Abstract
The RNA replication and transcription complex of coronaviruses is associated with an elaborate reticulovesicular network (RVN) of modified endoplasmic reticulum. Using cycloheximide and puromycin, we have studied the effect of translation inhibition on the RNA synthesis of severe acute respiratory syndrome coronavirus and mouse hepatitis virus. Both inhibitors prevented the usual exponential increase in viral RNA synthesis, with immunofluorescence and electron microscopy indicating that RVN development came to a standstill. Nevertheless, limited RNA synthesis was supported, implying that continued translation is not an absolute requirement and suggesting a direct link between RVN formation and accumulation of coronavirus proteins.
TEXT
Coronaviruses have a positive-strand, approximately 30-kb RNA genome of which the 5′ two-thirds encodes two large replicase polyproteins (pp1a and pp1ab) that are autoproteolytically cleaved to produce 15 or 16 mature nonstructural proteins (nsps) (3, 16, 21). The majority of nsps assemble into the viral replication and transcription complex (RTC), which includes the viral RNA-dependent RNA polymerase (nsp12) (17). The 3′-proximal part of the coronavirus genome encodes structural and accessory proteins, which are expressed from a nested set of subgenomic mRNAs (9, 12, 15).
Coronavirus infection induces elaborate membrane structures (5, 6, 18) that presumably serve as an RTC scaffold, a common feature among positive-strand RNA viruses (1, 2, 4, 8, 10, 11). This reticulovesicular membrane network (RVN) is derived from and continuous with the endoplasmic reticulum (ER) (6) and includes convoluted membranes (CMs) and a large number of unusual double-membrane vesicles (DMVs). The exact role in viral RNA synthesis of different RVN subdomains remains to be elucidated. Both viral nsps and double-stranded RNA (dsRNA) are associated with the RVN, but, surprisingly, they barely colocalize: whereas the bulk of viral replicase proteins are CM associated, dsRNA is mainly found inside DMVs (6).
In this study, we have addressed the relationship between coronavirus mRNA translation, RNA synthesis, and RVN development. Previously, the RNA synthesis of mouse hepatitis virus (MHV) was reported to decrease almost instantly after addition of the translation inhibitor cycloheximide (CHX) (14). It was suggested that the MHV RTC is structurally labile or contains an unstable protein factor. In an in vitro RTC activity assay for severe acute respiratory syndrome-related coronavirus (SARS-CoV), neither CHX nor the alternative translation inhibitor puromycin (PUR) directly affected RNA synthesis (19). To explore the impact of translation inhibition in more detail, we have now combined biochemical and cell biological approaches to analyze the effect of CHX and PUR treatment in MHV- and SARS-CoV-infected cells.
The effect of translation inhibition by CHX or PUR was first measured by metabolic labeling of viral RNA synthesis in infected cells, essentially as previously described (7). MHV-infected 17Clone-1 cells and SARS-CoV-infected Vero-E6 cells were compared, while taking into account the different infection kinetics of these two viruses. For each inhibitor, a dose was chosen that gave complete translation inhibition within 30 min after addition to the culture medium, which was verified by metabolic labeling of cellular protein synthesis (Fig. 1A). Cellular transcription was inhibited by using actinomycin D, and viral RNA synthesis was measured at different time points after translation inhibition by labeling with [3H]uridine for 1 h.
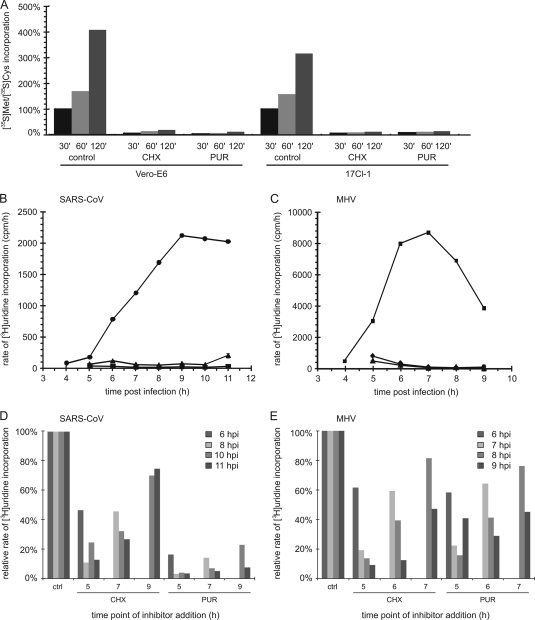
Fig. 1.
