Abstract
The middle T (MT) antigen of polyomavirus has provided fundamental insights into the regulation of mammalian cell growth in vitro and important animal models for the analysis of tumor induction. The mouse mammary tumor virus (MMTV)-MT model of breast cancer has been important for probing the cellular signaling pathways in mammary tumorigenesis. MT itself has no intrinsic enzymatic activity but, rather, transforms by binding to and activating key intracellular signaling molecules, phosphatidylinositol 3-kinase (PI3-kinase) being the best studied of these. Thus, MT mimics a constitutively activated receptor tyrosine kinase (RTK). Our recent work suggests that MT signaling, like that of RTKs, is often quite dependent on cellular context in vitro. Here, we examine contextual effects on signaling in animal models as well. In this study, we generated transgenic mice in which MT is expressed in the mouse prostate under the control of an (ARR)2-Probasin promoter. All male transgenic mice displayed mouse prostatic intraepithelial neoplasia (mPIN) in the ventral and dorsal/lateral prostate as early as 8 weeks of age. Notably, during the course of tumor development over time, invasive cancer, reactive stroma, and infiltration of inflammatory cells were seen. Transcriptional profiling analyses show regulation of multiple pathways, with marked upregulation of both the NF-κB and inflammatory pathways. Comparison of expression profiles of our MT prostate model with those from an MMTV-MT breast model (23) shows both tissue-specific and tissue-independent MT effects. The signature of genes regulated by MT in a tissue-independent manner may have prognostic value.
INTRODUCTION
In the male population of the United States, prostate cancer is the second most commonly diagnosed cancer after dermatologic cancer. Typically developing slowly with age, it ranks second among all cancers as a cause of mortality in men in the United States. Considerable insight concerning the effects of genetic alterations on prostate tissue has been gleaned from a number of genetically engineered mouse models (GEMs). For example, the activation of receptor tyrosine kinase (RTK) pathways has been modeled via transgenic mice expressing Fgf8b (15, 56), Neu (34), and Erbb2 (6). Loss-of-function models for tumor suppressor genes include Nkx3.1 (30) and PTEN (11, 61, 63, 68). Models of oncogene expression include the expression of Akt (40), PIK3CB (32), c-myc (14), H-ras (52), and two models expressing simian virus 40 (SV40) large T antigen (the transgenic adenocarcinoma of mouse prostate [TRAMP] model [22] and the large probasin-large T antigen [LPB-Tag] model [41]). The resulting lesions vary according to the target gene, displaying histological abnormalities ranging from hyperplasia to metastatic cancer. Tumor development in most transgenic mice created via the expression of a single gene is limited to hyperplasia or mouse prostatic intraepithelial neoplasia (mPIN), while high levels of expression of myc and SV40 T antigen resulted in metastatic cancers (14, 22). Gene deletion of Nkx3.1 or PTEN did not result in metastatic adenocarcinoma (11, 30, 61, 63) except in one PTEN study (68). However, double knockout models, such as the deletion of PTEN together with p53 (9), p27 (10), or Nkx3.1 (31), develop more aggressive phenotypes, suggesting that multiple genetic hits may be required to accelerate prostate cancer progression initiated by PTEN loss.
Polyomavirus (PyV) middle T antigen (MT) is one of three early gene products encoded by the single early mRNA of this small DNA tumor virus (21, 51). It is the only protein encoded by the PyV genome that induces transformation in vitro. MT utilizes a hydrophobic C-terminal membrane anchor to associate with the cytoplasmic surface of the plasma membrane (and other membranes in contact with the cytoplasm) and subsequently binds and activates a subset of the src tyrosine kinase family (src, yes, and fyn), resulting in the phosphorylation of several key tyrosine residues on MT. These phosphorylated residues, along with adjacent amino acids, provide docking sites for cellular signaling molecules bearing SH2 or PTB domains, such as Tyr 250 for SHC (5, 12), Tyr 315 for phosphatidylinositol 3-kinase (PI3-kinase) (29, 71), and Tyr 322 for phospholipase C gamma (PLC-γ) (58). This phenomenon is highly similar to signal transduction triggered through RTKs that are activated by ligand binding, leading to autophosphorylation of several tyrosine residues on the activated receptor and subsequent activation of downstream signaling. Thus, MT has provided considerable insight into the mechanism by which constitutively activated receptor tyrosine kinases can direct cellular transformation. In particular, much of the original work on PI3-kinase signaling was done using MT as a model (51).
MT has been used to create mouse models to investigate several different types of cancers. The most extensively studied model is the mammary tumor model of Guy and colleagues. When expressed in the mammary gland under the control of the mouse mammary tumor virus long terminal repeat (MMTV-LTR), MT rapidly induces multifocal mammary adenocarcinomas and subsequently induces metastasis in 3 months in MMTV-MT634 mice (23). In addition, the roles of several signaling cascades associated with MT have been investigated. Mice with mutant forms of MT defective for binding to Shc (replacement of Tyr 250 by phenylalanine) and/or PI3-kinase (replacement of Tyr 315 by phenylalanine) exhibit significantly delayed tumor onset and do not develop metastatic cancer (70). Another well-characterized MT model is the pancreatic tumor model, which uses the RCAS-TVA system to introduce MT into the pancreas (33). By histological analyses, the resulting tumors appeared to arise from either acinar or ductal cells. Finally, two MT-driven prostate cancer models have also been described. In the first study, the C-3 promoter was utilized to express MT in the mouse prostate, resulting in mPIN (62). However, this promoter also drives gene expression in other tissues, causing other types of cancers in various organs, which limited its usefulness in prostate cancer studies. The second system utilized the LoxP-stop-LoxP cassette system to limit the expression of an MT transgene to only those tissues in which cre was expressed via either a second transgene or a viral vector (7). Upon deletion of the stopper sequences through cre recombinase expression (Probasin-Cre) in the prostate, MT expression was induced, again ultimately resulting in mPIN. However, this study did not detail disease progression or the molecular characteristics of the mPIN. Therefore, these two models have, to date, provided only limited understanding of the molecular mechanisms of MT-driven prostate cancer.
In this study, we used a modified rat Probasin composite promoter containing two copies of androgen-responsive regions [(ARR)2PB] cassette to drive the expression of MT in the mouse prostate. MT expression produced progressive tumor development that ultimately resulted in an invasive tumor phenotype. We have explored this process in detail by standard histological techniques, as well as molecular methods.
MATERIALS AND METHODS
Plasmid construction and generation of transgenic mice.
The (ARR)2PB-Cre vector was obtained from William Sellers's laboratory. This clone includes a minimal promoter and two androgen receptor (AR) binding sites, a cDNA encoding the Cre recombinase and a poly(A) signal. The probasin promoter region was excised from (ARR)2PB-Cre with XhoI and NotI, while the poly(A) tail region was digested with EcoRI and BamHI. The excised fragments were subcloned into the corresponding sites of pcDNA 3.1(−) to generate (ARR)2PB-Poly(A). MT was PCR amplified with oligonucleotides containing EcoRV restriction enzyme recognition sites. The resulting PCR products were digested with EcoRV and then subcloned into (ARR)2PB-Poly(A) to generate (ARR)2PB-MT. A restriction fragment containing the whole (ARR)2PB-MT cassette was then isolated for pronuclear injection to generate transgenic animals by standard means in the DFCI Core Facility. All procedures were performed according to protocols approved by the Institutional Animal Care and Use Committee of the Dana-Farber Cancer Institute.
Genotyping.
Founders and positive transgenic animals were screened by genomic DNA PCR using genomic DNA prepared from tail clips. Oligonucleotides for PCR primers were from the probasin promoter region (5′GACACTGCCCATGCCAATCAT-3′) and an internal region of MT (5′-ATATACAACTACATGAGCTG-3′). DNA isolation and testing were performed as previously described (27).
IHC analysis.
