Abstract
Immaculate and complete palatal seam disintegration, which takes place at the last phase of palate development, is essential for normal palate development. And in absence of palatal medial edge seam (MES) disintegration, cleft palate may arise. It has been established that Transforming Growth Factor (TGF) β induces both Epithelial Mesenchymal Transition (EMT) and/or apoptosis during MES disintegration. It is likely that MES might cease cell cycle to facilitate cellular changes prior to undergoing transformation or apoptosis, which has never been studied before. This study was designed to explore whether TGFβ, which is crucial for palatal MES disintegration, is capable of inducing cell cycle arrest. We studied the effects of TGFβ1 and TGFβ3, potent negative regulators of the cell cycle, on p15ink4b activity in MES cells. We surprisingly found that TGFβ1, but not TGFβ3, plays a major role in activation of the p15ink4b gene. In contrast, following successful cell cycle arrest by TGFβ1, it is TGFβ3 but not TGFβ1 that causes later cellular morphogenesis, such as EMT and apoptosis. Since TGFβ signaling activates Smads, we analyzed the roles of three Smad binding elements (SBEs) on the p15ink4b mouse promoter by site specific mutagenesis and found that these binding sites are functional. The ChIP assay demonstrated that TGFβ1, not TGFβ3, promotes Smad4 binding to two 5’ terminal SBEs but not the 3’ terminal site. Thus, TGFβ1 and TGFβ3 play separate yet complimentary roles in achieving cell cycle arrest and EMT/apoptosis and cell cycle arrest is a prerequisite for later cellular changes.
Keywords: cell cycle, palate, TGFβ, apoptosis, EMT
INTRODUCTION
Cleft palate, a common craniofacial deformity, can be manifested by the failure of palatal midline epithelial seam (MES) to disintegrate during the late phase of palatogenesis following optimum palatal growth, elevation and fusion. The Transforming Growth Factor β (TGFβ) plays an essential role in all phases of palate development (Heldin et al., 2009; Murray and Schutte, 2004; Schutte and Murray, 1999). While TGFβ1 is important for palatal shelve growth (Mu et al., 2008; Yang and Kaartinen, 2007), it is TGFβ3 that plays the primary role in MES disintegration (Nawshad et al., 2004). It has been shown that TGFβ1 is strongly expressed in the medial edge epithelial (MEE) cells prior to adherence with the opposite palatal shelve (Martinez-Alvarez et al., 2004; Martinez-Alvarez et al., 2000). Following adherence of the opposite palatal shelves, TGFβ1 gradually ceases to express, while TGFβ3 begins to express strongly in the MEE cells. Therefore, TGFβ1 expression precedes TGFβ3 in time dependent fashion (Mu et al., 2008). Moreover, the importance of TGFβ3 in MES disintegration either by Epithelia Mesenchymal Transition (EMT) and/or by apoptosis or both has been well established (Ahmed et al., 2007; Nawshad, 2008). But cellular morphogenesis such as EMT and/or apoptosis warrant significant rearrangement of structural proteins to facilitate cellular changes during which period cells must undergo cell cycle arrest. Therefore, it is likely that induction of cell cycle arrest might be a prerequisite for EMT and apoptosis.
It has been recently shown that cell cycle phase is a key event in a determination of subsequent pathway – EMT or an apoptosis (Tian et al., 2009). (Yang et al., 2006a) demonstrated in several epithelial cell lines that, in response to TGFβ treatment, the cells in G2/M phase undergo apoptosis, while if they are in the G1/S phase they transfer to EMT. (Hannon and Beach, 1994) showed that TGF-β inhibits cell proliferation in epithelial cells through several pathways including upregulation of the (CDK) inhibitors p15ink4b and p21cip1/waf1. Subsequently, (Tremain et al., 2000) showed that increasing TGFβ1 expression leads to increasing levels of p15ink4b protein and inactivation of TGFβ1 signaling reduced induction of p15ink4b. p15ink4b, a member of the family of D cyclin-dependent kinase inhibitors (CDK1) is an inducer of G1 cell cycle arrest. This protein binds early G1 CDK 4 or 6 and prevents its interaction with the cyclin D (Hannon and Beach, 1994; Wolff et al., 2003). The p15ink4b expression is directly regulated by TGFβ1 through interaction of Smad at the Smad Binding Element (SBE) in its promoter (Seoane et al., 2001). Therefore, it is likely that activation of p15ink4b in response to TGFβ might precede EMT and/or apoptosis to ensure proper execution of cellular changes. TGFβ1 is a well known potent inducer of growth inhibition in several cell types, including epithelial cells (Derynck et al., 2001). In the liver, TGFβ1 is a negative regulator of hepatocyte proliferation (Bissell, 2001; Bissell et al., 2001).
While ample studies have established the role of TGFβ3 in palatal seam disintegration by EMT and or apoptosis, there are no studies describing the cell cycle status of palatal MEE cells prior to cellular changes in response to TGFβ. In this study, we undertook experiments to address the activation of p15ink4b by TGFβ in the palatal MEE cells. We report for the first time that both TGFβ1 and TGFβ3 are capable of inducing cell cycle arrest by activating p15ink4b. While TGFβ1 is a more potent regulator of cell cycle arrest by inducing p15ink4b, it is TGFβ3 that executes EMT and apoptosis following induction of cell cycle arrest by TGFβ1. The data indicates that cell cycle arrest by TGFβ1 is prerequisite for TGFβ3 induced EMT and or apoptosis.
MATERIALS AND METHODS
Cell culture and treatment conditions
The Medial Edge Epithelial (MEE) primary cells derived from mice embryonic palatal shelve epithelial cells have been used in previous experiments (Ahmed et al., 2007; Nawshad et al., 2007). The cells were maintained in Dulbeco’s Modified Eagle’s medium (DMEM) supplemented with 100U/ml penicillin, 100µg/ml streptomycin and 10% fetal bovine serum. To analyze the effect of TGFβ1 or TGFβ3 on the p15ink4b protein expression, the cells were treated with different doses (1, 2, 4, 6, 8, 10 and 15 ng/mL) of exogenous recombinant human TGFβ1 and/or TGFβ3 (R&D systems CA) for 6, 12, 24, 36 and 48hours (h). For FACSArray, MEE cells were treated with 5ng/mL of TGFβ1 and/or TGFβ3 for 24h. For luciferase assays the cells were incubated with TGFβ1 or TGFβ3 (2ng and 5ng/mL) for 24h, whereas for the chromatin immune precipitation (ChIP) the time of incubation was 60 minutes (min). For cell migration assay, MEE cells were treated with TGFβ1 and TGFβ3 (5ng/ml) for 48h. For TUNEL, using live and dead cell assays by FACSArray, MEE cells were treated with TGFβ1 and TGFβ3 (5ng/ml) for 72h. To activate p15ink4b, MEE cells were treated with murine full length (1.3kb) pBluescript-p15cDNA (generously provided by Dr. Linda Wolff, Center for Cancer Research, National Cancer Institute, NIH, Bethesda, MD). To block p15ink4b, MEE cells were treated with lentivirus vector encoding short hairpin (sh) p15ink4b (pLK0.1-shp15RNA) obtained from OpenBiosystems (AL, USA).
Cell cycle analysis
MEE cells were grown in 10% FBS containing DMEM in T25 flasks. Approximately 60% confluent cells were treated with 6.0 µM Aphidicholin (Sigma-Aldrich, MO) for 16h, washed with Hanks Buffered Saline Solution (HBSS) (Medoatech, VA), and released into complete medium for 30 min. Cells were then treated with a medium containing TGFβ1 (5ng/mL) and TGFβ3 (5ng/mL) for 24h. MEE cells were detached by trypsin and collected in 15mL tube after washing with 1×PBS. Next, cells were fixed by adding ice-cold 70% ethanol drop by drop while slowly vortexing and incubated for 24h at −20°C. Fixed cells were then washed once with 1×PBS and the cell concentrations were adjusted to 4×105 and stained with 1mg/mL of Propidium Iodide (PI) and RNase (Sigma-Aldrich, MO) for 30 min. at room temperature. Finally, these PI stained cells were analyzed by BD FACSArray Bioanalyzer (BD Biosciences, CA) using green laser at 532nm wavelength.
Immunohistochemistry and Western blot
To demonstrate the in vivo expression of p15ink4b protein in the palate in time dependent fashion, CF1 (Charles River Laboratory, MA) embryos were collected at 14.0, 14.5, 15.5 and 16.5 days post coitum (dpc), fixed in Buin's fixative and 7µm sectioned were cut. These sections underwent immunohistochemistry techniques as previously described by us in (Nawshad and Hay, 2003). Similarly, for western blot analysis, protein collected from primary MEE cells from CF1 embryos, as described in (Ahmed et al., 2007; Nawshad et al., 2007) were used. Commercially available murine polyclonal p15ink4b antibody (Novus Biologicals, CO) was used for both immunohistochemistry and western blot analysis.
Plasmid constructs and site-directed mutagenesis
For the luciferase activity assay, we used the plasmid construct pGL3-p15-Lux generously provided by Dr. Linda Wolff (Center for Cancer Research, National Cancer Institute, NIH, Bethesda, MD) (Fig. 3A). This plasmid, generated on the base of the pGL3 vector (Promega, WI), encodes the firefly luciferase gene under control of the human p15ink4b gene promoter. Sequence of p15ink4b promoter has been identified and detailed in (Haviernik et al., 2003). Since we were interested in three potential Smad Binding Elements, SBE in the promoter of p15ink4b gene (Fig. 3B), we generated plasmids carrying mutations in each of these promoter regions. The mutations were introduced into pGL3-p15wt plasmid using the Site-Directed Mutagenesis Kit (Stratagene, CA) according to the manufacturer protocol. The following primers were used:
- Smad-binding element A:
- forward primer - 5’ GTTACCACTTTCAGGCAGCTTCAGAAAGTAG 3’,
- reverse primer – 5’ CTACTTTCTGAAGCTGCCTGAAAGTGGTAAC-3’;
- Smad-binding element B:
- forward primer 5’GTTGTTTGGCAACGGGCATAATGTGTGCCTCTC 3’,
- reverse primer – 5’ GAGAGGCACACATTATGCCCGTTGCCAAACAAC 3’;
- Smad-binding element C:
- forward primer - 5’GTTTCACCCAGAAGCAGGCATCTAGTCTTCTG 3’,
- reverse primer – 5’ CAGAAGACTAGATGCCTGCTTCTGGGTGAAAC 3’.
