Abstract
Hepatitis E virus (HEV) is an important but extremely understudied human pathogen, and the mechanisms of HEV replication and pathogenesis are largely unknown. We previously identified an attenuated genotype 3 HEV mutant (pSHEV-1) containing three unique amino acid mutations (F51L, T59A, and S390L) in the capsid protein. To determine the role of each of these mutations, we constructed three HEV single mutants (rF51L, rT59A, and rS390L) which were all found to be replication competent in Huh7 liver cells. To determine the pathogenicities of the mutants, we utilized the specific-pathogen-free (SPF) pig model for HEV and a unique inoculation procedure that bypasses the need for propagating infectious HEV in vitro. A total of 60 pigs were intrahepatically inoculated, via an ultrasound-guided technique, with in vitro-transcribed full-length capped RNA transcripts from the infectious clones of each single mutant, the pSHEV-1 triple mutant, wild-type pSHEV-3, or phosphate-buffered saline (PBS) buffer (n = 10). The results showed that the F51L mutation partially contributed to virus attenuation, whereas the T59A and S390L mutations resulted in more drastic attenuation of HEV in pigs, as evidenced by a significantly lower incidence of viremia, a delayed appearance and shorter duration of fecal virus shedding and viremia, and lower viral loads in liver, bile, and intestinal content collected at three different necropsy times. The results indicate that the three mutations in the capsid protein collectively contribute to HEV attenuation. This study has important implications for developing a modified live-attenuated vaccine against HEV.
INTRODUCTION
Hepatitis E virus (HEV) is a major cause of enterically transmitted acute hepatitis in many developing countries in Asia, the Middle East, and Africa and in Mexico (2, 13, 40–42, 49), although the disease has also been reported in many industrialized countries, including the United States (53). HEV is transmitted primarily by the fecal-oral route through contaminated drinking water or food. The disease generally affects young adults and has a mortality rate of less than 1% in the general population, although a significantly higher rate of mortality, up to 30%, has been reported in infected pregnant women (47). HEV is a small nonenveloped virus, and its genome is a single-strand, positive-sense RNA molecule of approximately 7.2 kb in size. Currently, HEV is classified in the genus Hepevirus of the family Hepeviridae (9).
The genomic RNA of HEV contains a short 5′ noncoding region (NCR), three open reading frames (ORF1, -2, and -3), and a 3′ NCR (22). A cap structure has been identified at the 5′ end of the viral genome and is required for efficient virus replication in vivo (14). The ORF2 and ORF3 proteins are translated from a bicistronic subgenomic mRNA (18, 25). ORF1 encodes a nonstructural protein with multiple functional domains, including methyltransferase, papain-like protease, helicase, and RNA-dependent RNA polymerase (RdRp) (29). It remains unclear whether the ORF1 polyprotein functions as a single protein with multiple functional domains or as individually cleaved smaller proteins. Thus far, the functions of the helicase and methyltransferase domains in ORF1 have been experimentally confirmed (26–28, 36). A proline-rich hypervariable region in ORF1 was found to be dispensable for HEV replication both in vitro and in vivo (51), although the biological significance of this region remains unknown. ORF2, located at the 3′ end of the genome, encodes the viral capsid protein, which contains a signal sequence and three glycosylation sites (13). The ORF2 capsid protein is involved in virion assembly, immunogenicity, and host cell receptor binding (23, 30, 34, 39, 55). The capsid protein is cleaved between amino acids (aa) 111 and 112, resulting in a 55-kDa capsid protein without the signal sequence that can still form virus-like particles (VLPs) (13). It has been demonstrated that mutations within the glycosylation sites prevent the formation of infectious virus particles (19). It has been reported that homodimers of the truncated HEV capsid proteins, E2 (amino acid residues 394 to 606) and p239 (amino acid residues 368 to 606), contain dominant antigenic determinants and that rhesus monkeys immunized with the homodimers are protected against HEV infection (31). The ORF3 gene overlaps with ORF2 (18, 25) and encodes a small multifunctional protein (1, 5, 6, 11, 12, 18, 46, 57, 58, 61–63, 68). ORF3 is translated from the third in-frame AUG codon in the ORF1/ORF2 intergenic region and encodes a protein that is essential for virus infectivity in vivo (25), although the expression of ORF3 protein is not required for virus replication, virion assembly, or infection in vitro (10, 11). The N terminus of ORF3 binds to HEV RNA and forms a complex with the capsid protein. The C terminus of the ORF3 protein may be involved in virion egress from infected cells and virion morphogenesis (12, 58, 68).
At least four major genotypes of HEV have been identified in mammalian species (40, 41). Genotype 1 and 2 strains are restricted to humans (43), whereas genotype 3 and 4 strains have been identified in humans, pigs, and several other animal species and are known to be zoonotic (3, 33, 37, 40, 41, 44, 45, 70, 71). The mechanisms of HEV pathogenesis and replication are still not well understood. In a previous study, we identified three mutations (F51L, T59A, and S390L) in the capsid protein of a strain of genotype 3 HEV (pSHEV-1) (24). These three mutations are unique to pSHEV-1, as the residues F51, T59, and S390 are conserved among all other known mammalian HEV strains (24). In a pilot study with only two pigs, we found that both pigs infected with pSHEV-1 had delayed seroconversion and delayed fecal virus shedding with undetectable viremia (24), thus suggesting a potential attenuation. Therefore, the objectives of this study were to determine if all three mutations contribute to HEV attenuation in a pathogenicity study with a large number of specific-pathogen-free (SPF) pigs and to dissect the role of each of these three amino acid mutations in virus attenuation. Three HEV single mutants (rF51L, rT59A, and rS390L) were constructed, their viabilities in Huh7 liver cells were tested, and subsequently the pathogenicity of each mutant was compared to those of the wild-type pSHEV-3 virus and the other mutants in a unique SPF pig model for HEV.
MATERIALS AND METHODS
Cells and infectious cDNA clones.
A subclone of a human hepatoma cell line (Huh7), originally isolated in Japan, was maintained in Dulbecco's modified Eagle medium (DMEM) (GIBCO) supplemented with 10% fetal bovine serum under a 5% CO2 atmosphere at 37°C. Transfected cells were maintained under the same conditions except at 34.5°C. The infectious cDNA clone pSHEV-3 from a strain of genotype 3 HEV and its derived mutant infectious clone pSHEV-1 containing the three capsid mutations (F51L, T59A, and S390L) were described in a previous study (24).
Construction of HEV mutants.
The wild-type genotype 3 HEV pSHEV-3 was used as the backbone for the construction of three single mutants (rF51L, rT59A, and rS390L) by site-directed mutagenesis. Briefly, a partial fragment of ORF2 was subcloned into pCR2.1 vector (Invitrogen) and subsequently used to generate three single-amino-acid mutants. Overlapping primers were designed based on the genomic sequence of pSHEV-3, each containing the desired nucleotide change (underlined): for mutant rF51L, forward primer 51F (5′-GTTGATTCTCAGCCCCTCGCC-3′) and reverse primer 51R (5′-GGCGAGGGGCTGAGAATCAA-3′); for mutant rT59A, forward primer 59F (5′-CTATATTCATCCAGCCAAC-3′) and reverse primer 59R (5′-GTTGGCTGGATGAATATAG-3′); and for mutant rS390L, forward primer 390F (5′-GACAGAATTGATTTCGTTGGCC-3′) and reverse primer 390R (5′-CGGCCAACGAAATCAATTCTGT-3′). Mutagenic primers were used in combination with the vector pCR2.1-specific primers M13F (5′-GTAAAACGACGGCCAG-3′) and M13R (5′-CAGGAAACAGCTATGAC-3′) to generate two overlapping PCR products, which were subsequently assembled into a final product of 1,463 bp by overlap extension PCR. Each PCR product was gel purified, digested with AvrII and NdeI, and then cloned back into the corresponding region of the pSHEV-3 infectious clone to produce each of the three full-length single HEV mutants, rF51L, rT59A, and rS390L. The entire ORF2 capsid gene for each mutant was sequenced to verify the presence of the introduced mutations and the absence of other, unwanted mutations.
In vitro RNA transcription.
To generate full-length capped RNA transcripts for in vitro viability studies, the wild-type pSHEV-3 infectious cDNA clone and the three single mutant clones rF51L, rT59A, and rS390L were each linearized with XbaI. Linearized plasmids were extracted with phenol-chloroform, followed by ethanol precipitation. Capped RNA transcripts were synthesized by using a T7 riboprobe in vitro transcription system (Promega) as previously described (11). Each reaction was performed in a final volume of 25 μl containing 2.5 μg of linearized cDNA template, 5 μl of 5× transcription buffer, 2.5 μl of 10 mM dithiothreitol, 1 μl of 40 U/ml RNasin, 2.5 μl of nucleoside triphosphates (5 mM each ATP, CTP, and UTP and 0.5 mM GTP), 2.5 μl of 5 mM m7(3′-O-methyl)-G(5′)ppp(5′)G (Ambion), and 1 μl of 20-U/ml T7 polymerase. The reaction mixture was incubated for 1.5 h at 37°C. To evaluate the integrity and quantity of the RNA transcripts, a 2-μl aliquot of the transcription reaction mixture was analyzed in a 1% agarose gel. Transcription mixtures were cooled on ice and directly used for in vitro transfection of Huh7 cells to determine the replication competency of the wild-type and mutant viruses.
