Abstract
In Arabidopsis seedlings and cauliflower florets, Rpn6 (a proteasome non-ATPase regulatory subunit) was found in two distinct protein complexes of ∼800 and 500 kDa, respectively. The large complex likely represents the proteasome 19S regulator particle (RP) because it displays the expected subunit composition and all characteristics. The small complex, designated PR500, shares at least three subunits with the “lid” subcomplex of 19S RP and is loosely associated with an hsp70 protein. In Arabidopsis COP9 signalosome mutants, PR500 was specifically absent or reduced to an extent that correlates with the severity of the mutations. Furthermore, PR500 was also diminished in response to potential protein-misfolding stresses caused by the heat shock and canavanine treatment. Immunofluorescence studies suggest that PR500 has a distinct localization pattern and is enriched in specific nuclear foci. We propose that PR500 may be evolved in higher plants to cope with the frequently encountered environmental stresses.
INTRODUCTION
The 26S proteasome is a major proteolytic device for selective protein breakdown in eukaryotes (Hershko and Ciechanover, 1998). This targeted protein degradation is one of the most important means utilized by all eukaryotes to control cellular protein quality, transcription, signal transduction, cell cycle progression, and metabolic activities.
The 26S proteasome is composed of a 20S catalytic core and two 19S regulatory particles on both ends. The 20S core particle (20S CP) is a hollow cylinder with interior proteolytic sites. The 19S regulatory particle (19S RP) is composed of six ATPases and at least 11 non-ATPase subunits in yeast (Baumeister et al., 1998; Glickman et al., 1998b). ATP can stimulate association of 19S RP with the 20S CP to form the 26S proteasome that is capable of degrading ubiquitin-tagged proteins (Orino et al., 1991). The 19S RP is believed to direct substrate recognition and processing (ubiquitin binding, unfolding, and deubiquitination) before substrates are funneled into the 20S CP for destruction (Coux et al., 1996; Baumeister et al., 1998). The 19S particle of Saccharomyces cerevisiae can be divided into two subcomplexes, the base and the lid (Glickman et al., 1998a). The base subcomplex consists of all six ATPases and Rpn1 and Rpn2 (the two largest non-ATPase subunits), and is in direct contact with the 20S CP. The lid subcomplex contains eight non-ATPase subunits, Rpn3, Rpn5–9, Rpn11, and Rpn12. It covers one or both ends of the 26S proteasome cylinders by contacting the base subcomplex. Rpn10 seems to play a role in connecting the lid to the base, because the lid can be easily disassociated from the 26S proteasome when Rpn10 is absent (Glickman et al., 1998a). More recently, a lid-like complex was isolated as a proteasome purification by-product of human red blood cells (Braun et al., 1999; Henke et al., 1999). However, there is no report until now to indicate that a lid-like subcomplex is present in vivo under physiological conditions in any organism.
On the other hand, the COP9 signalosome, a highly conserved cellular regulator (Wei and Deng, 1999), exhibits a remarkable resemblance to the lid subcomplex of the proteasome (Glickman et al., 1998a; Seeger et al., 1998; Wei et al., 1998). The COP9 signalosome also consists of eight subunits, and each of the subunits shares significant sequence similarities with a particular lid subunit in a one-to-one correlation. This suggests that the COP9 signalosome and the lid have likely evolved from a common ancestor (Wei and Deng, 1999). Recently, structures of purified human COP9 signalosome and lid subcomplex were compared by electron microscopy imaging (Kapelari et al., 2000). The result indicates that both complexes lack any symmetry in subunit arrangement and exhibit a central groove, although they do not appear to have identical structure. Although the lid is conserved throughout eukaryotes (Glickman et al., 1998a), COP9 signalosome is found from fission yeast to plants and mammals but is apparently absent from S. cerevisiae (Deng et al., 2000).
The COP9 signalosome was initially defined as a light-inactivable repressor of photomorphogenesis in Arabidopsis (Wei et al., 1994; Chamovitz et al., 1996). Mutant seedlings lacking the complex exhibit a constitutive photomorphogenic phenotype, constitutive activation of light-responsive gene expression, and lethality after the seedling stage (Wei and Deng, 1996, 1999). In mammals, subunits of the COP9 signalosome have been implicated to regulate multiple cellular signaling pathways and cell cycle progression (Wei and Deng, 1999). Most recently, a role of the COP9 signalosome in the proteasome-mediated degradation of a key photomorphogenic regulator HY5 has been documented (Osterlund et al., 2000). Together, available data points to a functional relation between the COP9 signalosome and the proteasome. However, there is no information available yet regarding the nature of this relationship.
As an initial step in searching the possible relationship between those two complexes, we characterized the in vivo forms of the Rpn6 protein and its possible regulation by the COP9 signalosome. Here we report the identification and purification of am Rpn6-containing complex named PR500. PR500 stably exists in plant cells under physiological conditions and localizes to distinct nuclear foci in plant cell nuclei. We showed that PR500 is absent or significantly reduced in stress conditions in which abnormal proteins are expected to accumulate and in the COP9 signalosome mutants of Arabidopsis, suggesting a role of PR500 in cellular stress response in plants.
MATERIALS AND METHODS
Plant Materials and Arabidopsis Strains
The fus6–1, fus6-G236, fus6-T236, cop8-1, cop10-1, and fus11-U203 mutants have been described (Castle and Meinke, 1994; Wei et al., 1994; Kwok et al., 1996; Karniol et al., 1999; Serino et al., 1999). The det1-1 and det1-8 were described by Pepper et al. (1994). Seeds were planted on agar plates containing growth medium (GM) and 1% sucrose (Wei et al., 1994) and were cold treated at 4°C for 7–12 d before being transferred to growth chambers. The fluence rate of white light was 156 μmol m−2 s−1. Cauliflower heads used for gel filtration and complex purification were purchased fresh from a local market.
Heat Shock and Canavanine Treatments
Five-day-old Arabidopsis seedlings, grown continuously in white light on GM plates with 1% sucrose, were transferred from a 22°C growth chamber to a 42°C chamber for 3 h or the time period indicated. For the canavanine treatment experiment, cold-treated Arabidopsis seeds were germinated on GM medium for 3 d. Then, the seedlings were transferred to GM medium supplemented with 0 or 4 mg/l canavanine and grown for 2 d more under continuous white light.