Translation inhibition affects the rate of coronavirus RNA synthesis. (A) Translation inhibition by CHX or PUR in Vero-E6 cells (100 μg/ml CHX or 1 mg/ml PUR) and 17Clone-1 (17Cl-1) cells (100 μg/ml of both inhibitors). Protein synthesis was measured for 30 to 120 min after the simultaneous addition of 35S-labeled methionine/cysteine and inhibitor. (B to E) Time course of SARS-CoV (B and D) and MHV (C and E) RNA synthesis in Vero-E6 and 17Clone-1 cells, respectively, labeled with [3H]uridine for 1 h prior to lysis in the presence or absence (control [●]) of the translation inhibitors CHX (100 μg/ml [▴]) and PUR (1 mg/ml [panel B] or 100 μg/ml [panel C] [■]). Labeling was performed as described previously (7), and incorporation was quantified by liquid scintillation counting of aliquots of cell lysates corresponding to ∼3,500 cells. Cellular transcription was inhibited by giving 10 μg/ml of actinomycin D 1 h prior to labeling. CHX or PUR was added at 4 h p.i. to SARS-CoV-infected Vero-E6 cells (B) or MHV-infected 17Clone-1 cells (C), and viral RNA synthesis was measured each hour for the duration of the experiment. Subsequently, the effect of varying the time point of translation inhibition was studied by treating SARS-CoV-infected Vero-E6 cells with CHX or PUR at 5, 7, or 9 h p.i. (D) and MHV-infected 17Clone-1 cells at 5, 6, or 7 h p.i. (E), after which RNA synthesis was measured for 1 h at the indicated time points. The average of two experiments is shown.
SARS-CoV RNA synthesis became measurable at 5 h postinfection (p.i.) and peaked around 9 h p.i. (Fig. 1B). In agreement with previous reports (14), MHV RNA synthesis peaked around 7 h p.i. (Fig. 1C). Addition of CHX or PUR at 4 h p.i. blocked RNA synthesis of both viruses—immediately for SARS-CoV and somewhat more slowly for MHV (Fig. 1B and C), which may reflect differences in stability between the RTCs of the two viruses. For both coronaviruses studied here, the inhibitory effect of early PUR addition on viral RNA synthesis was similar to that of CHX. However, when the inhibitors were added at later time points, a difference became apparent. In the first hour of PUR treatment, SARS-CoV RNA synthesis dropped below 20% and subsequently declined to background levels (Fig. 1D). In the presence of CHX, however, RNA synthesis diminished rapidly to about 50% but then decreased only slowly during the hours that followed. For MHV, the latter trend was observed when either translation inhibitor was used (Fig. 1E), thus resulting in a less pronounced inhibition by CHX treatment than previously reported (14) and in longer-lasting residual viral RNA synthesis. In the case of SARS-CoV, the different inhibition profiles obtained with CHX and PUR suggest that the effect of the latter drug extends beyond the dissociation of ribosomes. Whereas CHX essentially “freezes” translating ribosomes on their mRNA, PUR treatment results in polysome disruption and affects the morphology of the ER (13), which may hamper RVN formation or function and, consequently, RNA synthesis. Recent studies using the drug brefeldin A also suggested a direct link between the integrity of the secretory pathway, RVN formation, and viral RTC activity (7, 20). Although the specific combination of cell lines and drugs used here may have influenced our analysis to a certain extent, our data do indicate that a limited level of coronavirus RTC activity can be maintained for several hours in the absence of de novo protein synthesis.