For the study of pathogenicity in pigs, capped RNA transcripts were synthesized using the mMessage mMachine T7 kit (Ambion) as described previously (24). Transcription reactions were performed for the wild-type pSHEV-3, the triple mutant pSHEV-1, and each of the three single mutant (rF51L, rT59A, and rS390L) infectious clones in a 200-μl reaction mixture containing 10 μg of linearized DNA plasmid, 20 μl of 10× buffer, 100 μl of 2× nucleoside triphosphate-Cap, 20 μl of enzyme mix, and an additional 10 μl of 30 mM GTP. After 1.5 h of incubation at 37°C, the RNA transcripts for each clone were pooled. The integrity of the RNA transcripts was determined by gel electrophoresis in a 1% agarose gel and their concentration measured with a NanoDrop DN-1000 instrument (Thermo Scientific). The capped RNA transcripts for each clone were diluted with 3 volumes of cold RNase-DNase-proteinase-free phosphate-buffered saline (PBS) (6 ml), divided into aliquots of 0.8 ml, immediately frozen on dry ice, and used for direct intrahepatic inoculation of pigs the next day.
In vitro transfection and immunofluorescence assay (IFA).
Huh7 cells were transfected with similar amounts of full-length capped RNA transcripts from the wild-type pSHEV-3 clone and each of the three single mutant clones rF51L, rT59A, and rS390L in order to determine their replication competencies in vitro. Monolayers of Huh7 cells were washed with Opti-MEM (Gibco) and transfected with RNA transcripts with DMRIE-C (Invitrogen) as previously described (11). Briefly, 25 μl of the transcription mixture was diluted in 1 ml of Opti-MEM containing 20 μl of DMRIE-C reagent, and the mixture was added to a T25 flask containing cells at 40 to 50% confluence. After incubation for 5 h at 34.5°C, the transfection mixture was replaced with fresh growth medium supplemented with 10% fetal bovine serum, and the cultures were maintained at 34.5°C. On day 3 posttransfection, cells were trypsinized, plated in eight-well LabTek chamber slides, and grown for an additional 3 days at 34.5°C. On day 6 posttransfection, cells were fixed with acetone for 2 min, washed with PBS, and incubated at room temperature for 30 min with a chimpanzee 1313 anti-HEV antiserum diluted 1:200 in a dilution buffer (0.5% bovine serum albumin [BSA], 0.5% milk, and 0.1% Triton X-100 in PBS). This chimpanzee anti-HEV antiserum was used because it has a very high titer and was shown to be specific for the HEV ORF2 protein (14, 24). Cells were washed with PBS and incubated with 1:2,000-diluted Alexa Fluor 488-conjugated goat anti-human IgG (Molecular Probes) for 30 min at room temperature. Slides were washed with PBS, covered with Vectashield fluorescent mounting medium (Vector Laboratories), and viewed under a fluorescence microscope (Nikon Eclipse TE300).
Experimental design for pathogenicity study of the HEV mutants in SPF pigs.
The study protocol was approved by the Institutional Animal Care and Use Committee (IACUC) of Iowa State University (approval number 3-09-6703-S) as well as by the IACUC of Virginia Polytechnic Institute and State University (approval number 09-058-CVM). Sixty 3-week-old, specific-pathogen-free (SPF) pigs were distributed randomly into six groups (groups A, B, C, D, E, and F) of 10 pigs each. Each group of pigs was housed separately in a biosafety level 2 (BSL2) facility. Prior to inoculation, all pigs tested negative for IgG anti-HEV antibodies by an enzyme-linked immunosorbent assay (ELISA) (43–45). Capped RNA transcripts were thawed and immediately inoculated by an ultrasound-guided percutaneous intrahepatic injection procedure into four different sites of the liver, with approximately 200 μl of capped RNA transcripts per injection site. Group A pigs were each injected intrahepatically with 0.8 ml of the capped RNA transcripts from the wild-type pSHEV-3 clone and served as positive controls. Pigs in groups B, C, and D were each injected intrahepatically with 0.8 ml of the capped RNA transcripts from the HEV single mutant clones rF51L, rT59A, and rS390L, respectively. Group E pigs were each similarly injected with 0.8 ml of the capped RNA transcripts from the triple mutant pSHEV-1 clone, and pigs in group F were injected with PBS and served as negative controls. All pigs were monitored daily for clinical signs of disease for a total of 8 weeks.
Serum samples and fecal swabs were collected prior to inoculation and weekly thereafter from each pig and were tested for the presence and the amount of HEV RNA. Serum samples were also tested for the presence of HEV antibodies. In order to evaluate and compare the levels of replication of different HEV mutants during the acute stages of infection, three pigs from each group were necropsied at 21 and 28 days postinoculation (dpi), and the remaining four pigs from each group were necropsied at 56 dpi. The viral loads (amounts of HEV RNA) in the bile, liver tissues, and intestinal content collected at each necropsy were quantified as well.
Serology.
Weekly serum samples were tested for IgG anti-HEV by an ELISA essentially as described previously (43–45). Briefly, a truncated recombinant ORF2 capsid protein containing the HEV immunodominant region from aa 452 to 617 (GenWay Biotech Inc., San Diego, CA) was used as the antigen. Horseradish peroxidase (HRP)-conjugated goat anti-swine IgG (Sigma) was used as the secondary antibody. Preimmune and hyperimmune anti-HEV-positive swine sera were included as negative and positive controls, respectively.
RNA extraction, cDNA synthesis, and nested reverse transcription-PCR (RT-PCR).
Samples of liver tissues (500 mg) collected at each necropsy were homogenized in 10% (wt/vol) sterile PBS. Homogenates were centrifuged at 3,000 rpm (Eppendorf centrifuge 5810) at 4°C for 15 min, and supernatants were used for the detection and quantification of HEV RNA. Intestinal content collected during each necropsy and weekly fecal swab materials from each pig were resuspended in PBS to make a 10% fecal suspension. Total RNA was extracted from 200 μl of serum, fecal suspension, liver homogenate, bile, and intestinal content samples using TRIzol reagent (GIBCO-BRL) and was resuspended in 30 μl of DNase-, RNase-, and proteinase-free water (Eppendorf Inc.). Synthesis of cDNA was performed for 60 min at 42°C using 1 μl of 10 μM R1 reverse primer (5′-CTACAGAGCGCCAGCCTTGATTGC-3′), 0.25 μl (20 U/μl) of Superscript II reverse transcriptase (Invitrogen), 1 μl of 0.1 M dithiothreitol, 4 μl of 5× RT buffer, 0.5 μl (40 U/μl) of RNase inhibitor (Promega), and 1 μl of 10 mM deoxynucleoside triphosphates (dNTPs).
A nested RT-PCR was used to detect HEV RNA in serum, fecal, bile, and liver samples. Primers used for RT-PCR were designed within the ORF2 capsid gene. Briefly, 5 μl of cDNA was amplified in a 50-μl volume in a nested PCR with AmpliTaq gold DNA polymerase (Applied Biosystems). The first round of PCR was performed using F1 (forward, 5′-AGCTCCTGTACCTGATGTTGACTC-3′) and R1 (reverse, 5′-CTACAGAGCGCCAGCCTTGATTGC-3′) primers to produce an expected fragment of 404 bp. A second round of PCR was carried out in the presence of the F2 (5′-GCTCACGTCATCTGTCGCTGCTGG-3′) and R2 (5′-GGGCTGAACCAAAATCCTGACATC-3′) primers to generate a product of 266 bp, using 5 μl of the first-round PCR products as the template. The PCR parameters for both rounds included an initial incubation at 95°C for 9 min, followed by 40 cycles of denaturation at 94°C for 1 min, annealing at 52°C for 1 min, and extension at 72°C for 1.5 min, with a final incubation at 72°C for 7 min. Amplified PCR products were analyzed by gel electrophoresis.
Quantification of HEV RNA by qRT-PCR.