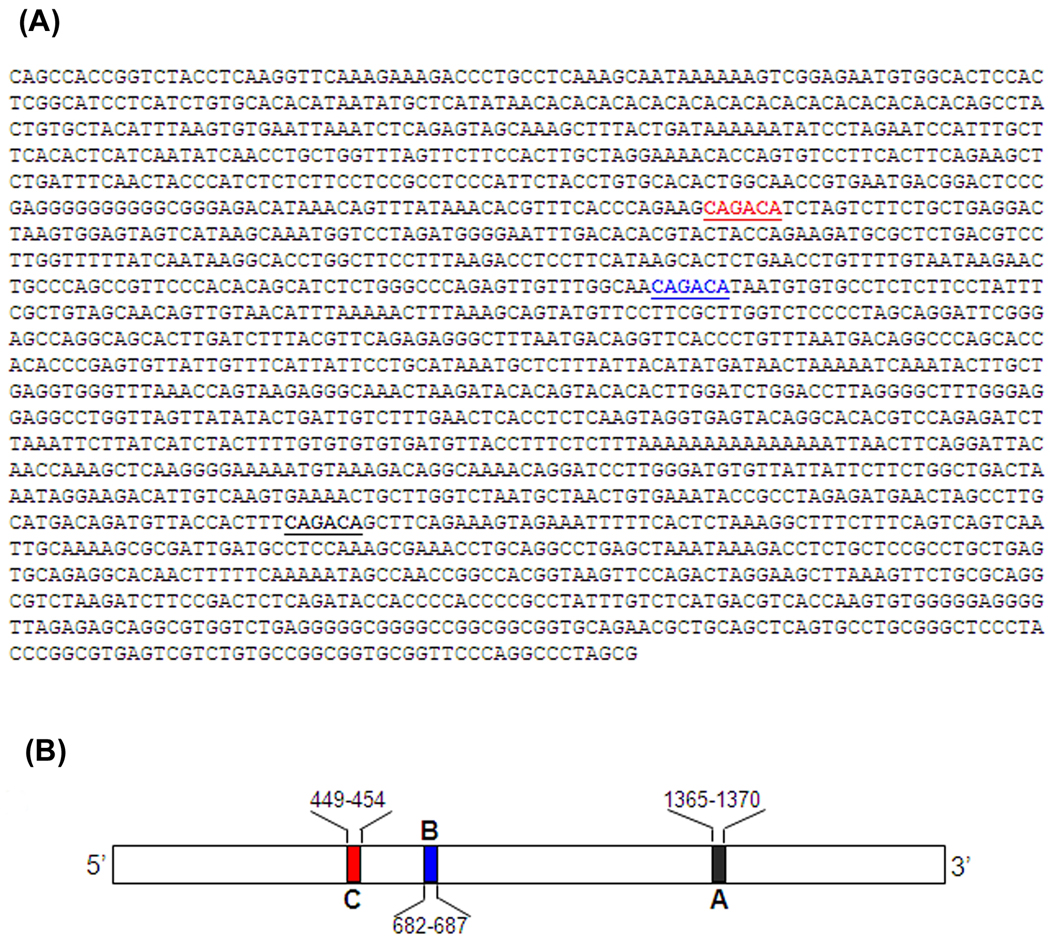
Fig. 3. Nucleotide sequence and a schematic diagram of the mouse p15ink4b promoter with potential Smad Binding Elements (SBEs).
A. Nucleotide sequence showing SBEs of the p15ink4b promoter −1.7 kb in obtained from Dr. Linda Wolff, NCI, NIH, Bethesda, MD, indicated by colored underlined sections (Red-C, Blue-B and Black- A).
B. Schematic diagram of the p15ink4b promoter −1.7 kb showing SBEs A, B, and C are shown as black, blue and red boxes respectively. SBE nucleotide positions are shown above or under diagram.
The PCR reaction included 18 cycles. Each cycle comprised 30 sec denaturation step at 95°C, 60 sec annealing at 55°C, and 7 min extension step at 68°C. All mutations were confirmed by DNA technique analysis of the plasmids called pGL3-p15mutA, pGL3-p15mutB, and pGL3-p15mutC respectively.
Transfection and luciferase assay
To measure the Smad-mediated effect of TGFβ1 and TGFβ3 on the p15ink4b promoter activity, we measured expression of the firefly luciferase in MEE cells transiently transfected with pGL3-p15-Lux plasmid or derivative plasmids containing mutations in the SBEs. The MEE cells were seeded in 12-well plate and incubated overnight to the final count of 2×105 cells per well. Then culture media was changed and the cells were transfected with pGL3p15wt plasmid or the identical plasmids containing mutations in Smad binding sites A, B or C as described above. The Lipofectamine 2000 transfection reagent (Invitrogen, CA) was used following the manufacturer’s protocol. Transfection of MEE cells with pGL3 vector without a p15ink4b promoter region (empty vector) was performed as a negative control. After 24h incubation, the cells were lysed in Passive Lysis Buffer (Promega, WI) adhering to manufacturer’s recommendations. Luciferase activity was detected using luminometer Sirius (Berthold Detection Systems, NC).
To eliminate background TGFβ activity, we blocked endogenous TGFβ1 and TGFβ3 by the treatment of the cells with respective antibodies. MEE cells were plated in 12-well tissue culture plates in concentrations of 0.5×105 cells per ml and then treated with antibodies against TGFβ1 and TGFβ3 (R&D systems, CA) twice. The first treatment was 8h after passage, anti TGFβ1 was used in concentrations of 1µg/ml, anti TGFβ3 - 1.4µg/ml. After 24h incubation with antibodies the antibody-containing media was removed, cells were washed with PBS and transfected with pGL3-p15wt plasmid as described above. After 4h incubation with transfection complexes, the culture media was changed. The same antibodies, using concentrations of 0.5µg/ml for anti TGFβ1 and 0.7µg/ml for anti TGFβ3, were added. The human recombinant (hr) TGFβ3 (2ng/ml or 5ng/ml) was added to the transfected cells treated with anti TGFβ1 antibody, while the hrTGFβ1 (2ng/ml or 5ng/ml) was added into the wells with the transfected anti TGFβ3 treated cells. The antibody treatment and the TGFβ1 and/or TGFβ3 treatment were repeated 24h later. The cells were split 24h after the last antibody treatment, washed with PBS and luciferase activity was analyzed as described above. The results are reported as the mean ± SD of at least three independent experiments.
Chromatin immunoprecipitation (ChIP) assay
Equal quantities (40×106 cells) of the control, TGFβ1 (5ng/ml) and TGFβ3 (5ng/ml) stimulated MEE cells in 70–80% monolayers were fixed by treatment with 1% formaldehyde (Electron Microscopy Sciences, CA) for 10 min at room temperature for the chromatin cross-linking. The reaction was stopped by adding glycine to a final concentration of 0.125M. Then cells were immediately washed twice with ice-cold PBS containing 0.5mM EDTA and harvested as described earlier in (Thillainadesan et al., 2008). Subsequent procedures were performed using SimpleChIP™ Enzymatic Chromatin IP kit (Magnetic beads) (Cell Signaling, MA) according to manufacturer’s protocol. Briefly, cell pellets were lysed in 10 ml of cell lysis buffer provided by the kit manufacturer and incubated on ice for 10 min. Cross-linked chromatin was isolated from the lysates by sonication 2 sets of 6- second at 4°C using Microson XL 2000 sonicator. Immune precipitation of the cross-linked chromatin with rabbit anti-Smad4 antibody (Cell Signaling, MA) was subsequently performed. For isotype control, rabbit IgG (Cell Signaling, MA) was used. The 100µl chromatin samples in binding buffer from Chromatin IP kit (Cell Signaling, MA), containing 15µg DNA were incubated overnight with antibodies at 4°C and slow shaking. The subsequent binding with magnetic beads, washing of immunocomplexes and DNA extraction were carried out following the manufacturer’s protocol. Purified DNA was analyzed by PCR with primers specific for Smad4-binding elements in p15ink4b promoter. The following primers were used:
- Smad-binding site A:
- forward 5’CTGCTTGGTCTAATGCTAACTGTG 3’
- reverse 5’GGTCTTTATTTAGCTCAGGCCTGC 3’;
- Smad-binding site site B:
- forward 5’ ACCTCCTTCATAAGCACTCTGAAC 3’
- reverse 5’GAGACCAAGCGAAGGAACATACTG 3’;
- Smad-binding site C:
- forward 5’ ATTCTACCTGTGCACACTGGCAAC 3’
- reverse 5’GGTAGTACGTGTGTCAAATTCCCC3’
Resulting DNA fragments were detected by electrophoresis in 1.5% agarose gel. To quantitatively analyze Smad4 binding with the respective A, B and C sites in the p15ink4b promoter under the TGFβ1 or TGFβ3 treatment, real-time PCR analysis of the DNA, purified from immunocomplexes with anti-Smad4 antibody was used. The same primers were applied. DNA samples were subjected to quantitation by real-time PCR using RealMasterMix Sybr Rox (5Prime, Hamburg, Germany). Reactions were performed in 20ul volume according to the manufacturer’s recommendations. A standard curve was generated on the base of serial dilutions of pGL3-p15wt plasmid. The reactions were carried out in triplicate using the iCycler with iQ Multicolor Real-time PCR Detection System (BioRad, CA) and iCycler software (BioRad, CA). The mean ± SD obtained from three independent experiments were compared.
Transwell cell migration assay
The cell migration assay was performed thrice as described in (Nawshad et al., 2007) using the Innocyte™Cell Migration Assay (EMD Biosciences, CA). Briefly, 8µm pore size transwell migration chambers in 96-well plates were used for migration analyses. Both treated and untreated MEE cells were allowed to migrate across the membrane insert toward media in the presence of serum for 48h at 37°C (chemotactic migration). Cells that migrate through the membrane and attach to the lower side of the cell culture insert were subsequently detached using a cell detachment buffer containing Calcein-AM fluorescent dye (excitation max.: 485nm; emission max.: 520nm). The data were obtained using a BD Bioanalyzer™ fluorescent plate reader (BD Biosciences, CA).
TUNEL assay
To identify the apoptotic cells via the distribution of DNA fragmentation, we performed TUNEL assay three times - which employs TdT to add dUTP to the fragmented DNA ends. We used the TUNEL Apoptosis Detection kit (Millipore, MA). The method has been previously described in (Ahmed et al., 2007; Nawshad et al., 2007).
