Abstract
How viruses evolve to select their receptor proteins for host cell entry is puzzling. We recently determined the crystal structures of NL63 coronavirus (NL63-CoV) and SARS coronavirus (SARS-CoV) receptor-binding domains (RBDs), each complexed with their common receptor, human angiotensin-converting enzyme 2 (hACE2), and proposed the existence of a virus-binding hot spot on hACE2. Here we investigated the function of this hypothetical hot spot using structure-guided biochemical and functional assays. The hot spot consists of a salt bridge surrounded by hydrophobic tunnel walls. Mutations that disturb the hot spot structure have significant effects on virus/receptor interactions, revealing critical energy contributions from the hot spot structure. The tunnel structure at the NL63-CoV/hACE2 interface is more compact than that at the SARS-CoV/hACE2 interface, and hence RBD/hACE2 binding affinities are decreased either by NL63-CoV mutations decreasing the tunnel space or by SARS-CoV mutations increasing the tunnel space. Furthermore, NL63-CoV RBD inhibits hACE2-dependent transduction by SARS-CoV spike protein, a successful application of the hot spot theory that has the potential to become a new antiviral strategy against SARS-CoV infections. These results suggest that the structural features of the hot spot on hACE2 were among the driving forces for the convergent evolution of NL63-CoV and SARS-CoV.
INTRODUCTION
Host receptor recognition by viruses is the first and essential step for viral infections. During the long history of evolutionary battles between viruses and hosts, viruses have evolved complex strategies for their receptor selections (2). Despite tremendous efforts to understand these strategies, the current picture of how viruses recognize their host receptors is still murky. Viruses exploit a wide variety of host cell surface molecules as their receptors. In addition to serving as receptors for viruses, these molecules are implicated in various host physiological functions such as cell adhesion, immune response, signaling pathways, proteolysis, and ion transport. On one hand, several viruses can share one host receptor. For example, coxsackievirus-adenovirus receptor, an immunoglobulin (Ig) superfamily member, is the receptor for both coxsackieviruses and adenoviruses (3). On the other hand, one virus can recognize several different host receptors. For example, herpes simplex viruses use one of at least three protein receptors: HVEM, which is a tumor necrosis factor receptor family member (23), and nectin-1 and nectin-2, both of which are Ig superfamily members (8, 31). Understanding the pattern of host receptor recognition by viruses has important implications for viral evolution, pathogenesis, host range, tropism, cross-species infections, emerging viral epidemics, and virus-mediated gene targeting.
A key question regarding the evolution of host receptor recognition by viruses is what features of these receptor molecules make them targeted by viruses. The receptors for viruses can be proteins, carbohydrates, or lipids (2). Compared with carbohydrates and lipids, protein receptors in general have more structural features and thus are engaged in more-specific and high-affinity binding interactions with viruses; they are the focus of this study. Among protein receptors, some (such as cell adhesion molecules) are more abundant than others (such as proteases and ion transporters). Although the availability of abundant protein receptors to viruses is probably one of the reasons why they were selected by viruses as receptors (30), it is not clear whether receptor proteins, especially nonabundant receptor proteins, contain any structural features that make them targeted by viruses.
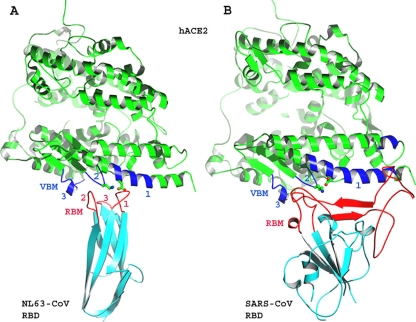
The structural features of receptor proteins can be identified from the atomic structures of virus/receptor interfaces. Defined structural and functional receptor-binding domains (RBDs) have been identified in many viral surface glycoproteins. One or more receptor-binding motifs (RBMs) on these viral RBDs mediate the interactions with their receptor proteins. To date several crystal structures are available for viral RBDs complexed with their receptor proteins (1, 4, 5, 13, 18, 32, 33). Among these structures, only two reveal how different viral RBDs can bind to their common receptor protein: the structures of NL63 coronavirus (NL63-CoV) and SARS coronavirus (SARS-CoV) RBDs, each complexed with their common receptor, human angiotensin-converting enzyme 2 (hACE2) (18, 32). Both NL63-CoV and SARS-CoV are important human viral pathogens. The former causes prevalent respiratory diseases (6, 29), whereas the latter was responsible for the worldwide epidemic of severe acute respiratory syndrome (SARS) diseases in 2002 to 2003 (12, 24). Coronavirus spike glycoproteins are envelope-anchored clove-shaped trimers (16). Each spike trimer contains three monomeric S1 heads, which function in receptor binding, and a trimeric S2 stalk, which functions in fusing the viral envelope and host membrane. NL63-CoV and SARS-CoV RBDs are located in the S1 heads of their respective spike proteins. There is no structural homology in their RBD core structures or RBMs (Fig. 1). The core structures of NL63-CoV and SARS-CoV RBDs are a two-layer β-sandwich and a single-layer β-sheet, respectively; the RBMs of NL63-CoV and SARS-CoV are three discontinuous short loops and one continuous long subdomain, respectively. Nevertheless, the two viral RBDs bind to the same three virus-binding motifs (VBMs) on hACE2 (18, 32).