All serum, liver tissue homogenate, bile, and intestinal content samples that were tested positive by the nested RT-PCR assay were retested by quantitative RT-PCR (qRT-PCR) to quantify the amounts of HEV RNA. Primers for the qRT-PCR were designed using the Beacon designer software (Premier Biosoft) to produce a 93-bp fragment within the ORF2 gene (HEV-forward, 5′-TTGGTGGCTATGCTATCT-3′; HEV-reverse, 5′-TGACATCAGTGGAGGTAA-3′). The HEV RNA standards were generated from the linearized clone pSHEV-3 using the mMessage mMachine T7 kit (Ambion) according to the manufacturer's protocol. Synthesized RNA transcripts were treated with 1 μl (2 U/μl) of Turbo DNase (Applied Biosystems) to remove the template DNA. The integrity of the RNA transcripts was confirmed by gel electrophoresis, and the RNA concentration was measured using a NanoDrop ND-1000 instrument (Thermo Scientific). Serial 10-fold dilutions of the in vitro-synthesized HEV RNA transcripts were used for determining the qRT-PCR sensitivity and detection limit. The qRT-PCR was performed in a 25-μl reaction mixture containing 12.5 μl of SensiMix SYBR & Fluorescein (Quantace Ltd.), 0.5 μl of each primer (10 μM), and 5 μl of cDNA template. The qRT-PCR parameters consisted of an initial PCR activation step of 95°C for 10 min followed by 40 cycles of denaturation at 95°C for 15 s, annealing at 59°C for 30 s, and extension at 72°C for 15 s. Amplification and data analysis were carried out on an iCycler thermal cycler (Bio-Rad). Samples were tested in triplicate, and a negative control with no template was included in every set of reactions.
Nucleotide sequencing of the viruses recovered from pigs inoculated with the HEV mutants.
Samples of bile, serum, feces, and intestinal content from selected pigs of each group inoculated with HEV mutants rF51L, rT59A, and rS390L were amplified by RT-PCR. The PCR products of the ORF2 gene region were subsequently sequenced to confirm that the viruses recovered from pigs originated from their respective inoculum. For the HEV mutants rF51L and rT59A, the reverse primer 5159R1 (5′-GGGCTGAACCAAAATCCTGACATC-3′) was used for cDNA synthesis, followed by a first-round PCR using the forward 5159F1 (5′-ATAACATGTCTTTTGCATCGCC-3′) and reverse 5159R1 primers and a second round of nested PCR using the forward 5159F2 (5′-CGTGCTTCTGCCTATGCTGC-3′) and reverse 5159R2 (5′-TCACGCCAAGCGGAGCCAAGG-3′) primers. For the HEV mutant rS390L, the reverse primer 390R1 (5′-GAGTAGGTCGGTCTTGCTCG-3′) was used for cDNA synthesis. The first-round PCR was performed using the forward 390F1 (5′-CAGCCACACGTTTCATGAAGG-3′) and reverse 390R1 primers, followed by a second round of nested PCR using the forward 390F2 (5′-CTTGCATTTCACCGGCACGAAC-3′) and reverse 390R2 (5′-GAATCACCCAGATCTATATCG-3′) primers.
Structural visualization of S390L mutant capsid.
All molecular graphic figures were produced with PyMOL 0.99 molecular graphics (7; http://www.pymol.org) on a Linux workstation. Coordinates were downloaded from Protein Data Bank (PDB) accession number 2ZTN (chain A, residues 129 to 606) and correspond to the X-ray structure of the genotype 3 HEV ORF2 (69). To build a backbone model from the alignment of the wild-type and mutant (S390L) forms of ORF2, we used the AL2TS service of the Protein Structure Prediction Center (Lawrence Livermore National Laboratory), which converts sequence structure alignment into standard PDB format.
Statistical analysis.
RT-PCR data (presence versus absence of HEV RNA) were analyzed using Fisher's exact test to simultaneously compare all the groups as well as for the two-way comparisons of interest. P values for the two-way comparisons after Fisher's exact test were adjusted using Bonferroni's procedure. Statistical significance was set to alpha = 0.05. All analyses were performed using commercially available software (version 9.2; SAS, Cary, NC).
RESULTS
The three HEV single mutants rF51L, rT59A, and rS390L are replication competent in Huh7 liver cells.
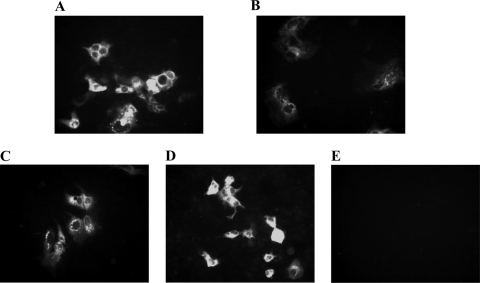
Capped RNA transcripts from the full-length cDNA clones of each of the three HEV single mutants (rF51L, rT59A, and rS390L) were transfected into Huh7 cells to determine their replication competence. The RNA transcripts from the wild-type pSHEV-3 clone were also included as a positive control. The viability of the triple mutant pSHEV-1 has been demonstrated in vitro previously (24) and thus was not tested in this study. The ORF2 viral antigens were detected by IFA in Huh7 cells transfected with the wild-type pSHEV-3, as well as in cells transfected with all three single mutants (rF51L, rT59A, and rS390L) (Fig. 1A, B, C, and D). A positive IFA signal was seen in the cytoplasm of Huh7 cells, whereas no detectable signal was found in mock-transfected cells (Fig. 1E). Since ORF2 protein is synthesized from subgenomic RNAs, production of ORF2 protein serves as a surrogate marker for viral RNA replication. Therefore, the number of positive cells was quantified by fluorescence-activated cell sorter (FACS) analysis of transfected cells stained for HEV ORF2 protein as previously described (56). No major differences in the production of ORF2 protein were observed between cells transfected with the mutants and the wild type. The mean percentage and standard deviation of ORF2-positive cells for the wild-type pSHEV-3 were 2.9% ± 2.2%, whereas the values for the single mutants rF51L, rT59A, and rS390L and the triple mutant pSHEV-1 were 4.2% ± 0.43%, 2.9% ± 0.40%, 1.9% ± 1%, and 2.4% ± 2.1%, respectively.
Fig. 1.
Immunofluorescence staining of a subclone of Huh7 cells transfected with similar amounts of capped full-length RNA transcripts. (A) pSHEV-3; (B) mutant rF51L; (C) mutant rT59A; (D) mutant rS390L; (E) mock transfection. Cells were stained for HEV ORF2 protein using chimpanzee 1313 anti-HEV immune serum.
Seroconversion to anti-HEV antibodies is delayed in SPF pigs infected with each of the three HEV single mutants compared to those infected with the wild-type pSHEV-3.
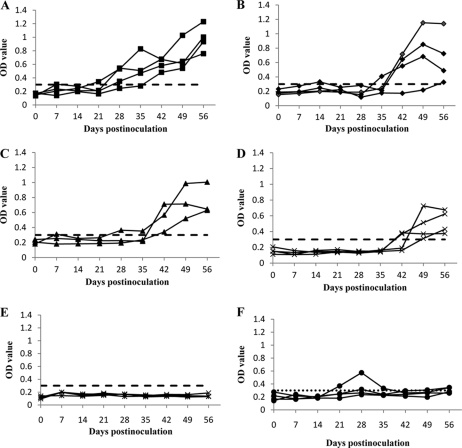
Seroconversion to IgG anti-HEV was observed in all three HEV single mutant groups as well as in the positive-control group (Fig. 2), indicating active HEV infection in the inoculated pigs. Pigs inoculated with the single mutants rF51L, rT59A, and rS390L had a 1- to 3-week-delayed seroconversion (Fig. 2B, C, and D) compared to those infected with the wild type (Fig. 2A), while pigs inoculated with the triple mutant pSHEV-1 failed to seroconvert by 56 dpi (Fig. 2E). As expected, all pigs from group F (PBS negative control) remained seronegative throughout the study, except for one pig at 21 to 35 dpi, which exhibited positive optical density (OD) values (Fig. 2F). Since this pig was negative for HEV RNA throughout the study and since the low OD values lasted for only 3 weeks, the transient elevation of OD values in this single pig is likely due to an unexplained nonspecific response.
Fig. 2.
Seroconversion to anti-HEV IgG in pigs inoculated with RNA transcripts from the wild-type pSHEV-3 and HEV mutants. (A) Positive-control pigs inoculated with the wild-type pSHEV-3; (B) pigs inoculated with the single mutant rF51L; (C) pigs inoculated with the single mutant rT59A (one pig from this group died of septicemia at 6 days after inoculation); (D) pigs inoculated with the single mutant rS390L; (E) pigs inoculated with the triple mutant pSHEV-1; (F) negative-control pigs inoculated with PBS. Only results for those pigs necropsied at 56 dpi are shown here. Anti-HEV IgG was plotted as the ELISA optical density (A405), and the ELISA cutoff value was set 0.3 as previously described (15, 24, 45).
