Abstract
RNAi interference (RNAi) is a powerful gene silencing technology that has immense potential for treating a vast array of human ailments, for which suppressing disease-associated genes may provide clinical benefit. Here, we review the development of RNAi as a therapeutic modality for neurodegenerative diseases affecting the central nervous system (CNS). We overview promising preclinical data for the application of RNAi in the CNS and discuss key challenges (e.g. delivery and specificity) that remain as these approaches transition to the clinic.
INTRODUCTION
Over a decade ago, Andrew Fire and Craig Mello described the capacity of double-stranded RNA molecules to inhibit gene expression in Caenorhabditis elegans, opening a new frontier in biological research (1). This highly specific and, at the time, puzzling observation was termed RNA interference (RNAi). In 2001, a group led by Thomas Tuschl discovered that small RNA duplexes (∼21 nt) can mediate RNAi in cultured mammalian cells; as such, they proposed that RNAi may be applicable as a gene-specific therapy for human disease (2). The following year, several groups achieved potent RNAi-mediated gene suppression in vivo, most notably, in adult mouse liver and brain (3,4). Together, these findings have ignited the development of RNAi into a therapeutic modality for a range of acquired and inherited human diseases for which repressing the expression of key genes may provide clinical benefit. Remarkably, just this past year, a study by Davis et al. (5) provided the first molecular evidence of successful RNAi-mediated silencing in humans.
The prospect of RNAi as a therapeutic intervention provides an opportunity to treat several diseases for which effective options are currently unavailable or limited. In particular, RNAi-based therapies are being investigated for diseases affecting the central nervous system (CNS), including sporadic and genetic neurological disorders which have posed challenges to translational scientists and clinicians for years. Recent studies using animal models have generated promising proof-of-concept data supporting that RNAi therapy can improve neuropathological and behavioral phenotypes. Here, we briefly overview the RNAi pathway and describe the various means by which it can be manipulated to achieve gene-specific silencing. In addition, we discuss the application of RNAi technologies in the CNS and highlight its potential to treat neurodegenerative disorders. Finally, we reflect on some of the key challenges that remain as RNAi transitions to the clinic.
THE WORKINGS OF RNAi
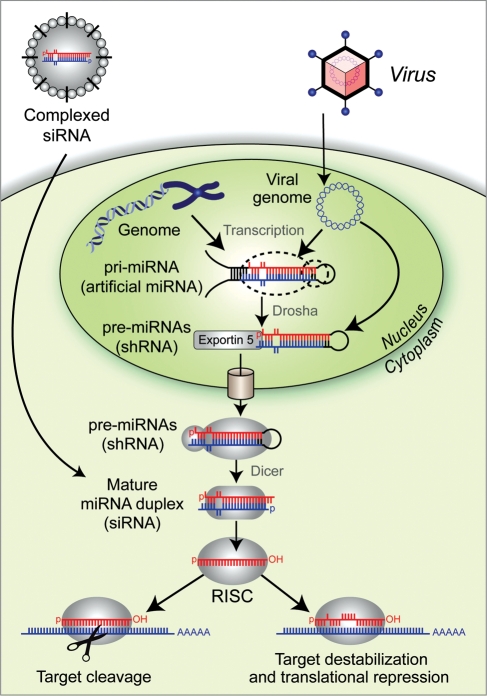
RNAi is an essential molecular mechanism governing cell fate determination, proliferation and many other biological processes (6). The cellular RNAi machinery works in conjunction with small RNAs to regulate gene expression and serve as an innate cellular response to viral invasion and transposable element activity (Fig. 1) (7). Endogenous RNAi for gene regulation occurs primarily via genomically encoded small RNAs known as microRNAs (miRNAs). Mature miRNAs (∼19–25nt) are processed from primary miRNA transcripts (pri-miRNAs), which contain stem-loop (i.e. hairpin) structures (8). Upon expression, pri-miRNAs are cleaved by the nuclear Drosha–DGCR8 microprocessor complex, producing intermediate hairpin RNAs (∼60–70nt) known as precursor-miRNAs (pre-miRNAs) (9,10). Pre-miRNAs are subsequently trafficked by Exportin-5 to the cytoplasm where they are further processed by a Dicer-containing complex, thus liberating the small miRNA duplex region (11,12). A single strand (the antisense ‘guide’ strand) of the resulting duplex is then incorporated into the RNA-induced silencing complex (RISC), which then binds to and silences target transcripts (13,14). The mode of target repression primarily depends upon the degree of complementarity; near perfect base-pairing is required for target transcript cleavage, while imperfect binding (typically in a target 3′-UTR) induces translational repression and mRNA decay. The latter scenario represents the canonical miRNA-based mechanism, and notably, short stretches of complementarity—as little as 6–7 bp—may be sufficient to trigger gene silencing (15).