Live and dead cell staining
As previously detailed in (Ahmed et al., 2007), MEE cells were grown in 10% FBS containing DMEM in T-25 flasks. Approximately 60% confluent cells were treated with 6.0µM Aphidicholin for 16h, washed with HBSS and released into complete medium for 30 min. Cells were then treated with complete medium containing 5ng/mL of TGFβ3 for 72h. Cells were collected every 24h for live and dead cell stain analysis by BD FACSArray Bioanalyzer (BD Biosciences, CA). Vibrant cell metabolic assay kit and Sytox red dead cell stain were purchased from Invitrogen (CA). Cells were stained according to the manufacturer's protocol. In brief, first, the floating cells were collected by spin down. 4µL of 1× PBS was added to the flask and 1mL of 1× PBS was added to dissolve the collected floating cells and then added this dissolved pellet to the flasks. Then 2µM concentration of C12-resazurin was added to those flasks and incubated for 15 min in 37°C. After this, cells were detached by trypsin and collected, dissolved the pellet in 5nM Sytox Red stain of 1× PBS, and incubated for a minimum of 15 min at room temperature in the dark. These stained cells were analyzed by BD FACSArray Bioanalyzer using green laser at 532nm wavelength and red laser at 635 nm wavelength for C12-resazurin and Sytox Red stain, respectively.
Statistical analysis
Quantitative data were analyzed by two-way ANOVA (OriginPro v. 8.0) and Student’s t test (Microsoft Excel). Standard deviation was calculated in all quantitative experiments for at least three independent preparations. The difference was considered to be statistically significant when P < 0.05.
RESULTS
TGFβ1 is a potent inducer of p15ink4b expression
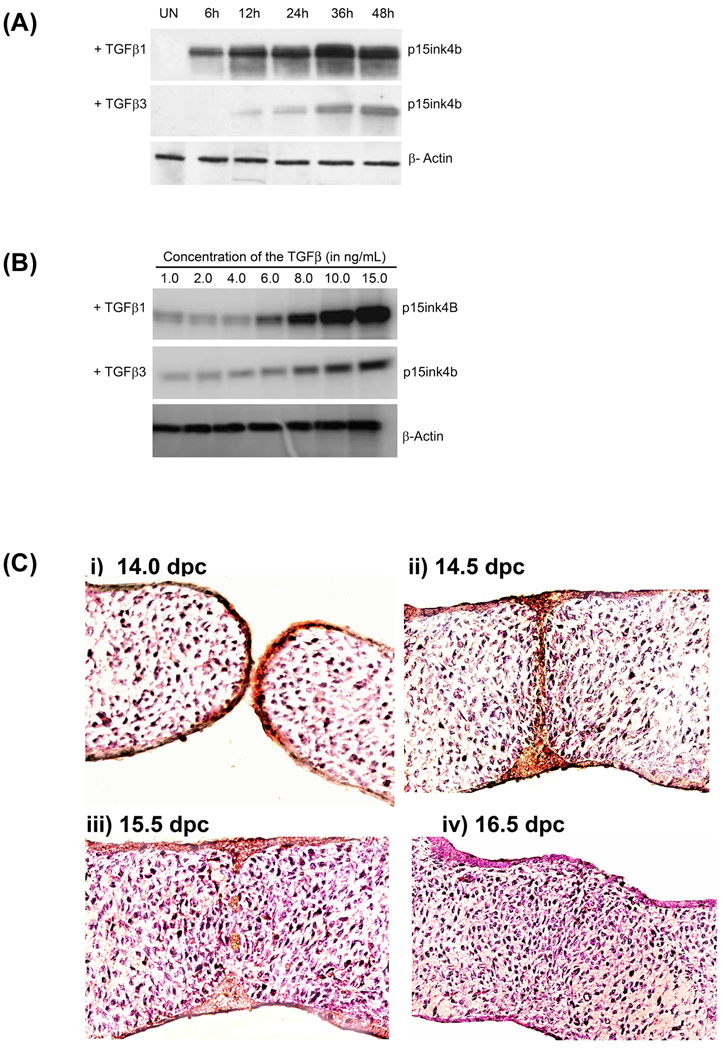
The first task was to establish the role of TGFβ1 and TGFβ3 separately in the medial edge epithelial (MEE) cells. It has been previously shown (Ahmed et al., 2007; Nawshad et al., 2007) that these homogenous primary medial edge epithelial cells behave in a near identical fashion as they would have in vivo. These results show that in response to TGFβ1 (5ng/mL), p15ink4b protein expression, as detected by western blot analysis, show significantly higher protein levels compared to TGFβ3 (5ng/mL) in a time dependent fashion (Fig. 1A). As time passes, so did the expression of p15ink4b protein levels in both treatment conditions. Moreover, expression p15ink4b was dependent on dose; as chronological increased doses of both TGFβ1 and TGFβ3 increased p15ink4b expression (Fig. 1B). These results demonstrated that while TGFβ1 had significantly more impact on p15ink4b expression compared to TGFβ3, they both increased the expression in a time and dose dependent manner (Fig. 1A and B). To confirm the MEE cells behavior and p15ink4b expressions, as shown in Fig. 1A and 1B, were in agreement with in vivo, we demonstrated that the embryonic palatal sections at 14.0 dpc express increased level of p15ink4b and continued to show higher expression till 14.5 dpc, however, the protein expression ceased by 15.5 dpc and showed no further expression at 16.5 dpc by which time palatal seam is completely disintegrated.
Fig. 1. p15ink4b expression in the MEE cells in response to TGFβ1 and TGFβ3.
A. Cell cycle arrest markers induced by TGFβ - upon TGFβ1 and TGFβ3 treatments (both 5ng/ml), protein expressions of the cell cycle arrest markers p15ink4b, gradually increased from 6h to strong expression at 48h. In response to TGFβ1, MEE cells showed higher expression compared to TGFβ3 treatment.
B. Both TGFβ1 and TGFβ3 treatment for 24h caused dose dependent increase of p15ink4b expression. All untreated control (UN) experiments were done using the same method as treated but the results shown are from 24h as this is the time point during which p15ink4b remain highest in vivo, (see Fig 1C).
C. In vivo, p15ink4b protein expression shown to be highest in the palatal epithelium immediately prior to embryonic palatal fusion at 14.0 dpc (i) and remained high when opposite palates are fused at 14.5 dpc (ii). But p15ink4b show very limited expression at 15.5 dpc (iii) when seam begin to disintegrate and expression is absent by 16.5 dpc (iv) when palatal seam is complete disintegrated
Cell cycle arrest is achieved by TGFβ1
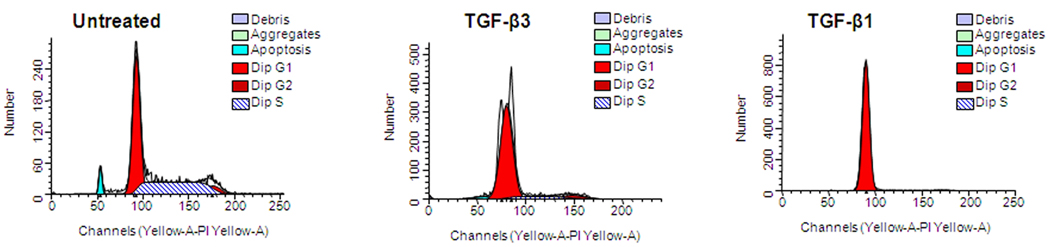
As TGFβ is a potent inducer of cell cycle arrest, we investigated whether TGFβ1 and TGFβ3 cause cell cycle arrest in the MEE cells. We found that MEE cells underwent cell cycle arrest almost immediately after TGFβ1 and TGFβ3 treatments, All MEE cells were synchronized at G1 phase (with Aphidicolin 6.0µM). G1 phase-synchronized MEE cells showed cessation of the cell cycle almost immediately in response to TGFβ1 and TGFβ3 signaling and nearly all (98%) MEE cells remain at the G1 state (Fig. 2). TGFβ1 completely halted MEE cell progression through the next phases of the cell cycle. At no point did MEE cells advance to the next (S, G2, M) phases of the cell cycle (Fig. 2). Nearly 79% of MEE cells treated with TGFβ3 also halted cells at G1 state. However, in response to TGFβ3, some cells progressed through the next phases of cell cycle (Fig. 2), unlike TGFβ1. In TGFβ untreated control cells, which had been treated with 10% FBS only, MEE cells successful progressed through all phases of the cell cycle (Fig. 2). These data support earlier findings that while both TGFβ1 and TGFβ3 are capable of inducing p15ink4b expression and cell cycle arrest, TGFβ1 has a more pronounced effect on p15ink4b expression and cell cycle arrest.
Fig. 2. Cell Cycle analysis by FACSArray in response to TGFβ1 and TGFβ3.
MEE cells undergo TGFβ1-induced cell cycle arrest: MEE cells grown in culture were treated with Aphidicolin (6 µM) for 16 h, which blocks DNA synthesis to synchronize all MEE cells at G1 phase. Once synchronized, MEE cells were released from the block for 30 min, treated with TGFβ1 and TGFβ3 (5ng/mL) for 24h and fixed with 70% ethanol (−20 °C), and flowcytometry on a FACSArray was done to evaluate cell cycle status. MEE cells treated with TGFβ1 failed to advance to the next phases (S, G2, and M) and remained stagnant at G1 (right panel). In contrast, although limited, some TGFβ3 treated cells were able to move to the next phases of cell cycle (middle panel). But untreated cells (control) progress through all phases of cell cycle (left panel)
p15ink4b promoter does harbor Smad binding regions
From earlier studies, it has been shown that p15ink4b has several potential Smad Binding Elements (SBEs). A Schematic diagram of the p15ink4b promoter −1.7 kb showing SBEs A, B, and C are shown as black, blue and red boxes respectively (Fig. 3). SBE nucleotide positions are shown above or under diagram.