Prostate tissues were isolated and fixed in 10% buffered formalin (Fisher Scientific) at 4°C overnight, followed by phosphate-buffered saline (PBS) washing. Tissues were then paraffin embedded, sectioned at 5 μm, and mounted on slides. Hematoxylin and eosin (H&E) staining and immunohistochemistry (IHC) assays were carried out as previously reported (27). In summary, slides were deparaffinized and immersed in 0.5% hydrogen peroxide in TBS (50 mM Tris and 150 mM NaCl). Next, antigens were retrieved with sodium citrate buffer (10 mM sodium citrate, 0.05% Tween 20, pH 6.0) in a microwave oven for 10 min and blocked in 3% goat serum diluted in TBS for 1 h. Primary antibodies were diluted in 3% goat serum in TBS as follows: anti-MT (1:10,000; polyclonal antibody 18-8 generated by the Roberts laboratory), Ki-67 (1:100; Dako), CK8/TROMA-1 (1:50; Developmental Studies Hybridoma Bank, The University of Iowa), anti-AR (1:50; Millipore), anti-phospho-AKT (Ser 473) (1:100; Cell Signaling Technology), anti-phospho-S6 ribosomal protein (Ser 240/244) (1:50; Cell Signaling Technology), anti-phospho-GSKβ (glycogen synthase kinase-β) (Ser 9) (1:400; Cell Signaling Technology), anti-phospho-ERK1/2 (extracellular signal-regulated kinase 1/2) (Thr 202/Tyr 204) (1:100; Cell Signaling Technology), anti-p65 (1:50; Cell Signaling Technology), anti-CXCL5 (1:250; R&D System), anti-p63 (1:100; Santa Cruz), anti-CD3 (1:1,000; Cell Marque), anti-B220 (1:200; BD Pharmingen), and anti-Mac2 (1:32,000; Cedarlane) antibodies. After either 1 h of incubation at room temperature or overnight incubation at 4°C, slides were washed with TBS three times, and secondary antibodies (1:200 diluted in 1.5% goat serum diluted in TBS) were added and incubated for 30 min at room temperature. After washing with TBS, slides were incubated with avidin biotin complex in TBS for 30 min at room temperature, and the stain was developed with 3,3′-diaminobenzidine (DAB) solution (Sigma). The slides were then washed and counterstained with Gill's hematoxylin solution (Sigma-Aldrich). The slides were washed again, and sequential dehydration steps were followed through 70%, 80%, 95%, and 100% ethanol. The slides were later soaked in xylene and mounted.
Masson's trichrome staining.
Paraffin-embedded sections were stained with Masson's trichrome (Accustain trichrome; Sigma-Aldrich). Staining procedures were carried out according to the manufacturer's protocol. In summary, sections were deparaffinized, incubated in preheated Bouin's solution (Sigma-Aldrich) for 15 min at 56°C, and then washed in running tap water. Then, sections were stained for 5 min each in the following staining solutions: Weigert's iron hematoxylin solution, Biebrich scarlet-acid fucshin, phosphotungstic-phosphomolybdic acid solution, aniline blue solution, and (for 2 min) 1% acetic acid solution. After each staining, sections were washed in running tap water for 5 min. After staining in phosphotungstic-phosphomolybdic acid solution, sections were directly stained in aniline blue solution. Finally, slides were dehydrated and mounted as described above.
Microarray analysis.
Total RNA samples were extracted from freshly isolated ventral prostate tissues of 8-week-old mice, four of the wild type and four (ARR)2PB-MT transgenic, and from freshly isolated mammary glands of 11- to 12-week-old mice, three of the wild type and three MMTV-MT transgenic, with Trizol (Invitrogen) and additionally cleaned using RNeasy MiniPrep columns (Qiagen). The integrity of the RNA was evaluated both by agarose gel visualization and by spectrophotometric measurement. RNA samples were submitted to the DFCI Microarray Core Facility for assays. DNA array experiments were conducted on mouse expression array 430A2.0 chips (Affymetrix). The gene expression cell files (.CEL) were normalized with the robust multiarray average (RMA) method implemented in the Limma package in the R/Bioconductor package. In addition, BRB Array Tools (developed by Richard Simon and the BRB-ArrayTools Development Team [http://linus.nci.nih.gov/BRB-ArrayTools.html]) were also used. Differential expression of genes was computed using the empirical Bayes method (eBayes), which was included in the Limma package in R (55). To compare pathways that could be differentially involved in tumorigenesis in prostate cancer and breast cancer in mice, we utilized Ingenuity Pathway Analysis software (Ingenuity) and Gene Set Enrichment Analysis (GSEA) (http://www.broad.mit.edu/gsea/) (59). Gene expression profile data were obtained from Gene Expression Omnibus (GEO); GEO accession numbers are as follows: MMTV-PyV MT (GSE3165) (26), TRAMP (GSE10525) (25), AKT transgenic mice (GSE1413) (39), and myristylated PI3-kinase p110β transgenic mice (GSE21543) (32). Myc data were from the authors' website (http://doe-mbi.ucla.edu/myc_driven_prostate_cancer/), and gene expression data of PTEN knockout and MMTV-MT mice are unpublished data from our laboratory. Each data set was separately processed to find differentially expressed genes by t test using BRB Array tools. Human clinical data were obtained from Glinsky et al. for prostate cancer (20) and from the Gene Expression Omnibus for breast cancer (GSE1456) (46). Genes consistently up- or downregulated in both prostate and breast tumors of mice were selected, and then human orthologs were identified from the Mouse Genome Informatics (MGI) database (http://www.informatics.jax.org/) and used for survival analyses. Kaplan-Meier plots were generated using GraphPad Prism (GraphPad Software). Microarray data were deposited in GEO under accession number GSE26234.
Real-time PCR.
All oligonucleotide pairs used for real-time PCR experiments were designed using the GenScript Real-Time PCR Primers Designs tool and are listed in Table S1 in the supplemental material. Total RNA was extracted from prostate lobes of both wild-type and transgenic mice using TRIzol reagent (Invitrogen). One microgram of total RNA was reverse transcribed using a SuperScript II RT-PCR kit (Invitrogen) with random hexamer primers according to the manufacturer's protocol. The resulting cDNA (25 ng) was used for PCRs with SYBR green master mix (Applied Biosystems). Thermocycling programs were 45 cycles of 95°C for 15 s, 60°C for 1 min, with an initial step of 95°C for 10 min. Expression levels were calculated according to the Pfaffl method (48). The levels of amplified transcripts were normalized to the levels of beta actin obtained.
RESULTS
An (ARR)2PB-MT transgene directs the expression of MT in the mouse prostate.
To effect prostate-specific expression of MT in the mouse, we utilized the well-known (ARR)2PB promoter cassette, which drives gene expression upon androgen stimulation. Within this cassette, there are two androgen-responsive regions (ARR) containing androgen-responsive elements (AREs), which drive hormone-dependent gene expression during mouse prostate development (73). A PCR-amplified cDNA encoding MT was subcloned into the (ARR)2PB cassette to produce the (ARR)2PB-MT construct (Fig. 1A). This plasmid was then linearized and microinjected into the pronucleus of fertilized mouse eggs to generate transgenic animals. PCR analyses of genomic DNA from tail clips were used to screen for founders, and representative results are shown in Fig. 1B. By this approach, three transgenic lines were obtained: GG982, FF23, and 36B. All three models ultimately displayed generally similar characteristics. However, there were certain phenotypic differences among them, presumably arising from prostate lobe-specific differences in MT expression, with the FF23 line expressing MT in all three lobes while the GG982 and 36B lines failed to express MT in the anterior prostate (see below). Since the GG982 and 36B lines were roughly equivalent, we used the GG982 and FF23 lines for subsequent experiments.
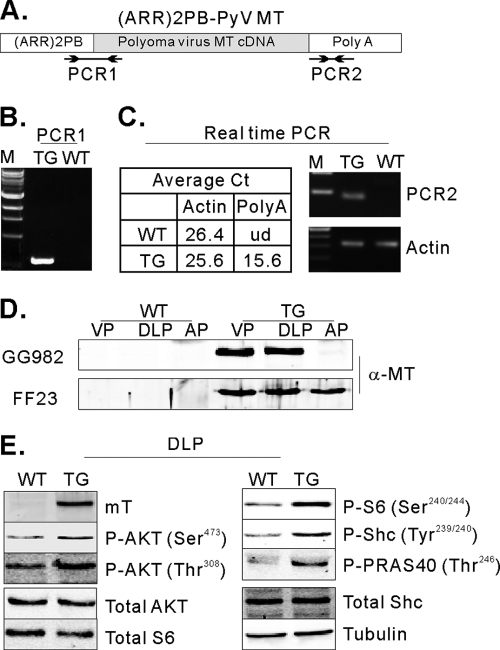
Fig. 1.