Fig. 1.
Overall structures of NL63-CoV and SARS-CoV RBDs, each complexed with their common receptor, human ACE2. (A) Crystal structure of NL63-CoV RBD complexed with hACE2 (PDB 3KBH). hACE2 is green, virus-binding motifs (VBMs) are blue, the NL63-CoV RBD core structure is cyan, and receptor-binding motifs (RBMs) are red. Lys353 and Asp38 in hACE2, which are critical for the RBD/hACE2 interactions, are shown in ball-and-stick format. (B) Crystal structure of SARS-CoV RBD complexed with hACE2 (PDB 2AJF).
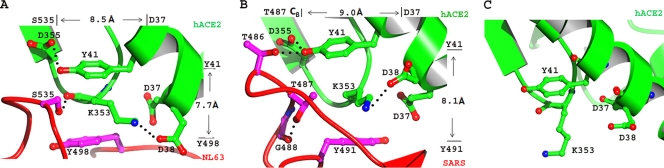
Our previous structural studies led to the hypothesis that a virus-binding hot spot exists on hACE2 and plays an important role in the binding of both NL63-CoV and SARS-CoV (32). This hypothetical hot spot consists of a critical Lys353-Asp38 salt bridge on hACE2, which is surrounded by four hydrophobic tunnel walls (Fig. 2A and B). Two of the tunnel walls, Tyr41 (top wall) and Asp37 (right wall), are contributed by hACE2, whereas the other two tunnel walls are contributed by the viruses: Tyr498 (bottom wall) and Ser535 (left wall) from NL63-CoV and Tyr491 (bottom wall) and Thr487 (left wall) from SARS-CoV. Details of how this hypothetical hot spot may contribute to the virus/receptor interactions are unknown. In this study we use structure-guided biochemical and functional approaches to investigate the role of each of the components of the hot spot structure in the virus/receptor interactions. We then apply the hot spot theory to the development of a potentially new antiviral strategy against SARS-CoV infections. We also discuss how the structural features of the hot spot drove the convergent evolution of two different viruses.
Fig. 2.
Detailed structure of a common virus-binding hot spot on human ACE2. (A) Hot spot structure at the NL63-CoV/hACE2 interface. (B) Hot spot structure at the SARS-CoV/hACE2 interface. (C) Conformation of Lys353 on the surface of unbound human ACE2 (PDB 1R42).
MATERIALS AND METHODS
Protein expression and purification.
Soluble proteins, including NL63-CoV RBD (residues 461 to 616), SARS-CoV RBD (residues 306 to 527), and hACE2 peptidase domain (residues 19 to 615), were expressed and purified as previously described (17, 18, 32). In brief, the proteins were expressed in insect cells using the Bac-to-Bac system (Life Technologies Inc.). Each expression construct (Invitrogen) contained an N-terminal honeybee melittin signal sequence and a C-terminal His tag sequence. Mutations were introduced by PCR site-directed mutagenesis to the expression constructs. Recombinant baculoviruses were generated and amplified in Sf9 insect cells. The protein to be purified was harvested from Sf9 cell supernatants, loaded onto a Ni-nitrilotriacetic acid (Ni-NTA) column, eluted from the Ni-NTA column with 0.5 M imidazole, and further purified by gel filtration chromatography on Superdex 200 (GE Healthcare). Fractions containing the purified protein were pooled together, loaded into an Amicon ultra-15 centrifugal filter unit (10,000-molecular-weight [MW] cutoff) (Millipore), and centrifuged at 10,000 rpm until the protein concentration reached 10 mg/ml.
RBD/hACE2 binding assays.