TGFβ1 induces Smad4 transactivation of the p15ink4b promoter
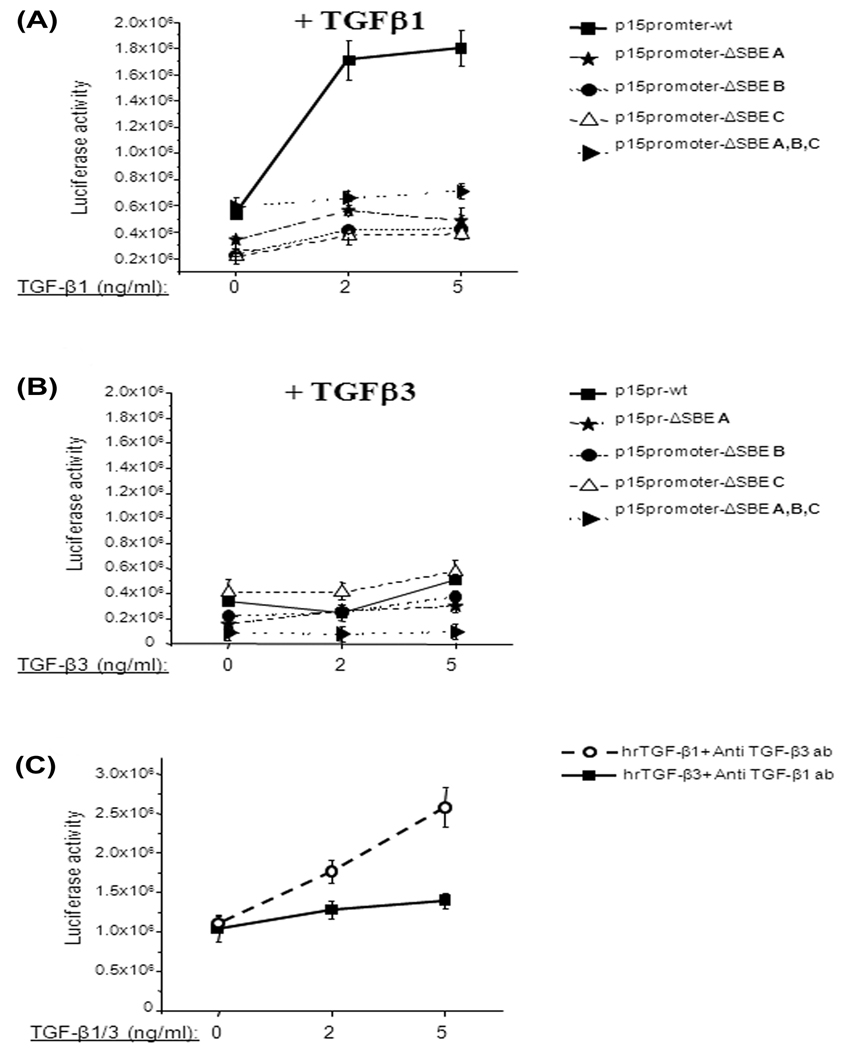
The effect of TGFβ1 on p15ink4b promoter activity was explored by transfecting MEE cells with pGL3-p15-Lux plasmid and, following incubation, the transfected cells with TGFβ1 (2 or 5ng/ml) for 24h. Luciferase assay showed a transactivation effect on the p15ink4b promoter in response to TGFβ1 (Fig. 4A; p15promoter-wt). Treatment with 2ng/ml of TGFβ1 resulted in ~3.4-folds increase of luciferase activity in cells transfected by pGL3-p15-Lux wild type (wt) plasmid (control). Increasing the TGFβ1 concentration to 5ng/ml did not significantly affect p15ink4b promoter activity in comparison to 2ng/ml (3.7 fold increase). However, mutations in the Smad binding elements (SBE) in p15ink4b promoter dramatically reduced TGFβ1 transactivating effect (Fig. 4A). TGFβ1 at 2ng/ml concentration reduced the luciferase activity 14 by ~2.3-folds, in comparison to wt plasmid, in constructs with mutations in the A or B of SBEs. However, mutations of all three (A, B and C sites) SBEs as well as only the C site, led to significant loss of TGFβ1 mediated transactivation effect compared to wt p15ink4b promoter activity. And the values of luciferase activity in these constructs (mutated A, B and C sites as well as only the C site) are similar to value of untreated activity. Interestingly, the independent mutations on the A and B sites have ~2.3 fold decrease in the gene activity compared to the wt. These data suggest that all three Smad4 binding elements are important for TGFβ1 induced p15ink4b gene activity. The results suggest that the C site of SBE is critical for the TGFβ1 effect, since mutation in this region completely reduced the effect of TGFβ1 mediated p15ink4b promoter activity.
Fig. 4. Effect TGFβ on the gene expression under control of p15ink4b promoter.
A. MEE cells were transiently transfected with pGL3-p15-Lux wild type plasmid and/or plasmids containing mutations in the Smad-binding sites. After transfection, the cells were incubated with TGFβ1 (2 or 5ng/ml) for 24 hours. Background luciferase activity (empty vector) was subtracted from all data. Error bars indicate standard deviation from at least three independent experiments. TGFβ1 increases p15ink4b promoter activity by threefold in MEE cells. TGFβ1 effect is Smad4-dependent, since addition of TGFβ1 to MEE cells transfected with plasmids containing mutations in Smad binding sites did not increase luciferase activity. All three SBEs in p15ink4b promoter are implicated in TGFβ1 induced promoter activation
B. MEE cells were transiently transfected as describe in figure legend 3A. After transfection, the cells were incubated with TGFβ3 (2 or 5ng/ml) for 24 hours. Background luciferase activity (empty vector) was subtracted from all data. Error bars indicate standard deviation from at least three independent experiments. Addition of TGFβ3 does not significantly increase luciferase activity in MEE cells transfected with pGL3-p15wt-Lux plasmid as well as with the plasmids containing mutations in Smad-binding sites.
C. Endogenous TGFβ1 and TGFβ3 were blocked using the antibody (anti TGFβ1 1µg/ml and anti TGFβ3 1.4µg/ml). Following blocking, the cells were transfected by pGL3-p15-Lux wild type plasmid. After transfection the cell were incubated with exogenous recombinant TGF-β1/TGFβ3 (2 or 5ng/ml) for 24 hours in presence of the antibody. Background luciferase activity (empty vector) was subtracted from all data. Error bars indicate standard deviation of three independent preparations. After blocking endogenous TGFβ3, addition of TGFβ1 increases p15ink4b promoter activity in dose-dependent manner in MEE cells. After blocking endogenous TGFβ1, addition of TGFβ3 does not have significant effect on p15ink4b promoter activity.
TGFβ3 does not have significant effect on Smad4-mediated activation of the p15ink4b promoter
The transfection method and luciferase assay are describe in a previous section. After transfection, the cells were incubated with TGFβ3. Treatment with 2ng/ml of TGFβ3 did not significantly increase luciferase activity in MEE cells transfected with pGL3-p15wt-Lux plasmid (Fig. 4B) whereas 5ng/ml of TGFβ3 increased luciferase activity 1.5-fold. The MEE cells produced similar results after transfection with plasmid containing the mutation in the SBE A (Fig. 4B) compared to the wild type plasmid. Treatment with TGFβ3 did not affect luciferase activity in MEE cells containing mutations in Smad-binding sites B and C as well as plasmid with all three mutated sites. These results indicate that unlike TGFβ1, TGFβ3 has limited to no effect on p15ink4b promoter activity.
TGFβ1 independently activates p15ink4b promoter when endogenous TGFβ3 is blocked
To eliminate effects from endogenous TGFβ1 or TGFβ3, MEE cells were pre-treated with antibodies against TGFβ1 and TGFβ3 for 24h. The cells were then transfected with pGL3-p15-Lux plasmid. After transfection, exogenous recombinant TGFβ1 or TGFβ3 were added and cells were then incubated for 24h in the presence of the antibodies. This approach made it possible to evaluate the effects of TGFβ1 and TGFβ3 on the p15ink4b promoter separately without the ambiguity imposed by endogenous TGFβ activity. The data indicate that human recombinant (hr) TGFβ1 significantly increased p15ink4b promoter activity in a dose-dependent manner (Fig. 4C) when TGFβ3 was inhibited, whereas exogenous TGFβ3 did not affect p15ink4b promoter activity, when TGFβ1 was blocked. Therefore, we conclude that TGFβ1 is more potent in activating p15ink4b promoter activity compared to TGFβ3.
Smad4 bind to all the Smad-binding elements in p15ink4b promoter
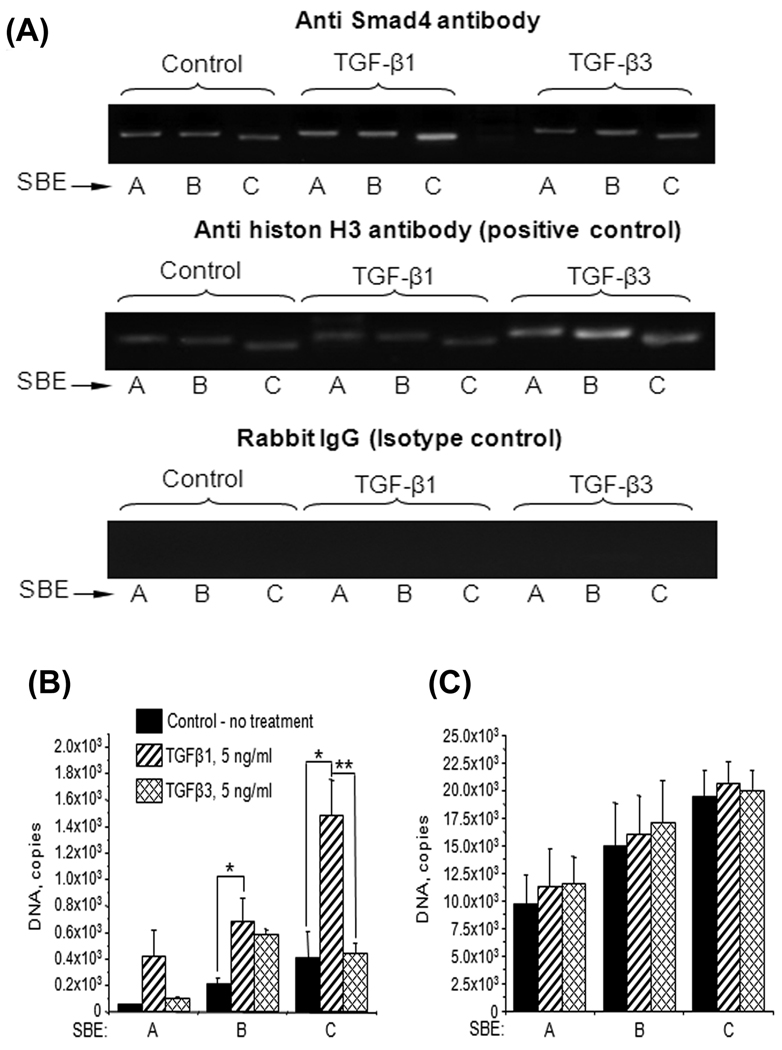
Since the SBEs are functional and important to the activation of p15ink4b promoter as shown earlier, we wanted to identify if these SBEs are harboring Smad proteins in the p15ink4b promoter. We employed Chromatin immunoprecipitation assay to identify whether or not Smad4 was bound to the SBEs (A, B and C). The PCR analysis of the chromatin immunoprecipitated with anti-Smad4 antibody from TGFβ1 and TGFβ3 treated or untreated cells displayed that all 3 SBEs bound to Smad4 protein (Fig. 5A, top panel). The specificity of the bindings was confirmed by the positive (Histon H3) and negative (Rabbit Ig G) controls (Fig. 5A lower two panels). Quantitative real-time PCR analysis demonstrates the selective binding of Smad4 with different SBEs in the p15ink4b promoter depending on the treatment with TGFβ1 or TGFβ3 (Fig. 5B). We found that TGFβ1 significantly increased binding of Smad4 with all three SBEs in the p15ink4b promoter, whereas TGFβ3 significantly increased only binding with SBE B. SBE C bound most strongly with Smad4 protein. Comparison of qPCR results of the analysis of immunoprecipitated chromatin with the real-time PCR data of the total purified but not immunoprecipitated chromatin from the same cells (input control, Fig. 5C) confirms that the revealed differences in Smad4 binding with SBE in p15ink4b promoter induced by TGFβ1 and TGFβ3 signaling are statistically significant. Since mutation SBE C decreased luciferase expression most (Fig. 4A) it appears that site C is the most important region in the p15ink4b promoter for Smad4 mediated transactivation induced by TGFβ1 signaling. The differences in response to TGFβ are significant as there are no statistically significant differences between treated and untreated samples in non-IP chromatin. It is normally that total DNA quantity is 1–2 log higher in input control than in IP chromatin, because usually 90–95% of DNA is lost during IP and subsequent purification.