Expression of functional polyomavirus middle T in the mouse prostate. (A) Schematic diagram of the DNA construct used for the creation of transgenic mice. PCR1 represents the expected PCR product from genomic DNA for genotyping. PCR2 represents the PCR product of real-time PCR for measuring expression levels. Arrows indicate oligonucleotides used for PCRs. (B) An example of a genotyping PCR. TG, transgenic mouse, WT, wild type. (C) The table summarizes the average real-time cycle threshold (Ct) values. ud, undetected. The real-time PCR products were run on an agarose gel. (D) Detection of MT expression in the mouse prostate. Cellular lysates were prepared from the anterior (AP), ventral (VP), and dorsal/lateral prostate (DLP) from 10-week-old TG (GG982) and WT littermates and 30-week-old TG (FF23) and WT littermates. Transgene expression was confirmed by immunoblotting experiments using an anti-MT antibody. (E) MT induces functional signal transduction. Total lysates from VP and DLP from WT and TG mice were analyzed by immunoblotting using antibodies directed against anti-phospho-AKT (Ser473), anti-phospho-AKT (Thr308), anti-phospho-S6 (Ser240/244), anti-phospho-Shc (Tyr239/240), anti-PRAS40 (Thr246), anti-AKT, anti-S6, and antitubulin antibodies.
First, we confirmed the expression of MT in the transgenic mouse prostates. Total RNA was purified from the ventral prostates of 10-week-old mice, and real-time PCR was performed with oligonucleotides targeted to the poly(A) signal region (Fig. 1A). As shown in Fig. 1C, MT expression was detected in transgenic mice but not in wild-type animals. We next confirmed protein expression. Total cellular lysates were prepared from the ventral prostate (VP), dorsal/lateral prostate (DLP), and anterior prostate (AP) lobes of transgenic and control mice, and immunoblotting experiments were performed using an anti-MT antibody. As shown in Fig. 1D, MT expression was detected in the ventral and dorsal/lateral lobes of the transgenic prostates in all lines but not in the anterior lobes of line GG982 (Fig. 1D) or line 36B3 (data not shown). However, in the FF23 line, MT is also expressed in the anterior lobe (Fig. 1D).
MT transduces oncogenic signals via interactions between several phosphorylated tyrosine residues on MT and SH2 or phosphotyrosine binding (PTB) domain-containing proteins, such as SHC, PI3-kinase, and PLC-γ. These associations lead to clear activation of PI3-kinase and, in some cases, RAS pathways, while activation of PLC-γ is usually difficult to detect. To verify the molecular function of MT in mouse prostate cells, we performed a series of immunoblotting experiments, examining the dorsal/lateral prostate, where MT is consistently expressed. We first probed with anti-phospho-AKT (Ser 473) and (Thr 308) antibodies to see if the PI3-kinase pathway was activated. As shown in Fig. 1E, phospho-AKT signals were marginally but reproducibly increased in dorsal/lateral lobes (Fig. 1E). Since phospho-AKT levels are only weakly increased, we also examined the phosphorylation of AKT substrates. Phosphorylation of both a proximal AKT target, PRAS40, and a downstream target, S6 ribosomal protein (S6RP) (Ser 240/244), was clearly increased in transgenic mice, further supporting the activation of the PI3K/AKT/mTOR pathway (Fig. 1E). In addition, we examined the activation of the ras pathway, measuring tyrosine phosphorylation of SHC (Tyr 239), which was also significantly increased compared with that of controls (Fig. 1E).
MT-induced lesions exhibit features of adenocarcinomas characterized by luminal cell markers.
We next sacrificed mice at various ages to assess whether MT drives tumorigenesis in the mouse prostate. At 4 weeks, the prostates of both control and transgenic mice were histologically normal (data not shown). By 10 weeks of age, however, the ventral and dorsal/lateral prostate lobes of all transgenic mice (but not control mice) displayed features of mPIN. Specifically, we observed intraductal proliferation of epithelial cells with atypical hyperchromatic nuclei (Fig. 2A). (In contrast, the FF23 line developed mPIN in all lobes where MT was expressed, including the AP.) As a point of comparison, transgenic mice expressing activated PI3-kinase p110β or activated Akt in the prostate also display mPIN at this age (39, 40). However, the mPIN induced by MT tends to involve a larger proportion of the glands than the activated-PI3-kinase p110β and activated-AKT models. We further analyzed the mitotic index by performing IHC staining for the proliferation marker KI-67. As shown in Fig. 2B, significantly more Ki-67-positive cells were detected in the prostates of transgenic mice than in those of wild-type mice.
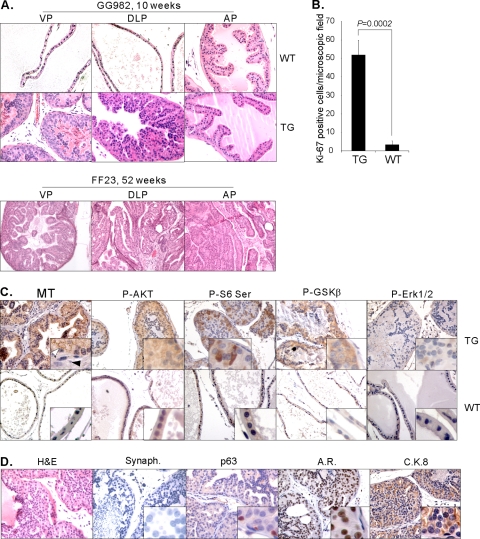
Fig. 2.
Development of adenocarcinoma upon expression of MT in the mouse prostate. (A) H&E staining displays normal prostate from a WT mouse (10 weeks old) and characteristics of mPIN in the VP and DLP of a TG mouse of the GG982 line (10 weeks old). For comparison, H&E staining on sections of an FF23 transgenic mouse prostate (52 weeks old) demonstrates the development of lesions in all prostate lobes where MT is expressed. (B) High proliferation index in the MT-induced lesions. IHC analysis was conducted with an anti-KI-67 antibody, and KI-67-positive cells were counted from five microscopic fields and averaged. The P value was calculated using the t test. Error bars show standard deviations. (C) IHC analysis was conducted with anti-MT, anti-phospho-AKT (Ser 473), anti-phospho-S6 (Ser 240/244), anti-phospho-GSKβ (Ser 9), and anti-phospho-ERK1/2 (Thr 202/Tyr 204) antibodies on DLPs of WT and TG mice (12 weeks old). Expression of MT in the transgenic mouse is detected in epithelial cells (white arrowhead) but not in stromal cells (black arrowhead). Notably, membrane-localized signals are detected. (D) Paraffin-embedded sections prepared from DLP of 10-week-old TG mice were subjected to IHC with anti-Synaptophysin (Synaph.), anti-p63, anti-androgen receptor, and anti-CK8 antibody. Images were taken at 40× magnification except for H&E staining of a sample from an FF23 transgenic mouse, which was taken at 20× magnification.
Next, we examined which cell types feature MT expression by IHC staining with anti-MT antibody. As shown in Fig. 2C, the expression of MT was clearly evident in epithelial cells but was not observed in stromal cells. We then determined whether activation of the PI3-kinase pathway is detected in PIN lesions. Increased levels of phosphorylation were detected for AKT and its substrate GSKβ, as well as S6RP (Fig. 2C). These results, together with those of the immunoblotting experiments shown in Fig. 1E, suggest that activation of the PI3-kinase pathway does indeed occur in the prostates of the transgenic mice. In contrast, phosphorylation of the downstream target Erk1/2 was not significantly altered in transgenic mice compared with that in wild-type mice, as is often seen in MT-transformed cells (51).
Mouse prostate epithelial cells consist of three major cell types: neuroendocrine cells, secretory luminal cells, and basal cells. IHC analysis experiments were performed to identify the nature of the neoplastic cells. We analyzed the expression of the following markers specific for the various cell types: an antisynaptophysin antibody for neuroendocrine cells, anti-p63 antibody for basal cells, and anti-Ck8/18 antibody and anti-androgen receptor (AR) antibody for luminal cells. As shown in Fig. 2D, abnormal cells within the lesions express AR and CK8/18 but are negative for synaptophysin and p63 expression (normal basal cells surrounding the lesions can be p63 positive). We examined even older mice, but they did not express synaptophysin in the mPIN (data not shown). These results indicate that, similar to human PIN, the prostate lesions caused by MT overexpression display a luminal phenotype.