Surface plasmon resonance assays were carried out using a Biacore 2000 as previously described (32). In brief, to measure the affinities for binding between mutant viral RBDs and wild-type hACE2, hACE2 was immobilized on a C5 sensor chip through direct covalent coupling via amine groups. The surface of the sensor chip was activated with N-hydroxysuccinimide (NHS), the receptor was injected and immobilized to the surface of the chip, and the remaining activated surface of the chip was blocked with ethanolamine. Soluble RBDs were introduced at a flow rate of 20 μl/min at different concentrations (62.5 nM, 125 nM, and 250 nM). The on rate (kon), the off rate (koff), and the dissociation constant (Kd) were determined for the RBD/receptor binding interactions using BIA-EVALUATIONS software. To measure the affinities for binding between mutant hACE2 and wild-type viral RBDs, RBDs were immobilized on the sensor chip and hACE2 was the soluble analyte. As negative controls, soluble RBDs or hACE2 was passed through an empty sensor chip and buffer alone was passed through sensor chips containing RBDs or hACE2 as immobilized ligands.
Transduction assays with pseudotyped virus.
Transduction was assayed using murine leukemia viruses (MLVs) expressing β-galactosidase and pseudotyped with NL63-CoV or SARS-CoV spike protein. To prepare pseudotyped viruses, HEK293T cells were cotransfected with spike protein-encoding pcDNA3.1 and MLV β-galactosidase-transducing vector pBAG (25). At 2 days posttransfection, viral supernatants were harvested and concentrated in a spin concentrator. Approximately 4 ml of supernatant was typically concentrated (10,000-MW cutoff) to between 100 to 200 μl. HEK293T cells transiently expressing hACE2 in pcDNA3.1 were inoculated in 96-well dishes by adding 5 μl of concentrated viral supernatant to 50 μl cell culture medium per well. Transduction efficiency was quantified 2 days later by measuring β-galactosidase activity. The inoculated cells were lysed in phosphate-buffered saline (PBS) containing 0.5% NP-40 and 3 mg/ml o-nitrophenyl-β-d-glucopyranoside and monitored by spectrometry (optical density at 410 nm [OD410]). The intracellular C termini of the spike protein and hACE2 contained a C9 tag and a hemagglutinin (HA) tag, respectively. The concentrations of the spike protein packaged in pseudotyped viruses and of hACE2 expressed on the HEK293T cell surface were detected by Western blotting using anti-C9 and anti-HA antibodies, respectively. As a negative control, the plasmid expressing the spike protein was replaced by a plasmid that does not express any protein.
RESULTS
To investigate the role of the hot spot structure in the virus/receptor binding interactions, we mutated each of the components of the hot spot structure. We then examined how the mutations affect the affinities for binding between RBDs and hACE2 using surface plasmon resonance Biacore assays. We also investigated how the mutations impact the interactions between spike proteins and hACE2 by transduction assays using pseudotyped virus.
For Biacore assays, we first measured the affinities for binding between the wild-type hACE2 peptidase domain and prototypic NL63-CoV RBD (strain Amsterdam 1) and between the wild-type hACE2 peptidase domain and prototypic SARS-CoV RBD (strain Tor2, which was isolated during the 2002 to 2003 SARS epidemic). hACE2 was immobilized on the Biacore sensor chip through direct covalent coupling via amine groups, and NL63-CoV or SARS-CoV RBD was injected over the chip as the soluble analyte. The measured Kd for SARS-CoV RBD and hACE2 binding was 20.8 nM (Fig. 3A), consistent with the Kd of 16.2 nM measured in a previous study (19). The measured Kd for NL63-CoV RBD and hACE2 binding was 34.9 nM (Fig. 3A), the first reported Kd for binding between the two proteins. The same RBD fragment used in this study (residues 461 to 616) also bound to hACE2 with high affinity in a previous study using a coimmunoprecipitation analysis (21). Interestingly, although SARS-CoV and NL63-CoV RBDs had similar Kds for binding with hACE2, NL63-CoV RBD bound to hACE2 with significantly lower koff and kon. It has been shown that koff and kon are dictated by short-range van der Waals interactions and long-range electrostatic interactions between the proteins, respectively (26). Therefore, the lower koff and kon of the NL63-CoV-RBD/hACE2 complex likely reflected a less electrostatic and more hydrophobic interface between the two proteins.
Fig. 3.
Surface plasmon resonance Biacore analyses of the binding interactions between viral RBDs and human ACE2. Each experiment was repeated 5 times at three different protein concentrations. The corresponding standard errors are shown. (A) Kinetics of the binding interaction between wild-type hACE2 and wild-type RBDs. (B) Biacore analyses of NL63-CoV RBD and hACE2. Single mutations were introduced to hACE2 or RBD to modify every component of the hot spot structure. Ka, association constant. (C) Biacore analyses of SARS-CoV RBD and hACE2.