Next, for both SARS-CoV- and MHV-infected cells, we analyzed the distribution and accumulation of nonstructural proteins and viral RNA by performing dual-immunofluorescence (IF) microscopy as described previously (6, 7). We employed rabbit antisera recognizing the transmembrane replicase subunit nsp4 and a mouse monoclonal antibody labeling double-stranded RNA, with the latter presumably representing the replicative intermediates of the viral RNA. Although variable between individual cells, on average the numbers of small nsp4- and dsRNA-containing foci present at 4 h p.i. remained essentially the same during subsequent CHX or PUR treatment, in contrast to the significant proliferation of the signal observed in untreated control cells (Fig. 2). Still, in SARS-CoV-infected cells, the customary development of the early, small foci into a perinuclear collection of larger foci was not prevented by translation inhibition (Fig. 2A). In MHV-infected cells, such a migration to the perinuclear area did not occur during CHX or PUR treatment (Fig. 2B). In addition, when cultures infected with SARS-CoV or MHV at a multiplicity of infection of 1 were given translation inhibitors early in infection (4 or 2 h p.i.), the percentage of IF-positive cells essentially remained stable for the next 3 h (Fig. 2C). In the same time frame (4 to 7 or 2 to 5 h p.i., respectively), the number of positive cells tripled in untreated control cultures infected with SARS-CoV or MHV. In terms of RVN development, these IF data suggested that translation inhibition leads to a “status quo,” with complexes formed prior to drug treatment remaining stable and, according to the metabolic labeling data, able to sustain a modest level of viral RNA synthesis (Fig. 1).
Fig. 2.
Subcellular localization of nsp4 and dsRNA following translation inhibition. SARS-CoV-infected Vero-E6 cells (A) and MHV-infected 17Clone-1 cells (B) were treated with CHX or PUR from 4 to 7 h p.i. (SARS-CoV) or 2 to 5 h p.i. (MHV). Cells were then fixed and analyzed by dual-labeling IF microscopy for nsp4 (red) and dsRNA (green). Nuclei were visualized by Hoechst 33258 staining (blue). The bar represents 25 μm. (C) Microscopic images containing between 300 and 450 cells were analyzed for nsp4 and dsRNA labeling and scored “IF positive” when either of these was visible. The average and standard error of the mean (SEM) of all cells per sample are shown.
Recently, the morphogenesis and ultrastructure of the SARS-CoV-induced RVN were documented in detail by transmission electron microscopy (TEM) and electron tomography (6). Using TEM, we now compared the RVN ultrastructures in SARS-CoV-infected Vero cells fixed at either 4 or 7 h p.i., including cultures that had been treated with either CHX or PUR from 4 to 7 h p.i. (Fig. 3). Whereas CHX-treated and untreated mock-infected cells were morphologically indistinguishable, PUR treatment resulted in the previously described strong reduction of polysome numbers and an increase of tubular ER (data not shown) (13). Between 4 and 7 h p.i., the RVN developed from mostly clusters of individual DMVs into the vesicle packets (VPs) that are typical of the later stages of infection (Fig. 3A and B) (6). Following CHX or PUR treatment, RVN development was significantly hampered, and only rarely did single DMVs transform into the larger VPs (Fig. 3C and D). In line with the IF data presented in Fig. 2, however, translation inhibition did not induce degradation of virus-induced membrane structures, which likely continued to support limited viral RNA synthesis (Fig. 1).