The mPIN induced by MT progresses to invasive cancer with prominent reactive stroma and inflammation.
To investigate disease progression further, mice were sacrificed at more advanced ages (30 weeks to 2 years old). The sizes of prostates of transgenic mice and wild-type littermates were compared by macroscopic examination. In general, the prostates of transgenic mice gradually enlarged over the course of time. As shown in Fig. 3A, the sizes of the prostates of transgenic mice are increased compared to those of wild-type mice at the age of 80 to 90 weeks. The VP and the LP often become joined with connective tissue as mice age, so it is difficult to measure to weigh them separately. Since MT induces mPIN in both lobes, we combined the weights of the VP and DLP to avoid errors during tissue isolation. Examination of tumors harvested from mice at different ages suggests that the tumors grew slowly and continuously.
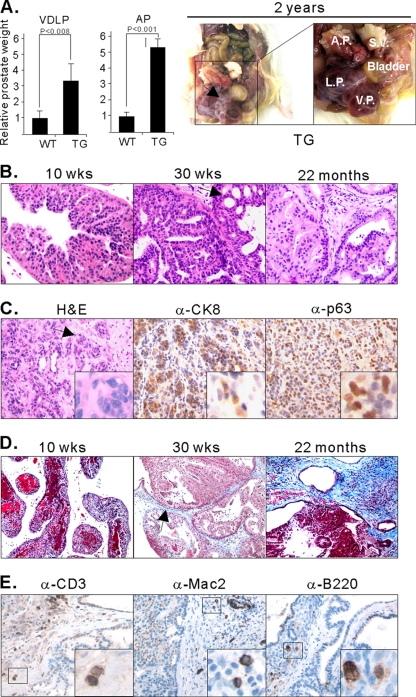
Fig. 3.
The lesions induced by MT resulted in severe phenotypes, invasion, and reactive stroma. (A) Enlargement of prostate lobes of TG mice. The weights of the VP/DLP (VDLP) were combined to avoid possible errors during tissue isolation. The weights of prostate tissues were normalized with the body weights. The graph was then standardized to the weight of VP/DLP of the wild type, and the relative differences between the weights of tissues from wild-type and transgenic mice are presented. Error bars show standard deviations. Prostates of WT mice were obtained from mice 91 to 92 weeks old, and prostates of TG mice were isolated from mice 81 to 96 weeks old. Frank tumors developed, as seen in an approximately 2-year-old FF23 line mouse (right panels). S.V., seminal vesicle. (B) H&E staining of DLP isolated from GG982 line mice at 10 weeks, 30 weeks, and 2 years of age. The black arrow indicates the cribriform phenotype. At least 5 mice were examined at each specific age. (C) H&E staining of the AP of a 2-year-old FF23 line mouse exhibits extensive invasive prostate cancer. IHC assays were conducted with anti-CK8 and anti-p63 antibodies. The black arrow indicates irregular nests of atypical epithelial cells penetrating the stroma surrounding the prostatic ducts, indicating invasion. (D) Masson's trichrome staining shows the development of reactive stroma in blue color in old mice (>30 weeks old). The black arrow indicates blue staining with Masson's Trichrome. (E) IHC experiments with anti-CD3, anti-B220, and anti-MAC2 antibody show infiltration of inflammatory cells.
Along with the increase in size, prostates of older transgenic mice presented with severe histopathological abnormalities. For example, cribriform intraductal lesions—an indication of high-grade PIN in human prostate cancer—were clearly observed in the ventral and dorsal/lateral lobes (Fig. 3B, indicated with a black arrow) in the GG982 line and in all three lobes in the FF23 line (Fig. 2A). In general, more advanced adenocarcinomas were seen in the DLP in both lines. More strikingly, 50% (5/10) of mice of the FF23 line older than 20 months developed invasive carcinomas in the AP (Fig. 3C). As shown in Fig. 3C, irregular nests of atypical epithelial cells penetrate the stroma surrounding the prostate ducts, a clear indication of invasion. To confirm the invasion of prostate cancer cells, we performed IHC assays with anti-CK8/18 and anti-p63. As shown in Fig. 3C, the expression of p63 is no longer confined to the basement membrane area (compare with Fig. 2D) but is now randomly dispersed within the stromal compartment. In addition, the invasive tumor component contained cells expressing CK8 markers, as well as cells expressing p63. Increased expression of p63 has been previously reported in the study of mouse prostate cancers driven by PTEN deletion (69). In addition, the expression of both a luminal marker (CK8) and a basal cell marker (p63) were reported in Nkx3.1+/−;PTEN+/− mice (1).
Another characteristic of the prostate lesions resulting from the expression of MT is the presence of abundant stromal cells, which is often observed in the progression of human cancers (28). Thickened stroma was observed among relatively old mice (>30 weeks of age) and was more prominent in the DLP than in the VP. To test whether these cells could be classed as reactive stroma, we stained with an anti-KI-67 antibody, which demonstrated that these stromal cells had a high proliferative rate (data not shown). To further evaluate the reactive nature of the stroma, we performed Masson's trichrome staining that allows the differentiation of smooth muscle cells (red staining) from collagen (blue staining). As shown in Fig. 3D, blue staining is readily detected in aged mice (>30 weeks old), indicating that the stroma consists mostly of fibrous tissue. Thus, the stroma in the prostate tumors induced by the expression of MT displayed striking similarity to the reactive stroma seen in human prostate cancer (64).
Prostates from mice 1 year of age or older frequently showed the presence of inflammatory infiltrations within the reactive stroma. To further characterize the type of inflammatory cells, we performed additional IHC experiments with anti-CD3 (pan-T lymphocyte marker), anti-MAC2 (macrophage marker), and anti-B220 (B cell marker) antibodies (Fig. 3E). Most inflammatory cells were either CD3 positive or MAC2 positive. A smaller population of B220-positive cells was also observed. These results indicate that the inflammatory cells were composed of a mixture of T cells, B cells, and macrophages.
Comparison of molecular signatures among various murine prostate cancer models suggests that MT directs a particularly inflammatory disease phenotype.
To shed further light on the molecular mechanisms of prostatic tumorigenesis caused by MT expression, we conducted DNA microarray analyses. Using the empirical Bayes method (13), 1,028 probes out of 22,690 probes were identified as differentially regulated by at least 2-fold by MT. Approximately 58% of these genes (583) were upregulated in tumors. A list of all the differentially expressed genes is presented in Fig. 4A and Table S2 in the supplemental material. Real-time PCR validated some of the microarray results, as shown in Fig. 4B. Gene Ontology analysis identified genes on this list that were involved in a variety of processes, including cell cycle, immune response, cell proliferation, apoptosis, and metabolism (see Tables S3 and S4 in the supplemental material). As will be discussed below, chemokines were the single most prominently overexpressed group of genes.
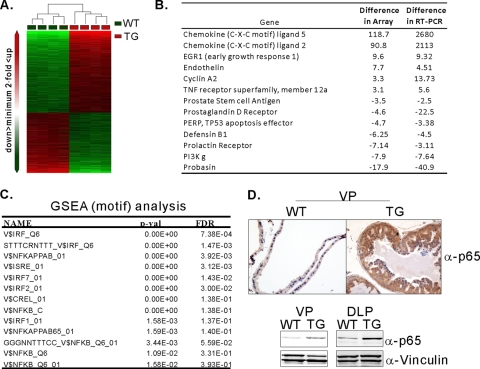
Fig. 4.
Analyses of DNA microarrays identify enrichment of genes associated with the NF-κB signaling pathway. (A) Hierarchical clustering of gene expression data from WT and TG prostate tissues. Genes differentially expressed by at least 2-fold were selected for hierarchical clustering. The heat map depicts differential gene expression between the WT and TG groups. Each row represents an individual gene, and each column represents an individual tissue sample. The red and green colors represent relatively higher and lower expression levels, respectively. (B) Summary of real-time PCR data of selected genes identified from DNA array analyses. (C) GSEA with the C3 motif database identified gene sets enriched in the NF-κB pathway and the interferon signaling cascade. FDR, false discovery rate. (D) IHC assays and immunoblotting experiments with anti-p65 antibodies demonstrate increased levels of p65 in the prostate of transgenic mice.