Using Biacore, we also measured the affinities for binding between hACE2 and NL63-CoV RBD and between hACE2 and SARS-CoV RBD in a reverse way: NL63-CoV or SARS-CoV RBD was immobilized on the sensor chip, and hACE2 was injected over the chip as the soluble analyte. The measured Kds were 68.0 nM for NL63-CoV RBD and hACE2 and 137 nM for SARS-CoV RBD and hACE2, both of which were higher than when hACE2 was immobilized (Fig. 3A). Such discrepancies in measured Kd were mostly due to the differences in measured kon. No matter whether hACE2 or RBDs were immobilized, koffs remained similar. When hACE2 was immobilized, however, kon was significantly higher. Why did kon increase when hACE2, instead of RBDs, was immobilized? This is because hACE2 has a larger molecular weight than either of the RBDs, and thus when immobilized, hACE2 can provide more accessible surface area for complex formation, leading to higher kon. Therefore, the surface accessibility of the immobilized protein, but not the dissociation rate, accounted for the discrepancies in measured Kd.
To evaluate how mutations of the hot spot structure affect the affinities for binding between RBDs and hACE2, we introduced single mutations to either RBDs or hACE2 that modified every component of the hot spot structure. These mutations were K353A, D38A, D37A, Y41A, and Y41F in hACE2, Y498A, S535A, and S535T in NL63-CoV RBD, and Y491A, T487A, and T487S in SARS-CoV RBD (Fig. 2A and B). We expressed and purified each of the 11 hACE2 and RBD mutants. To measure the affinities for binding between mutant RBDs and wild-type hACE2, hACE2 was immobilized on the sensor chip and mutant RBDs were the soluble analytes. To measure the affinities for binding between wild-type RBDs and mutant hACE2, NL63-CoV or SARS-CoV RBD was immobilized on the sensor chip and mutant hACE2 was the soluble analyte. The results were then compared with the affinities for binding between wild-type hACE2 and wild-type RBDs (Fig. 3B and C).
In this study we not only measured direct interactions between viral RBDs and hACE2 using recombinant proteins but also examined the spike/receptor interactions using functional assays. To this end, we carried out transduction assays with pseudotyped virus to investigate whether changes in RBD/hACE2 interactions could lead to corresponding changes in viral entry and membrane fusion in the context of the full-length spike proteins and their receptor protein. We prepared retroviral MLVs expressing β-galactosidase and pseudotyped with NL63-CoV or SARS-CoV spike protein. These MLVs were incubated with hACE2-expressing HEK293T cells. The transduction efficiency of the pseudotyped viruses was measured by determining β-galactosidase activity of inoculated cell lysate. To measure the interactions between wild-type hACE2 and mutant spike proteins or between mutant hACE2 and wild-type spike proteins, we introduced single mutations to hACE2 or the spike proteins that were the same as the mutations used for Biacore assays (Fig. 4A and B). The expression levels of the spike proteins in pseudotyped viruses and of hACE2 molecules on HEK293T cells were detected by Western blotting using antibodies against their intracellular C-terminal C9 and HA tags, respectively. The Western blotting results showed that all of the mutant spike proteins and mutant hACE2 molecules were well expressed, and the expression levels of these mutant proteins were quantified and calibrated against those of the wild-type proteins (Fig. 4C). Finally, the measured transduction efficiencies for mutant spike proteins and mutant hACE2 were normalized against the transduction efficiency of viruses pseudotyped with wild-type spike proteins in cells expressing wild-type hACE2 (Fig. 4).
Fig. 4.
Transduction assays with pseudotyped virus of the interactions between viral spike proteins and human ACE2. Retroviral MLVs expressing β-galactosidase and pseudotyped with the NL63-CoV or SARS-CoV spike protein were used to infect hACE2-expressing HEK293T cells. Transduction efficiency of the pseudotyped viruses was measured by β-galactosidase assays. After mutations were introduced into the spike proteins or hACE2, the corresponding transduction efficiency was normalized against the transduction efficiency of viruses pseudotyped with wild-type spike proteins in cells expressing wild-type hACE2. Each experiment was repeated 6 times. The corresponding standard errors are shown. (A) Transduction of MLVs pseudotyped with NL63-CoV spike protein in hACE2-expressing cells. (B) Transduction of MLVs pseudotyped with SARS-CoV spike protein in hACE2-expressing cells. (C) Western blotting of coronavirus spike proteins and hACE2. The NL63-CoV and SARS-CoV spike proteins packaged in pseudotyped retroviruses both contained a C-terminal C9 tag, and the hACE2 expressed on the HEK293T cell surface contained a C-terminal HA tag. The expression levels of the spike proteins and hACE2 were detected by Western blotting using anti-C9 and anti-HA antibodies, respectively. The protein bands were quantified using software Image J (version 1.6).