Protein Extraction and Gel Filtration Chromatography
Arabidopsis seedlings or cauliflower tissue were homogenized in liquid nitrogen (Arabidopsis) or in cold buffer (cauliflower) containing 50 mM NaCl, 10 mM MgCl2, 5 mM EDTA, 2 mM dithiothreitol (DTT), 10% glycerol, protease inhibitor cocktail (Boehringer Mannheim, Indianapolis, IN) and 25 (cauliflower) or 50 (Arabidopsis) mM Tris-HCl, pH 7.5, with phenylmethylsulfonyl fluoride and β-mercaptoethanol added to 5 mM, respectively, just before use. The homogenate was centrifuged for 10 min, and the supernatant was filtered through a 0.2-μm filter (Gelman Sciences, Ann Arbor, MI) before loading onto a Superose 6 (HR 10/30) gel filtration column (Pharmacia, Piscataway, NJ). The column was equilibrated with a PBSM buffer (1.76 mm KH2PO4, 10 mm Na2HPO4, 136 mm NaCl, 2.6 mm KCl, 2 mm MgCl2, and 10% glycerol) or Tris buffer (50 mM Tris, pH 7.5, 50 mM NaCl, 5 mM EDTA, 10 mM MgCl2, 10% glycerol, and 1 mM DTT). Proteins were eluted in the same buffer for column equilibration at a flow rate of 0.2 ml/min. All manipulations were carried out at 4°C. Fractions of 0.25 or 0.5 ml were collected starting from the onset of the column void volume (7.5 ml) and concentrated using StrataClean Resin (Stratagene, La Jolla, CA) as described before (Kwok et al., 1998). Equal volumes of each fraction were subjected to SDS-PAGE followed by immunoblot analysis. The molecular mass standards were described previously (Kwok et al., 1999). When ATP was included, it was 10 mM in the homogenization buffer and 2 mM in subsequent gel filtration buffers, or as indicated in the text.
Immunoblot Analysis and Immunoprecipitation
Western blot and immunoprecipitation analyses were carried out as previously described (Staub et al., 1996), except that Tween-20 was not included in the PBS buffer during immunoprecipitation, and a final concentration of 0.55 M NaCl was included in the PBS washing buffer. Rabbit polyclonal antibodies to Rpn6 and Rpt5 were described previously (Kwok et al., 1999). The polyclonal antibodies against Arabidopsis Mbp1/Rpn10 and Rpt1 (subunits of 19S RP) were reported before (van Nocker et al., 1996). The monoclonal antibodies against 21D7/Rpn3 was described by Smith et al. (1997). The antibodies against human Rpt2 (S4) were kindly provided by Dr. Carlos Gorbea (University of Utah Medical School, Salt Lake City, UT). The rabbit polyclonal antibodies against hsp70 were made against purified wheat germ hsp70 protein (Crookes and Olsen, 1998). The antibodies against Rpn5a will be described separately (Kurepa and Vierstra, unpublished data).
Arabidopsis Protoplast Immunofluorescence Staining
The procedure for protoplast preparation and immunofluorescence was as described previously (Matsui et al., 1995; Kwok et al., 1998). Fresh seedlings were picked and immediately chopped with razor blades to small pieces. The pieces were added to the solution containing cell-wall–digesting enzymes as described by Matsui et al. (1995). Purified rabbit polyclonal antisera against Rpn6, Rpt5, or COP9 signalosome subunits were used for staining the protoplasts after affinity purification (Kwok et al., 1998). Secondary antibody was a goat anti-rabbit antibody conjugated to fluorescein isothiocyanate (Sigma, St. Louis, MO) and was used at a dilution of 1:400. The samples were double stained for nuclei with 1 mg/ml 4′,6-diamidino-2-phenylindole.
Purification of the Cauliflower 19S and PR500 Complexes
The surface layer of cauliflower heads was shaved and homogenized with a commercial blender in buffer A (25 mM Tris-HCl, pH 7.5, 20 mM NaCl, 6 mM MgCl2, 5 mM EDTA, 5 mM β-mercaptoethanol, 2 mM DTT, and 10% glycerol) containing 2 mM phenylmethylsulfonyl fluoride and the protease inhibitor cocktail (Boehringer Mannheim). Homogenates were filtered through eight layers of cheesecloth and centrifuged for 30 min at 13,000 × g at 4°C. Ammonium sulfate fine powder was added slowly to the supernatant to bring the ammonium sulfate concentration to 30% saturation. The mixture was incubated at 4°C for 1 h and centrifuged for 30 min at 13,000 × g. The supernatant was filtered through a 2-μm pore size nylon filter and was applied to a 300 ml phenyl Sepharose (Pharmacia) column. The column was initially equilibrated with buffer B (25 mM Tris-HCl, pH 7.5, 2 mM MgCl2, 10% glycerol, and 1 mM DTT) containing ammonium sulfate to 30% saturation and 10 mM NaCl. The column was then washed with (≥3 volumes) buffer B containing a 15% saturation of ammonium sulfate. The Rpn6-enriched fractions were eluted with buffer B supplemented with 10 mM NaCl. The eluted fraction was passed through a P-DG gel desalting column (Bio-Rad, Hercules, CA) to completely remove trace amounts of ammonium sulfate. The sample was loaded onto a Q-Sepharose ion exchange column and was washed (≥3 volume) with buffer B containing 150 mM NaCl. The complexes were eluted with 350 mM NaCl in buffer B. The eluted fraction was then adjusted to a NaCl concentration of 100 mM with buffer B before separation on a heparin affinity column (Pharmacia). The heparin column was washed with 4 volumes of the loading buffer (buffer B with 100 mM NaCl), and the Rpn6-associated complexes were eluted with 350 mM NaCl in buffer B. The Rpn6-containing fraction was adjusted to a NaCl concentration of 200 mM and loaded onto a 1 ml Mono-Q column (Pharmacia). A linear 200–400 mM NaCl gradient elution (50 ml total) was used to separate the PR500 complex (eluted at 240 mM) from the 19S complex (eluted at 270 mM). The eluted fractions (1 ml each) containing PR500 and 19S particles were pooled separately, and each of the complexes was further purified on a Superose 6 HR columns in the PBSM buffer (1.76 mM KH2PO4, 10 mM Na2HPO4, 136 mM NaCl, 2.6 mM KCl, 2 mM MgCl2 and 10% glycerol). The proteins were collected in 0.5-ml fractions, and each fraction was concentrated by Strataclean beads (Stratagene). The proteins were eluted from the beads by mixing with 2× SDS gel-loading buffer and boiling for 3 min, separated by a 10% SDS-PAGE, and visualized by silver or Coomassie blue staining. In large-scale purification, the peak fractions of PR500 from the Superose 6 HR gel filtration column were pooled and applied onto a 1-ml heparin column for a gradient elution after the NaCl concentration was adjusted. The linear gradient from 100–300 mM NaCl was carried out over 20 ml in buffer B plus 0.25% Nonidet P-40 without glycerol.
