p65 DNA binding activity through an ELISA-based kit showed a 71% reduction in p65 activation and DNA binding in cells that received Ad-Cav-1 compared to the activity in cells transduced with Ad-GFP, confirming that Cav-1 modulates HIV-1 transcription through NF-κB in primary CD4+ T cells.
Fig. 7.
Cav-1 modulates HIV gene expression in primary CD4+ T lymphocytes. (A and B) Primary CD4+ T lymphocytes were isolated from human peripheral blood. Cells were dually infected with VSV-G-pseudotyped NL4-3.Luc.R−E− and adenovirus vector expressing GFP (Ad-GFP) or Cav-1 (Ad-Cav-1). The expression levels of GFP and Cav-1 are shown. (C) The levels of viral gene expression in the presence and absence of Cav-1 were measured by luciferase activity and determined relative to the levels in cells transduced with Ad-GFP. (D) p65 phosphorylation in the presence or absence of Cav-1 was evaluated by Western blot analysis. (E) The effect of Cav-1 on viral gene expression was measured in the presence of peptide SN50, an inhibitor of NF-κB cytoplasm-to-nucleus translocation, or the mutant SN50M. (F) Viral gene expression was measured by luciferase activity in one round of replication. NF-κB binding to target DNA in the presence and absence of Cav-1 was determined using an ELISA-based kit. All experiments were performed in triplicates, and results shown are means ± SD with P values.
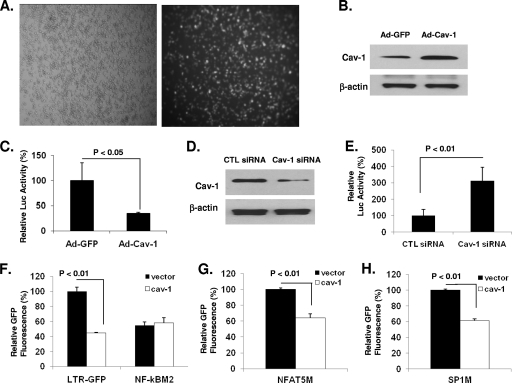
Since macrophages express endogenous Cav-1, we tested transcriptional inhibition of viral gene expression using two approaches. First, we introduced exogenous Cav-1 into MDMs using Ad-Cav-1 in a manner similar to that described for the CD4+ T cells and determined the level of viral gene expression in one round of replication by measuring luciferase activity. As expected, Western blot analysis of MDMs transduced with Ad-GFP revealed endogenous Cav-1 expression, with increased amounts of Cav-1 in MDMs treated with Ad-Cav-1 (Fig. 8B). The luciferase activity in cells transduced with Ad-Cav-1 and infected with pseudotyped NL4-3.Luc.R−E− virus showed a reduction of 3-fold compared to the activity in MDMs transduced with Ad-GFP (Fig. 8C). In a second approach, the endogenous Cav-1 expression was knocked down by small interfering RNA (siRNA) treatment in MDMs infected with the pseudotyped HIV (Fig. 8D), and luciferase activity was measured to determine the level of viral gene expression. As shown in Fig. 8E, the luciferase expression increased dramatically (3-fold) in MDMs treated with siRNA compared to the activity in cells receiving control siRNA, suggesting a role for Cav-1 in repressing viral gene expression and subsequently inhibiting viral replication in macrophages.
Fig. 8.
Cav-1 modulates HIV-1 gene expression in physiologically relevant primary monocyte-derived macrophages (MDMs). (A and B) MDMs were isolated from human peripheral blood. Cells were dually infected with VSV-G-pseudotyped NL4-3.Luc.R−E− and Ad-Cav-1 or Ad-GFP. The expression levels of GFP and Cav-1 are shown. (C) The level of viral gene expression in the presence of overexpressed or endogenous Cav-1 was measured by luciferase activity in one round of replication and determined relative to the levels in cells transduced with Ad-GFP. (D) To determine the physiological relevance of the inhibition of HIV gene expression by Cav-1, MDMs infected with VSV-G-pseudotyped NL4-3.Luc.R−E− were transfected with specific siRNA that targets Cav-1 or control (CTL) siRNA using Fugene 6. (E) The level of viral gene expression in one round of replication was measured by luciferase activity under the viral LTR promoter. (F, G, and H) In separate experiments, MDMs were transfected with pLTR-GFP or NF-κBM2 (F), NFAT5M (G), or Sp1M (H) along with pCZ-Cav-1 or pCZ-vector to examine the role of NF-κB in Cav-1-mediated inhibition of HIV gene expression in macrophages. The cells were harvested 48 h posttransfection and subjected to flow cytometry to measure GFP fluorescence intensity. All experiments were performed in triplicates, and results shown are means ± SD with P values. CTL, control.