Fig. 5. TGFβ treatment increase binding Smad4 protein with p14ink4b promoter.
A. Chromatin was isolated from TGFβ1 (5ng/ml) or TGFβ3 (5ng/ml) treated/untreated MEE cells, immunoprecipitated with anti Smad4 antibody (overnight) and analyzed by quantitative real-time PCR with primers recognizing A, B and C Smad4 binding elements in p15ink4b promoter. Serial dilutions of pGL3-p15wild type plasmid were used as quantitative standards. Both TGFβ1 and TGFβ3 increase binding of Smad4 with SBE in the p15ink4b promoter. TGFβ1 induces binding of Smab4 with all three SBE in variable degree, whereas TGFβ3 increases only binding with SBE B.
B. Chromatin was isolated from TGFβ1 (5ng/ml) or TGFβ3 (5ng/ml) treated/untreated MEE cells, immunoprecipitated with anti Smad4 antibody (overnight) and analyzed by quantitative real-time PCR with primers recognizing A, B and C Smad4 binding elements in p15ink4b promoter. Serial dilutions of pGL3-p15wild type plasmid were used as quantitative standards. Both TGFβ1 and TGFβ3 increase binding of Smad4 with SBE in the p15ink4b promoter. TGFβ1 induces binding of Smab4 with all three SBE in variable degree, whereas TGFβ3 increases only binding with SBE B. The results are shown as a mean ± SD obtained from three independent chromatin preparations. Asterisk indicates p ≤ 0.05, double asterisk – p ≤ 0.005.
C. Input control for quantitative real-time PCR of immunoprecipitated chromatin. Chromatin was isolated as described in A and analyzed by quantitative PCR with primers recognizing A, B and C Smad4 binding elements in p15ink4b promoter with DNA standards described in A. Primers recognizing B and C Smad4 binding elements display higher efficiency than primers specific for SBE A, however statistically significant differences between TGFβ1-, TGFβ3-treated or untreated samples are not detected (p ≥ 0.05). Error bars show the standard deviation of three independent chromatin preparations.
While TGFβ3 alone facilitates cell migration but TGFβ1 and TGFβ3 both can induce apoptosis
To establish the roles of TGFβ1 and TGFβ3 on cell migration and apoptosis, we undertook Transwell cell migration assay (Fig. 6A) and TUNEL staining (Fig. 6B) using a combination of treatment conditions. The results demonstrate that TGFβ3 causes increased cell migration compared to TGFβ1. Further, addition of TGFβ1 with TGFβ3 significantly impact cell migration. However, inducing cell proliferation with 20% FBS, in presence of both TGFβ1 and TGFβ3, reduce cell migration. And cell migration can be halted with blocking p15ink4b (with pLK0.1-shp15) when cell are treated with both TGFβ1 and TGFβ3, but the effect of halting p15 in presence of either/or TGFβ1 and TGFβ3 alone on cell migration was not significant. The cell migration can be facilitated by TGFβ3, but not TGFβ1, with increasing p15ink4b (with full length ~1.3kb murine p15ink4b cDNA, pBluescript-p15ink4b). Such independent functions by these two isoforms of TGFβ ligand, where 1 is important for cell cycle arrest and 3 for migration, suggest that TGFβ1 induced p15ink4b activation may be a prerequisite for cell migration by TGFβ3.
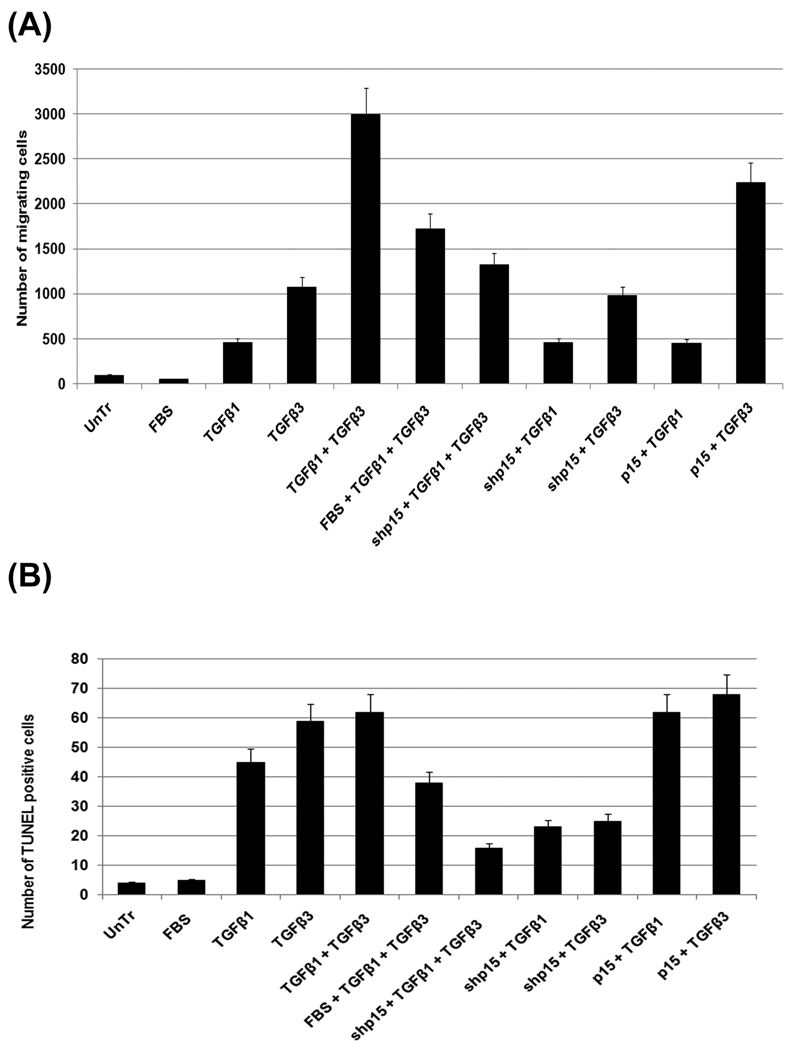
Fig. 6. MEE cell migration and apoptotic assays in response to TGFβ.
The roles of TGFβ1 and TGFβ3 on cell migration and apoptosis are shown here with Transwell cell migration assay, TUNEL assay as well as FACSArray.
A. Our transwell cell migration assay results show that showed lack of cell migration in untreated (control) MEE cells (mean ±SD; n=3; <p0.05;), but, when MEE cells treated with TGFβ3 (5ng/mL) for 48h, they become motile. In combination of treatments, such as, inducing cell proliferation with 20% FBS (FBS), inducing TGFβ1 and TGFβ3 with exogenous recombinant TGFβ1 (5ng/mL) and TGFβ3 (5ng/mL), lentivirus vector encoding short hairpin (sh) p15ink4b (pLK0.1-shp15RNA) was used to block p14ink4b and murine full length (1.3kb) pBluescript-p15cDNA (p15) to was used to induce p15ink4b, our transwell cell migration assay results demonstrate that TGFβ3 increases cell migration when compared to TGFβ1 and in combination of both the isoforms, they have additive effect on cell migration. Moreover, blocking cell cycle with shp15 in the presence of either TGFβ1 or TGFβ3 alone does not affect cell migration. But when cells are treated with in both TGFβ1 and TGFβ3, blockage of p15, significantly reduces cell migration. Induction of p15 significantly increases cell migration when treated with TGFβ3 but has no effect when treated with TGFβ1 indicating that while TGFβ3 alone is a better promoter of cell migration, it needs the induction of p15 and cell cycle arrest by TGFβ1 to have full effect.
B. With 16 Biotin UTP labelled TUNEL positive cells in MEE culture for determining apoptosis, our results show that TGFβ1 (5ng/mL) and TGFβ3 (5ng/mL) for 72h have similar effect on apoptosis and both are capable of inducing apoptosis. TGFβ3 has slightly more effect than TGFβ1 and but together they do not have an additive effect such seen for migration assay (A). As seen in cell migration, apoptosis also can be blocked when p15ink4b is blocked (with shp15) and can be enhanced when induced p15 (with full length p15cDNA) indicating induction of cell cycle arrest by p15ink4b may be a requirement for apoptosis.