RESULTS
Rpn6 Is Present in 19S RP as Well as a Smaller Protein Complex
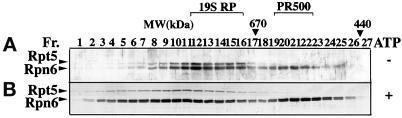
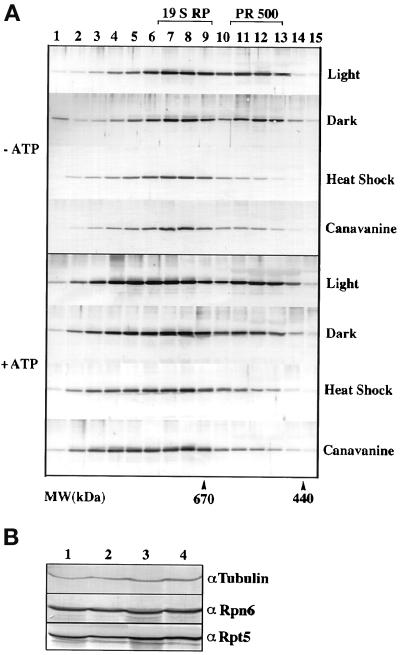
We recently reported the molecular characterization of Rpn6 (AtS9) and Rpt5 (AtS6A), a non-ATPase and an ATPase subunit of the Arabidopsis 19S RP (Kwok et al., 1999). To further examine their functional forms, we tested whether Rpn6 is also present in any form other than in the 19S RP. Gel filtration chromatography was used to size fractionate crude plant extracts prepared from Arabidopsis seedlings or cauliflower florets. In both cases, Rpn6 was eluted in two separate complexes with estimated molecular masses of ∼800 and 500 kDa in all buffers tested (Figure 1A). In contrast, Rpt5 was eluted in only a single 800-kDa complex cofractionating with Rpn6 and was not found in any other major form in the absence of ATP. The 800-kDa complex size is similar to the reported molecular size of the mammalian 19S RP (Chu-ping et al., 1994; Sawada et al., 1997). Consistent with Rpn6 and Rpt5 being subunits of the 19S RP, Rpn6 and Rpt5 not only cofractionated but also coimmunoprecipitated each other from plant extract (Kwok et al., 1999). These results led us to hypothesize that the 800-kDa Rpn6- and Rpt5-containing complex is the Arabidopsis and cauliflower 19S RP.
Figure 1.
Rpn6 is present in two distinct protein complexes in plants. Cell extract from cauliflower head was fractionated in a Superose 6 gel filtration column. Selected elution fractions (Fr.) of 0.25 ml each were analyzed by immunoblot for Rpn6 and Rpt5. The elution positions of molecular mass markers are shown (in kDa) above the gel blots. (A) Cell extraction and subsequent size fractionation were conducted under standard condition (see MATERIALS AND METHODS) in which no ATP was present. (B) ATP was included in the extraction buffer, the column equilibration buffer, and elution buffer in otherwise identical experimental conditions as in A. Note that Rpn6 is eluted in a peak at ∼500 kDa in the presence and absence of ATP and that both Rpn6 and Rpt5 cofractionate in an 800-kDa peak in the absence of ATP and in fractions of larger molecular mass in the presence of ATP.
One characteristic of the 19S particle is its ATP-dependent association with the 20S CP to form the 26S proteasome. Therefore, we tested whether Rpn6 and Rpt5 could be incorporated into larger complexes in the presence of ATP. As shown in Figure 1B, both Rpn6 and Rpt5 shifted from the 800-kDa complex toward larger molecular mass complexes when ATP was present in the extraction and the subsequent gel filtration buffers. Thus, the 800-kDa complex is likely to be the 19S RP, and the higher molecular mass complexes that we observed in the presence of ATP are the 26S proteasome with one or two 19S RP. In contrast, the 500-kDa Rpn6-containing complex is not affected by ATP (Figure 1B). We tentatively designate this 500-kDa complex as PR500 for proteasome-related 500-kDa complex. To further confirm that PR500 indeed exists in vivo, different buffer strengths (25, 50, and 200 mM Tris), different ion strengths (6 or 10 mM Mg2+, and 20, 100, or 200 mM NaCl), and different buffer pH (6–8.5) were also used to extract proteins and elute proteins from gel filtration column. PR500 peak was constantly observed (unpublished results).
PR500 Contains a Subset of the 19S RP Subunits
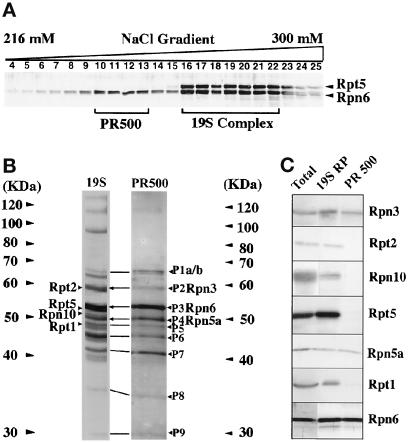
To ascertain whether the 800-kDa complex is the 19S regulator and to reveal the composition of PR500, we purified both complexes to near homogeneity from cauliflower (see MATERIALS AND METHODS). As shown in Figure 2A, the two complexes could be separated from each other in a Mono-Q column by a fine salt gradient elution. Peak fractions containing PR500 (fractions 10–13) or 19S RP (fractions 16–22) from the Mono-Q column were pooled. Each of the complexes was further purified by Superose 6 size fractionation, which also ensured that the complexes of native sizes were isolated.
Figure 2.
Purification and characterization of PR500 and 19S RP from cauliflower. (A) PR500 was separated from the 19S RP in an NaCl linear gradient on a Mono-Q column. Fractions (1 ml each) taken from 216 mM (fraction 4) to 300 mM NaCl (fraction 25) were analyzed for Rpn6 and Rpt5 by immunoblot. Fractions 10–13 were pooled as PR500 and fractions 16–22 were pooled as 19S RP. These two pooled samples were further purified by gel filtration as detailed in MATERIALS AND METHODS. (B) Compositional comparison between the purified PR500 and 19S RP preparations. The silver-stained SDS-PAGE lanes for each complex are shown, with positions of molecular size markers indicated. The identities of the seven subunits, three subunits shared between the two complexes and the four subunits present in only 19S RP, were confirmed by Western blot and are marked. Note that the corresponding protein bands between 19S RP and PR500 are indicated by a line or arrow (only the three with know identities). (C) Western blot analysis of the two purified complexes. All seven antibodies against distinct proteasome 19S RP subunits reacted with specific bands from the purified 19S RP preparation as well as the total cell extracts, whereas only three of them reacted with the purified PR500 preparation.
The purified 800-kDa complex contained ∼19 major protein bands (Figure 2B). Its protein profile resembled that of 19S RP purified from animal sources (Chu-ping et al., 1994). Its identity as the 19S RP was confirmed by immunoblot analysis using seven antibodies that recognize four non-ATPase (Rpn3, Rpn6, Rpn10, and Rpn5a) and three ATPase subunits of the 19S RP (Rpt1, Rpt2, and Rpt5). All of these antibodies specifically reacted with proteins of the expected sizes (Figure 2C).
Because the PR500 preparation following the gel filtration column still contained a few minor protein bands that did not comigrate with PR500, an additional purification step by heparin gradient chromatography in the presence of 0.25% Nonidet P-40 was carried out (see MATERIALS AND METHODS). This resulted in nine distinct cofractionating bands, ranging from 30–63 kDa (Figure 2B). A direct comparison of the protein profiles of 19S RP and PR500 complexes (Figure 2B) indicated that essentially all proteins in the PR500 complex might also be present in the 19S RP. The only exception was the largest band (P1), which was a doublet in PR500, whereas the corresponding protein of P1 in 19S RP preparation was not (Figure 2B). The nature of this difference is not yet clear. The P1 doublet in PR500 could be due to partial degradation, novel modification, or the presence of a new protein. Assuming all of the cofractionated proteins were components of PR500, we further examined the molecular identity of PR500 components by a Western blot using available antibodies against seven 19S proteasome subunits. Among the seven tested antibodies, three of them (Rpn6, Rpn3, and Rpn5a) recognized a protein in the purified PR500 (Figure 2C). The remaining four proteins, one non-ATPase (Rpn10) and three ATPases (Rpt1, Rpt2, and Rpt5), were not present in the PR500 complex. Because the yeast lid subcomplex is composed of eight non-ATPase subunits including Rpn3, Rpn5, and Rpn6 (Glickman et al., 1998a), PR500 could potentially contain all subunits of the lid subcomplex with room for one or two extra subunits.