Macrophages have different transcription factor profiles than lymphocytes, and therefore, we tested whether the inhibition of HIV gene expression by Cav-1 involves the NF-κB pathway. MDMs were transfected with wild-type pLTR-GFP or LTR with mutants (NF-κBM2, MFAT5M, or Sp1M) along with a Tat-encoding plasmid (pTatZ) and pCZ-Cav-1. As shown in Fig. 8F to H, Cav-1 was still able to significantly inhibit LTR-driven GFP expression in cells transfected with NFAT5M and Sp1M. Only mutation of the two NF-κB sites abolished the influence of Cav-1 on HIV-1 LTR-driven GFP expression, establishing that Cav-1 repression of HIV gene expression in macrophages also involves the modulation of the NF-κB pathway.
DISCUSSION
Cav-1 has been shown to inhibit HIV replication in 293T and U87 cells, as well as showing enhanced expression of Cav-1 in HIV-infected macrophages (31, 34). These observations, along with the lack of Cav-1 in CD4+ T cells and the capability of macrophages to express this protein, supports the hypothesis in which Cav-1 contributes to HIV's persistent infection of macrophages. Previously, we reported that Cav-1 binding to HIV Env gp41 blocks hemifusion and subsequently inhibits bystander apoptosis (58). Based on this indirect information, we proposed a potential mechanism for Cav-1's ability to inhibit HIV replication that involves this association of Cav-1 and envelope. In the same study, we demonstrated that a Cav-1 peptide containing the scaffold domain for interacting with other proteins, including Env gp41, showed inhibition of virus replication. However, there is no clear and direct evidence as to whether the Cav-1 binding to the HIV Env influences viral replication. In the current study, all the experiments were performed in the absence of envelope to exclude the potential confounding factor of Cav-1 and gp41's association in virus replication. In this study, we have investigated the mechanism by which Cav-1 inhibits HIV replication at the level of gene expression. We have established for the first time that Cav-1 represses viral gene expression and that this suppression involves the NF-κB pathway. Inhibition of viral gene expression is evident in all of the cell types we examined, including primary CD4+ T cells and macrophages, as shown by luciferase expression under the control of the HIV LTR in a provirus context. Our results reveal that increasing amounts of Cav-1 inhibit viral gene expression in macrophages. The endogenous Cav-1 expression knockdown experiment using specific siRNA in primary macrophages, with its resulting upregulation of viral gene expression, establishes with no ambiguity that Cav-1 regulates HIV replication in macrophages.