Figure 1.
Diagram of the endogenous RNAi pathway and means to co-opt the machinery.
We refer the reader to a recent more in-depth review of miRNA biogenesis and function (7).
EXPLOITING THE RNAi PATHWAY
Over the past decade, our increased understanding of miRNA biogenesis and gene-silencing mechanisms has promoted the development of various strategies for co-opting the cellular RNAi machinery to direct specific silencing of virtually any gene. These RNAi-based technologies have become invaluable molecular tools to study gene function and are being investigated as therapeutic reagents for many human diseases.
The potential to artificially induce gene silencing depends on our ability to design inhibitory RNAs that properly engage the RNAi machinery and to introduce them into target cells or tissues. The central RNAi effectors, known as small-interfering RNAs (siRNAs), are essentially designed to mimic mature miRNA duplexes, but with the guide strand exhibiting perfect complementarity to the intended target transcript to trigger the potent cleavage-based silencing mechanism (Fig. 1). Notably, selecting potent and highly specific siRNA sequences is not exactly trivial, and numerous empirical evaluations of large-scale siRNA knock-down studies have allowed researchers to define several siRNA design guidelines (14,16). For example, a key consideration is that siRNA sequences should be selected or manipulated to promote accurate loading of the antisense guide strand into RISC, leaving the sense strand to be degraded (17,18). Furthermore, GC-content and positional preferences of certain nucleotides also influence siRNA efficacy. Given the multifaceted nature of designing optimal siRNAs, we direct the reader to additional literature on the subject (19,20).
SiRNAs may be chemically synthesized using modified bases for improved stability and specificity and reduced immunostimulatory properties (21). Upon entering the cell by endosomal uptake and escape or by electroporation, siRNAs engage the RNAi pathway at the Dicer-to-RISC stage (Fig. 1). Alternatively, siRNAs can be incorporated into expression-based systems by embedding the sequences into stem-loop structures designed to mimic pri-miRNAs (artificial miRNAs) or pre-miRNAs (short-hairpin RNAs or shRNAs) (22,23). These RNAi triggers may be integrated into a variety of expression systems; shRNAs are classically transcribed from Pol III promoters (e.g. U6 and H1) which provide strict control of the start and stop transcription sites, whereas artificial miRNAs are more amenable to Pol II-based systems, which enable tissue-specific and regulated expression strategies (24). Following expression, these hairpin-based RNAi substrates are processed via the RNAi pathway, thus releasing their siRNA sequence. Expression-based RNAi vectors afford unique opportunities for employment of viral-based delivery systems, stable long-term gene suppression and finer control of spatiotemporal silencing, among other related advantages associated with transgenic approaches.
To date, both non-viral and viral approaches have been employed to successfully achieve RNAi-mediated gene silencing in a variety of tissues, including the brain and spinal cord. For therapeutic development, the delivery modality depends largely upon the targeted tissue or cell population and desired interval of silencing. Focal delivery of synthetic siRNAs can be accomplished by direct intraparenchymal injection (25). This approach is restricted by the capacity of small RNAs to diffuse throughout the tissue and infiltrate target cells. Furthermore, the duration of gene silencing is limited by the relatively short half-life of siRNAs, and chronic knock-down requires repeated injections or indwelling devices for continued or pulsed infusion. Alternatively, long-term gene suppression is attainable following a single injection of viruses engineered to express stem-loop RNAs (26,27). As with non-viral strategies, the ability of viruses to diffuse within tissue and transduce target cells likewise limits their RNAi delivery capacity (28). Lentiviruses, capable of integrating into the genome, are advantageous when targeting proliferating cells, so that therapeutic gene expression can be maintained through cell division, if desired. In contrast, adeno-associated viruses (AAVs) may be preferred for targeting non-dividing cells (e.g. neurons). AAVs typically do not integrate, but rather, persist episomally, and are thus less prone to causing insertional mutagenesis within the host genome (29). Viral-based delivery systems are further discussed in the accompanied article on gene therapy in this issue.