Next, we performed Gene Set Enrichment Analysis (GSEA), which helps to identify gene sets enriched in relevant biological processes. We used the C2 (curated gene sets) and C3 (motif gene sets) gene sets (59). Among the gene sets enriched, both curated and motif analyses identified NF-κB target genes and inflammatory genes as consistently enriched in MT-induced prostate cancer (Fig. 4C; also see Tables S5 and S6 in the supplemental material). As shown in Fig. 4D, IHC experiments corroborated the array results showing increased levels of the p65 subunit of the NF-κB complex and increased levels of p65 in the transgenic mouse (Fig. 4D).
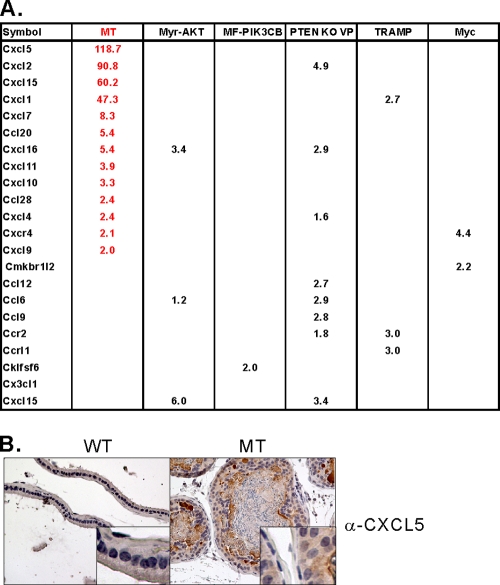
Finally, we compared the molecular signatures obtained from the MT transgenic mice to those from previously reported prostate models. Notably, the various mouse models result in diseases of distinctly different severities. The lesions developed in the PIK3CB (32) and Akt mice (40) are characterized as mPIN, which never proceed to invasive prostate cancer, in contrast to the PTEN null tumors used in our laboratory; furthermore, the MT and PTEN null tumors in other backgrounds (37, 63, 68) invade, and the myc (14) and TRAMP mice (22) progress from invasive tumors to frankly metastatic lesions. While this comparison provides some insight into the differences in tumor severity phenotypes, another aspect of gene expression proved to be most striking. Notably, the microarray data identified chemokines as the gene family most highly overexpressed in MT tumors when compared with the other models (Fig. 5A). Principal among these are CXCL5, CXCL2, CCL5, and CXCL15. The overexpression of CXCL5 and CXCL2 was confirmed by real-time PCR (Fig. 4B). We further examined the expression of one of the chemokine family proteins, CXCL5, by IHC assay (Fig. 5B). IHC analysis shows high signals for CXCL5 in an MT transgenic mouse. Recent studies have suggested that extracellular matrix genes (versican and tanscin) and chemokines (BDFN, CCL5, CXCL5, and CXCL16) may be induced by the interaction between stromal and epithelial cells during human cancer progression (60, 66). The upregulation of chemokine families in this highly stromal mouse prostate cancer model further suggests a possible link between overexpression of chemokines and the development of proliferative stromal cells. Chemokine expression may also explain the infiltration of inflammatory cells seen in our histological studies (Fig. 3E).
Fig. 5.
Expression of chemokine family genes is predominantly induced in transgenic mice arisen by expression of MT. (A) Comparison of chemokine gene expression in mouse prostate cancers among different genetically engineered models (GEMs): MT transgenic mice, myr-AKT transgenic mice, myr-flag-PIK3CB transgenic mice, VP and AP of PTEN knockout mice, TRAMP transgenic mice, and high-Myc transgenic mice. Differential expression of chemokines was obtained from the results of each study separately by a t test (P < 0.001) between wild types and tumor models using BRB tools, and the fold differences are summarized. (B) Paraffin-embedded sections were prepared from WT and MT transgenic mice, and IHC assays with anti-CXCL5 antibody were conducted.
Comparisons of molecular signatures of breast cancer and prostate cancer driven by MT expression.
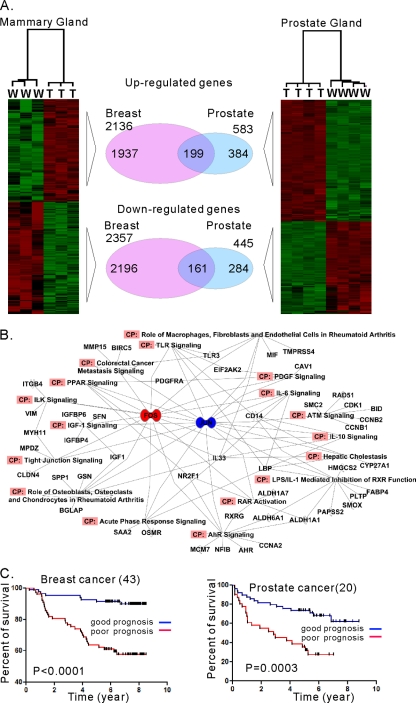
We previously reported that activation of the PI3-kinase pathway is required for MT-mediated transformation in both human and mouse mammary epithelial cells but that the mechanism of action could vary in a cell type-dependent manner (65). We wanted to determine whether there might be a core signature of MT transformation common to different cellular contexts. We compared gene expression data obtained from breast and prostate tumors induced by transgenic expression of MT. As shown in Fig. 6A, we identified 360 genes that were consistently up- or downregulated in both tissues. These include SAA1, Stathmin, Jun, Fos, CD44, Ki67, and others (see Table S7 in the supplemental material). However, many genes are regulated by MT in a tissue-specific manner (see Tables S8 and S9 in the supplemental material). Interestingly, chemokine genes are almost exclusively upregulated in prostate cancer (see Table S9). Notably, downregulation of chemokine genes (except CXCL5) was also observed in MMTV-MT transgenic mice in a previous gene expression study using a different platform of arrays (GSE3165) (26). Likewise, some genes, including Car6, Car12, Skp2, and FoxC1, were uniquely identified in breast tumors. This result indicates that the molecular mechanisms of tumorigenesis by MT expression may differ depending on the cellular context. Notably, a number of the commonly regulated genes, including SAA1/2, Tacstd2, Jun, CD24a, Tspan8, and TMPRSS4, were also identified as PI3-kinase pathway-dependent genes (32), consistent with the idea that activation of the PI3-kinase pathway is intrinsically important in MT transformation. To delineate the possible pathways associated with MT expression, we used Ingenuity Pathway Analysis software and sought canonical pathways. As shown in Fig. 6B, the AP1 complex (Jun/Fos) was mapped at the center of the pathways. This confirms that MT activates the AP1 pathway (57). In addition, AP1 has been used a prognostic marker for advanced types of breast cancer (67) and prostate cancer (45).
Fig. 6.
Comparison between gene expression profiles of mammary tumors and prostate tumors induced by MT expression. (A) Gene expression profiles demonstrate that both common and differential molecular mechanisms may drive tumor formation in a tissue-dependent manner. (B) Ingenuity Pathway analysis with genes common to breast and prostate cancers induced by MT suggests that AP1 (Jun/Fos) is one of the most significant factors in several canonical pathways. CP, canonical pathway. (C) Kaplan-Meier analyses of human breast and prostate cancers were conducted with genes consistently found to be differentially expressed in both mouse tissues.
A previous study using comparative analysis of MMTV-MT models demonstrated that a key gene set for predicting metastatic human breast cancers was also found in MT-associated cancer genes identified from mouse models (49). This result demonstrated the utility of the signature of MT-associated cancer. We therefore wondered whether the common gene set representing the molecular signature of the intrinsic MT function (Fig. 6A; also see Table S7 in the supplemental material) might be clinically correlated in breast and prostate cancers. Using human orthologs of the signature genes, we applied unsupervised clustering to both human prostate cancer data (20) and human breast cancer data (46). In both cases, human cancer samples were separated into two clusters. We then performed Kaplan-Meier plots to see whether these two clusters have different survival or recurrent disease patterns. As shown in Fig. 6C, for breast cancer cohorts, the common signature is significantly associated with survival, which is consistent with the results of previous studies (49). Furthermore, the signature can also be used for predicting the outcome of human prostate cancers. This indicates that molecular mechanisms associated with MT in developing cancers might have relevance to clinical outcomes, although MT is not directly involved in human cancers.