Unrelated to HIV, the modulation of NF-κB transcriptional activation by Cav-1 has been reported by several groups in different cell types and physiological experimental approaches, yielding conflicting results. In endothelial cells, the loss of Cav-1 results in decreased activation of NF-κB in response to lipopolysaccharide challenge (38) and linoleic acid treatment (62). The knockdown of Cav-1 using siRNA in human endothelium-derived EA.hy926 cells had no effect on NF-κB activation by TNF (13). In a monocrotaline-induced pulmonary hypertension model, decreased Cav-1 in rat lung tissue is accompanied by inhibited IκBα expression and increased NF-κB activation (22). The overexpression of Cav-1 is reported to suppress NF-κB activation in murine macrophages (57). In three human cancer cell lines, upregulation of Cav-1 was observed together with downregulation of NF-κB after treatment with growth hormone-releasing hormone (GHRH) antagonist MIA-602 (4). These conflicting results may have to do with the use of different stimuli and the experimental approaches taken subsequently, influencing Cav-1's relationship with factors prior to NF-κB activation. Our results with HIV infection convincingly show that Cav-1 significantly reduces viral gene expression by inhibiting NF-κB activation. The reduction in phosphorylation of IKKα and IKKβ that we observe in the presence of Cav-1 suggests that the influence of Cav-1 on NF-κB is upstream from the IKK activation step. IKKα and IKKβ are part of a larger multiprotein complex, upstream from the activation of NF-κB, whose activation leads to the phosphorylation of IκB and its release from the NF-κB complex (25, 37). The reduction of the subsequent steps of NF-κB activity, such as the decrease we saw in IκBα and p65 phosphorylation, inhibition of the translocation of the NF-κB complex, and binding to cis-acting elements, are therefore related to Cav-1's action upstream of the IKK activation steps. There are several upstream activators and regulators of IKK (25), and defining the upstream element that Cav-1 associates with in affecting the NF-κB pathway will be critical for complete understanding of Cav-1-mediated inhibition of HIV replication.
One of the major hurdles in AIDS therapy is the difficulty in eradicating cellular reservoirs of persistent and latent HIV infections (8). HIV-1 latency and persistent infection is tightly regulated by viral and host factors that control the silencing and initiation of viral transcription (30, 35). Additionally, AIDS disease progression correlates closely with HIV virus replication in infected patients, which is also linked to the regulation of viral genome transcription (10). Among a variety of host transcriptional factor binding sites, the regulatory region between nucleotides −104 and −80 of the HIV LTR, which harbors two consensus NF-κB binding sites, is the most conserved and extensively characterized (41). The sequences of the κB sites and the 4-nucleotide spacer are highly conserved on most isolates of HIV (24, 56), which suggests that NF-κB is a key factor in HIV transcription. The deletion or site-directed mutation of these NF-κB sites diminishes HIV-1 LTR transcription (41). Physical interactions of NF-κB and the HIV-1 LTR have also been demonstrated, both in vitro and in vivo (40, 59). Our finding that Cav-1 inhibits HIV gene expression mediated by the NF-κB pathway suggests that Cav-1 can be one of the key cellular regulators of HIV replication in macrophages. Cav-1's upregulation during HIV infection, which is mediated by Tat (31), only strengthens the notion that Cav-1 plays a key role in establishing the persistent infection of macrophages in a feedback loop by the repression of viral gene expression. Several restriction factors that impede HIV replication in macrophages have been described (reviewed in references 5 and 30). HIV replication can be restricted at different steps of the virus life cycle. These include restrictions at the early preintegration, the transcriptional, and the late posttranscriptional stages of virus replication. Cav-1 is, therefore, a new addition to the growing number of factors which contribute to the restriction of HIV replication in macrophages in order for the virus to establish a chronic infection and form a viral reservoir.
ACKNOWLEDGMENT
This research was supported by a grant from the National Institutes of Health (AI39126) to A. Mergia.
Footnotes
Published ahead of print on 23 March 2011.