Both Biacore assays and transduction assays with pseudotyped virus yielded results that were highly consistent with each other (Fig. 3 and 4; Tables 1 and 2). It is worth noting that recombinant SARS-CoV and NL63-CoV RBDs are both monomers in solution, whereas the full-length spike proteins are trimers on virus surfaces (14). Thus, the good correlation between the RBD/hACE2 binding affinities and the spike-mediated transduction efficiency strongly suggests that the measured RBD/hACE2 binding activities reflect the native states of the proteins. Our results showed that all of the targeted mutations produced significantly reduced RBD/hACE2 binding affinities and spike-guided transduction compared with those for the corresponding wild-type proteins (t test; P < 0.01 for both Ka and transduction), with the exception of D37A in hACE2 (P > 0.075 for transduction) and Y498 in NL63-CoV RBD (P > 0.10 for both Ka and transduction). Here we combine these biochemical and functional data with our previous structural data and discuss molecular and structural features of the virus-binding hot spot that make hACE2 a common target by two different viruses.
Table 1.
| Form of hACE2 | Dataa for interaction with RBD of: |
|||
|---|---|---|---|---|
| NL63-CoV |
SARS-CoV |
|||
| Ka (10−3/nM) | Transduction (%) | Ka (10−3/nM) | Transduction (%) | |
| WTb | 14.7 (±1.5) | 100 (±2.3) | 7.30 (±1.2) | 100 (±4.8) |
| K353A | Too lowc | 19.3 (±2.9) | Too low | 13.5 (±2.2) |
| D38A | 3.37 (±0.49) | 47.2 (±2.3) | 2.44 (±0.37) | 59.3 (±4.5) |
| D37A | 8.77 (±0.77) | 87.6 (±6.8) | 2.72 (±0.46) | 85.1 (±2.4) |
| Y41A | 0.69 (±0.14) | 28.5 (±2.0) | 0.63 (±0.21) | 24.0 (±2.8) |
| Y41F | 0.96 (±0.33) | 35.7 (±2.1) | 0.69 (±0.24) | 25.9 (±4.3) |
Values are means (±standard errors). Ka, association constant.
WT, wild type.
Too low, too low for measurement.
Table 2.
Summary of Biacore and pseudotyped-virus transduction data from Fig. 3 and 4, with hACE2 immobilized
| Virus and form of RBD | Dataa for interaction with hACE2 |
|
|---|---|---|
| Ka (10−3/nM) | Transduction (%) | |
| NL63-CoV | ||
| WTb | 28.7 (±3.8) | 100 (±2.3) |
| Y498A | 24.3 (±2.7) | 91.9 (±3.4) |
| S535A | 1.25 (±0.32) | 64.9 (±2.4) |
| S535T | 0.43 (±0.11) | 39.9 (±1.8) |
| SARS-CoV | ||
| WT | 48.1 (±4.0) | 100 (±4.8) |
| Y491A | Too lowc | 50.4 (±2.4) |
| T487A | Too low | 57.7 (±6.1) |
| T487S | Too low | 50.9 (±3.8) |
Values are means (±standard errors). Ka, association constant.
WT, wild type.
Too low, too low for measurement.
The hot spot structures at the NL63-CoV/hACE2 and SARS-CoV/hACE2 interfaces have many common features. The Lys353-Asp38 salt bridge plays a central role in the hot spot structure at both of the interfaces. Because of the hydrophobic environment, the salt bridge not only provides a significant amount of energy to the virus/receptor binding interactions but also fills a critical void in the hydrophobic stacking interactions at the virus/receptor interfaces. Correspondingly, alanine substitution for either Lys353 or Asp38 in hACE2 significantly decreased the RBD/hACE2 binding affinities and viral transductions (Fig. 3 and 4). The hydrophobic tunnel walls of the hot spot structure also make important contributions to the virus/receptor binding interactions; they not only support the side chain of Lys353 to form the salt bridge but also provide hydrophobic stacking interactions at the virus/receptor interfaces. Some hydrophobic tunnel walls contribute more energy to the virus/receptor binding interactions than others (Fig. 3 and 4). For example, Tyr41 in hACE2 (top wall) is more important than Asp37 in hACE2 (right wall), probably because Tyr41 functions better as a tunnel wall with its aromatic ring. Alanine substitution for Tyr41 significantly decreased RBD/hACE2 binding affinities and viral transductions, suggesting that the stacking interaction between Tyr41 and Lys353 is essential for the hot spot structure. Interestingly, although a phenylalanine at the 41 position can potentially function as a tunnel wall with its aromatic ring, the Y41F mutation also significantly decreased RBD/hACE2 binding affinities and viral transductions. Detailed structural analysis reveals that the hydroxyl group of Tyr41 forms a hydrogen bond with receptor Asp355 at the NL63-CoV/hACE2 interface and two hydrogen bonds with receptor Asp355 and RBD Thr486 at the SARS-CoV/hACE2 interface (Fig. 2A and B). Thus, the side chain of Tyr41 needs to be firmly anchored in order for it to function properly as a tunnel wall. Residue 41 is a histidine in the ACE2 proteins from several bat species (10). Not only is His41 a poor hydrophobic stacker, but also it cannot be anchored properly to function as a tunnel wall. As a result, these bat ACE2 proteins were poor receptors for human SARS-CoV strains unless an H41Y mutation was introduced (10). Overall, the salt bridge and many of the tunnel walls of the hot spot structure contribute energy to the virus/receptor binding interactions.