DISCUSSION
For the studies described here, we have created transgenic mice using polyomavirus MT under the control of an androgen receptor-driven promoter. MT expressed in the mouse prostate drives the slow development of invasive cancer, mimicking the slow progression of the human disease. Ordinarily, the majority of tumors arising from epithelial cells are caused by the cumulative effects of multiple genetic or epigenetic alterations (24). These alterations result in gain of function of oncogenes, loss of function of tumor suppressors, or epigenetic changes. These changes disturb the cellular homeostasis and eventually induce diseases, including cancers. MT affects multiple signaling pathways directly, in large part through tyrosine kinase activation. Most importantly, the MT model also displays significant histophysiological similarities with human prostate cancers, including a large component of actively dividing stromal cells and infiltration by inflammatory cells.
Activation of RTKs is often seen in human prostate cancer, although the correlation of insulinlike growth factor (IGF) levels in plasma and clinical outcomes remains controversial (8, 50). However, several clinical studies demonstrated that the level of ErbB2 expression is often elevated in advanced prostate cancer patients, especially those who have undergone radiation or hormone deprivation therapies (18, 42, 44, 54). Modeling of RTK activation in the prostate has been carried out via transgenic expression of Erbb2-delta (a C-terminal truncation mutant) (6), Neu (34), or FGF8b (15, 56). Although these models developed disease of different severities, the RTK models displayed some similarities, including relatively slow progression of mPIN to microinvasion, proliferation of stromal cells, and infiltration of inflammatory cells. Notably, our current MT model is similar to these models in a number of characteristics. Although mPIN was observed at an early age, the disease progressed slowly to invasive lesions over 1.5 years. In addition, extensive development of the stromal compartment was also seen in MT animals. Presumably, many of these similarities are explained mechanistically by the similarity of the signaling pathways, such as the AKT and Raf pathways, activated by both MT and RTKs.
The infiltration of inflammatory cells seen in the histological studies recalls the importance of interactions of tumor cells with the underlying stromal cells seen in the MT breast model, where the stroma contributes to the migratory and invasive behavior of MT tumor cells (17). Tumor-associated proteases, which are important for metastasis, come mostly from the breast stroma (47). Particular attention has been paid to macrophages, and loss of macrophage infiltration in CSF-1 null mice results in tumors that do not invade well and results in a dramatic decrease in metastasis (35). These studies prove that the tumor microenvironment in the MT-driven breast tumors can promote tumor progression to invasion or metastasis. Since similar stromal phenotypes were observed in the MT-driven prostate tumor model described here, further studies of this model may help in understanding the mechanisms of invasive prostate cancer.
The high levels of chemokine production seen in our MT prostate tumor model make it a unique animal model to study the function of chemokines in prostate cancer development.
DNA microarray experiments identified chemokine ligands as the most highly expressed class of genes in MT transgenic tumors. These ligands act through G protein-coupled receptors, and their signaling cascades are known to be involved in promoting angiogenesis and to affect the constitution of tumor microenvironments (2). Increased expression of CXCL5 was previously associated with macrophage and myeloid cell infiltration in MMTV-MT tumors (4, 72). The addition of CCL5 increased the migratory behavior of tumor-associated myeloid cells, while antagonists of CXCR2 reduced metastasis (72). The expression levels of chemokines, such as CXCL5, CCL5, or CXCL16, are significantly associated with prostate cancer progression, including invasive and bone metastatic prostate tumors (3, 36, 60). It is quite possible that chemokine production by the MT-driven tumor epithelia may be driving infiltration of both stromal and immune cells. It is also known that MT-transformed mouse mammary epithelial cells produce CCL2 and CCL5 (Y. Zhu and B. Schaffhausen, unpublished results). It is also possible that the stromal production of chemokines subsequently leads to infiltration by inflammatory cells. Notably, a recent study from the Hanahan laboratory showed that cancer-associated fibroblasts can play a key role in tumor inflammation by expressing inflammatory genes, such as chemokines (16). Perhaps MT elicits stromal infiltration independently of chemokines and it is the stroma that produces the chemokines that subsequently lead to infiltration by inflammatory cells.
There are several possible origins for the stromal cells that are so prevalent in these tumors. Since prostate stromal cells also express substantial levels of androgen receptor, transgene expression from the (ARR)2-probasin promoter in those cells is possible. However, MT expression was seen only in luminal epithelial cells by IHC experiments, as shown in Fig. 2C. Therefore, the development of the cancer-associated stromal phenotype, as shown in Fig. 3, is more likely being induced by paracrine factors arising in epithelial cells expressing MT. Indeed, the observation that the abnormal cells appear only within the prostate lumen, with no increased stromal involvement at young ages, is consistent with the hypothesis that MT acts solely in the prostate epithelium. However, we cannot completely rule out the possibility that some of the activated stroma may have arisen from cells expressing low levels of MT, which could be below the detection level in IHC. Alternatively, the reactive stroma could have arisen by epithelial-mesenchymal transition (EMT) in MT-expressing epithelial cells late in tumor progression.
Regardless of which cells are expressing the chemokines and driving infiltration of both stromal cells and white cells, the question arises as to what aspect of MT signaling initiates these events. Most of MT's effects on host cells are thought to arise via two pathways: first, the PI3-kinase pathway, which is activated by binding of the p85 adaptor subunit of PI3-kinase to the phospho-YMPM motif at Tyr 315, and second, the RAS/RAF/ERK pathway, activated by binding of the RAS regulator, Shc, to the NPXY motif at Tyr 250. Since prostate tumors driven by p110β and AKT transgenes do not express chemokines abundantly, it seems unlikely that the PI3-kinase pathway is driving their expression. However, both MT expression and, to a lesser degree, ablation of PTEN activate the RAS/RAF/MEK/ERK pathway, making it a reasonable candidate for chemokine induction. Another key tyrosine residue in the interaction with cellular signaling molecules is Tyr 322, which is the site for PLC-γ association. Notably, previous work pointed to the importance of PLC-γ in metastasis in MMTV-MT for mammary tumors and TRAMP for prostate tumors (53), further suggesting a potential role for PLC-γ in MT-driven prostate cancer. However, since we did not observe metastasis in the MT prostate cancer model, the role of Tyr 322 here might be different from the breast case. We are currently generating transgenic animals expressing MT mutant alleles at Tyr 250, Tyr 315, or Tyr 322 to test these hypotheses.
To explore the common molecular mechanisms in tumorigenesis induced by MT expression, we compared the gene expression data from breast and prostate cancers. The expression of the majority of genes was regulated in a tissue-dependent manner. This suggests that viral oncogenes activate potentially different molecular pathways in different cell types. This is not surprising, since it is well known that the importance of individual MT signaling pathways differs among tissues. There are also differences in gene expression profiles obtained for the MMTV-MT model when MMTV-MT634 tumors are derived in different mouse strain backgrounds and, indeed, even in passage of different MMTV-MT634-derived tumors in FVB mice (38, 43, 49). Such differences could arise as a function of aging, mouse strain differences, gender differences, and disease-specific stages. Our current study did not address these variances. Differences could also result from the differential contribution of stromal components. Indeed, the MMTV-MT model displayed a greater ratio of a luminal cell marker (Krt8) and a stromal cell marker (Desmin) than of (ARR)2PB-MT (data not shown). Taken together, these constraints limit our ability to interpret the tissue-specific differences we observed to be shared by the two MT models. However, this same logic adds increased importance to the features that we observed. Among the commonly regulated genes were genes present in PI3-kinase pathways, suggesting that MT utilized this pathway for inducing cancer. This is consistent with the observation that polyomavirus that is unable to activate PI3-kinase is defective in tumorigenesis in many tissues (19). Functional genomic annotation analyses identified many commonly upregulated genes involved in cell cycle regulation and AP1 signaling, whereas downregulated genes are negatively associated with RXR signaling. Consistent with these findings, higher AP1 levels were clinically correlated with poorer prognosis in breast (67) and prostate cancer (45). Indeed, analysis of commonly expressed genes allowed us to construct a gene signature which appeared to have prognostic utility in a retrospective examination of a limited set of prostate tumors. Therefore, molecular pathways inducing the expression of these genes would be potential targets for therapeutic applications, even though MT itself does not induce breast or prostate cancer in humans.