REFERENCES
- 1. Adachi A., et al. 1987. Productive, persistent infection of human colorectal cell lines with human immunodeficiency virus. J. Virol. 61:209–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Asin S., Bren G. D., Carmona E. M., Solan N. J., Paya C. V. 2001. NF-κB cis-acting motifs of the human immunodeficiency virus (HIV) long terminal repeat regulate HIV transcription in human macrophages. J. Virol. 75:11408–11416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Asin S., Taylor J. A., Trushin S., Bren G., Paya C. V. 1999. Iκκ mediates NF-κB activation in human immunodeficiency virus-infected cells. J. Virol. 73:3893–3903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bellyei S., et al. 2010. GHRH antagonists reduce the invasive and metastatic potential of human cancer cell lines in vitro. Cancer Lett. 293:31–40 [DOI] [PubMed] [Google Scholar]
- 5. Bergamaschi A., Pancino G. 2010. Host hindrance to HIV-1 replication in monocytes and macrophages. Retrovirology 7:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Burgermeister E., Liscovitch M., Rocken C., Schmid R. M., Ebert M. P. 2008. Caveats of caveolin-1 in cancer progression. Cancer Lett. 268:187–201 [DOI] [PubMed] [Google Scholar]
- 7. Burgermeister E., et al. 2007. Differential expression and function of caveolin-1 in human gastric cancer progression. Cancer Res. 67:8519–8526 [DOI] [PubMed] [Google Scholar]
- 8. Coiras M., Lopez-Huertas M. R., Perez-Olmeda M., Alcami J. 2009. Understanding HIV-1 latency provides clues for the eradication of long-term reservoirs. Nat. Rev. Microbiol. 7:798–812 [DOI] [PubMed] [Google Scholar]
- 9. Coiras M., Lopez-Huertas M. R., Rullas J., Mittelbrunn M., Alcami J. 2007. Basal shuttle of NF-kappaB/I kappaB alpha in resting T lymphocytes regulates HIV-1 LTR dependent expression. Retrovirology 4:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Coombs R. W., et al. 1989. Plasma viremia in human immunodeficiency virus infection. N. Engl. J. Med. 321:1626–1631 [DOI] [PubMed] [Google Scholar]
- 11. Couet J., Li S., Okamoto T., Ikezu T., Lisanti M. P. 1997. Identification of peptide and protein ligands for the caveolin-scaffolding domain. Implications for the interaction of caveolin with caveolae-associated proteins. J. Biol. Chem. 272:6525–6533 [DOI] [PubMed] [Google Scholar]
- 12. Cullen B. R. 1991. Regulation of HIV-1 gene expression. FASEB J. 5:2361–2368 [DOI] [PubMed] [Google Scholar]
- 13. D'Alessio A., et al. 2010. Targeting of tumor necrosis factor receptor 1 to low density plasma membrane domains in human endothelial cells. J. Biol. Chem. 285:23868–23879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dupree P., Parton R. G., Raposo G., Kurzchalia T. V., Simons K. 1993. Caveolae and sorting in the trans-Golgi network of epithelial cells. EMBO J. 12:1597–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Frank P. G., Pavlides S., Cheung M. W., Daumer K., Lisanti M. P. 2008. Role of caveolin-1 in the regulation of lipoprotein metabolism. Am. J. Physiol. Cell Physiol. 295:C242–C248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Frank P. G., Pavlides S., Lisanti M. P. 2009. Caveolae and transcytosis in endothelial cells: role in atherosclerosis. Cell Tissue Res. 335:41–47 [DOI] [PubMed] [Google Scholar]
- 17. Gargalovic P., Dory L. 2003. Caveolins and macrophage lipid metabolism. J. Lipid Res. 44:11–21 [DOI] [PubMed] [Google Scholar]
- 18. Goetz J. G., Lajoie P., Wiseman S. M., Nabi I. R. 2008. Caveolin-1 in tumor progression: the good, the bad and the ugly. Cancer Metastasis Rev. 27:715–735 [DOI] [PubMed] [Google Scholar]
- 19. Harris J., et al. 2002. Expression of caveolin by bovine lymphocytes and antigen-presenting cells. Immunology 105:190–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. He J., et al. 1995. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J. Virol. 69:6705–6711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hovanessian A. G., et al. 2004. The caveolin-1 binding domain of HIV-1 glycoprotein gp41 is an efficient B cell epitope vaccine candidate against virus infection. Immunity 21:617–627 [DOI] [PubMed] [Google Scholar]