An hsp70 Chaperone Protein May Associate with the PR500 Complex
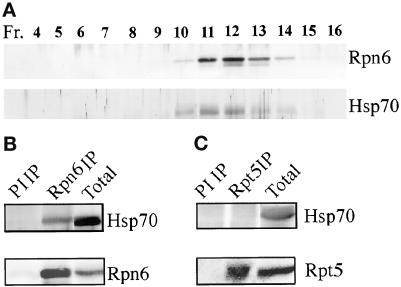
During the purification of the PR500 complex, we noted that a 70-kDa protein cofractionated with PR500 even after the Superose-6 gel filtration step but was lost in the final heparin chromatography step. Because the hsp70 chaperones have recently been shown to be required for the proteasome function (Luders et al., 2000; Pirkakala et al., 2000), we tested whether this 70-kDa protein is related to hsp70 chaperones. Indeed, as shown in Figure 3A, this protein was recognized by polyclonal antibodies against hsp70. To test whether this hsp70 is physically associated with the PR500 complex, we attempted to coimmunoprecipitate hsp70 with either Rpn6 or Rpt5. Whereas Rpn6 antibody could immunoprecipitate hsp70, the Rpt5 antibody could not (Figure 3, B and C). Therefore, we conclude that Arabidopsis hsp70 is specifically associated with PR500 but not with the 19S RP.
Figure 3.
hsp70 is physically associated with PR500. (A) Rpn6 and hsp70 cofractionation profile on Superose 6 HR gel filtration column before the last step of purification (see MATERIALS AND METHODS). The fractions from the gel filtration column were separated by SDS-PAGE and immunoblot analyzed. (B and C) Coimmunoprecipitation with Rpn6 or Rpt5 antibodies, respectively. Proteins partially purified by phenyl Sepharose high performance and Q-Sepharose fast flow columns were used for the immunoprecipitation (IP) experiments. The precipitants were subjected to Western blot analysis with antibodies against hsp70 and Rpn6 or Rpt5. PI IP, preimmune serum immunoprecipitation.
The COP9 Signalosome Is Required for PR500 Accumulation
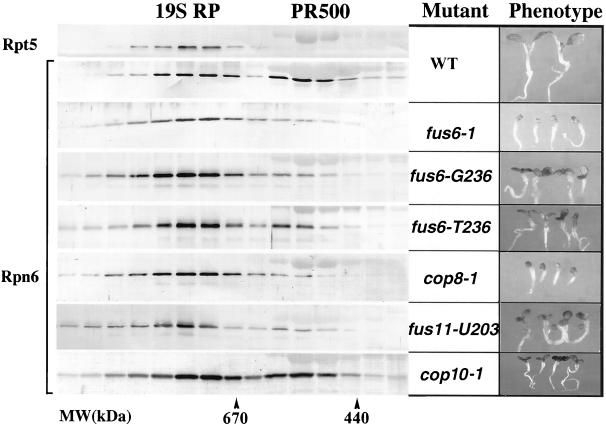
To investigate the possible role of COP9 signalosome in regulating Rpn6-containing complexes, we examined the effect of the COP9 signalosome mutations on the 19S RP and PR500 accumulation. In Arabidopsis, FUS6/COP11, COP8, and FUS11 encode the CSN1, CSN4, and CSN3 subunits of the COP9 signalosome, respectively (Chamovitz et al., 1996; Serino et al., 1999; Deng et al., 2000), and mutations in any of these genes disrupt COP9 signalosome assembly. As shown in Figure 4, a series of Rpn6 gel filtration analyses using equal amounts of wild-type and mutant cell extracts was carried out to examine the levels of the Rpn6-containing complexes. In wild type, whereas Rpt5 was exclusively fractionated with 19S RP, ∼30–40% of Rpn6 was present in PR500. Rpn6 was used as an indication of the PR500 abundance, and it is evident that the PR500 complex was either significantly reduced (fus6-T236, 15%; fus6-G236, 8%; and fus11-U203, 9%) or essentially absent (fus6-1 and cop8-1, <5%) in the different COP9 signalosome mutants (Figure 4). Furthermore, the level of PR500 accumulation correlates with the severity of the molecular lesions as well as with the severity of the phenotype (Figure 4; Peng, Staub, Serino, Kwok, Kurepa, Bruce, Vierstra, Wei, and Deng, unpublished results. In contrast, in det1-8, cop1-5, and cop10-1 mutants, which affect photomorphogenesis to a similar degree as the severe COP9 signalosome mutants but do not affect the level or structural integrity of COP9 signalosome (Kwok et al., 1998), their abundance of PR500 was ∼30–40%, similar to that of wild type (Figure 4; Peng, Staub, Serino, Kwok, Kurepa, Bruce, Vierstra, Wei, and Deng, unpublished results). Furthermore, the presence of ATP in the extraction and gel filtration buffers did not affect PR500 levels observed in any mutant strains, although in these conditions a good portion of the 19S were incorporated into higher molecular mass complexes (Peng, Staub, Serino, Kwok, Kurepa, Bruce, Vierstra, Wei, and Deng, unpublished results).
Figure 4.
The COP9 signalosome is required for PR500 complex accumulation in Arabidopsis. Total cell extracts from light-grown wild-type or mutant seedlings were fractionated on a Superose 6 column. Selected fractions (0.5 ml per fraction) were analyzed for Rpn6 or Rpt5 (top lane) by immunoblot. The positions of 19S RP and PR500 are indicated on top, and the molecular mass standards are indicated at the bottom. Gel filtration was performed as in Figure 1 except that the buffers were modified as described in MATERIALS AND METHODS. Seedlings of Arabidopsis COP9 signalosome mutants and wild-type (WT) are shown on the right for phenotype comparison. The mutant and wild-type seedlings were grown in constant white light for 5 d. All images were taken with the same magnification.
Under our experimental conditions, the distribution of Rpt5 was unaffected by the various mutations examined. It was present only in the fractions containing the 19S RP or putative 26S proteasome in all the mutants described above (Peng, Staub, Serino, Kwok, Kurepa, Bruce, Vierstra, Wei, and Deng, unpublished results) as in wild-type Arabidopsis seedlings (Figure 4, top). Therefore, we concluded that the COP9 signalosome is essential for the accumulation of PR500 but not for the accumulation of 19S RP and for the assembly of the 26S proteasome.