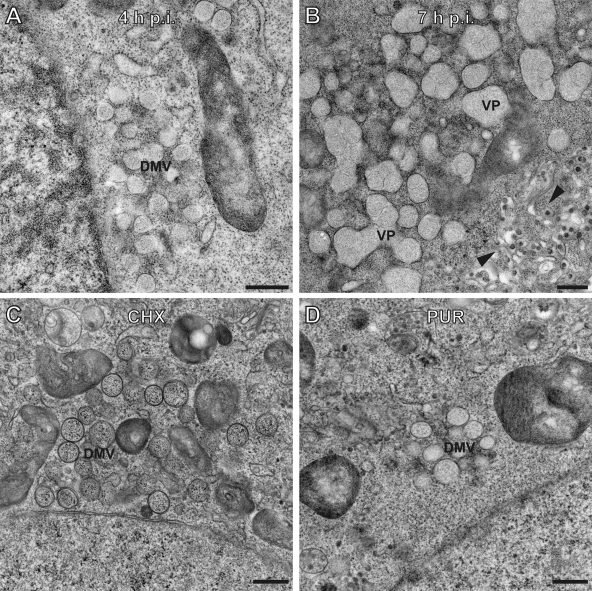
Fig. 3.
Development of SARS-CoV double-membrane vesicles into vesicle packets is hampered when translation is inhibited. Electron micrographs of SARS-CoV-infected Vero-E6 cells showing normal development of double-membrane vesicles (DMVs) at 4 h p.i. (A) and vesicle packets (VPs) at 7 h p.i. (B). Treatment with CHX (C) or PUR (D) starting at 4 h p.i. prevented the customary development of the virus-induced RVN, resulting in the prolonged presence of DMV clusters and a lack of VPs at 7 h p.i. Bars represent 500 nm.
Next, a quantitative DMV analysis was performed by cutting thin EM sections in a plane parallel with the cell monolayer and counting inner vesicles present in DMVs and VPs. As described previously (6), counting was facilitated by stitching electron micrographs covering a complete slice through the center of the cell into large mosaics, each composed of 25 to 100 images. Per condition, 32 cells from three independent experiments were analyzed to calculate the average number of SARS-CoV-induced vesicles per μm2 of cytoplasm. Unfortunately, the analysis was hampered by the fact that DMVs are still relatively rare at 4 h p.i. (i.e., cells may be infected, but random EM images may not reveal any DMVs) (Fig. 2). Also the relative asynchronicity of infection between cells in the same culture contributed to significant variation in DMV counts. Still, as documented previously (6), average DMV numbers clearly increased between 4 and 7 h p.i. Upon translation inhibition with either CHX or PUR, this trend was clearly lacking (Fig. 4), suggesting a direct connection between RVN development and the accumulation of coronavirus proteins.
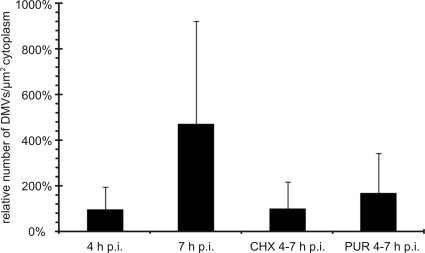
Fig. 4.
The number of RVN inner vesicles is stable when translation is inhibited. Thin sections of SARS-CoV-infected Vero-E6 cells were used to count RVN inner vesicles in DMVs or VPs. For this purpose, images covering a complete slice through the center of the cell were stitched into large mosaics to facilitate the analysis. Per condition, 32 cells from three independent experiments were analyzed. The graph shows the average number of inner vesicles per square micrometer of cytoplasm and illustrates how translation inhibition by either CHX or PUR prevented expansion of the number of inner vesicles as it normally occurs between 4 and 7 h p.i. DMVs were never observed in mock-infected cells. Error bars represent the standard deviations of the averages from three independent experiments.
Taken together, our data suggest that the coronavirus RVN-forming membrane alterations, which are likely triggered by the synthesis of increasing amounts of transmembrane proteins like nsp3, nsp4, and nsp6, contribute to the efficiency of the replicative cycle, rather than being the mere consequence of cellular responses to infection. Our study of RNA synthesis and RVN development early in infection revealed that coronavirus RTCs (i) are stable even when viral protein synthesis is stalled, (ii) remain capable of limited but sustained RNA synthesis under these conditions, and (iii) can be active in the absence of RVN expansion, although it is clear that the normal development of replication structures likely contributes to the rapid increase in viral RNA synthesis that is typical of the exponential phase of infection.