- 22. Huang J., et al. 2008. Pyrrolidine dithiocarbamate restores endothelial cell membrane integrity and attenuates monocrotaline-induced pulmonary artery hypertension. Am. J. Physiol. Lung Cell. Mol. Physiol. 294:L1250–L1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huang J. H., et al. 2007. Identification of the HIV-1 gp41 core-binding motif in the scaffolding domain of caveolin-1. J. Biol. Chem. 282:6143–6152 [DOI] [PubMed] [Google Scholar]
- 24. Jeeninga R. E., et al. 2000. Functional differences between the long terminal repeat transcriptional promoters of human immunodeficiency virus type 1 subtypes A through G. J. Virol. 74:3740–3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Karin M., Ben-Neriah Y. 2000. Phosphorylation meets ubiquitination: the control of NF-[kappa]B activity. Annu. Rev. Immunol. 18:621–663 [DOI] [PubMed] [Google Scholar]
- 26. Kilareski E. M., Shah S., Nonnemacher M. R., Wigdahl B. 2009. Regulation of HIV-1 transcription in cells of the monocyte-macrophage lineage. Retrovirology 6:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kogo H., Fujimoto T. 2000. Caveolin-1 isoforms are encoded by distinct mRNAs. Identification of mouse caveolin-1 mRNA variants caused by alternative transcription initiation and splicing. FEBS Lett. 465:119–123 [DOI] [PubMed] [Google Scholar]
- 28. Kurzchalia T. V., et al. 1992. VIP21, a 21-kD membrane protein is an integral component of trans-Golgi-network-derived transport vesicles. J. Cell Biol. 118:1003–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kurzchalia T. V., Parton R. G. 1999. Membrane microdomains and caveolae. Curr. Opin. Cell Biol. 11:424–431 [DOI] [PubMed] [Google Scholar]
- 30. Le Douce V., Herbein G., Rohr O., Schwartz C. 2010. Molecular mechanisms of HIV-1 persistence in the monocyte-macrophage lineage. Retrovirology 7:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lin S., Wang X. M., Nadeau P. E., Mergia A. 2010. HIV infection upregulates caveolin 1 expression to restrict virus production. J. Virol. 84:9487–9496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lisanti M. P., et al. 1995. Caveolae, transmembrane signalling and cellular transformation. Mol. Membr. Biol. 12:121–124 [DOI] [PubMed] [Google Scholar]
- 33. Liu P., Rudick M., Anderson R. G. 2002. Multiple functions of caveolin-1. J. Biol. Chem. 277:41295–41298 [DOI] [PubMed] [Google Scholar]
- 34. Llano M., et al. 2002. Blockade of human immunodeficiency virus type 1 expression by caveolin-1. J. Virol. 76:9152–9164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Margolis D. M. 2010. Mechanisms of HIV latency: an emerging picture of complexity. Curr. HIV/AIDS Rep. 7:37–43 [DOI] [PubMed] [Google Scholar]
- 36. Mercier I., et al. 2009. Clinical and translational implications of the caveolin gene family: lessons from mouse models and human genetic disorders. Lab Invest. 89:614–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mingyan Y., Xinyong L., De Clercq E. 2009. NF-kappaB: the inducible factors of HIV-1 transcription and their inhibitors. Mini Rev. Med. Chem. 9:60–69 [DOI] [PubMed] [Google Scholar]
- 38. Mirza M. K., et al. 2010. Caveolin-1 deficiency dampens Toll-like receptor 4 signaling through eNOS activation. Am. J. Pathol. 176:2344–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Molina J. M., Scadden D. T., Byrn R., Dinarello C. A., Groopman J. E. 1989. Production of tumor necrosis factor alpha and interleukin 1 beta by monocytic cells infected with human immunodeficiency virus. J. Clin. Invest. 84:733–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mukerjee R., Sawaya B. E., Khalili K., Amini S. 2007. Association of p65 and C/EBPbeta with HIV-1 LTR modulates transcription of the viral promoter. J. Cell. Biochem. 100:1210–1216 [DOI] [PubMed] [Google Scholar]
- 41. Nabel G., Baltimore D. 1987. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature 326:711–713 [DOI] [PubMed] [Google Scholar]
- 42. Okamoto T., Schlegel A., Scherer P. E., Lisanti M. P. 1998. Caveolins, a family of scaffolding proteins for organizing “preassembled signaling complexes” at the plasma membrane. J. Biol. Chem. 273:5419–5422 [DOI] [PubMed] [Google Scholar]