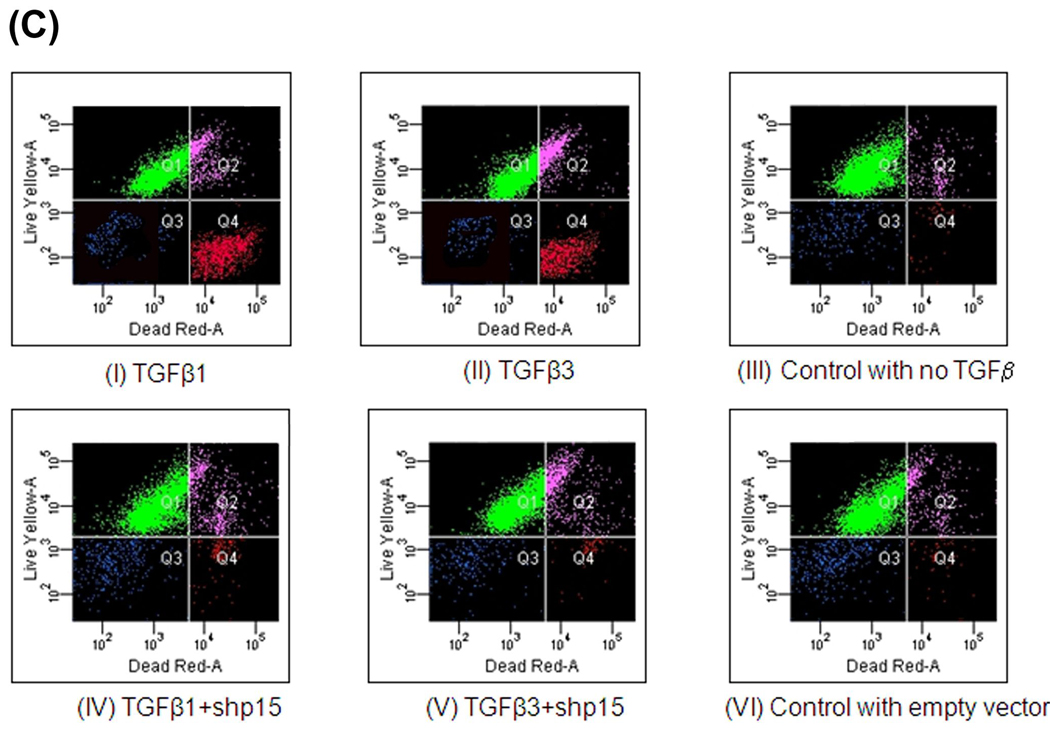
C. TGFβ3 or TGFβ1 promote MEE cell to undergo apoptosis: MES cells grown in culture to 80% confluency were treated with. TGFβ3 (5ng/mL) or TGFβ1 (5ng/mL). Nucleic acid dyes, C12-resazurin at 488 nm, and Sytox Red at 633 nm excitation show “Live” (Q1), “viable” cells (Q2) and “dead” (Q4) cells, respectively and “debris” in blue (Q3). Within 72h of TGFβ1 and TGFβ3 treatment the number of “dead cells” in Q4 reached nearly 33% and 25% respectively for TGFβ3 and TGFβ1 treated total MEE cells - with the remaining being “live” and “viable” cells. As shown in TUNEL assay (B), TGFβ1 and TGFβ3 induced apoptotic can be blocked by inhibiting p15ink4b with shp15 as demonstrated by reduced number of “dead” cells in Q4 similar to control, when compared with TGFβ1 and TGFβ3 treated “dead” cells.
Similarly for TUNEL positive cells (Fig. 6B) in MEE culture for determining apoptosis, our results showed that unlike cell migration, the ligands, TGFβ1 and TGFβ3, affect apoptosis. While both are capable of inducing apoptosis, TGFβ3 has slightly more effect than TGFβ1 and have no significant additive effect when they function jointly. Similar to the cell migration results, apoptosis can also be blocked when p15ink4b is blocked (with shp15), but unlike, cell migration, both TGFβ1 and TGFβ3, by itself, enhanced apoptosis. And both TGFβ1 and TGFβ3 had similar effect on apoptosis when induced by p15ink4b (with full length cDNA), whereas in cell migration, the effect was noticeable only in case of TGFβ3, not TGFβ1. This indicates that induction of cell cycle arrest by p15ink4b may be a requirement for apoptosis. These TUNEL staining results were confirmed by the Live and dead cell analysis using nucleic acid dyes, C12-resazurin at 488 nm, and Sytox Red at 633 nm excitation show “Live”, “viable” and “dead” cells. Our FACSArray results show that within 72h of TGFβ1 and TGFβ3 treatment the number of “dead cells” in Q4 reached nearly 33% and 25% respectively for TGFβ3 and TGFβ1 treated MEE cells (Fig. 6C). As shown in TUNEL assay (Fig. 6B), TGFβ1 and TGFβ3 induced apoptotic can be blocked by inhibiting p15ink4b with shp15 as demonstrated by reduced number of “dead” cells in Q4 similar to control, when compared with TGFβ1 and TGFβ3 treated “dead” cells.
DISCUSSION
Palatogenesis is an important event during the craniofacial development of the group of higher vertebrates known as amniotes. The stages of palatal development traditionally have been defined by the position of the palatal shelves in the oral cavity and the opposing palatal shelves’ level of union at the midline (Ferguson, 1988). Formation of the medial epithelial seam (MES) by palatal shelf fusion is a crucial step of palate development. Complete disintegration of the MES is the final essential phase of palatal confluency with surrounding mesenchymal cells. The cellular mechanism underlying seam degeneration and the fate of MES cells has long been a major focus of the field; however, several controversies still surround these topics. For the past 50 years, scientists have used the latest, most sophisticated methods available to them, ranging from basic ultra structural cell biology to cell labeling using genetic lineage markers. Two major models have been proposed for seam degeneration: Epithelial Mesenchymal Transition (EMT) and/or Apoptosis.
TGFβ have multiple effects, including apoptosis and regulation of cell proliferation, involving EMT (Nawshad et al., 2004; Tian and Schiemann, 2009). It has been established that TGFβ induces both EMT and apoptosis during palatal seam disintegration (Ahmed et al., 2007; Nawshad, 2008; Valdes et al., 2002). TGFβ-induced apoptosis and EMT are mediated by some of the same signaling molecules, most of which are activated during an early stage of TGFβ treatment (Yang et al., 2006a; Yang et al., 2006b). While a role for TGFβ in palatal EMT and apoptosis has been well established, the prerequisite and preparation for MEE cells to undergo such cellular changes has never been addressed. It is likely that cells undergoing drastic morphological and functional alterations cease cell proliferation to facilitate the transition, so that these dramatic changes can be adapted. Therefore, cessation of cell proliferation and activation of cell cycle arrest might be a prerequisite for cells undergoing differentiation and/or transition. It has been well established that the process of TGFβ induced changes in morphology and phenotype are coincident with cell survival, when cells elicit an epithelial-fibroblastic transition (Sanchez et al., 1999). Proliferation control by TGFβ is an important modulator of cell migration and a key regulator of EMT (Caja et al., 2007). Caja et al., (2007) (Caja et al., 2007) suggests that when hepatoma cells escape from the suppressive effects of TGFβ on cell proliferation the same cytokine also induces EMT. It was recently shown that cell cycle phase is a key event in determining the subsequent pathway – EMT or an apoptosis; the growth arrest is required for cells to undergoing differentiation (Yang et al., 2006b). It was shown in hepatocytes, that as a response to TGFβ treatment, the cells undergo an apoptosis if they are in G2/M phase, while if they are in the G1/S phase they transfer to EMT (Yang et al., 2006b).
There is ample evidence to suggest a direct role of TGFβ to inhibit cell cycle progression by blocking a number of steps involved in cdk activation (Bhowmick et al., 2003; Satterwhite and Moses, 1994), TGFβ is a representative growth inhibitor for much kind of epithelial cells (Moses et al., 1990; Pietenpol et al., 1990). TGFβ signaling leads to transcriptional activation of the CDK inhibitors p21WAF1/CIP1 and p15ink4b (Massague, 1998; Massague and Wotton, 2000). p15ink4b has been shown to be a direct target gene for TGFβ1 (Hannon and Beach, 1994). Exposure of TGFβ1 in human keratiocytes, results in induction of p21WAF1/CIP1 and p15ink4b (Sakaguchi et al., 2004). When primary mouse keratinocytes are cultured in the presence of a neutralizing antibody to TGFβ1, the senescence response is suppressed and the level of the TGFβ1 target, p15ink4b, increases as does association of this kinase inhibitor with cyclinD/cdk4 complexes (Tremain et al., 2000). The p15ink4b activity is not limited to cell cycle arrest but it also induces cell differentiation in granulocyte/macrophage differentiation (Amanullah et al., 2000).
Both TGFβ induced apoptosis and EMT are mediated by some of the same signaling molecules, such as TGFβ receptors, Smads, p38 MAPK, PKA and STAT3, most of which are activated at an early stage after TGFβ treatment (Tian et al., 2009). In response to TGFβ stimulation, Smad2 and Smad3 are phosphorylated. The phosphorylated Smad2/3 are engaged in a complex with Smad4 and then translocated into the nucleus. In nuclear compartment this protein complex directly binds to the Smad binding element (SBE), the specific sequence in promoters of target genes, including p15ink4B.
Studies showed that multiple factors including Smads, Sp1 and Myc bind to different DNA elements of the p15ink4b promoter to mediate TGFβ-induced gene expression (Guo et al., 2009). Promoter studies in keratinocytes revealed that TGFβ stimulatory effects on p15ink4b gene are mediated through the Smad pathway and the transcription factor Sp1(Ho et al., 2004). (Peng et al., 2002) found two Smad4 binding elements, located in the region between − 359 and −329 bp and between −95 and 46bps of the p15ink4b promoter. However, SBE between −359 and −329bp is more active than the later region. The p15ink4b promoter reporter gene assays revealed that Smad4-dependent transcriptional activation is mediated by this SBE, which indicates that p15ink4b, is one of the down-stream target genes regulated by Smad.
Moreover, (Gomis et al., 2006) found that the induction of p15ink4b involves two separate SBEs, CAGACA (between −431 and −425bp) and AAGA (between −525 and −521bp) and both are responsive to TGFβ. When the primary MEE cells from mouse embryos are collected at 14.0 days post coitum (dpc) and treated with TGFβ1 and TGFβ3 for 48h, we were able to demonstrate that TGFβ induces the cell cycle arrest through activation of p15ink4b protein expression in a time and dose dependent manner (Fig. 1A and B). However, TGFβ1 has a more pronounced effect on p15ink4b expression than TGFβ3 (Fig. 1A and B). Moreover, p15ink4b expression is highest in vivo when opposite palatal shelves about to fuse at 14.0 dpc (Fig. 1C, i) and when the fuse at 14.5 dpc (Fig. 1C, ii). However, the p15ink4b expression decreased immediately following palatal fusion and show very limited at 15.5 dpc (Fig. 1C, iii) or no expression at 16.5 dpc (Fig. 1C, iv) by which time palatal seam is completely disintegrated. Interestingly, changes in the p15ink4b expression are in accord with the TGFβ expression in vivo. It has been previously shown that TGFβ1 and TGFβ3 expression are confined mostly (in the palatal medial edge epithelial cells) to at time just prior to palatal fusion. As palates fuse, the TGFβ1 expression ceases but TGFβ3 continues to maintain increased expression indicating two separate yet complimentary roles of TGFβ1 and TGFβ3. Since TGFβ1 is a more potent inducer of p15ink4b expression of palatal MEE cells in culture condition (Fig. 1 and B), it is therefore, likely that the higher expression of TGFβ1 in palates in vivo, prior palatal fusion, is related to cell cycle arrest. TGFβ3 may have additive effects on cell cycle arrest concurrent with TGFβ1 suggesting that they may be complimentary in vivo.