Pigs inoculated with HEV mutants have delayed and shorter viremia and fecal virus shedding and lower serum viral loads than those inoculated with wild-type pSHEV-3.
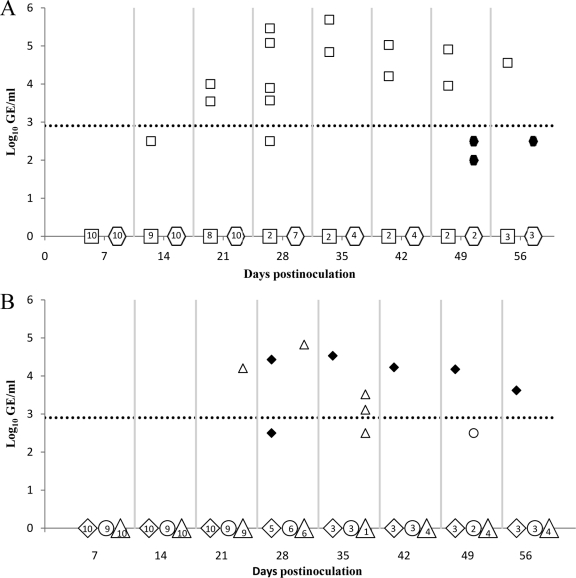
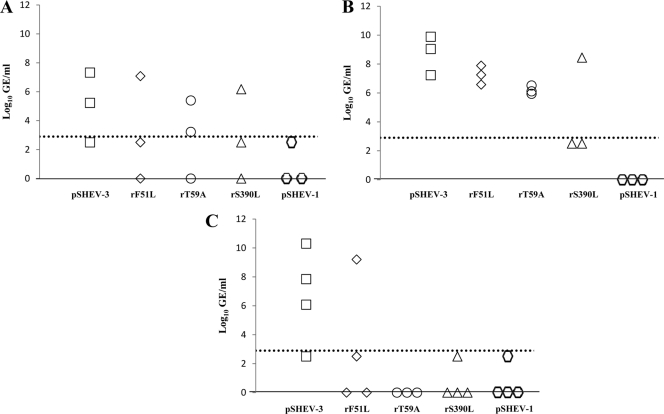
Weekly serum samples from all pigs were tested for the presence of HEV viremia (Table 1) and serum viral RNA loads (Fig. 3) by nested RT-PCR and quantitative real-time RT-PCR (qRT-PCR), respectively. Pigs inoculated with PBS showed no detectable HEV RNA, while those inoculated with the wild-type pSHEV-3 had viremia starting at 14 dpi, with an average duration of continuous viremia of 3.2 weeks (Table 1), and serum viral loads ranging from 3.5 to 5.7 log10 genomic equivalent copies (GE) per ml (Fig. 3A). Pigs inoculated with the mutant viruses had a delayed and transient viremia that was of generally lower magnitude than that in the wild-type-virus-inoculated pigs (Table 1; Fig. 3A and B).
Table 1.
HEV RNA in feces and sera of specific-pathogen-free pigs intrahepatically inoculated with capped RNA transcripts from wild-type pSHEV-3 and mutant rF51L, rT59A, rS390L, and pSHEV-1 infectious clones
| Group | Inoculum | Sample | No. of positive pigs/no. tested at dpi: |
Total no. positive/no. tested | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 7 | 14 | 21 | 28 | 35 | 42 | 49 | 56 | ||||
| A | pSHEV-3 | Feces | 0/10 | 3/10 | 8/10 | 10/10 | 7/7 | 4/4 | 4/4 | 4/4 | 4/4 | 10/10 |
| Serum | 0/10 | 0/10 | 1/10 | 2/10 | 6/7 | 2/4 | 2/4 | 2/4 | 1/4 | 7/10 | ||
| B | rF51L | Feces | 0/10 | 2/10 | 2/10 | 5/10 | 5/7 | 4/4 | 3/4 | 3/4 | 4/4 | 7/10 |
| Serum | 0/10 | 0/10 | 0/10 | 0/10 | 2/7 | 1/4 | 1/4 | 1/4 | 1/4 | 2/10 | ||
| C | rT59A | Feces | 0/10 | 0/9 | 0/9a | 3/9b | 5/6 | 3/3 | 0/3 | 0/3 | 0/3 | 8/10 |
| Serum | 0/10 | 0/9 | 0/9 | 0/9 | 0/6d | 0/3 | 0/3 | 0/3 | 1/3 | 1/10 | ||
| D | rS390L | Feces | 0/10 | 0/10 | 0/10a | 3/10b | 7/7 | 4/4 | 4/4 | 0/4 | 0/4 | 8/10 |
| Serum | 0/10 | 0/10 | 0/10 | 1/10 | 1/7 | 3/4 | 0/4 | 0/4 | 0/4 | 5/10 | ||
| E | pSHEV-1 | Feces | 0/10 | 0/10 | 0/10a | 1/10b | 1/7 | 0/4 | 0/4 | 0/4 | 0/4 | 1/10 |
| Serum | 0/10 | 0/10 | 0/10 | 0/10 | 0/7d | 0/4 | 0/4 | 2/4 | 1/4 | 2/10 | ||
| F | PBS | Feces | 0/10 | 0/10 | 0/10a | 0/10b | 0/7c | 0/4 | 0/4 | 0/4 | 0/4 | 0/10 |
| Serum | 0/10 | 0/10 | 0/10 | 0/10 | 0/7 | 0/4 | 0/4 | 0/4 | 0/4 | 0/10 | ||
Group with a statistically significant difference in the incidence of fecal virus shedding compared to the wild-type group at 14 dpi (P < 0.05).
Group with a statistically significant difference in the incidence of fecal virus shedding compared to the wild-type group at 21 dpi (P < 0.05).
Group with a statistically significant difference in the incidence of fecal virus shedding compared to the wild-type group at 28 dpi (P < 0.05).
Group with a statistically significant difference in the incidence of viremia compared to the wild-type group at 21 dpi (P < 0.05).
Fig. 3.
Detection and quantification of viral RNA loads in sera of pigs inoculated with the wild-type pSHEV-3 and HEV mutants at the different days postinoculation (dpi). (A) HEV genomic equivalent (GE) titers in sera from pigs inoculated with wild-type pSHEV-3 (□) and triple mutant pSHEV-1 ( ). (B) HEV GE titers in sera from pigs inoculated with the single mutants rF51L (♦), rT59A (○), and rS390L (▵). The numbers inside the symbol on the x axis represent the number of pigs in each group that were negative for viremia at the respective dpi. The HEV genomic copy numbers are presented as the log10 of GE per ml of serum (y axis). The dotted line represents the limit of the sensitivity of the qRT-PCR assay (2.90 log10 GE/ml).
). (B) HEV GE titers in sera from pigs inoculated with the single mutants rF51L (♦), rT59A (○), and rS390L (▵). The numbers inside the symbol on the x axis represent the number of pigs in each group that were negative for viremia at the respective dpi. The HEV genomic copy numbers are presented as the log10 of GE per ml of serum (y axis). The dotted line represents the limit of the sensitivity of the qRT-PCR assay (2.90 log10 GE/ml).
Weekly fecal samples were collected from all pigs and tested for the presence of HEV RNA by nested RT-PCR (Table 1). All fecal samples collected from PBS-inoculated pigs were negative for HEV RNA. Pigs inoculated with wild-type pSHEV-3 or mutant rF51L shed virus in feces as early as 7 dpi, and the average durations of continuous fecal viral shedding were 7.8 and 5.2 weeks, respectively (Table 1). Pigs inoculated with mutant rT59A or rS390L shed virus in feces by 21 dpi, and the average durations of continuous fecal virus shedding were reduced to 1.7 and 3.2 weeks, respectively (Table 1). Only one of the pigs inoculated with the triple mutant pSHEV-1 had detectable HEV RNA in feces at 21 and 28 dpi (Table 1).
Pigs inoculated with HEV mutants have lower viral loads in liver tissues, bile, and intestinal content collected from each necropsy than those inoculated with the wild-type pSHEV-3.