With various RNAi triggers and delivery options available, researchers must decide which combination will yield a suitable inhibitory RNA dose to achieve potent and specific silencing in their experimental setting. Dose optimization remains a crucial consideration given the potential for RNAi treatments to induce cellular toxicity. High levels of exogenously supplied RNAi substrates may perturb cellular function by saturating the RNAi machinery, thus disrupting natural miRNA-mediated gene regulation (30–32). Also, artificial inhibitory RNAs have the potential to bind to and regulate unintended mRNA targets, an effect known as off-target gene silencing (33). Off-targeting primarily occurs when the seed region (nucleotides 2–8 of the siRNA) pairs with 3′-UTR sequences of unintended mRNAs and directs translational repression and destabilization of those transcripts, similar to the canonical miRNA-based silencing mechanism (34). Together, these potential side-effects may have severe consequences; for example, Grimm et al. (30) reported that high-level shRNA expression, typical from strong Pol III promoters, in mouse liver caused fatality. Subsequent work from our laboratory demonstrated that artificial miRNAs may have lower toxicity potentials. In comparison studies, we found that shRNAs are more potent but induce toxicity in vitro and in vivo, whereas artificial miRNAs are expressed at tolerably lower levels, yet maintain potent gene-silencing capacities (32,35). Together, these results highlight the need to consider and monitor dosing in RNAi experiments. Dosing of synthetic siRNAs is rather straightforward. However, the final inhibitory RNA levels resulting from expression-based systems depends upon many factors (e.g. vector platforms, delivery modalities, promoter selection, hairpin structure and availability of RNAi pathway components) which are likely to be unique to each experimental setting. However, an extensive RNAi toolbox provides scientists with the means to identify suitable combinations to pursue their research objectives.
SILENCING CNS DISEASE
For a host of neurodegenerative disorders [e.g. Alzheimer's (AD), Parkinson's (PD) and polyglutamine (polyQ) repeat diseases], aberrant accumulation of misfolded proteins appears to play a central role in disease onset and progression (36). Thus, a modest reduction in the levels of neurotoxic proteins is expected to provide significant therapeutic relief. Using RNAi-based approaches, investigators have successfully inhibited the expression of disease-causing proteins in cell and animal models of neurodegeneration, in many cases, improving neuropathological and behavioral phenotypes.
Alzheimer's disease (AD)
AD, the most common neurodegenerative disorder and a leading cause of dementia worldwide, is characterized by the presence of amyloid plaques and neurofibrillary tangles (NFTs) within the brain (37). Amyloid plaques are composed of the misfolded Aβ peptide, which is produced by proteolytic processing of the amyloid precursor protein (APP). RNAi strategies have been used to reduce Aβ peptides in vivo by targeting the expression of enzymes (e.g. BACE1) required for the proteolytic processing of APP, or by directly targeting the expression of APP (38,39). In each case, measurable reductions in Aβ peptide levels correlated with improved neuropathology and memory-related phenotypes. The other pathological hallmark of AD, NFTs, is composed of hyperphosphorylated Tau (40), another therapeutic target for AD (41). Tau does not appear to be essential for mammalian brain function and is implicated in a number of neurodegenerative diseases (42). Notably, tau knockout mice are resistant to human Aβ-induced brain dysfunction (43,44). Although no studies have directly targeted Tau expression in mouse models of AD, Piedrahita et al. (41) recently blocked the production of NFTs in triple transgenic AD mice by silencing cyclin-dependent kinase 5 (CDK5) which is required for Tau hyperphosphorylation. Thus, this study validates CDK5 and, indirectly, Tau as viable RNAi targets for AD.
Parkinson's disease (PD)
PD is the second most common neurodegenerative disease, and patients’ brains are often littered with Lewy bodies, which are protein aggregates comprised primarily of alpha-synuclein (α-syn) (45,46). Interestingly, single-point mutations in α-syn as well as genetic duplication or triplication of the α-syn gene (SCNA) are linked to hereditary parkinsonism (47). These data have led investigators to target α-syn expression with RNAi as a plausible therapeutic approach to PD (48–50). To date, studies have yielded conflicting results regarding the effectiveness and tolerability of this strategy, with at least one group reporting nigrostriatal degeneration after depleting α-syn levels in the rat brain (51). Whether this is a model- or sequence-specific effect remains unknown. Meanwhile, as genetic studies uncover additional putative therapeutic targets (i.e. leucine-rich repeat kinase-2), RNAi approaches can be used to validate them in genetic and sporadic models of PD.