Our cell cycle arrest study using a FACSArray showed that G1 synchronized MEE cells, treated with TGFβ1 and TGFβ3 induced cell cycle arrest and remain at G1 phase. But this effect is more prominent with TGFβ1 than TGFβ3 (Fig. 2A and B) Both TGFβ1 and TGFβ3 have similar effects in the expression of p15ink4b and in cell cycle arrest; however, our studies indicate that the role of TGFβ1 in cell cycle arrest is more pronounced than TGFβ3.
Our experiments into gene activity using luciferase assays in MEE cells transfected with pGL3-p15-Lux plasmid demonstrated that TGFβ1 had higher gene activity (three folds increase) effect on the p15ink4b promoter, compared to TGFβ3 (Fig. 4A and B). It is noteworthy that the effect of TGFβ3 in p15ink4b gene activity is very similar to wild type, making TGFβ3 less likely to have a primary role in p15ink4b gene activity. The three SBEs have variable roles in the p15ink4b gene activity, since the mutation of three SBEs (Fig. 4A, B and C) effect p15ink4b gene activity significantly in both TGFβ1 and TGFβ3 treated MEE cells. To demonstrate which isoform of TGFβ has the most effect on p15ink4b, we blocked TGFβ1 and TGFβ3 using specific antibodies. Blocking endogenous TGFβ3 followed by the addition of TGFβ1 led to an increase in p15ink4b promoter activity by two folds in a dose-dependent manner in MEE cells (Fig. 4C). But addition of TGFβ3, when TGFβ1 was blocked, did not have a significant effect. While the luciferase data clearly support a more prominent role of TGFβ1 in p15ink4b gene activity over TGFβ3, it is noteworthy, that when endogenous TGFβ3 was not blocked, addition of the TGFβ1, activates p15ink4b promoter by threefolds (Fig. 4A) indicating that TGFβ1 and TGFβ3 are complimentary and act synergistically. These data clearly attest to our findings both in protein expression (Fig, 1) and cell cycle arrest FACSArray (Fig. 2) demonstrating that TGFβ1 promotes cell cycle arrest by inducing p15ink4b more than TGFβ3 does. By aggregating p15ink4b protein expression, p15ink4b gene activity and cell cycle arrest data by FACSArray, we can suggest that TGFβ1 may be the major causative factor for the induction of cell cycle arrest in the palatal MEE cells just prior to palatal fusion at 14.5 dpc.
Our data also indicate that TGFβ effect on p15ink4b induced cell-cycle arrest is Smad-dependent. Analysis of Smad binding with SBEs in p15ink4b promoter showed that all three elements are involved in interaction with Smad protein (Fig. 5A) when cells are activated with TGFβ1 and TGFβ3. While both TGFβ1 and TGFβ3 increased binding of Smad4 protein with the p15ink4b promoter, the effect of TGFβ1 was significantly higher (Fig. 5B) which correlates with its stronger p15ink4b gene activity and protein expression (Fig. 1 and Fig. 3). Moreover, our data demonstrate that site C of the SBEs has higher binding capacity as shown by increased DNA quantities indicting that activated Smads, in response to TGFβ1, binds both to site C and site B in response to TGFβ3. However the binding interaction of TGFβ1, demonstrated by DNA amount, is much higher to site C than site B. (Fig. 5B).
We were intrigue by the expression pattern on TGFβ1 and TGFβ3 in palatal MEE cells and their role in cell cycle arrest. Two questions still remained unanswered whether TGFβ1 is inducing cell cycle arrest by activating p15ink4b, and what are the sequential roles of TGFβ1 and TGFβ3 in palatal seam disintegration. While others have shown that TGFβ3 plays crucial role in palatal seam disintegration by EMT and/or Apoptosis (Ahmed et al., 2007; Nawshad, 2008), we undertook the functional assays to demonstrate the roles of TGFβ1 and TGFβ3 in cell migration and apoptosis. Our data overwhelmingly support a major role of TGFβ3 in cell migration compared to TGFβ1 (Fig. 6A), however, the functional acquisition of cell cycle arrest by inducing p15ink4b by TGFβ1 is important for TGFβ3 induced cell migration. The results also indicate that cell migration is dependent on the status cell cycle arrest since the blockage of cell cycle arrest by p15ink4b inhibitor, shp15ink4b, and decreased cell migration in presence of both the isoforms. And addition of p15ink4b along with exogenous TGFβ3, not TGFβ1, increased cell migration. Blocking p15ink4b or adding p15ink4b, along with exogenous TGFβ1, did not significantly affect cell migration suggesting that TGFβ3 is a potent inducer of cell migration and its effect is dependent on the cell cycle status that can be introduced by TGFβ1, a potent inducer of cell cycle arrest marker, p15ink4b, as shown in Fig1A and B. These data clearly indicate two separate yet complimentary roles of TGFβ1 and TGFβ3. Whereas, TGFβ1 ensures immediate cell cycle arrest by activating p15ink4b and preparing MEE cells to undergo morphological changes, TGFβ3 promotes cell migration, suggesting that these two isoforms (1 and 3) of TGFβ play a synergistic role in MEE cell behavior. Interestingly, unlike cell migration, in apoptosis, these two isoforms (1 and 3) can both induce apoptosis unilaterally in a near identical fashion as demonstrated by TUNEL positive cells as well as FACSArray for live and dead cell analysis (Fig. 6 B and C). Our results suggest that TGFβ3 is a more potent promoter of cell migration; however, both TGFβ1 and TGFβ3 are capable of inducing apoptosis. Since the expression of TGFβ1 is either limited or no longer present in the palatal seam in vivo upon palatal fusion, it is likely that the induction of apoptosis in vivo is facilitated by TGFβ3 - as its expression following fusion increases dramatically and is the preferentially remaining TGFβ isoform in the palatal seam at that stage. Some research groups suggested a functional connection between TGFβ1 and TGFβ3 during development based on a shared mechanism of activation (Mu et al., 2008; Yang and Kaartinen, 2007).
Although increasing evidence indicates that cell cycle state is an important factor for cellular response to extra cellular stimuli, very little attention has been given to the two distinct cellular responses induced by cell stimuli for the same cells, for example in the case of TGFβ induced concomitant apoptosis and EMT. The observation that TGFβ induced caspase activity in cells synchronized at G2/M phase was significantly higher than cells at G1/S phase provided an explanation on how TGFβ induced very strong apoptosis in these cells. It has been widely known that growth arrest is a prerequisite for cells undergoing differentiation. As TGFβ induced EMT occurred in the G1/S phase, TGFβ induced G1/S phase growth arrest may provide cells a precondition for undergoing EMT. TGFβ induced EMT of cells at G1/S phase explained how cells survived the TGFβ treatment under the same condition. It is obvious that in regular cell cultures without synchronization, cells are highly heterogeneous in terms of the cell cycle phases and will therefore respond differentially to TGFβ, leading to a variety of cell fates. These data not only explained why only some cells undergo apoptosis within a defined relatively short period of TGFβ treatment but they also provided a new insight into the mechanism through which same signal molecule (TGFβ1) induces concomitantly different cellular fates in the same cell type. This explanation could be important in the understanding of multifunctional effects of TGFβ in some physiological and pathological processes. It has been reported that the EMT conferred a degree of resistance to TGFβ induced apoptosis in fetal rat hepatocytes (Valdes et al., 2002). This phenomenon may not be restricted to the TGFβ induced apoptosis and EMT, since Snail induced EMT has also been associated with the blockage of cell cycle and the resistance to different pro apoptotic stimuli-induced apoptosis (Vega et al., 2004). Although it has been known that there is a relationship between the cell cycle, TGFβ induced apoptosis and EMT, little is known about cellular changes which control cell cycle-dependent apoptosis or EMT. Most of these proteins were key players in a wide variety of cellular processes, including cell growth and death, regulation of cytoskeleton arrangement and signaling pathways.
EMT and apoptosis are highly conserved, fundamental processes that govern morphogenesis in development. The key to understanding EMT and apoptosis lies not only in the instructions the MES cells carry with them, but also within the characteristics of the landscape that determine the way cells behave during development. Our and others evidences suggest that TGFβ can promote both to contribute during palatogenesis, especially during palatal seam disintegration. In this study, for the first time, we suggest that cell cycle arrest is a key phase that takes place immediate following palatal fusion. And induction of p15ink4b by TGFβ is the first and foremost phase following palatal fusion and without cell cycle arrest, EMT and or apoptosis cannot proceed.
ACKNOWLEDGMENT
We thank Dr. Linda Wolff (Center for Cancer Research, National Cancer Institute, NIH, Bethesda, MD) for providing us with murine p15ink4b promoter and p15ink4b full length cDNA. This research is supported by NIDCR, NIH grant (R01DE017986) to Dr. Ali Nawshad.