- 43. Palade G. E. 1953. Fine structure of blood capillaries. J. Appl. Phys 24:1424 [Google Scholar]
- 44. Parent M., et al. 2005. Poly(ADP-ribose) polymerase-1 is a negative regulator of HIV-1 transcription through competitive binding to TAR RNA with Tat.positive transcription elongation factor b (p-TEFb) complex. J. Biol. Chem. 280:448–457 [DOI] [PubMed] [Google Scholar]
- 45. Park J., Nadeau P. E., Mergia A. 2009. Activity of TAR in inducible inhibition of HIV replication by foamy virus vector expressing siRNAs under the control of HIV LTR. Virus Res. 140:112–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Parton R. G., Simons K. 2007. The multiple faces of caveolae. Nat. Rev. Mol. Cell. Biol. 8:185–194 [DOI] [PubMed] [Google Scholar]
- 47. Pelkmans L., Kartenbeck J., Helenius A. 2001. Caveolar endocytosis of simian virus 40 reveals a new two-step vesicular-transport pathway to the ER. Nat. Cell Biol. 3:473–483 [DOI] [PubMed] [Google Scholar]
- 48. Poli G., et al. 1990. Tumor necrosis factor alpha functions in an autocrine manner in the induction of human immunodeficiency virus expression. Proc. Natl. Acad. Sci. U. S. A. 87:782–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pomerantz R. J., Feinberg M. B., Trono D., Baltimore D. 1990. Lipopolysaccharide is a potent monocyte/macrophage-specific stimulator of human immunodeficiency virus type 1 expression. J. Exp. Med. 172:253–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Quest A. F., Gutierrez-Pajares J. L., Torres V. A. 2008. Caveolin-1: an ambiguous partner in cell signalling and cancer. J. Cell. Mol. Med. 12:1130–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Quest A. F., Leyton L., Parraga M. 2004. Caveolins, caveolae, and lipid rafts in cellular transport, signaling, and disease. Biochem. Cell Biol. 82:129–144 [DOI] [PubMed] [Google Scholar]
- 52. Sen J., et al. 1995. Expression and induction of nuclear factor-kappa B-related proteins in thymocytes. J. Immunol. 154:3213–3221 [PubMed] [Google Scholar]
- 53. Shin J. S., Gao Z., Abraham S. N. 2000. Involvement of cellular caveolae in bacterial entry into mast cells. Science 289:785–788 [DOI] [PubMed] [Google Scholar]
- 54. Stan R. V. 2005. Structure of caveolae. Biochim. Biophys. Acta 1746:334–348 [DOI] [PubMed] [Google Scholar]
- 55. Stan R. V., Tkachenko E., Niesman I. R. 2004. PV1 is a key structural component for the formation of the stomatal and fenestral diaphragms. Mol. Biol. Cell 15:3615–3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Verhoef K., Sanders R. W., Fontaine V., Kitajima S., Berkhout B. 1999. Evolution of the human immunodeficiency virus type 1 long terminal repeat promoter by conversion of an NF-kappaB enhancer element into a GABP binding site. J. Virol. 73:1331–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang X. M., Kim H. P., Song R., Choi A. M. 2006. Caveolin-1 confers antiinflammatory effects in murine macrophages via the MKK3/p38 MAPK pathway. Am. J. Respir. Cell Mol. Biol. 34:434–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wang X. M., Nadeau P. E., Lo Y. T., Mergia A. 2010. Caveolin-1 modulates HIV-1 envelope-induced bystander apoptosis through gp41. J. Virol. 84:6515–6526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Williams S. A., Kwon H., Chen L. F., Greene W. C. 2007. Sustained induction of NF-kappa B is required for efficient expression of latent human immunodeficiency virus type 1. J. Virol. 81:6043–6056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Williams T. M., Lisanti M. P. 2005. Caveolin-1 in oncogenic transformation, cancer, and metastasis. Am. J. Physiol. Cell Physiol. 288:C494–C506 [DOI] [PubMed] [Google Scholar]
- 61. Yamada E. 1955. The fine structure of the gall bladder epithelium of the mouse. J. Biophys. Biochem. Cytol. 1:445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zheng Y., et al. 2009. Role of caveolin-1 in EGCG-mediated protection against linoleic-acid-induced endothelial cell activation. J. Nutr. Biochem. 20:202–209 [DOI] [PMC free article] [PubMed] [Google Scholar]