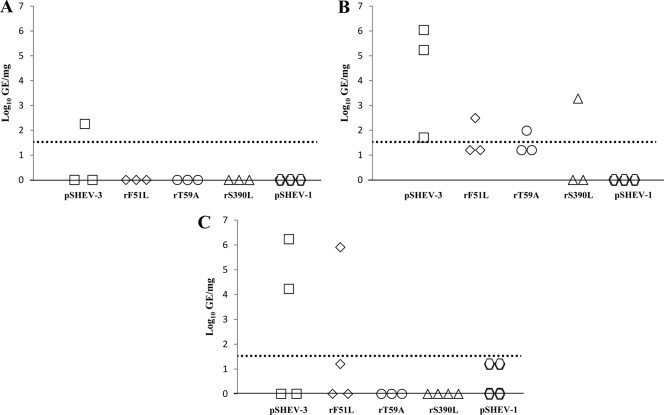
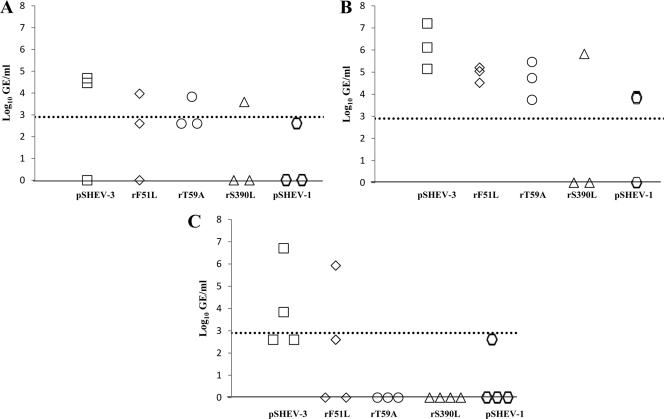
Pigs inoculated with PBS were negative for HEV RNA in liver, bile, and intestinal content collected during all three different necropsies at 21, 28, and 56 dpi. HEV RNA was detected in the livers of 1/3, 3/3, and 2/4 wild-type-virus-inoculated pigs at 21, 28, and 56 dpi, respectively (Fig. 4A, B, and C). Pigs inoculated with each of the single mutants (rF51L, rT59A, and rS390L) had detectable HEV RNA in the liver, with relatively lower viral GE titers at 28 dpi (Fig. 4B). HEV RNA was detected in 2/4 pigs inoculated with the wild type, rF51L, and pSHEV-1 at 56 dpi, though the titers of the mutant viruses were generally lower than that of the wild-type virus (Fig. 4C).
Fig. 4.
Detection and quantification of viral RNA loads in livers of pigs inoculated with the wild-type pSHEV-3 and HEV mutants. HEV GE titers were determined from samples harvested at necropsies on days 21 (A), 28 (B), and 56 (C). Data are shown as log10 of GE per mg of liver. The dotted line represents the limit of the sensitivity of the qRT-PCR assay (1.53 log10 GE/mg).
Viral RNA was detected in bile samples collected from each group of infected pigs at 21 dpi, with lower titers observed in triple mutant pSHEV-1-infected pigs (Fig. 5A). Viral RNA was consistently detected in the wild-type- and single mutant-infected pigs at 28 dpi, with no viral RNA detected in the triple mutant-infected pigs (Fig. 5B). At the necropsy at 56 dpi, high levels of viral RNA were still detectable in the bile of pigs infected with the wild-type virus, but only low levels of virus were detectable in the bile of most pigs infected with the single and triple mutants (Fig. 5C).
Fig. 5.
Detection and quantification of viral RNA loads in bile samples from pigs inoculated with the wild-type pSHEV-3 and HEV mutants. HEV GE titers were determined from samples harvested at necropsies on days 21 (A), 28 (B), and 56 (C). Data are shown as log10 of GE per ml of bile. The dotted line represents the limit of the sensitivity of the qRT-PCR assay (2.90 log10 GE/ml).
Viral RNA was detected in intestinal content collected from each group of infected pigs at 21 and 28 dpi (Fig. 6A and B). At 56 dpi, HEV RNA was detected consistently in the intestinal contents of wild-type virus-infected pigs and also in 2/4 and 1/4 of the pigs inoculated with rF51L and pSHEV-1, respectively (Fig. 6C). No pigs inoculated with mutant rT59A or rS390L had detectable viral RNA in the intestinal content at 56 dpi (Fig. 6C).
Fig. 6.
Detection and quantification of viral RNA loads in intestinal contents of pigs inoculated with the wild-type pSHEV-3 or HEV mutants. HEV GE titers were determined from samples harvested at necropsies on days 21 (A), 28 (B), and 56 (C). Data are shown as log10 of GE per ml of intestinal content suspension. The dotted line represents the limit of the sensitivity of the qRT-PCR assay (2.90 log10 GE/ml).
Mutations F51L and T59A are genetically stable in vivo, whereas mutation S390L reverts to the wild-type phenotype at later stages of infection in pigs.
The viruses recovered from pigs inoculated with the HEV mutants rF51L, rT59A, and rS390L were partially sequenced to determine the in vivo stabilities of the three single mutations introduced in the viral capsid protein. Sequence analyses revealed that mutation F51L was retained in the viruses recovered from feces, bile, and intestinal contents collected at 28 and 56 dpi from pigs inoculated with the mutant rF51L. Similarly, the mutation T59A was also retained in the viruses recovered from bile and intestinal contents collected at 28 dpi from pigs inoculated with the mutant rT59A. However, sequence analyses of viruses recovered from feces, serum, intestinal contents, and bile collected at 21, 28, 35, and 42 dpi from six different pigs inoculated with the single mutant rS390L revealed a mixture of both the mutant and the wild-type phenotypes. The revertant population was detected as early as 21 dpi and increasingly became the predominant phenotype over the course of infection, with no detection of the mutant rS390L in some viruses recovered from pigs at 42 dpi.
3-D structural location of the S390L mutation in the viral capsid.
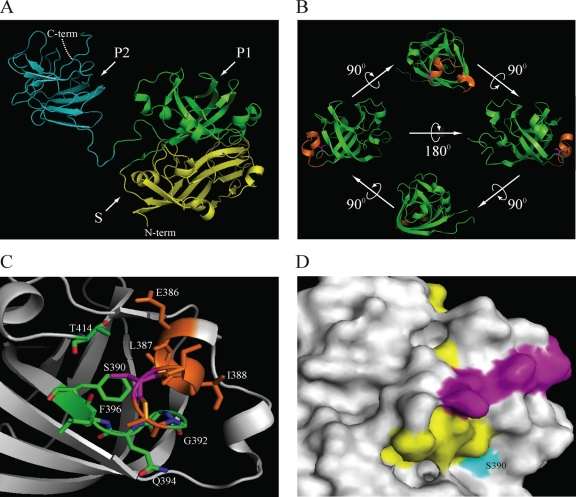
Three-dimensional (3-D) modeling of the HEV capsid structure revealed that the S390L mutation is localized within a putative sugar binding sequence (376ADTLLGGLPTELISSA391) within the P1 linear domain (residues 314 to 453) (20) of the C terminus of the capsid protein that is strictly conserved among all four HEV genotypes. This potential sialic acid binding site maps to a helix-turn-helix motif (Fig. 7) and forms a hidden pocket that may be responsible for binding to cell receptors (20).
Fig. 7.
Structural location of S390 within the capsid protein of HEV. (A) Crystal structure of the truncated HEV capsid protein (residues 129 to 606) (PDB accession no. 2ZTN) (69). Domains are indicated as follow: shell (S) (residues 118 to 313) in yellow and subdomains P1 (residues 314 to 453) in green and P2 (residues 454 to 606) in cyan of the protruding (P) domain. (B) Ribbon diagram of the ternary P1 domain, showing the putative glycosylation site (376ADTLLGGLPTELISSA391, in orange). The S390 residue is shown in magenta and maps within a helix-turn-helix motif. (C) Schematic presenting the residues within a 4-Å distance from S390. Those residues that are part of the putative glycosylation site are in orange (386ELISSA391). Residues in green are part of the loop and β-sheet and comprise residues 392GGQLF396. The side chain of an additional residue, T414, located within a 4-Å distance is also indicated. (D) Surface map showing the location of residue S390 and linear epitopes of the 12A10 and 16D7 (purple) and 15B2 (yellow) monoclonal antibodies (23). Images were generated using PyMOL (7).
DISCUSSION
In this study, we report the identification of three amino acid mutations in the HEV capsid protein that collectively contributed to virus attenuation in vivo. The three amino acid mutations F51L, T59A, and S390L in the ORF2 capsid were identified in the genome of a genotype 3 HEV infectious clone (pSHEV-1) from a previous study (24). ORF2 partially overlaps ORF3; however, the three mutations in the ORF2 capsid do not change the amino acid sequence of the ORF3 protein. In this study, three HEV mutants with a single amino acid change (rF51L, rT59A, and rS390L) were constructed, and all were shown to be replication competent and viable after transfection into a subclone of Huh7 human liver cells. No major difference was observed in the production of HEV ORF2 protein in cells transfected with the wild type and the mutants. The pig model system has been used to understand various aspects of HEV replication, pathogenesis, and cross-species infection (16, 44, 45, 51, 66). Due to limited resources and restricted procedures for nonhuman primates, it is not possible to use a large number of nonhuman primates for HEV pathogenicity studies. Therefore, we conducted a pathogenicity study of these three single mutants using a large number of SPF pigs in each group (n = 10). We showed that in the positive-control pigs inoculated with the wild-type pSHEV-3, fecal virus shedding preceded the onset of viremia, and the patterns of viremia and fecal virus shedding in pSHEV-3-infected pigs were consistent with those observed in previous studies (21, 43). An IgG anti-HEV response was detected 1 to 2 weeks after the onset of viremia and remained detectable until the end of the 8-week study. Peak viral loads in serum, liver, bile, and intestinal content were detected at 28 dpi, with a temporal decrease of viral genome titers concurrent with the increase of HEV antibodies. In pigs inoculated with the wild-type pSHEV-3, the virus was not fully cleared by the immune response by 56 dpi, as evidenced by continuous fecal shedding in all pigs and the presence of high viral loads in liver, bile, and intestinal content at the end of the 8-week study.