In summary, we report a new mouse model of prostatic adenocarcinoma that shows several interesting features, including invasiveness and infiltration of inflammatory cells. These features are also found in human prostate cancers. In the most recent decade, several chemical compounds have been synthesized to target PI3-kinase and MAP kinase pathways, probably the most activated pathways in cancer. Since MT effectively activates these pathways, our MT transgenic mice can be utilized as an appropriate model for studying advanced prostate cancers and for evaluating therapeutic chemical compounds.
Supplementary Material
ACKNOWLEDGMENTS
We thank A. Upadhyaya and L. Clayton for helpful comments on the manuscript and E. Li and J. Horner for creating transgenic mice.
This work was supported in part by grants from the National Institutes of Health (T.M.R., B.S., and M.L.), the Prostate Cancer Foundation (M.L.), the Linda and Arthur Gelb Center for Translational Research (M.L.), and the DFHCC Prostate SPORE (T.M.R. and M.L.). In compliance with Harvard Medical School guidelines, we disclose the consulting relationships: Novartis Pharmaceuticals, Inc. (T.M.R. and M.L.).
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 16 March 2011.
REFERENCES
- 1. Abate-Shen C., et al. 2003. Nkx3.1; Pten mutant mice develop invasive prostate adenocarcinoma and lymph node metastases. Cancer Res. 63:3886–3890 [PubMed] [Google Scholar]
- 2. Balkwill F. 2004. Cancer and the chemokine network. Nat. Rev. Cancer 4:540–550 [DOI] [PubMed] [Google Scholar]
- 3. Begley L. A., et al. 2008. CXCL5 promotes prostate cancer progression. Neoplasia 10:244–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bierie B., et al. 2008. Transforming growth factor-beta regulates mammary carcinoma cell survival and interaction with the adjacent microenvironment. Cancer Res. 68:1809–1819 [DOI] [PubMed] [Google Scholar]
- 5. Campbell K. S., et al. 1994. Polyoma middle tumor antigen interacts with SHC protein via the NPTY (Asn-Pro-Thr-Tyr) motif in middle tumor antigen. Proc. Natl. Acad. Sci. U. S. A. 91:6344–6348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Casimiro M., et al. 2007. ErbB-2 induces the cyclin D1 gene in prostate epithelial cells in vitro and in vivo. Cancer Res. 67:4364–4372 [DOI] [PubMed] [Google Scholar]
- 7. Cecena G., Wen F., Cardiff R. D., Oshima R. G. 2006. Differential sensitivity of mouse epithelial tissues to the polyomavirus middle T oncogene. Am. J. Pathol. 168:310–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chan J. M., et al. 1998. Plasma insulin-like growth factor-I and prostate cancer risk: a prospective study. Science 279:563–566 [DOI] [PubMed] [Google Scholar]
- 9. Chen Z., et al. 2005. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature 436:725–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Di Cristofano A., De Acetis M., Koff A., Cordon-Cardo C., Pandolfi P. P. 2001. Pten and p27KIP1 cooperate in prostate cancer tumor suppression in the mouse. Nat. Genet. 27:222–224 [DOI] [PubMed] [Google Scholar]
- 11. Di Cristofano A., Pesce B., Cordon-Cardo C., Pandolfi P. P. 1998. Pten is essential for embryonic development and tumour suppression. Nat. Genet. 19:348–355 [DOI] [PubMed] [Google Scholar]
- 12. Dilworth S. M., et al. 1994. Transformation by polyoma virus middle T-antigen involves the binding and tyrosine phosphorylation of Shc. Nature 367:87–90 [DOI] [PubMed] [Google Scholar]
- 13. Efron B., Tisbshirani R., Storey J. D., Tusher V. 2001. Empirical Bayes analysis of a microarray experiment. J. Am. Stat. Assoc. 96:1151–1160 [Google Scholar]
- 14. Ellwood-Yen K., et al. 2003. Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer Cell 4:223–238 [DOI] [PubMed] [Google Scholar]
- 15. Elo T. D., et al. 2010. Stromal activation associated with development of prostate cancer in prostate-targeted fibroblast growth factor 8b transgenic mice. Neoplasia 12:915–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Erez N., Truitt M., Olson P., Arron S. T., Hanahan D. 2010. Cancer-associated fibroblasts are activated in incipient neoplasia to orchestrate tumor-promoting inflammation in an NF-kappaB-dependent manner. Cancer Cell 17:135–147 [DOI] [PubMed] [Google Scholar]
- 17. Fluck M. M., Schaffhausen B. S. 2009. Lessons in signaling and tumorigenesis from polyomavirus middle T antigen. Microbiol. Mol. Biol. Rev. 73:542–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fossa A., et al. 2002. Independent prognostic significance of HER-2 oncoprotein expression in pN0 prostate cancer undergoing curative radiotherapy. Int. J. Cancer 99:100–105 [DOI] [PubMed] [Google Scholar]
- 19. Freund R., Dawe C. J., Carroll J. P., Benjamin T. L. 1992. Changes in frequency, morphology, and behavior of tumors induced in mice by a polyoma virus mutant with a specifically altered oncogene. Am. J. Pathol. 141:1409–1425 [PMC free article] [PubMed] [Google Scholar]
- 20. Glinsky G. V., Glinskii A. B., Stephenson A. J., Hoffman R. M., Gerald W. L. 2004. Gene expression profiling predicts clinical outcome of prostate cancer. J. Clin. Invest. 113:913–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gottlieb K. A., Villarreal L. P. 2001. Natural biology of polyomavirus middle T antigen. Microbiol. Mol. Biol. Rev. 65:288–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Greenberg N. M., et al. 1995. Prostate cancer in a transgenic mouse. Proc. Natl. Acad. Sci. U. S. A. 92:3439–3443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guy C. T., Cardiff R. D., Muller W. J. 1992. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol. Cell. Biol. 12:954–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hahn W. C., Weinberg R. A. 2002. Modelling the molecular circuitry of cancer. Nat. Rev. Cancer. 2:331–341 [DOI] [PubMed] [Google Scholar]
- 25. Haram K. M., et al. 2008. Gene expression profile of mouse prostate tumors reveals dysregulations in major biological processes and identifies potential murine targets for preclinical development of human prostate cancer therapy. Prostate 68:1517–1530 [DOI] [PubMed] [Google Scholar]
- 26. Herschkowitz J. I., et al. 2007. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 8:R76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jia S., et al. 2008. Essential roles of PI(3)K-p110beta in cell growth, metabolism and tumorigenesis. Nature 454:776–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kalluri R., Zeisberg M. 2006. Fibroblasts in cancer. Nat. Rev. Cancer 6:392–401 [DOI] [PubMed] [Google Scholar]
- 29. Kaplan D. R., et al. 1986. Phosphatidylinositol metabolism and polyoma-mediated transformation. Proc. Natl. Acad. Sci. U. S. A. 83:3624–3628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim M. J., et al. 2002. Nkx3.1 mutant mice recapitulate early stages of prostate carcinogenesis. Cancer Res. 62:2999–3004 [PubMed] [Google Scholar]
- 31. Kim M. J., et al. 2002. Cooperativity of Nkx3.1 and Pten loss of function in a mouse model of prostate carcinogenesis. Proc. Natl. Acad. Sci. U. S. A. 99:2884–2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee S. H., et al. 2010. A constitutively activated form of the p110beta isoform of PI3-kinase induces prostatic intraepithelial neoplasia in mice. Proc. Natl. Acad. Sci. U. S. A. 107:11002–11007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lewis B. C., Klimstra D. S., Varmus H. E. 2003. The c-myc and PyMT oncogenes induce different tumor types in a somatic mouse model for pancreatic cancer. Genes Dev. 17:3127–3138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li Z., Szabolcs M., Terwilliger J. D., Efstratiadis A. 2006. Prostatic intraepithelial neoplasia and adenocarcinoma in mice expressing a probasin-Neu oncogenic transgene. Carcinogenesis 27:1054–1067 [DOI] [PubMed] [Google Scholar]