The polyQ repeat disease family
The polyQ repeat disease family consists of nine dominantly inherited monogenic disorders: Huntington's disease (HD), dentatorubral–pallidoluysian atrophy, spinal bulbar muscular atrophy and six of the spinocerebellar ataxias (SCA1–3, 6, 7 and 17) (reviewed in 52–55). In each case, otherwise unrelated genes harbor within their coding regions an expanded CAG trinucleotide repeat stretch which, upon expression, yields mutant proteins containing an expanded polyQ stretch. The polyQ expansion confers a toxic gain of function to the respective mutant polyQ proteins, which over time wreak havoc in distinct neuronal populations, fatally disrupting numerous key cellular pathways. The exact mechanism underlying polyQ-mediated pathogenesis remains unclear; however, for each disorder, the expression of mutant polyQ-expanded proteins drives disease manifestation, thus making them obvious single-gene targets for silencing-based treatments. As such, this family of neurodegenerative diseases has been at the forefront of RNAi therapeutic development targeting the CNS, and encouraging proof-of-concept results have been observed in several instances. Given that such studies have been extensively reviewed elsewhere (56,57), we focus here on highlighting some recent advances.
In 2004, Xia et al. (58) first published their pioneering work demonstrating the therapeutic potential of RNAi in a transgenic mouse model for SCA1. Since then, numerous laboratories have used various RNAi strategies to inhibit mutant polyQ protein expression in rodent models (59–65). In each study, investigators achieved measurable phenotypic correction, lending strong support to the clinical application of RNAi as novel therapeutic modality for polyQ diseases. Building on early successes, investigators are now focused on optimizing delivery, potency and safety of RNAi triggers and evaluating the tolerability of target gene knock-down. The latter remains an important consideration if the target gene provides a vital cellular function. Knockout mice provide key evidence regarding the requirement of genes for normal CNS development and function. However, RNAi-based strategies rarely achieve 100% target gene suppression, as observed in knockouts, and current delivery limitations prevent knock-down throughout the entire brain. Indeed, recent independent reports have shown that non-allele-specific silencing of wild-type and mutant huntingtin in striata of adult rodent HD models is well-tolerated and provides therapeutic benefit to the animals (64,65), even though huntingtin knock-out mice are non-viable (66,67), and removal of neuronally expressed huntingtin in the early post-natal period causes late-onset neurodegeneration (68). This example highlights the need to evaluate the effects of partial reduction in target gene expression in a spatiotemporal manner relevant to the therapeutic strategy.
Researchers are also working to develop RNAi reagents that can preferentially silence mutant alleles (i.e. allele-specific silencing), with the goal of reducing mutant protein toxicity while maintaining tolerable levels of the wild-type protein. For some of the polyQ diseases (e.g. SCA3, SCA7 and HD), investigators have achieved allele-specific silencing using RNAi sequences directed at single nucleotide polymorphisms (SNPs) in linkage disequilibrium with CAG expanded alleles (69–72). While this approach holds promise, the prevalence of targetable disease-associated SNPs among patient populations presents a key limiting factor. For HD, researchers have identified several SNPs that may allow coverage of ∼85% of the Euro-Caucasian HD population (71,72); however, the relevant siRNAs remain to be evaluated for therapeutic efficacy and safety in animal models. Nevertheless, their initial findings warrant additional SNP typing in other HD populations and for the other polyQ diseases. Through an alternative approach, Hu et al. (73) recently reported preferential silencing of polyQ-expanded huntingtin using miRNA mimics which bind to the CAG expansion imperfectly and impede protein production, presumably by interfering with ribosomal processivity. This method relies on the increased probability of the inhibitors to bind the expanded CAG tract and requires a discernable difference between wild-type and mutant CAG repeat lengths. From a drug development perspective, this result raises the possibility of using a single allele-specific RNAi reagent to treat all nine polyQ disorders, and we anticipate future studies testing the efficacy and safety of this strategy in rodents. Finally, a third approach for allele-selective inhibition involves targeting polyQ-associated splice-isoforms. This tactic is clearly applicable to SCA6, where two fully functional Cav2.1 isoforms (one lacking and one containing the polyQ domain) are expressed in the affected cerebellar Purkinje neurons (74). Together, these allele-specific silencing approaches and current accompanying results are promising; however, the ability to control inhibitory RNA dosing in vivo will be crucial for achieving sufficient selectivity. Also, whether allele-specific silencing will be required for these and other diseases remains under scrutiny and debate, thus warranting continued development of both selective and non-selective silencing strategies to ensure that all avenues are tested in our efforts to develop effective and safe RNAi-based treatments.