In a previous pilot study with only two pigs (24), we demonstrated the viability of the triple mutant pSHEV-1 in vivo. In this study, we confirmed that the triple mutant pSHEV-1 is infectious in vivo, as HEV RNA was detected in serum and fecal samples as well as in liver, bile, and intestinal contents of inoculated pigs. However, the pSHEV-1 with the triple mutations in the capsid protein was clearly attenuated compared to the wild-type pSHEV-3, as fewer pigs were viremic or shed virus in feces. For those pigs that did have detectable viremia and fecal virus shedding, viremia was delayed by 5 weeks, serum viral loads were drastically lower, and fecal virus shedding was also delayed and had a shorter duration. In addition, none of the pigs inoculated with pSHEV-1 seroconverted by 56 dpi, which is consistent with our pilot study, in which the two pigs inoculated with pSHEV-1 did not seroconvert until 63 dpi (24).
To determine which of the three amino acid mutations were responsible for the observed attenuation of the pSHEV-1 virus in pigs, we constructed three HEV single mutants (rF51L, rT59A, and rS390L), each containing a single amino acid mutation, and compared the in vivo pathogenicities of the three HEV single mutants in pigs to those of each other and the wild-type pSHEV-3. For the rF51L mutant, seroconversion and viremia were delayed 2 to 3 weeks compared to those with the wild-type pSHEV-3, and the incidence of viremia was lower. However, the serum viral loads were similar for the rF51L mutant and the wild-type pSHEV-3, and like the wild-type pSHEV-3, the mutant rF51L virus was also not fully cleared in pigs at the end of the 8-week study, as evidenced by viremia and fecal virus shedding at 56 dpi. Inefficient viral clearance for the rF51L mutant-inoculated pigs was also observed in liver, bile, and intestinal content, although viral loads were lower than those in the pigs inoculated with wild-type pSHEV-3. The results suggest that the F51L mutation in the capsid protein only partially contributes to virus attenuation.
For the rT59A mutant, seroconversion in pigs was delayed 1 to 3 weeks. Viral RNA was detected in the liver, bile, intestinal content, and fecal materials at 21 to 35 dpi, but viremia was not detected in any of the rT59A-inoculated pigs until 56 dpi. Unlike the wild-type pSHEV-3 and mutant rF51L, the mutant rT59A virus was completely cleared from feces, liver, bile, and intestinal content at 56 dpi. Also, the viral loads in bile, liver, and intestinal contents collected at the necropsy at 28 dpi were lower than those of the wild-type pSHEV-3. Therefore, the T59A mutation in the capsid is important for virus attenuation in pigs.
For mutant rS390L, the inoculated pigs had a more drastic delay in seroconversion (3 to 4 weeks later than seroconversion with the wild-type pSHEV-3). Similarly, viremia and fecal virus shedding were also delayed compared to those with the wild-type pSHEV-3 virus. Among the three single mutants, mutant rS390L had the lowest viral loads in liver and bile of pigs necropsied at 21, 28, and 56 dpi. Pigs infected with mutant rS390L completely cleared the virus from serum and feces by 42 and 49 dpi, respectively, and the liver, bile, and intestinal content collected at the end of the 8-week study had no detectable virus. Therefore, the results indicate that the S390L mutation in the HEV capsid protein is critical for virus attenuation in pigs.
Mutations F51L and T59A are both located in the N-terminal region of the HEV capsid protein, which contains a signal peptide (aa 1 to 22) involved in the translocation of the protein across the endoplasmic reticulum membrane (72), followed by an arginine-rich domain (aa 23 to 111) that may be involved in RNA encapsidation (59, 72). Therefore, it is possible that the F51L and T59A mutations contribute to virus attenuation by causing a defect in viral genomic RNA packaging, thus resulting in a lower level of infectious virus production in vivo. The observed reduction of viral loads in bile, liver, and intestinal content and the lower incidence of viremia and fecal virus shedding with both mutants are consistent with this explanation. The T59A mutation appears to have a greater impact on viral replication than the F51L mutation, as no viremia was detected and viral loads and fecal virus shedding were greatly reduced. It is possible that the polarity change of the mutation T59A more drastically affects the function of the N-terminal domain. This would explain the observed difference in the level of attenuation between the two mutants, since the F51L mutation did not result in a polarity change.
The C terminus of the HEV capsid protein is the major structural domain responsible for virion assembly, immunogenicity, and host cell receptor binding (23, 30, 34, 39, 55). Constructs (aa 112 to 607) lacking the N terminus have been shown to self-assemble into virus-like particles (VLPs) in a baculovirus expression system (34), and a truncated C-terminal peptide, p239 (aa 368 to 606), of the HEV capsid protein induced protective immunity against HEV challenge in nonhuman primates (32). This p239 peptide has also been reported to bind and penetrate cell lines that are susceptible to HEV, suggesting the presence of HEV receptor binding sites in this C-terminal region (23). The crystal structure of genotype 3 HEV VLPs assembled from a truncated version of the ORF2 capsid protein (residues 129 to 606) was determined at 3.5-Å resolution (69). Three structural domains have been defined within ORF2: S (residues 118 to 313), P1 (residues 314 to 453), and P2 (residues 454 to 606). P1 and S are closely associated and exposed on the surface of the capsid around the icosahedral 3-fold axis, and P1 and P2 are connected through a flexible proline-rich hinge that spans residues 445 to 467 (69) (Fig. 7A). The S domain forms a continuous capsid shell, whereas P2 is highly exposed and is largely responsible for antigenicity determination and virus neutralization (20, 69). Although there is no direct evidence regarding glycosylation of the HEV VLP capsid, consensus sites have been located within the S (Asn137-Leu-Ser and Asn130-Leu-Thr) and P2 (Asn562-Thr-Thr) domains (69, 72), with evidence pointing toward a direct role of Asn137 and Asn130 in virus infectivity and virion assembly (49). Structural analysis of the P1 domain, an antiparallel β-barrel composed of six β-strands and four short helices (20, 69) (Fig. 7B), reveals the presence of a putative sugar binding site (376ADTLLGGLPTELISSA391) that resembles the one site identified in the endosialidase of the K1F phage (PDB accession no. 1VOE), which maps within a helix-turn-helix motif of P1 (Fig. 7B and C) and may be responsible for binding to cell receptors (20). Amino acid position 390, which is conserved among all four major genotypes of mammalian HEV, is localized within this potential sialic acid binding site in the P1 linear domain. Although the HEV dimer shows a crossing topology of the C-terminal P2 domain versus the P1 and S domains, stability of the dimer is ensured by the interaction between the P2 domain of one monomer and the P2 and P1 domains of the other (69), with the sugar binding site exposed in the opposite interface of P1 (Fig. 7D). Interestingly, He et al. showed that various monoclonal antibodies directed against conformational and linear epitopes in P1 and P2 were able to block the binding of a truncated peptide (p239, residues 368 to 606) of ORF2, which occurs as 23-nm particles, to various cell lines (23). Largely based on selective blocking by monoclonal antibodies, He et al. concluded that the region comprising residues 368 to 606 should contain at least two distinct sites, located separately in the monomeric and dimeric domains, involved in binding. Remarkably, two of the monoclonal antibodies against linear epitopes in P1 (residues 423 to 438 [antibody 12A10] and 423 to 443 [antibody Ab 16D7]) that neutralize the binding of p239 particles to Huh7 cells map on the same interface as Ser390 (23) (Fig. 7D), suggesting a potential site of contact within that region. Therefore, the mutation from a polar residue (Ser) to a nonpolar residue (Leu) at position 390 may prevent the rS390L mutant from efficiently interacting with the host cell receptor. Crystal structure studies have shown that this putative sugar binding motif may be responsible for destabilization of HEV capsid trimers during uncoating (20). Defects in receptor binding and uncoating would both result in a lower level of virus replication in the host and thus would explain why mutant rS390L appeared to contribute the most to virus attenuation among the three single HEV mutants.