Heat Shock and Canavanine Treatments Down-Regulate PR500
Because the cop1-5, cop10-,1 and det1-8 mutants did not affect PR500 level, it is unlikely that the defective photomorphogenic development in the COP9 signalosome mutants is responsible for the observed reduction of PR500 level. Because the COP9 signalosome is required for regulated protein degradation mediated by the 26S proteasome (Osterlund et al., 2000), it is possible that the defect in proteasome-mediated protein degradation in the COP9 signalosome mutants might be related to the observed PR500 level change. To test the possibility, we examined PR500 levels under conditions elevating the production of misfolded proteins, thus increasing demand of the proteasome activity. First, the arginine analogue canavanine was used to supplement Arabidopsis GM. The incorporation of canavanine instead of arginine into cellular proteins causes protein misfolding, thus generating a large amount of substrate for the ubiquitin-proteasome pathway (Rosenthal et al., 1989; Rosenthal, 1992). Furthermore, heat shock is well known to cause misfolding of cellular proteins, thus increasing the accumulation of substrates for the ubiquitin-proteasome pathway (Alberts et al., 1994). As shown in Figure 5A, when 3-d-old seedlings were transferred to a plate supplemented with 4 mg/l canavanine for 2 d, PR500 peak diminished whether ATP was included in the buffers or not. Furthermore, when 5-d-old seedlings were heat shock treated for 3 h at 42°C, the PR 500 peak also diminished in the two buffer systems tested (Figure 5A). In contrast, light- and dark-grown seedlings had no detectable difference in PR500 abundance. These results suggested that cellular level of PR500 was decreased under stresses where higher proteasome activity is expected.
Figure 5.
Effect of heat shock and canavanine treatments on PR500 accumulation. Gel filtrations were conducted as in Figure 4 and selected fractions were analyzed. (A) Gel filtration profiles of Rpn6 from seedlings grown under indicated conditions. The positions of PR500 and 19S RP are indicated on the top. Molecular size markers are indicated at the bottom. “−ATP, protein extraction and gel filtration were conducted in buffers without ATP. +ATP, protein extraction and gel filtration were conducted in buffers with 10 and 2 mM ATP each. (B) The protein levels of Rpn6 and Rpt5 in the controls and stress-treated Arabidopsis seedlings. Lanes 1 and 3, the protein extracts from the control and heat shock-treated (42°C for 3 h) seedlings. Lanes 2 and 4, the protein extracts from the seedlings grown in the absence or presence of 4 mM canavanine. The tubulin antibodies were used as a control for equal loading.
To evaluate whether PR500 was degraded under the stress treatments, we examined the overall cellular levels of Rpn6 and Rpt5 in the controls and the stress-treated plants. Because PR500 contains a significant portion (30–40%) of the total cellular Rpn6 protein, a net loss of PR500 would result in a noticeable decrease of the Rpn6 relative to that of tubulin, a control for loading, in the total protein Western blot. As shown in Figure 5B, neither canavanine nor heat shock treatments caused detectable changes in the level of Rpn6 and Rpt5 (Figure 5B). Therefore, our result is consistent with the notion that PR500 was not lost under these treatments. Because there is no Rpn6 monomer before or after the treatments, the Rpn6-containing PR500 likely shifted to incorporate into the 26S proteasome or other complexed forms. However, we cannot rule out a possibility that the Rpn6 in PR500 was degraded, and at the same time increased synthesis of Rpn6 was able to compensate this loss of Rpn6.
PR500 Has a Nuclear Localization Pattern Distinct from the COP9 Signalosome and the Proteasome
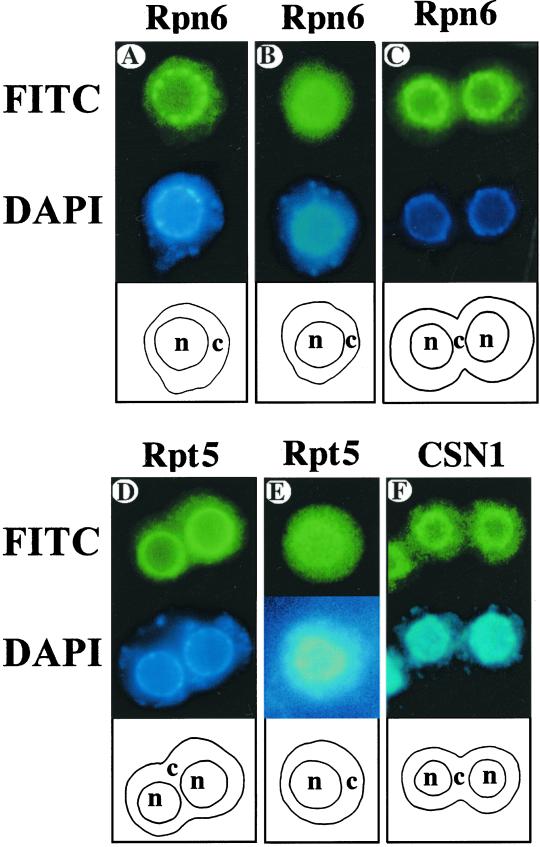
In wild-type Arabidopsis protoplasts, both Rpn6 and Rpt5 antibodies predominantly stained the nucleoplasm (Figure 6; Kwok et al., 1998, 1999). However, antibodies against Rpn6 (Figure 6A), but not Rpt5 (Figure 6D), intensely stained numerous discrete speckles (20–40 per nucleus) within the nucleoplasm in all cell types examined (see Figure 6, A and D). Consistent staining patterns were observed in 100 individual cells of each cell type. Because Rpn6 is a component of both 19S RP and PR500 and it is present in these speckles, whereas Rpt5 is only in the 19S RP and does not stain any speckle, it is possible that the bright speckles represent the localization of PR500. The uniform nuclear-enriched staining patterns of Rpn6 and Rpt5 could represent the localization of 19S RP alone or the 26S proteasome.
Figure 6.
Subcellular localization of proteasome, PR500, and the COP9 signalosome in Arabidopsis protoplasts. Protoplasts were prepared from 7-d-old light-grown wild-type (A, D, and F), mutant fus6-1 (B and E), and cop10-1 (C) seedlings. The protoplasts (top of each panel) were stained with purified Rpn6, Rpt5, and CSN1 antibodies and decorated with a fluorescein (FITC) secondary antibody. The corresponding labeling of nuclei by 4′,6-diamidino-2-phenylindole (DAPI) is shown in the middle of each panel. A diagram of the cell (c) and nucleus (n) outlines is shown at the bottom of each panel. The fus6-1 mutant is defective in the COP9 signalosome, whereas the cop10-1 mutant accumulates a normal level of the COP9 signalosome.
To substantiate this conclusion, we took advantage of the fact that PR500 complex is significantly diminished in the COP9 signalosome mutants but not in det1 and cop10 mutants of Arabidopsis. Protoplasts were prepared from roots of 7-d-old mutant seedlings that were either completely lacking PR500 (fus6-1 and fus12-R380)) or had normal PR500 accumulation (cop10-1 and det1-8). These protoplasts were then probed with the Rpt5 and Rpn6 antibodies. As shown in Figure 6, B and E, the Rpn6-associated speckles were not detected in cells prepared from the COP9 signalosome-deficient fus6-1 mutants. In contrast, the staining patterns of Rpn6 in the mutants of cop10-1 (Figure 6C) and det1-8 (Peng, Staub, Serino, Kwok, Kurepa, Bruce, Vierstra, Wei, and Deng, unpublished results) were essentially the same as that of wild-type seedlings. Therefore, the absence of the bright speckles specifically correlates with the loss of PR500 in the COP9 signalosome mutants and not in other cop/det/fus mutants. A distinct localization in specific nuclear foci of PR500 also strongly supports the conclusion that PR500 complex exists in vivo.