CHALLENGES FOR THERAPEUTIC RNAi
With the enthusiasm surrounding the discovery of RNAi, there was anticipation that RNAi-based therapeutics would rapidly reach the clinic, reminiscent of early days in gene-based medicine research. However, the expectation of early success has since been mellowed by numerous unresolved challenges faced previously by other nucleic acid-based technologies. To realize the therapeutic potential of RNAi, strategies are being devised to circumvent natural barriers to delivery, avoid immune/non-immune toxicities and monitor delivery and therapeutic indices in real-time.
Delivery of inhibitory RNAs to the CNS is a formidable task due, in part, to the blood–brain barrier. As previously noted, the most suitable RNAi delivery modality will be dictated by our understanding of disease pathogenesis (e.g. onset, progression and affected tissue/cell type) and the desired duration of gene silencing. Non-viral-delivered nucleic acids (e.g. ‘naked’ or complexed synthetic siRNAs) may access the CNS using three major entry routes: through the vasculature, cerebrospinal fluid or by direct intraparenchymal delivery into the brain. For this, the limiting factors include stability of the siRNA complexes and their capacity to penetrate target cells without stimulating immune responses. Initial efforts to address some of these challenges focused on incorporating chemical modifications into the sugars, backbone or bases of siRNA duplexes (21). Certain modifications led to increased stability, which effectively lowered the dose needed to achieve measurable and reproducible gene silencing. However, internal modifications failed to improve CNS entry and uptake after systemic delivery. Rather, new efforts have moved towards testing liposomes, nanoparticles and cell-penetrating peptides, among others, to stabilize and navigate siRNAs into and throughout the brain (62,75,76). Investigators have also conjugated ‘brain homing’ peptides or antibodies to liposomes and nanoparticles that increased brain uptake after systemic delivery (77–80). Future studies are likely to focus on exploiting the presence of disease-related epitopes as a means to increase further the efficacy, specificity and potency of non-viral siRNA delivery to the brain. Finally, the potential for an adverse immune response to RNAi therapy is an important consideration, particularly in neurodegenerative diseases where the affected brain is already in a ‘heightened state of alert’ as it deals with chronic pro-inflammatory signaling cascades. In general, innate immune responses to non-viral-delivered siRNAs are mediated by members of the toll-like receptor family or by two different dsRNA-sensing proteins: retinoic acid-inducible gene-1 or dsRNA-binding protein kinase (81). These interactions can occur during internalization (endosomal or lysosomal compartments) or intracellular release of the siRNA molecule and are dose- and sequence-dependent. Importantly, the use of chemically modified or nanoparticle-encased siRNA duplexes avoids stimulation of these pathways.
The challenges faced by viral delivery methods currently being evaluated in the laboratory and in the neurology clinic are discussed in the article on gene therapy in this issue.
Future discoveries in RNAi biology will continue to guide development of RNAi-based therapies. One exciting recent finding is the observation that some miRNAs reside in plasma and other extracellular body fluids (82–84). These studies build on the work of Valadi and colleagues (83) who first reported on the exchange of mRNA and miRNAs between cells via exosomes (microvesicles released by many cell types including neurons, glia and cancer cells). Recent data show that ‘secreted’ miRNAs can silence gene expression after uptake by neighboring cells (85,86). Some groups have identified individual components of the proposed pathway, but the mechanism and enzymatic complexes involved in cell-to-cell miRNA trafficking remain largely unknown (85,87). Nevertheless, the therapeutic prospect of ‘secreted’ RNAi-based strategies is significant and warrants further investigation.
Another challenge facing neuroscientists using RNAi is our current inability to monitor, in real-time, the delivery, activity and specificity of an RNAi molecule in the brain. Post-treatment sampling is impractical in the brain, unlike for skin diseases, infections and many tumors. Model systems are thus required to establish correlations between gene-silencing potency and dose-specific toxicity. Biomarkers particular to the neurodegenerative disease under study offer the best way to monitor a coincident response to therapeutic RNAi, although for many disorders the validity of a given biomarker to represent a particular disease stage is far from known. Finally, advances in imaging techniques to track RNA in vivo with quantum dots are showing promise (88).
Conflict of Interest statement. None declared.
FUNDING
This work was supported by the National Institutes of Health (NS050210, NS067111, and HD44093), the Hereditary Disease Foundation, and the Roy J. Carver Trust.