It has been well documented that one or more amino acid changes in capsid or envelope proteins can contribute to the attenuation phenotype of viruses (8, 50, 52, 64, 65). Single point mutations in the capsids of poliovirus (65), murine norovirus (4), infectious bursal disease virus (64), and adeno-associated viruses (67) have all been demonstrated to have significant effects on both in vitro and in vivo growth characteristics of the viruses. Reversion of an attenuated phenotype to a pathogenic wild type has also been reported for many animal viruses, such as poliovirus (35), porcine reproductive and respiratory syndrome virus (48), and infectious bursal disease virus (54). In the current study, the F51L and T59A mutants are genetically stable in pigs, as the viruses recovered from the inoculated pigs over the course of infection all retained these two mutations. However, in pigs inoculated with the mutant rS390L, a mixture of both the wild-type and attenuated mutant phenotype viruses was recovered from feces, serum, bile, and intestinal content as early as 21 dpi. As the course of infection progressed, the wild-type phenotype virus population increasingly became predominant, and by 42 dpi, the rS390L mutant virus was no longer detectable in some infected pigs. The fact that the mutant rS390L had mutated back to the wild-type sequence at later stages of virus infection indicates a strong biological importance of this amino acid residue in the functionality of the HEV capsid protein. It took at least 21 days of replication in pigs before the wild-type phenotype population reached a level similar to that of the mutant virus population in infected pigs (data not shown). The lower populations of the wild-type virus, during the first few weeks of infection contribute to the observed attenuation in pigs inoculated with the mutant rS390L. However, in our previous pilot study with only two pigs (24), the three nonsilent mutations F51L, T59A, and S390L in the capsid gene of the triple mutant pSHEV-1 were retained in the genome for up to 70 dpi. Therefore, reversion to the wild-type phenotype appears to be greater for the single mutant (such as rS390L) than the triple mutant pSHEV-1, since only one nucleotide change is required to restore the original sequence. All three mutants (F51L, T59A, and S390L) are created by a single nucleotide change, and therefore it will be interesting to see in a future animal study if the attenuation phenotype of mutant S390L can be stabilized by changing two nucleotides of codon 390.
Taken together, the results from this study indicated that the three amino acid mutations in the HEV capsid protein collectively contribute to virus attenuation. The F51L mutation resulted in only partial attenuation, whereas the T59A and S390L mutations attenuated the virus more drastically. The significant attenuation for the triple mutant pSHEV-1 was likely due to the additive effect of the three individual amino acid mutations. Additional phenotypic markers of HEV attenuation need to be identified and characterized for the development of a modified live-attenuated vaccine when HEV can be efficiently propagated in vitro.
ACKNOWLEDGMENTS
We thank Stephen Werre of the Statistical Services Lab for assistance with statistical analysis of the data. We also thank Shayleen Schalk and Matt Umphress for assistance with sample collection and animal manipulations and Barbara Dryman for technical assistance.
This study was supported by grants from the National Institutes of Health (R01AI050611 and R01AI074667).
Footnotes
Published ahead of print on 30 March 2011.
REFERENCES
- 1. Agrawal S., Gupta D., Panda S. K. 2001. The 3′ end of hepatitis E virus (HEV) genome binds specifically to the viral RNA-dependent RNA polymerase (RdRp). Virology 282:87–101 [DOI] [PubMed] [Google Scholar]
- 2. Arankalle V. A., et al. 1994. Seroepidemiology of water-borne hepatitis in India and evidence for a third enterically-transmitted hepatitis agent. Proc. Natl. Acad. Sci. U. S. A. 91:3428–3432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arankalle V. A., Chobe L. P., Chadha M. S. 2006. Type-IV Indian swine HEV infects rhesus monkeys. J. Viral Hepat. 13:742–745 [DOI] [PubMed] [Google Scholar]
- 4. Bailey D., Thackray L. B., Goodfellow I. G. 2008. A single amino acid substitution in the murine norovirus capsid protein is sufficient for attenuation in vivo. J. Virol. 82:7725–7728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chandra V., Kalia M., Hajela K., Jameel S. 2010. The ORF3 protein of hepatitis E virus delays degradation of activated growth factor receptors by interacting with CIN85 and blocking formation of the Cbl-CIN85 complex. J. Virol. 84:3857–3867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chandra V., Kar-Roy A., Kumari S., Mayor S., Jameel S. 2008. The hepatitis E virus ORF3 protein modulates epidermal growth factor receptor trafficking, STAT3 translocation, and the acute-phase response. J. Virol. 82:7100–7110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. DeLano W. L. 2002. The PyMOL molecular graphics system. DeLano Scientific, San Carlos, CA [Google Scholar]
- 8. Dietzschold B., et al. 1983. Characterization of an antigenic determinant of the glycoprotein that correlates with pathogenicity of rabies virus. Proc. Natl. Acad. Sci. U. S. A. 80:70–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Emerson S. U., et al. 2004. Hepevirus. Elsevier/Academic Press, London, United Kingdom [Google Scholar]
- 10. Emerson S. U., et al. 2004. In vitro replication of hepatitis E virus (HEV) genomes and of an HEV replicon expressing green fluorescent protein. J. Virol. 78:4838–4846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Emerson S. U., Nguyen H., Torian U., Purcell R. H. 2006. ORF3 protein of hepatitis E virus is not required for replication, virion assembly, or infection of hepatoma cells in vitro. J. Virol. 80:10457–10464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Emerson S. U., et al. 2010. Release of genotype 1 hepatitis E virus from cultured hepatoma and polarized intestinal cells depends on open reading frame 3 protein and requires an intact PXXP motif. J. Virol. 84:9059–9069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Emerson S. U., Purcell R. H. 2003. Hepatitis E virus. Rev. Med. Virol. 13:145–154 [DOI] [PubMed] [Google Scholar]
- 14. Emerson S. U., et al. 2001. Recombinant hepatitis E virus genomes infectious for primates: importance of capping and discovery of a cis-reactive element. Proc. Natl. Acad. Sci. U. S. A. 98:15270–15275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Feagins A. R., et al. 2011. Intergenotypic chimeric hepatitis E viruses (HEV) with the genotype 4 human HEV capsid gene in the backbone of genotype 3 swine HEV are infectious in pigs. Virus Res. 156:141–146, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Feagins A. R., Opriessnig T., Huang Y. W., Halbur P. G., Meng X. J. 2008. Cross-species infection of specific-pathogen-free pigs by a genotype 4 strain of human hepatitis E virus. J. Med. Virol. 80:1379–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Reference deleted.
- 18. Graff J., Torian U., Nguyen H., Emerson S. U. 2006. A bicistronic subgenomic mRNA encodes both the ORF2 and ORF3 proteins of hepatitis E virus. J. Virol. 80:5919–5926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Graff J., et al. 2008. Mutations within potential glycosylation sites in the capsid protein of hepatitis E virus prevent the formation of infectious virus particles. J. Virol. 82:1185–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guu T. S., et al. 2009. Structure of the hepatitis E virus-like particle suggests mechanisms for virus assembly and receptor binding. Proc. Natl. Acad. Sci. U. S. A. 106:12992–12997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Halbur P. G., et al. 2001. Comparative pathogenesis of infection of pigs with hepatitis E viruses recovered from a pig and a human. J. Clin. Microbiol. 3:918–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Harrison T. J. 1999. Hepatitis E virus—an update. Liver 19:171–176 [DOI] [PubMed] [Google Scholar]
- 23. He S., et al. 2008. Putative receptor-binding sites of hepatitis E virus. J. Gen. Virol. 89:245–249 [DOI] [PubMed] [Google Scholar]
- 24. Huang Y. W., et al. 2005. Capped RNA transcripts of full-length cDNA clones of swine hepatitis E virus are replication competent when transfected into Huh7 cells and infectious when intrahepatically inoculated into pigs. J. Virol. 79:1552–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang Y. W., Opriessnig T., Halbur P. G., Meng X. J. 2007. Initiation at the third in-frame AUG codon of open reading frame 3 of the hepatitis E virus is essential for viral infectivity in vivo. J. Virol. 81:3018–3026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kabrane-Lazizi Y., Meng X. J., Purcell R. H., Emerson S. U. 1999. Evidence that the genomic RNA of hepatitis E virus is capped. J. Virol. 73:8848–8850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Karpe Y. A., Lole K. S. 2010. NTPase and 5′ to 3′ RNA duplex-unwinding activities of the hepatitis E virus helicase domain. J. Virol. 84:3595–3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Karpe Y. A., Lole K. S. 2010. RNA 5′-triphosphatase activity of the hepatitis E virus helicase domain. J. Virol. 84:9637–9641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Koonin E. V., et al. 1992. Computer-assisted assignment of functional domains in the nonstructural polyprotein of hepatitis E virus: delineation of an additional group of positive-strand RNA plant and animal viruses. Proc. Natl. Acad. Sci. U. S. A. 89:8259–8263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li F., et al. 2000. Recombinant subunit ORF2.1 antigen and induction of antibody against immunodominant epitopes in the hepatitis E virus capsid protein. J. Med. Virol. 60:379–386 [DOI] [PubMed] [Google Scholar]
- 31. Li S. W., et al. 2005. Mutational analysis of essential interactions involved in the assembly of hepatitis E virus capsid. J. Biol. Chem. 280:3400–3406 [DOI] [PubMed] [Google Scholar]
- 32. Li S. W., et al. 2005. A bacterially expressed particulate hepatitis E vaccine: antigenicity, immunogenicity and protectivity on primates. Vaccine 23:2893–2901 [DOI] [PubMed] [Google Scholar]
- 33. Li T. C., et al. 2005. Hepatitis E virus transmission from wild boar meat. Emerg. Infect. Dis. 11:1958–1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li T. C., et al. 1997. Expression and self-assembly of empty virus-like particles of hepatitis E virus. J. Virol. 71:7207–7213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Macadam A. J., et al. 1989. Reversion of the attenuated and temperature-sensitive phenotypes of the Sabin type 3 strain of poliovirus in vaccinees. Virology 172:408–414 [DOI] [PubMed] [Google Scholar]
- 36. Magden J., et al. 2001. Virus-specific mRNA capping enzyme encoded by hepatitis E virus. J. Virol. 75:6249–6255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Matsubayashi K., et al. 2004. Transfusion-transmitted hepatitis E caused by apparently indigenous hepatitis E virus strain in Hokkaido, Japan. Transfusion 44:934–940 [DOI] [PubMed] [Google Scholar]
- 38. Reference deleted.