List of abbreviations and acronyms
- TGFβ
Transforming Growth Factor β
- EMT
Epithelial Mesenchymal Transition
- MES
Midline Epithelial Seam
- SBE
Smad Binding Element
- CDK
Cyclin Dependent Kinase
- MEE
Medial Edge Epithelial
- ChIP
Chromatin Immune Precipitation
REFERENCES
- Ahmed S, Liu CC, Nawshad A. Mechanisms of palatal epithelial seam disintegration by transforming growth factor (TGF) beta3. Developmental biology. 2007;309(2):193–207. doi: 10.1016/j.ydbio.2007.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanullah A, Hoffman B, Liebermann DA. Deregulated E2F-1 blocks terminal differentiation and loss of leukemogenicity of M1 myeloblastic leukemia cells without abrogating induction of p15(INK4B) and p16(INK4A) Blood. 2000;96(2):475–482. [PubMed] [Google Scholar]
- Bhowmick NA, Ghiassi M, Aakre M, Brown K, Singh V, Moses HL. TGF-beta-induced RhoA and p160ROCK activation is involved in the inhibition of Cdc25A with resultant cell-cycle arrest. Proc Natl Acad Sci U S A. 2003;100(26):15548–15553. doi: 10.1073/pnas.2536483100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissell DM. Chronic liver injury, TGF-beta, and cancer. Experimental & molecular medicine. 2001;33(4):179–190. doi: 10.1038/emm.2001.31. [DOI] [PubMed] [Google Scholar]
- Bissell DM, Roulot D, George J. Transforming growth factor beta and the liver. Hepatology (Baltimore, Md. 2001;34(5):859–867. doi: 10.1053/jhep.2001.28457. [DOI] [PubMed] [Google Scholar]
- Caja L, Ortiz C, Bertran E, Murillo MM, Miro-Obradors MJ, Palacios E, Fabregat I. Differential intracellular signalling induced by TGF-beta in rat adult hepatocytes and hepatoma cells: implications in liver carcinogenesis. Cell Signal. 2007;19(4):683–694. doi: 10.1016/j.cellsig.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Derynck R, Akhurst RJ, Balmain A. TGF-beta signaling in tumor suppression and cancer progression. Nat Genet. 2001;29(2):117–129. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
- Ferguson MW. Palate development. Development. 1988;103 Suppl:41–60. doi: 10.1242/dev.103.Supplement.41. [DOI] [PubMed] [Google Scholar]
- Gomis RR, Alarcon C, Nadal C, Van Poznak C, Massague J. C/EBPbeta at the core of the TGFbeta cytostatic response and its evasion in metastatic breast cancer cells. Cancer Cell. 2006;10(3):203–214. doi: 10.1016/j.ccr.2006.07.019. [DOI] [PubMed] [Google Scholar]
- Guo P, Zhao KW, Dong XY, Sun X, Dong JT. Acetylation of KLF5 alters the assembly of p15 transcription factors in transforming growth factor-beta-mediated induction in epithelial cells. J Biol Chem. 2009;284(27):18184–18193. doi: 10.1074/jbc.M109.007096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon GJ, Beach D. p15INK4B is a potential effector of TGF-beta-induced cell cycle arrest. Nature. 1994;371(6494):257–261. doi: 10.1038/371257a0. [DOI] [PubMed] [Google Scholar]
- Haviernik P, Schmidt M, Hu X, Wolff L. Consistent inactivation of p19(Arf) but not p15(Ink4b) in murine myeloid cells transformed in vivo by deregulated c-Myc. Oncogene. 2003;22(11):1600–1610. doi: 10.1038/sj.onc.1206268. [DOI] [PubMed] [Google Scholar]
- Heldin CH, Landstrom M, Moustakas A. Mechanism of TGF-beta signaling to growth arrest, apoptosis, and epithelial-mesenchymal transition. Curr Opin Cell Biol. 2009;21(2):166–176. doi: 10.1016/j.ceb.2009.01.021. [DOI] [PubMed] [Google Scholar]
- Ho J, de Guise C, Kim C, Lemay S, Wang XF, Lebrun JJ. Activin induces hepatocyte cell growth arrest through induction of the cyclin-dependent kinase inhibitor p15INK4B and Sp1. Cell Signal. 2004;16(6):693–701. doi: 10.1016/j.cellsig.2003.11.002. [DOI] [PubMed] [Google Scholar]
- Martinez-Alvarez C, Blanco MJ, Perez R, Rabadan MA, Aparicio M, Resel E, Martinez T, Nieto MA. Snail family members and cell survival in physiological and pathological cleft palates. Dev Biol. 2004;265(1):207–218. doi: 10.1016/j.ydbio.2003.09.022. [DOI] [PubMed] [Google Scholar]
- Martinez-Alvarez C, Tudela C, Perez-Miguelsanz J, O'Kane S, Puerta J, Ferguson MW. Medial edge epithelial cell fate during palatal fusion. Dev Biol. 2000;220(2):343–357. doi: 10.1006/dbio.2000.9644. [DOI] [PubMed] [Google Scholar]
- Massague J. TGF-beta signal transduction. Annu Rev Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- Massague J, Wotton D. Transcriptional control by the TGF-beta/Smad signaling system. Embo J. 2000;19(8):1745–1754. doi: 10.1093/emboj/19.8.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses HL, Yang EY, Pietenpol JA. TGF-beta stimulation and inhibition of cell proliferation: new mechanistic insights. Cell. 1990;63(2):245–247. doi: 10.1016/0092-8674(90)90155-8. [DOI] [PubMed] [Google Scholar]
- Mu Z, Yang Z, Yu D, Zhao Z, Munger JS. TGFbeta1 and TGFbeta3 are partially redundant effectors in brain vascular morphogenesis. Mech Dev. 2008;125(5–6):508–516. doi: 10.1016/j.mod.2008.01.003. [DOI] [PubMed] [Google Scholar]
- Murray JC, Schutte BC. Cleft palate: players, pathways, and pursuits. J Clin Invest. 2004;113(12):1676–1678. doi: 10.1172/JCI22154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawshad A. Palatal seam disintegration: to die or not to die? that is no longer the question. Dev Dyn. 2008;237(10):2643–2656. doi: 10.1002/dvdy.21599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawshad A, Hay ED. TGFbeta3 signaling activates transcription of the LEF1 gene to induce epithelial mesenchymal transformation during mouse palate development. J Cell Biol. 2003;163(6):1291–1301. doi: 10.1083/jcb.200306024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawshad A, LaGamba D, Hay ED. Transforming growth factor beta (TGFbeta) signalling in palatal growth, apoptosis and epithelial mesenchymal transformation (EMT) Archives of oral biology. 2004;49(9):675–689. doi: 10.1016/j.archoralbio.2004.05.007. [DOI] [PubMed] [Google Scholar]
- Nawshad A, Medici D, Liu CC, Hay ED. TGFbeta3 inhibits E-cadherin gene expression in palate medial-edge epithelial cells through a Smad2-Smad4-LEF1 transcription complex. J Cell Sci. 2007;120(Pt 9):1646–1653. doi: 10.1242/jcs.003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng B, Fleming JB, Breslin T, Grau AM, Fojioka S, Abbruzzese JL, Evans DB, Ayers D, Wathen K, Wu T, Robertson KD, Chiao PJ. Suppression of Tumorigenesis and Induction of p15ink4b by Smad4/DPC4 in Human Pancreatic Cancer Cells. Clin Cancer Res. 2002;8(11):3628–3638. [PubMed] [Google Scholar]
- Pietenpol JA, Stein RW, Moran E, Yaciuk P, Schlegel R, Lyons RM, Pittelkow MR, Munger K, Howley PM, Moses HL. TGF-beta 1 inhibition of c-myc transcription and growth in keratinocytes is abrogated by viral transforming proteins with pRB binding domains. Cell. 1990;61(5):777–785. doi: 10.1016/0092-8674(90)90188-k. [DOI] [PubMed] [Google Scholar]
- Sakaguchi M, Miyazaki M, Sonegawa H, Kashiwagi M, Ohba M, Kuroki T, Namba M, Huh NH. PKCalpha mediates TGFbeta-induced growth inhibition of human keratinocytes via phosphorylation of S100C/A11. J Cell Biol. 2004;164(7):979–984. doi: 10.1083/jcb.200312041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez A, Alvarez AM, Lopez Pedrosa JM, Roncero C, Benito M, Fabregat I. Apoptotic response to TGF-beta in fetal hepatocytes depends upon their state of differentiation. Exp Cell Res. 1999;252(2):281–291. doi: 10.1006/excr.1999.4624. [DOI] [PubMed] [Google Scholar]
- Satterwhite DJ, Moses HL. Mechanisms of transforming growth factor-beta 1-induced cell cycle arrest. Invasion & metastasis. 1994;14(1–6):309–318. [PubMed] [Google Scholar]
- Schutte BC, Murray JC. The many faces and factors of orofacial clefts. Hum Mol Genet. 1999;8(10):1853–1859. doi: 10.1093/hmg/8.10.1853. [DOI] [PubMed] [Google Scholar]
- Seoane J, Pouponnot C, Staller P, Schader M, Eilers M, Massague J. TGFbeta influences Myc, Miz-1 and Smad to control the CDK inhibitor p15INK4b. Nat Cell Biol. 2001;3(4):400–408. doi: 10.1038/35070086. [DOI] [PubMed] [Google Scholar]
- Thillainadesan G, Isovic M, Loney E, Andrews J, Tini M, Torchia J. Genome analysis identifies the p15ink4b tumor suppressor as a direct target of the ZNF217/CoREST complex. Mol Cell Biol. 2008;28(19):6066–6077. doi: 10.1128/MCB.00246-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian HY, Zhang KH, Gao X, Lei WW, Zhang L, Yu ML, Song JG, Zhao FK. Comparative proteomic analysis of cell cycle-dependent apoptosis induced by transforming growth factor-beta. Biochim Biophys Acta. 2009;1794(10):1387–1397. doi: 10.1016/j.bbapap.2009.05.003. [DOI] [PubMed] [Google Scholar]
- Tian M, Schiemann WP. The TGF-beta paradox in human cancer: an update. Future Oncol. 2009;5(2):259–271. doi: 10.2217/14796694.5.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremain R, Marko M, Kinnimulki V, Ueno H, Bottinger E, Glick A. Defects in TGF-beta signaling overcome senescence of mouse keratinocytes expressing v-Ha-ras. Oncogene. 2000;19(13):1698–1709. doi: 10.1038/sj.onc.1203471. [DOI] [PubMed] [Google Scholar]
- Valdes F, Alvarez AM, Locascio A, Vega S, Herrera B, Fernandez M, Benito M, Nieto MA, Fabregat I. The epithelial mesenchymal transition confers resistance to the apoptotic effects of transforming growth factor Beta in fetal rat hepatocytes. Mol Cancer Res. 2002;1(1):68–78. [PubMed] [Google Scholar]
- Vega S, Morales AV, Ocana OH, Valdes F, Fabregat I, Nieto MA. Snail blocks the cell cycle and confers resistance to cell death. Genes Dev. 2004;18(10):1131–1143. doi: 10.1101/gad.294104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff L, Garin MT, Koller R, Bies J, Liao W, Malumbres M, Tessarollo L, Powell D, Perella C. Hypermethylation of the Ink4b locus in murine myeloid leukemia and increased susceptibility to leukemia in p15(Ink4b)-deficient mice. Oncogene. 2003;22(58):9265–9274. doi: 10.1038/sj.onc.1207092. [DOI] [PubMed] [Google Scholar]
- Yang LT, Kaartinen V. Tgfb1 expressed in the Tgfb3 locus partially rescues the cleft palate phenotype of Tgfb3 null mutants. Dev Biol. 2007;312(1):384–395. doi: 10.1016/j.ydbio.2007.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Pan X, Lei W, Wang J, Shi J, Li F, Song J. Regulation of Transforming Growth Factor-{beta}1-Induced Apoptosis and Epithelial-to-Mesenchymal Transition by Protein Kinase A and Signal Transducers and Activators of Transcription 3. Cancer Res. 2006a;66(17):8617–8624. doi: 10.1158/0008-5472.CAN-06-1308. [DOI] [PubMed] [Google Scholar]
- Yang Y, Pan X, Lei W, Wang J, Song J. Transforming growth factor-beta1 induces epithelial-to-mesenchymal transition and apoptosis via a cell cycle-dependent mechanism. Oncogene. 2006b;25(55):7235–7244. doi: 10.1038/sj.onc.1209712. [DOI] [PubMed] [Google Scholar]