The hot spot structures at the NL63-CoV/hACE2 and SARS-CoV/hACE2 interfaces differ in a subtle but functionally important way. The tunnel structure at the NL63-CoV/hACE2 interface is more compact than that at the SARS-CoV/hACE2 interface (Fig. 2A and B). At the NL63-CoV/hACE2 interface, the closest distances between the two pairs of opposing tunnel walls, Ser535-Asp37 (left to right) and Tyr41-Tyr498 (top to bottom), are 8.5 Å and 7.7 Å, respectively. At the SARS-CoV/hACE2 interface, if a serine replaces threonine at the 487 position in SARS-CoV RBD, these distances become 9.0 Å and 8.1 Å, respectively. Because of the compactness of the tunnel structure at the NL63-CoV/hACE2 interface, S535T mutation in NL63-CoV RBD decreased the tunnel space and was energetically unstable (Fig. 3B and 4A). In contrast, because of the extra space of the tunnel structure at the SARS-CoV/hACE2 interface, T487S mutation in SARS-CoV RBD increased the tunnel space but was also energetically unstable (Fig. 3C and 4B). Indeed, residue 487 was a serine in RBDs of some low-pathogenicity SARS-CoV strains and was largely responsible for the lack of human-to-human transmission of these viral strains (18, 19, 27). Thus, although S535T mutation in NL63-CoV RBD and T487S mutation in SARS-CoV RBD exerted opposite effects on the same left tunnel wall of the hot spot structure, they both reduced RBD/hACE2 binding affinities and viral transductions. For similar reasons, compared with Tyr498 in NL63-CoV RBD, Tyr491 in SARS-CoV RBD provides more support to the hot spot structure as the bottom tunnel wall in a more spacious tunnel space, and hence alanine substitution for Tyr491 decreased RBD/hACE2 binding affinities and viral transductions (Fig. 3 and 4). Therefore, the seemingly small differences in the hot spot structure at the two virus/receptor interfaces not only have significant impacts on virus/receptor binding interactions but also have important epidemic implications.
One of the direct implications of our study is the possibility of using NL63-CoV RBD as an inhibitor to block SARS-CoV infections, because NL63-CoV RBD can compete with SARS-CoV for the common virus-binding hot spot on hACE2. To test this possibility, we inoculated MLVs pseudotyped with SARS-CoV spike protein onto hACE2-expressing HEK293T cells in the presence of various concentrations of purified NL63-CoV RBD or SARS-CoV RBD (Fig. 5). Transduction was shown as a percentage of β-galactosidase activity observed in the absence of any inhibitor. The results showed that NL63-CoV RBD indeed inhibited SARS-CoV spike-mediated transductions. At 10 μg/ml (0.47 μM), NL63-CoV RBD inhibited SARS-CoV spike-mediated transductions by over 80%. This method has the potential to become a new antiviral strategy against SARS-CoV infections, as it represents the first case in which SARS-CoV infection can be inhibited by a protein from a different virus. It also represents a successful application of the common virus-binding hot spot theory derived from the present study.
Fig. 5.
Inhibition of SARS-CoV spike-mediated transduction by NL63-CoV RBD. MLVs pseudotyped with SARS-CoV spike protein were used to infect hACE2-expressing HEK293T cells in the presence of various concentrations of purified NL63-CoV RBD, SARS-CoV RBD, SARS-CoV RBD containing the T487S mutation, hACE2 (positive control), and bovine serum albumin (BSA; negative control). Transduction is shown as a percentage of β-galactosidase activity observed in the absence of any inhibitor. Each experiment was repeated 5 times. The corresponding standard errors are shown. The results show that NL63-CoV can efficiently inhibit SARS-CoV spike-mediated transduction.