DISCUSSION
PR500 Is a Freely Existing 19S RP-Related Protein Complex
The purified PR500 complex contains nine distinct proteins, most of which appear to be components of the 19S RP (Figure 2C). This complex is similar, but not identical, to the recently reported proteasome lid subcomplex observed in S. cerevisiae and human (Glickman et al., 1998a; Braun et al., 1999; Henke et al., 1999). PR500 clearly shares common features in its composition with the lid subcomplex. For example, it contains at least three non-ATPase subunits (Rpn6, Rpn5a, and Rpn3) that are also present in the lid subcomplex. Furthermore, PR500 does not contain Rpn10, the two largest 19S RP subunits, Rpn1 and Rpn2, and the three ATPase subunits, all of which are not part of the lid subcomplex either. Thus, it is possible that PR500 contains the entire lid subcomplex (eight subunits) plus one or two additional components.
It is important to note that the PR500 complex is itself a freely existing structure apart from the 19S particles in normal plant cells, whereas the yeast lid is a stable subcomplex that can be detached in vitro only from the 26S proteasome purified from the Rpn10 deletion strain. Although a lid-like subcomplex can be isolated as a by-product during the COP9 signalosome purification from human cells, its presence under physiological conditions has not been demonstrated (Braun et al., 1999; Henke et al., 1999). Four major lines of evidence support the conclusion that PR500 exists under physiological conditions. First, PR500 was consistently observed by gel filtration of crude cell extracts under different buffer conditions with or without ATP. Second, the PR500 complex appears to have a distinct nuclear localization pattern. Third, the mutations of COP9 signalosome subunits resulted in specific loss of PR500, as demonstrated by both gel filtration and immunolocalization studies. Fourth, both heat shock and canavanine treatments can specifically reduced PR500 levels.
A Possible Role of PR500 in Plant Stress Response
Our studies identified three conditions in which PR500 accumulation was diminished. PR500 was diminished in COP9 signalosome mutants, in Arabidopsis seedlings grown on the arginine analogue canavanine and subjected to heat shock treatment. The later two treatments are known to elevated accumulation of misfolded proteins that are substrates for the 26S proteasome, thereby increasing the workload of the ubiquitin-proteasome pathway. Interestingly, the COP9 signalosome was recently shown to be essential for the proteasome-mediated degradation of a key transcription factor (Osterlund et al., 2000). Furthermore, in the COP9 signalosome mutants, we recently observed that there is a dramatic increase in the amount of cellular ubiquitinated proteins (Peng and Deng, unpublished data). This potential defect of the ubiquitin-proteasome pathway specific to the COP9 signalosome mutants is likely sensed and interpreted by Arabidopsis cells as an increase in demand for the ubiquitin-proteasome pathway. Thus, in all three cases in which we observed a decrease of PR500 levels share a common link, a high demand for the proteasome activity. It is also worth noting that, although our data support an indirect role of the COP9 signalosome in regulating the abundance of PR500, it does not exclude an additional and more direct role of the COP9 signalosome in regulating PR500 function or association with proteasome (as discussed below).
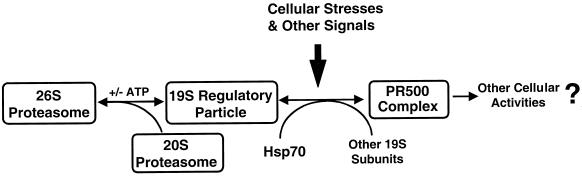
Because our result suggests that PR500 under those stresses was not degraded, it is likely incorporated into 26S or other forms of proteasome. It is interesting to speculate about the biochemical nature of the association of PR500 to the proteasome. As diagramed in Figure 7, one of several alternative possibilities could occur. First and the simplest, PR500 may be a cellular reservoir of 19S components for either assembly or disassociation. For example, upon high demand of the cellular proteasome activity, PR500 could be rapidly mobilized to assemble into 19S RP and then the 26S proteasome. In this case, any components present in PR500 but not in the final 19S RP will be discarded during the assembly process. This hypothesis would be consistent with an association with an hsp70 chaperone, which is known to help multisubunit protein complex assembly and ordered disassociation (Crookes and Olsen, 1998; Luders et al., 2000). However, because the 26S proteasome is rather abundant in most cells, it has only rather limited value in storing some extra lid-like complex just for rapid 26S proteasome assembly in response to the stresses. Second, PR500 could be used as a special lid subcomplex. During stress situations, this special lid subcomplex may be used to replace the regular lid subcomplex in a portion of the proteasome pool. This type of the proteasome with PR500 as lid may thus possess special characteristics that may better suit the stress situation. However, no evidence is available at present to suggest that PR500 is a distinct lid for 26S proteasome. Third, PR500 may physically associate with a normal and intact 26S proteasome and modify its activity or specificity to better suit stress environments.
Figure 7.
A model showing the dynamic relationships among PR500, 19S regulatory particle, and other proteasome structures. Under this model, 19S regulatory particle and PR500 are both present in the cell, and cellular stresses and other signals can modulate their equilibrium. Note that the presence of ATP (+) will help assembly of the 26S proteasome, whereas absence of ATP (−) will cause the disassociation of the 26S proteasome into 20S and 19S components. Free subunits and subcomplexes apart from the 19S particle may also participate in other unknown cellular activities. It is not clear whether the incorporation of PR500 into 19S or 26S proteasome would constitute a normal or distinct lid subcomplex, which would have different functional implications. See DISCUSSION for more detail.
Although our current data cannot differentiate among these possibilities, there are precedents that the activity and specificity of the proteasome can be modulated by additional regulatory proteins, including activators, inhibitors, and modulators. For example, a heterotrimeric modulator, containing two ATPase subunits (Rpt5/S6/Tbp1 and Rpt4/S10b/Sug2) in common with the 19S RP and one novel protein (p27), has been reported (DeMartino et al., 1996). This modulator facilitates the ATP-dependent association of the 19S and 20S particles. A 220-kDa activator containing S10b/Rpt4 and S6′ (TBP1/Rpt5) ATPases has also been isolated (Hastings et al., 1999). Recently, a novel 530-kDa protein complex distinct from PR500 has been shown to be associated with the 20S proteasome core (Tanaka et al., 2000). Moreover, functionally distinct forms of 26S proteasome have also been reported (Kawahara et al., 2000). PR500 could act according to one of the above-proposed ways to enhance the capacity of plant cells to deal with stress situations.