- 39. Meng J., et al. 2001. Identification and characterization of the neutralization epitope(s) of the hepatitis E virus. Virology 288:203–211 [DOI] [PubMed] [Google Scholar]
- 40. Meng X. J. 2010. Hepatitis E virus: animal reservoirs and zoonotic risk. Vet. Microbiol. 140:256–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Meng X. J. 2010. Recent advances in hepatitis E virus. J. Viral Hepat. 17:153–161 [DOI] [PubMed] [Google Scholar]
- 42. Meng X. J. 2003. Swine hepatitis E virus: cross-species infection and risk in xenotransplantation. Curr. Top. Microbiol. Immunol. 278:185–216 [DOI] [PubMed] [Google Scholar]
- 43. Meng X. J., et al. 1998. Experimental infection of pigs with the newly identified swine hepatitis E virus (swine HEV), but not with human strains of HEV. Arch. Virol. 143:1405–1415 [DOI] [PubMed] [Google Scholar]
- 44. Meng X. J., et al. 1998. Genetic and experimental evidence for cross-species infection by swine hepatitis E virus. J. Virol. 72:9714–9721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Meng X. J., et al. 1997. A novel virus in swine is closely related to the human hepatitis E virus. Proc. Natl. Acad. Sci. U. S. A. 94:9860–9865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Moin S. M., Panteva M., Jameel S. 2007. The hepatitis E virus Orf3 protein protects cells from mitochondrial depolarization and death. J. Biol. Chem. 282:21124–21133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Navaneethan U., Al Mohajer M., Shata M. T. 2008. Hepatitis E and pregnancy: understanding the pathogenesis. Liver Int. 28:1190–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nielsen H. S., et al. 2001. Reversion of a live porcine reproductive and respiratory syndrome virus vaccine investigated by parallel mutations. J. Gen. Virol. 82:1263–1272 [DOI] [PubMed] [Google Scholar]
- 49. Okamoto H. 2007. Genetic variability and evolution of hepatitis E virus. Virus Res. 127:216–228 [DOI] [PubMed] [Google Scholar]
- 50. Polo J. M., Johnston R. E. 1990. Attenuating mutations in glycoproteins E1 and E2 of Sindbis virus produce a highly attenuated strain when combined in vitro. J. Virol. 64:4438–4444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pudupakam R. S., et al. 2009. Deletions of the hypervariable region (HVR) in open reading frame 1 of hepatitis E virus do not abolish virus infectivity: evidence for attenuation of HVR deletion mutants in vivo. J. Virol. 83:384–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pugachev K. V., Galinski M. S., Frey T. K. 2000. Infectious cDNA clone of the RA27/3 vaccine strain of rubella virus. Virology 273:189–197 [DOI] [PubMed] [Google Scholar]
- 53. Purcell R. H., Emerson S. U. 2001. Hepatitis E virus, p. 3051–3061 In Knipe D. M., Howley P. M. (ed.), Fields virology, 4th ed., vol. 2 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 54. Raue R., et al. 2004. Reversion of molecularly engineered, partially attenuated, very virulent infectious bursal disease virus during infection of commercial chickens. Avian Pathol. 33:181–189 [DOI] [PubMed] [Google Scholar]
- 55. Riddell M. A., Li F., Anderson D. A. 2000. Identification of immunodominant and conformational epitopes in the capsid protein of hepatitis E virus by using monoclonal antibodies. J. Virol. 74:8011–8017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shukla P., et al. 2011. Cross-species infections of cultured cells by hepatitis E virus and discovery of an infectious virus-host recombinant. Proc. Natl. Acad. Sci. U. S. A. 108:2438–2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Surjit M., Jameel S., Lal S. K. 2004. The ORF2 protein of hepatitis E virus binds the 5′ region of viral RNA. J. Virol. 78:320–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Takahashi M., et al. 2008. Monoclonal antibodies raised against the ORF3 protein of hepatitis E virus (HEV) can capture HEV particles in culture supernatant and serum but not those in feces. Arch. Virol. 153:1703–1713 [DOI] [PubMed] [Google Scholar]
- 59. Tam A. W., et al. 1991. Hepatitis E virus (HEV): molecular cloning and sequencing of the full-length viral genome. Virology 185:120–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Reference deleted.
- 61. Tyagi S., Korkaya H., Zafrullah M., Jameel S., Lal S. K. 2002. The phosphorylated form of the ORF3 protein of hepatitis E virus interacts with its non-glycosylated form of the major capsid protein, ORF2. J. Biol. Chem. 277:22759–22767 [DOI] [PubMed] [Google Scholar]
- 62. Tyagi S., Surjit M., Lal S. K. 2005. The 41-amino-acid C-terminal region of the hepatitis E virus ORF3 protein interacts with bikunin, a kunitz-type serine protease inhibitor. J. Virol. 79:12081–12087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tyagi S., Surjit M., Roy A. K., Jameel S., Lal S. K. 2004. The ORF3 protein of hepatitis E virus interacts with liver-specific alpha1-microglobulin and its precursor alpha1-microglobulin/bikunin precursor (AMBP) and expedites their export from the hepatocyte. J. Biol. Chem. 279:29308–29319 [DOI] [PubMed] [Google Scholar]
- 64. van Loon A. A., de Haas N., Zeyda I., Mundt E. 2002. Alteration of amino acids in VP2 of very virulent infectious bursal disease virus results in tissue culture adaptation and attenuation in chickens. J. Gen. Virol. 83:121–129 [DOI] [PubMed] [Google Scholar]
- 65. Westrop G. D., et al. 1989. Genetic basis of attenuation of the Sabin type 3 oral poliovirus vaccine. J. Virol. 63:1338–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Williams T. P., et al. 2001. Evidence of extrahepatic sites of replication of the hepatitis E virus in a swine model. J. Clin. Microbiol. 39:3040–3046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wu Z., et al. 2006. Single amino acid changes can influence titer, heparin binding, and tissue tropism in different adeno-associated virus serotypes. J. Virol. 80:11393–11397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Yamada K., et al. 2009. ORF3 protein of hepatitis E virus is essential for virion release from infected cells. J. Gen. Virol. 90:1880–1891 [DOI] [PubMed] [Google Scholar]
- 69. Yamashita T., et al. 2009. Biological and immunological characteristics of hepatitis E virus-like particles based on the crystal structure. Proc. Natl. Acad. Sci. U. S. A. 106:12986–12991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yazaki Y., et al. 2003. Sporadic acute or fulminant hepatitis E in Hokkaido, Japan, may be food-borne, as suggested by the presence of hepatitis E virus in pig liver as food. J. Gen. Virol. 84:2351–2357 [DOI] [PubMed] [Google Scholar]
- 71. Yoo D., et al. 2001. Prevalence of hepatitis E virus antibodies in Canadian swine herds and identification of a novel variant of swine hepatitis E virus. Clin. Diagn. Lab. Immunol. 8:1213–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zafrullah M., Ozdener M. H., Kumar R., Panda S. K., Jameel S. 1999. Mutational analysis of glycosylation, membrane translocation, and cell surface expression of the hepatitis E virus ORF2 protein. J. Virol. 73:4074–4082 [DOI] [PMC free article] [PubMed] [Google Scholar]