DISCUSSION
Binding to the same hot spot on hACE2 was likely an outcome of convergent evolution by NL63-CoV and SARS-CoV. Our study provides several lines of evidence to support this notion. First, despite no structural homology in their RBDs, NL63-CoV and SARS-CoV both bind to the hot spot region and form highly similar and energetically stable tunnel structures (Fig. 1 and 2). Second, despite no structural homology in their RBMs, both viruses insert an RBM loop between VBM2 and VBM3 on hACE2 (Fig. 1). Third, despite being presented by nonhomologous RBM loops, Ser535 in NL63-CoV RBD and Thr487 in SARS-CoV RBD (Ser487 in RBDs of some low-pathogenicity SARS-CoV strains) occupy identical positions as the left tunnel wall in the hot spot structure (Fig. 2A and B) and contribute energy to the virus/receptor binding interactions (Fig. 3 and 4). Last, despite pointing in opposite directions, Tyr498 in NL63-CoV RBD and Tyr491 in SARS-CoV RBD also occupy identical positions as the bottom tunnel wall in the hot spot structure (Fig. 2A and B). These data suggest that NL63-CoV and SARS-CoV likely evolved independent strategies to achieve the same functional goal, supporting a convergent evolutionary relationship between the two viruses.
The likely convergent evolution of NL63-CoV and SARS-CoV was at least partly driven by the structural features of the virus-binding hot spot on hACE2. The general structural features of the hot spot favor virus binding: it is located in a region on hACE2 that is furthest from the membrane, relatively flat, free of glycosylation, and thereby easily accessible to viruses (Fig. 1). The detailed structural features of the hot spot, such as its potential to form the energetically stable tunnel structure, also favor virus binding (Fig. 2A and B). In the unbound hACE2 structure, where structural restraints from viruses are absent, Lys353 projects into solution; it does not form a salt bridge with Asp38 or stack with Tyr41 or Asp37 (Fig. 2C) (15, 28). Thus, the virus-binding hot spot is not preexistent or preorganized on hACE2; instead, it is induced to form by virus binding. Therefore, while the hot spot is mainly an intrinsic property of hACE2, it is also a dynamic structure and receives structural contributions from both hACE2 and the viruses, although the contributions from hACE2 are more pronounced. The virus-binding hot spot on hACE2 is likely different from the hot spots for host protein/protein interactions. Host protein partners have coevolutionary relationships (9, 11), and hence hot spots for host protein/protein interactions are usually preexistent and preorganized in unbound host protein structures (20, 22). Viruses and receptors, however, do not usually have such coevolutionary relationships; viruses evolve to adapt to host receptors, but receptors do not evolve to adapt to viruses. Occasionally, however, if a virus exerts a large enough impact on a host, the host receptor may evolve away from virus binding (7). So far the virus-binding hot spot on hACE2 is not known for its interaction with any other host proteins. Overall, the potential of the hot spot region on hACE2 to form energetically stable tunnel structures and some general structural features of this region on the receptor surface were among the possible reasons why the hot spot was exploited by two different viruses.
ACKNOWLEDGMENTS
We thank Lorraine Albritton for the pBAG vector and Michael Farzan for spike protein genes.
This work was supported by NIH grant R01AI089728 (to F.L.) and by a University of Minnesota AHC Faculty Research Development Grant (to F.L. and L.M.M.).
Footnotes
Published ahead of print on 16 March 2011.
REFERENCES
- 1. Abraham J., Corbett K. D., Farzan M., Choe H., Harrison S. C. 2010. Structural basis for receptor recognition by New World hemorrhagic fever arenaviruses. Nat. Struct. Mol. Biol. 17:438–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baranowski E., Ruiz-Jarabo C. M., Domingo E. 2001. Evolution of cell recognition by viruses. Science 292:1102–1105 [DOI] [PubMed] [Google Scholar]
- 3. Bergelson J. M., et al. 1997. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science 275:1320–1323 [DOI] [PubMed] [Google Scholar]
- 4. Bewley M. C., Springer K., Zhang Y. B., Freimuth P., Flanagan J. M. 1999. Structural analysis of the mechanism of adenovirus binding to its human cellular receptor, CAR. Science 286:1579–1583 [DOI] [PubMed] [Google Scholar]
- 5. Carfi A., et al. 2001. Herpes simplex virus glycoprotein D bound to the human receptor HveA. Mol. Cell 8:169–179 [DOI] [PubMed] [Google Scholar]
- 6. Fouchier R. A. M., et al. 2004. A previously undescribed coronavirus associated with respiratory disease in humans. Proc. Natl. Acad. Sci. U. S. A. 101:6212–6216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Galvani A. P., Slatkin M. 2003. Evaluating plague and smallpox as historical selective pressures for the CCR5-Delta 32 HIV-resistance allele. Proc. Natl. Acad. Sci. U. S. A. 100:15276–15279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Geraghty R. J., Krummenacher C., Cohen G. H., Eisenberg R. J., Spear P. G. 1998. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science 280:1618–1620 [DOI] [PubMed] [Google Scholar]