A proposed functional role of PR500 in plant stress responses could fit well with the immobile life style of higher plants. Unlike animals, plants cannot escape and need to endure and cope with the environmental stresses. For example, direct exposure of a leaf to sunlight can cause a rise of temperature of 4–5°C above ambient temperature when water is not abundantly available (Taiz and Zeiger, 1991). Thus, it is rather frequent that plant leaves encounter temperatures above 40°C during the year. This stress is likely to increase misfolding or denaturing of proteins. To survive, plants would need to effectively deal with those situations. The proteasome was demonstrated to be critical in dealing with this type of stress. For example, several knock-out mutants of the 19S RP subunits have been shown to be hypersensitive to heat shock stress and canavanine treatments in both yeast and plants (Finley et al., 1987; Lambertson et al., 1999; Takeuchi et al., 1999; Vierstra, 2000). Regardless of the specific mechanism of PR500 action, it is not unreasonable to suggest that this free pool of PR500 may be a unique feature of higher plants evolved to cope with the frequent protein-misfolding stresses and their immobile life style.
ACKNOWLEDGMENTS
We are grateful to Maria W. Smith, Carlos Gorbea, Mina Kurepa, and Steve van Nocker for kindly providing the monoclonal antibodies against 21D7/Rpn3 (subunit 3 of 19S RP), polyclonal antibodies against human Rpt2, Rpt1, and Rpn10, respectively. We are also grateful to Magnus Holm and Claus Swechheimer for comments and critical reading of this manuscript. This research is supported by a National Science Foundation grants (MCD-9513366 and MCD-0077217) to X.-W.D., a grant from Council for Tobacco Research, United States of America, to N.W., and an RDV-United States Department of Agriculture-National Research Initiative Competitive Grants Program grant (97-35301-4218) to R.D.V. X.-W.D. is a National Science Foundation Presidential Faculty Fellow. Z.P. is, in part, supported by a National Institutes of Health postdoctoral fellowship. G.S is a recipient of an Institute Pasteur Fondazione Cenci-Bologneti Fellowship.
REFERENCES
- Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson JD. Protein function. In: Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson JD, editors. Molecular Biology of the Cell. New York: Garland Press; 1994. pp. 214–215. [Google Scholar]
- Baumeister W, Walz J, Zuhl F, Seemuller E. The proteasome: paradigm of a self-compartmentalizing protease. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- Braun BC, Glickman M, Kraft R, Dahlmann B, Kloetzel PM, Finley D, Schmidt M. The base of the proteasome regulatory particle exhibits chaperone-like activity. Nat Cell Biol. 1999;1:221–226. doi: 10.1038/12043. [DOI] [PubMed] [Google Scholar]
- Castle L, Meinke D. A FUSCA gene of Arabidopsis encodes a novel protein essential for plant development. Plant Cell. 1994;6:25–41. doi: 10.1105/tpc.6.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamovitz DA, Wei N, Osterlund MT, von Arnim AG, Staub JM, Matsui M, Deng X-W. The COP9 complex, a novel multisubunit nuclear regulator involved in light control of a plant developmental switch. Cell. 1996;86:115–121. doi: 10.1016/s0092-8674(00)80082-3. [DOI] [PubMed] [Google Scholar]
- Chu-ping M, Vu JH, Proske RJ, Slaughter CA, DeMartino G. Identification, purification, and characterization of a high molecular weight, ATP-dependent activator (PA700) of the 20S proteasome. J Biol Chem. 1994;269:3539–3547. [PubMed] [Google Scholar]
- Coux O, Tanaka K, Goldberg AL. Structure and function of 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- Crookes WJ, Olsen LJ. The effects of chaperones and the influence of protein assembly on peroxisomal protein import. J Biol Chem. 1998;273:17236–17242. doi: 10.1074/jbc.273.27.17236. [DOI] [PubMed] [Google Scholar]
- DeMartino GN, Proske RJ, Moomaw CR, Strong AA, Song X, Hisamatsu H, Tanaka K, Slaughter CA. Identification, purification, and characterization of a PA700-dependent activator of the proteasome. J Biol Chem. 1996;271:3112–3118. doi: 10.1074/jbc.271.6.3112. [DOI] [PubMed] [Google Scholar]
- Deng XW, Dubiel W, Wei N, Hofmann K, Mundt K, Colicelli J, Kato JY, Naumann M, Segal D, Seeger M, Glickman M, Carr A, Chamovitz DA. Unified nomenclature for the COP9 signalosome: an essential regulator of development. Trends Genet. 2000;16:289. doi: 10.1016/s0168-9525(00)02071-0. [DOI] [PubMed] [Google Scholar]
- Finley D, Ozkaynak E, Varshavsky A. The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stress. Cell, 1987;48:1035–1046. doi: 10.1016/0092-8674(87)90711-2. [DOI] [PubMed] [Google Scholar]
- Glickman MH, Rubin DM, Coux O, Wefes I, Pfeifer G, Cjeka Z, Baumeister W, Fried VA, Finley D. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to COP9 signalosome and eIF3. Cell. 1998a;94:615–623. doi: 10.1016/s0092-8674(00)81603-7. [DOI] [PubMed] [Google Scholar]
- Glickman MH, Rubin DM, Fried VA, Finley D. The regulatory particle of the Saccharomyces cerevisiae proteasome. Mol Cell Biol. 1998b;18:3149–3162. doi: 10.1128/mcb.18.6.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings RA, Eyheralde I, Dawson SP, Walker G, Reynolds SE, Billett MA, Mayer JR. A 220-kDa activator complex of the 26S proteasome in insects and humans. J Biol Chem. 1999;274:25691–25700. doi: 10.1074/jbc.274.36.25691. [DOI] [PubMed] [Google Scholar]
- Henke W, Ferrell K, Bech-Otschir D, Seeger M, Schade R, Jungblut P, Naumann M, Dubiel W. Comparison of human COP9 signalosome and 26S proteasome lid. Mol Biol Rep. 1999;26:29–34. doi: 10.1023/a:1006991419464. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Kapelari B, Bech-Otschir D, Hegerl R, Schade R, Dumdey R, Dubiel W. Electron microscopy and subunit-subunit interaction studies reveal a first architecture of COP9 signalosome. J Mol Biol. 2000;300:1169–1178. doi: 10.1006/jmbi.2000.3912. [DOI] [PubMed] [Google Scholar]