- 35. Lin E. Y., Nguyen A. V., Russell R. G., Pollard J. W. 2001. Colony-stimulating factor 1 promotes progression of mammary tumors to malignancy. J. Exp. Med. 193:727–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lu Y., et al. 2008. CXCL16 functions as a novel chemotactic factor for prostate cancer cells in vitro. Mol. Cancer Res. 6:546–554 [DOI] [PubMed] [Google Scholar]
- 37. Ma X., et al. 2005. Targeted biallelic inactivation of Pten in the mouse prostate leads to prostate cancer accompanied by increased epithelial cell proliferation but not by reduced apoptosis. Cancer Res. 65:5730–5739 [DOI] [PubMed] [Google Scholar]
- 38. Maglione J. E., et al. 2004. Polyomavirus middle T-induced mammary intraepithelial neoplasia outgrowths: single origin, divergent evolution, and multiple outcomes. Mol. Cancer Ther. 3:941–953 [PubMed] [Google Scholar]
- 39. Majumder P. K., et al. 2004. mTOR inhibition reverses Akt-dependent prostate intraepithelial neoplasia through regulation of apoptotic and HIF-1-dependent pathways. Nat. Med. 10:594–601 [DOI] [PubMed] [Google Scholar]
- 40. Majumder P. K., et al. 2003. Prostate intraepithelial neoplasia induced by prostate restricted Akt activation: the MPAKT model. Proc. Natl. Acad. Sci. U. S. A. 100:7841–7846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Masumori N., et al. 2001. A probasin-large T antigen transgenic mouse line develops prostate adenocarcinoma and neuroendocrine carcinoma with metastatic potential. Cancer Res. 61:2239–2249 [PubMed] [Google Scholar]
- 42. Myers R. B., Srivastava S., Oelschlager D. K., Grizzle W. E. 1994. Expression of p160erbB-3 and p185erbB-2 in prostatic intraepithelial neoplasia and prostatic adenocarcinoma. J. Natl. Cancer Inst. 86:1140–1145 [DOI] [PubMed] [Google Scholar]
- 43. Namba R., et al. 2006. Heterogeneity of mammary lesions represent molecular differences. BMC Cancer 6:275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Osman I., et al. 2005. Serum levels of shed Her2/neu protein in men with prostate cancer correlate with disease progression. J. Urol. 174:2174–2177 [DOI] [PubMed] [Google Scholar]
- 45. Ouyang X., et al. 2008. Activator protein-1 transcription factors are associated with progression and recurrence of prostate cancer. Cancer Res. 68:2132–2144 [DOI] [PubMed] [Google Scholar]
- 46. Pawitan Y., et al. 2005. Gene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohorts. Breast Cancer Res. 7:R953–R964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pedersen T. X., et al. 2005. Extracellular protease mRNAs are predominantly expressed in the stromal areas of microdissected mouse breast carcinomas. Carcinogenesis 26:1233–1240 [DOI] [PubMed] [Google Scholar]
- 48. Pfaffl M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Qiu T. H., et al. 2004. Global expression profiling identifies signatures of tumor virulence in MMTV-PyMT-transgenic mice: correlation to human disease. Cancer Res. 64:5973–5981 [DOI] [PubMed] [Google Scholar]
- 50. Schaefer C., Friedman G. D., Quesenberry J. C. P. 1998. IGF-1 and prostate cancer. Science 282:199a [Google Scholar]
- 51. Schaffhausen B. S., Roberts T. M. 2009. Lessons from polyoma middle T antigen on signaling and transformation: a DNA tumor virus contribution to the war on cancer. Virology 384:304–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Scherl A., Li J. F., Cardiff R. D., Schreiber-Agus N. 2004. Prostatic intraepithelial neoplasia and intestinal metaplasia in prostates of probasin-RAS transgenic mice. Prostate 59:448–459 [DOI] [PubMed] [Google Scholar]
- 53. Shepard C. R., Kassis J., Whaley D. L., Kim H. G., Wells A. 2007. PLC gamma contributes to metastasis of in situ-occurring mammary and prostate tumors. Oncogene 26:3020–3026 [DOI] [PubMed] [Google Scholar]
- 54. Signoretti S., et al. 2000. Her-2-neu expression and progression toward androgen independence in human prostate cancer. J. Natl. Cancer Inst. 92:1918–1925 [DOI] [PubMed] [Google Scholar]
- 55. Smyth G. K. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl. Genet. Mol. Biol. 3:Article3 [DOI] [PubMed] [Google Scholar]
- 56. Song Z., et al. 2002. Fibroblast growth factor 8 isoform B overexpression in prostate epithelium: a new mouse model for prostatic intraepithelial neoplasia. Cancer Res. 62:5096–5105 [PubMed] [Google Scholar]
- 57. Srinivas S., Schonthal A., Eckhart W. 1994. Polyomavirus middle-sized tumor antigen modulates c-Jun phosphorylation and transcriptional activity. Proc. Natl. Acad. Sci. U. S. A. 91:10064–10068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Su W., Liu W., Schaffhausen B. S., Roberts T. M. 1995. Association of polyomavirus middle tumor antigen with phospholipase C-gamma 1. J. Biol. Chem. 270:12331–12334 [DOI] [PubMed] [Google Scholar]
- 59. Subramanian A., et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102:15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sung S. Y., et al. 2008. Coevolution of prostate cancer and bone stroma in three-dimensional coculture: implications for cancer growth and metastasis. Cancer Res. 68:9996–10003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Suzuki H., et al. 1998. Interfocal heterogeneity of PTEN/MMAC1 gene alterations in multiple metastatic prostate cancer tissues. Cancer Res. 58:204–209 [PubMed] [Google Scholar]
- 62. Tehranian A., et al. 1996. Neoplastic transformation of prostatic and urogenital epithelium by the polyoma virus middle T gene. Am. J. Pathol. 149:1177–1191 [PMC free article] [PubMed] [Google Scholar]
- 63. Trotman L. C., et al. 2003. Pten dose dictates cancer progression in the prostate. PLoS Biol. 1:E59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tuxhorn J. A., Ayala G. E., Rowley D. R. 2001. Reactive stroma in prostate cancer progression. J. Urol. 166:2472–2483 [PubMed] [Google Scholar]
- 65. Utermark T., Schaffhausen B. S., Roberts T. M., Zhao J. J. 2007. The p110alpha isoform of phosphatidylinositol 3-kinase is essential for polyomavirus middle T antigen-mediated transformation. J. Virol. 81:7069–7076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Vindrieux D., Escobar P., Lazennec G. 2009. Emerging roles of chemokines in prostate cancer. Endocr. Relat. Cancer 16:663–673 [DOI] [PubMed] [Google Scholar]
- 67. Vleugel M. M., Greijer A. E., Bos R., van der Wall E., van Diest P. J. 2006. c-Jun activation is associated with proliferation and angiogenesis in invasive breast cancer. Hum. Pathol. 37:668–674 [DOI] [PubMed] [Google Scholar]
- 68. Wang S., et al. 2003. Prostate-specific deletion of the murine Pten tumor suppressor gene leads to metastatic prostate cancer. Cancer Cell 4:209–221 [DOI] [PubMed] [Google Scholar]
- 69. Wang S., et al. 2006. Pten deletion leads to the expansion of a prostatic stem/progenitor cell subpopulation and tumor initiation. Proc. Natl. Acad. Sci. U. S. A. 103:1480–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Webster M. A., et al. 1998. Requirement for both Shc and phosphatidylinositol 3′ kinase signaling pathways in polyomavirus middle T-mediated mammary tumorigenesis. Mol. Cell. Biol. 18:2344–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Whitman M., Kaplan D. R., Schaffhausen B., Cantley L., Roberts T. M. 1985. Association of phosphatidylinositol kinase activity with polyoma middle-T competent for transformation. Nature 315:239–242 [DOI] [PubMed] [Google Scholar]
- 72. Yang L., et al. 2008. Abrogation of TGF beta signaling in mammary carcinomas recruits Gr-1+CD11b+ myeloid cells that promote metastasis. Cancer Cell 13:23–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zhang J., Thomas T. Z., Kasper S., Matusik R. J. 2000. A small composite probasin promoter confers high levels of prostate-specific gene expression through regulation by androgens and glucocorticoids in vitro and in vivo. Endocrinology 141:4698–4710 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.