Acknowledgments
This work was supported by the Council for Chemical Sciences of the Netherlands Organization for Scientific Research (NWO-CW) through ECHO grant 700.55.002 and TOP grant 700.57.301.
Footnotes
Published ahead of print on 23 March 2011.
REFERENCES
- 1. Carette J. E., Stuiver M., Van Lent J., Wellink J., Van Kammen A. 2000. Cowpea mosaic virus infection induces a massive proliferation of endoplasmic reticulum but not Golgi membranes and is dependent on de novo membrane synthesis. J. Virol. 74:6556–6563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Egger D., Teterina N., Ehrenfeld E., Bienz K. 2000. Formation of the poliovirus replication complex requires coupled viral translation, vesicle production, and viral RNA synthesis. J. Virol. 74:6570–6580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gorbalenya A. E., Enjuanes L., Ziebuhr J., Snijder E. J. 2006. Nidovirales: evolving the largest RNA virus genome. Virus Res. 117:17–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gosert R., et al. 2003. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J. Virol. 77:5487–5492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gosert R., Kanjanahaluethai A., Egger D., Bienz K., Baker S. C. 2002. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 76:3697–3708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Knoops K., et al. 2008. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 6:e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Knoops K., et al. 2010. Integrity of the early secretory pathway promotes, but is not required for, severe acute respiratory syndrome coronavirus RNA synthesis and virus-induced remodeling of endoplasmic reticulum membranes. J. Virol. 84:833–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Miller D. J., Schwartz M. D., Dye B. T., Ahlquist P. 2003. Engineered retargeting of viral RNA replication complexes to an alternative intracellular membrane. J. Virol. 77:12193–12202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Narayanan K., Huang C., Makino S. 2008. SARS coronavirus accessory proteins. Virus Res. 133:113–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pedersen K. W., van der Meer Y., Roos N., Snijder E. J. 1999. Open reading frame 1a-encoded subunits of the arterivirus replicase induce endoplasmic reticulum-derived double-membrane vesicles which carry the viral replication complex. J. Virol. 73:2016–2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Peranen J., Kaariainen L. 1991. Biogenesis of type I cytopathic vacuoles in Semliki Forest virus-infected BHK cells. J. Virol. 65:1623–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Perlman S., Netland J. 2009. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 7:439–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Puhka M., Vihinen H., Joensuu M., Jokitalo E. 2007. Endoplasmic reticulum remains continuous and undergoes sheet-to-tubule transformation during cell division in mammalian cells. J. Cell Biol. 179:895–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sawicki S. G., Sawicki D. L. 1986. Coronavirus minus-strand RNA synthesis and effect of cycloheximide on coronavirus RNA synthesis. J. Virol. 57:328–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sawicki S. G., Sawicki D. L., Siddell S. G. 2007. A contemporary view of coronavirus transcription. J. Virol. 81:20–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Snijder E. J., et al. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331:991–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Te Velthuis A. J. W., Arnold J. J., Cameron C. E., van den Worm S. H. E., Snijder E. J. 2010. The RNA polymerase activity of SARS-coronavirus nsp12 is primer dependent. Nucleic Acids Res. 38:203–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ulasli M., Verheije M. H., de Haan C. A., Reggiori F. 2010. Qualitative and quantitative ultrastructural analysis of the membrane rearrangements induced by coronavirus. Cell. Microbiol. 12:844–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. van Hemert M. J., et al. 2008. SARS-coronavirus replication/transcription complexes are membrane-protected and need a host factor for activity in vitro. PLoS Pathog. 4:e1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Verheije M. H., et al. 2008. Mouse hepatitis coronavirus RNA replication depends on GBF1-mediated ARF1 activation. PLoS Pathog. 4:e1000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ziebuhr J., Snijder E. J., Gorbalenya A. E. 2000. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 81:853–879 [DOI] [PubMed] [Google Scholar]