- Karniol B, Malec P, Chamovitz DA. Arabidopsis FUSCA5 encodes a novel phosphoprotein that is a component of COP9 complex. Plant Cell. 1999;11:839–848. doi: 10.1105/tpc.11.5.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara H, Kasahara M, Nishiyama A, Ohsumi K, Goto T, Kishimoto T, Saeki Y, Yokosawa H, Shimbara N, Murata S, Chiba T, Suzuki K, Tanaka K. Developmentally regulated, alternative splicing of the Rpn10 gene generates multiple forms of 26S proteasome. EMBO J. 2000;19:4144–4153. doi: 10.1093/emboj/19.15.4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok SF, Piekos B, Misera S, Deng X-W. A complement of ten essential and pleiotropic Arabidopsis COP/DET/FUS genes is necessary for repression of photomorphogenesis in darkness. Plant Physiol. 1996;110:732–742. doi: 10.1104/pp.110.3.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok SF, Solano R, Tsuge T, Chamovitz DA, Matsui M, Ecker JR, Deng X-W. Arabidopsis homologs of a c-Jun coactivator are present both in monomeric form and in the COP9 complex and their abundance is differentially affected by the pleiotropic cop/det/fus mutations. Plant Cell. 1998;10:1779–1790. doi: 10.1105/tpc.10.11.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok SF, Staub JM, Deng X-W. Characterization of two subunits of Arabidopsis 19S proteasome regulatory complex and its possible interaction with the COP9 complex. J Mol Biol, 1999;285:85–95. doi: 10.1006/jmbi.1998.2315. [DOI] [PubMed] [Google Scholar]
- Lambertson D, Chen L, Madura K. Pleiotropic defects caused by loss of the proteasome-interaction factors Rad23 and Rpn10 of Saccharomyces cerevisiae. Genetics, 1999;153:69–79. doi: 10.1093/genetics/153.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luders J, Demand J, Hohfeld J. The ubiquitin-related BAG-1 provides a link between the molecular chaperones Hsc70/hsp70 and the proteasome. J Biol Chem. 2000;275:4613–4617. doi: 10.1074/jbc.275.7.4613. [DOI] [PubMed] [Google Scholar]
- Matsui M, Stoop CD, von Arnim AG, Wei N, Deng X-W. Arabidopsis COP1 protein specifically interacts in vitro with a cytoskeleton-associated protein, CIP1. Proc Natl Acad Sci USA. 1995;92:4239–4243. doi: 10.1073/pnas.92.10.4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orino E, Tanaka K, Tamura T, Sone S, Ogura T, Ichihara A. ATP-dependent reversible association of proteasomes with multiple protein components to form 26S complexes that degrade ubiquitinated proteins in human HL-60 cells. FEBS Lett. 1991;284:206–210. doi: 10.1016/0014-5793(91)80686-w. [DOI] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CH, Wei N, Deng X-W. Targeted destabilization of HY5 during light-regulated Arabidopsis development. Nature, 2000;405:402–466. doi: 10.1038/35013076. [DOI] [PubMed] [Google Scholar]
- Pepper A, Delaney T, Washburn T, Poole D, Chory J. DET1, a negative regulator of light-mediated development and gene expression in Arabidopsis, encodes a novel nuclear-localized protein. Cell. 1994;78:109–116. doi: 10.1016/0092-8674(94)90577-0. [DOI] [PubMed] [Google Scholar]
- Pirkakala L, Alastalo TP, Zuo X, Benjamin IJ, Sistonen L. Disruption of heat shock factor 1 reveals an essential role in the ubiquitin proteolytic pathway. Mol Cell Biol. 2000;20:2670–2675. doi: 10.1128/mcb.20.8.2670-2675.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal GA. The biochemical basis for the insecticidal properties of l-canavanine, a higher plant protective allelochemical, In: Otto D, Weber B, editors. Insectcides: Mechanism of Action and Resistance. Andover, England: Intercept Ltd.; 1992. pp. 35–46. [Google Scholar]
- Rosenthal GA, Reichart JM, Hoffman JA. l-Canavanine incorporation into vitellogenin and macromolecular conformation. J Biol Chem. 1989;264:13693–13696. [PubMed] [Google Scholar]
- Sawada H, Akiaishi T, Katsu M, Yokosawa H. Difference between PA700-like proteasome activator complex and the regulatory complex dissociated from the 26S proteasome implies the involvement of modulating factor in the 26S proteasome assembly. FEBS Lett. 1997;412:521–525. doi: 10.1016/s0014-5793(97)00851-x. [DOI] [PubMed] [Google Scholar]
- Seeger M, Kraft R, Ferrell K, Bech-Otschir D, Dumdey R, Schade R, Gordon C, Naumann M, Dubiel WR. A novel protein complex involved in signal transduction possessing similarities to 26S proteasome subunits. FASEB J. 1998;12:469–478. [PubMed] [Google Scholar]
- Serino G, Tsuge T, Kwok SF, Matsui M, Wei W, Deng XW. Arabidopsis cop8 and fus4 mutations define the same locus that encodes subunit 4 of the COP9 signalosome. Plant Cell. 1999;11:1967–1980. doi: 10.1105/tpc.11.10.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MW, Ito M, Miyawaki M, Sato S, Yoshikawa Y, Wada S, Maki H, Nakagawa H, Komamine A. Plant 21D7 protein, a nuclear antigen associated with cell division, is a component of the 26S proteasome. Plant Physiol. 1997;113:281–291. doi: 10.1104/pp.113.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staub JM, Wei N, Deng X-W. Evidence for FUS6 as a component of the nuclear-localized COP9 complex in Arabidopsis. Plant Cell. 1996;8:2047–2056. doi: 10.1105/tpc.8.11.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taiz L, Zeiger E. Heat stress and heat shock. In: Taiz L, Zeiger E, editors. Plant Physiology. 1991. pp. 360–361. [Google Scholar]
- Takeuchi J, Fujimuro M, Yokosawa H, Tanaka K, Toh-e A. Rpn9 is required for efficient assembly of the yeast 26S proteasome. Mol Cell Biol. 1999;19:6575–6584. doi: 10.1128/mcb.19.10.6575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka E, Sawada MT, Morinaga C, Yokosawa H, Sawada H. Isolation and characterization of a novel 530-kDa protein complex (PC530) capable of associating with the 20S proteasome from starfish oocytes. Arch Biochem Biophys. 2000;374:181–188. doi: 10.1006/abbi.1999.1584. [DOI] [PubMed] [Google Scholar]
- van Nocker S, Sadis S, Rubin DM, Glickman M, Fu H, Coux O, Wefes I, Finley D, Vierstra RD. The multiubiquitin-chain-binding protein Mcb1 is a component of the 26S proteasome in Saccharomyces cerevisiae and plays a nonessential, substrate-specific role in protein turnover. Mol Cell Biol. 1996;16:6020–6028. doi: 10.1128/mcb.16.11.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra RD. Program and Abstracts of the 11th International Conference on Arabidopsis Research. 2000. Organization and function of the ubiquitin/26S proteasome proteolytic pathway in Arabidopsis. , 18. [Google Scholar]
- Wei N, Chamovitz DA, Deng X-W. Arabidopsis COP9 is a component of a novel signaling complex mediating light control of development. Cell. 1994;78:117–124. doi: 10.1016/0092-8674(94)90578-9. [DOI] [PubMed] [Google Scholar]
- Wei N, Deng X-W. The role of pleiotropic COP/DET/FUS genes in light control of Arabidopsis seedling development. Plant Physiol. 1996;112:871–878. doi: 10.1104/pp.112.3.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei N, Deng XW. Making sense of the COP9 signalosome, a conserved regulatory protein complex from Arabidopsis to human. Trends Genet. 1999;15:98–103. doi: 10.1016/s0168-9525(98)01670-9. [DOI] [PubMed] [Google Scholar]
- Wei N, Tomohiko T, Serino G, Dohmae N, Takio K, Matsui M, Deng X-W. The COP9 complex is conserved between plants and mammals and is related to the 26S proteasome regulatory complex. Curr Biol. 1998;8:919–922. doi: 10.1016/s0960-9822(07)00372-7. [DOI] [PubMed] [Google Scholar]