- 9. Goh C. S., Cohen F. E. 2002. Co-evolutionary analysis reveals insights into protein-protein interactions. J. Mol. Biol. 324:177–192 [DOI] [PubMed] [Google Scholar]
- 10. Hou Y. X., et al. 2010. Angiotensin-converting enzyme 2 (ACE2) proteins of different bat species confer variable susceptibility to SARS-CoV entry. Arch. Virol. 155:1563–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jothi R., Cherukuri P. F., Tasneem A., Przytycka T. M. 2006. Co-evolutionary analysis of domains in interacting proteins reveals insights into domain-domain interactions mediating protein-protein interactions. J. Mol. Biol. 362:861–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ksiazek T. G., et al. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953–1966 [DOI] [PubMed] [Google Scholar]
- 13. Kwong P. D., et al. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lai M. M. C., Holmes K. V. 2001. Coronaviridae: the viruses and their replication, p. 1163–1186 In Knipe D. M., et al. (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 15. Li F. 2008. Structural analysis of major species barriers between humans and palm civets for severe acute respiratory syndrome coronavirus infections. J. Virol. 82:6984–6991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li F., et al. 2006. Conformational states of the severe acute respiratory syndrome coronavirus spike protein ectodomain. J. Virol. 80:6794–6800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li F., Li W. H., Farzan M., Harrison S. C. 2006. Interactions between SARS coronavirus and its receptor. Adv. Exp. Med. Biol. 581:229–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li F., Li W. H., Farzan M., Harrison S. C. 2005. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science 309:1864–1868 [DOI] [PubMed] [Google Scholar]
- 19. Li W. H., et al. 2005. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J. 24:1634–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li X., Keskin O., Ma B. Y., Nussinov R., Liang J. 2004. Protein-protein interactions: hot spots and structurally conserved residues often locate in complemented pockets that pre-organized in the unbound states: implications for docking. J. Mol. Biol. 344:781–795 [DOI] [PubMed] [Google Scholar]
- 21. Lin H. X., et al. 2008. Identification of residues in the receptor-binding domain (RBD) of the spike protein of human coronavirus NL63 that are critical for the RBD-ACE2 receptor interaction. J. Gen. Virol. 89:1015–1024 [DOI] [PubMed] [Google Scholar]
- 22. Ma B. Y., Wolfson H. J., Nussinov R. 2001. Protein functional epitopes: hot spots, dynamics and combinatorial libraries. Curr. Opin. Struct. Biol. 11:364–369 [DOI] [PubMed] [Google Scholar]
- 23. Montgomery R. I., Warner M. S., Lum B. J., Spear P. G. 1996. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell 87:427–436 [DOI] [PubMed] [Google Scholar]
- 24. Peiris J. S. M., et al. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361:1319–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Price J., Turner D., Cepko C. 1987. Lineage analysis in the vertebrate nervous system by retrovirus-mediated gene transfer. Proc. Natl. Acad. Sci. U. S. A. 84:156–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schreiber G., Fersht A. R. 1996. Rapid, electrostatically assisted association of proteins. Nat. Struct. Biol. 3:427–431 [DOI] [PubMed] [Google Scholar]
- 27. Song H. D., et al. 2005. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc. Natl. Acad. Sci. U. S. A. 102:2430–2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Towler P., et al. 2004. ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis. J. Biol. Chem. 279:17996–18007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. van der Hoek L., et al. 2004. Identification of a new human coronavirus. Nat. Med. 10:368–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang J. H. 2002. Protein recognition by cell surface receptors: physiological receptors versus virus interactions. Trends Biochem. Sci. 27:122–126 [DOI] [PubMed] [Google Scholar]
- 31. Warner M. S., et al. 1998. A cell surface protein with herpesvirus entry activity (HveB) confers susceptibility to infection by mutants of herpes simplex virus type 1, herpes simplex virus type 2, and pseudorabies virus. Virology 246:179–189 [DOI] [PubMed] [Google Scholar]
- 32. Wu K., Li W. K., Peng G., Li F. 2009. Crystal structure of NL63 respiratory coronavirus receptor-binding complexed with its human receptor. Proc. Natl. Acad. Sci. U. S. A. 106:19970–19974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xu K., et al. 2008. Host cell recognition by the henipaviruses: crystal structures of the Nipah G attachment glycoprotein and its complex with ephrin-B3. Proc. Natl. Acad. Sci. U. S. A. 105:9953–9958 [DOI] [PMC free article] [PubMed] [Google Scholar]