REFERENCES
- 1.Fire A., Xu S.Q., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Elbashir S.M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 3.McCaffrey A.P., Meuse L., Pham T.T., Conklin D.S., Hannon G.J., Kay M.A. RNA interference in adult mice. Nature. 2002;418:38–39. doi: 10.1038/418038a. [DOI] [PubMed] [Google Scholar]
- 4.Xia H., Mao Q., Paulson H.L., Davidson B.L. siRNA-mediated gene silencing in vitro and in vivo. Nat. Biotechnol. 2002;20:1006–1010. doi: 10.1038/nbt739. [DOI] [PubMed] [Google Scholar]
- 5.Davis M.E., Zuckerman J.E., Choi C.H., Seligson D., Tolcher A., Alabi C.A., Yen Y., Heidel J.D., Ribas A. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464:1067–1070. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carrington J.C., Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336–338. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- 7.Krol J., Loedige I., Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 8.Lee Y., Jeon K., Lee J.T., Kim S., Kim V.N. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J., Lee J., Provost P., Radmark O., Kim S., et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 10.Gregory R.I., Yan K.P., Amuthan G., Chendrimada T., Doratotaj B., Cooch N., Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 11.Yi R., Qin Y., Macara I.G., Cullen B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Provost P., Dishart D., Doucet J., Frendewey D., Samuelsson B., Radmark O. Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J. 2002;21:5864–5874. doi: 10.1093/emboj/cdf578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwarz D.S., Hutvagner G., Du T., Xu Z., Aronin N., Zamore P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 14.Khvorova A., Reynolds A., Jayasena S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 15.Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 16.Matveeva O., Nechipurenko Y., Rossi L., Moore B., Saetrom P., Ogurtsov A.Y., Atkins J.F., Shabalina S.A. Comparison of approaches for rational siRNA design leading to a new efficient and transparent method. Nucleic Acids Res. 2007;35:e63. doi: 10.1093/nar/gkm088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matranga C., Tomari Y., Shin C., Bartel D.P., Zamore P.D. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 18.Leuschner P.J., Ameres S.L., Kueng S., Martinez J. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep. 2006;7:314–320. doi: 10.1038/sj.embor.7400637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Birmingham A., Anderson E., Sullivan K., Reynolds A., Boese Q., Leake D., Karpilow J., Khvorova A. A protocol for designing siRNAs with high functionality and specificity. Nat. Protoc. 2007;2:2068–2078. doi: 10.1038/nprot.2007.278. [DOI] [PubMed] [Google Scholar]
- 20.Jackson A.L., Linsley P.S. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nat. Rev. Drug Discov. 2010;9:57–67. doi: 10.1038/nrd3010. [DOI] [PubMed] [Google Scholar]
- 21.Behlke M.A. Chemical modification of siRNAs for in vivo use. Oligonucleotides. 2008;18:305–319. doi: 10.1089/oli.2008.0164. [DOI] [PubMed] [Google Scholar]
- 22.Paul C.P., Good P.D., Winer I., Engelke D.R. Effective expression of small interfering RNA in human cells. Nat. Biotechnol. 2002;20:505–508. doi: 10.1038/nbt0502-505. [DOI] [PubMed] [Google Scholar]
- 23.Zeng Y., Wagner E.J., Cullen B.R. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol. Cell. 2002;9:1327–1333. doi: 10.1016/s1097-2765(02)00541-5. [DOI] [PubMed] [Google Scholar]
- 24.Boudreau R.L., Mas Monteys A., Davidson B.L. Minimizing variables among hairpin-based RNAi vectors reveals the potency of shRNAs. RNA. 2008;14:1834–1844. doi: 10.1261/rna.1062908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolff J.A., Budker V. The mechanism of naked DNA uptake and expression. Adv. Genet. 2005;54:3–20. doi: 10.1016/S0065-2660(05)54001-X. [DOI] [PubMed] [Google Scholar]
- 26.Rubinson D.A., Dillon C.P., Kwiatkowski A.V., Sievers C., Yang L., Kopinja J., Rooney D.L., Ihrig M.M., McManus M.T., Gertler F.B., et al. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat Genet. 2003;33:401–406. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- 27.Davidson B.L., Harper S.Q. Viral delivery of recombinant short hairpin RNAs. Methods Enzymol. 2005;392:145–173. doi: 10.1016/S0076-6879(04)92009-5. [DOI] [PubMed] [Google Scholar]
- 28.Davidson B.L., Breakefield X.O. Viral vectors for gene delivery to the nervous system. Nat. Rev. Neurosci. 2003;4:353–364. doi: 10.1038/nrn1104. [DOI] [PubMed] [Google Scholar]
- 29.Heilbronn R., Weger S. Viral vectors for gene transfer: current status of gene therapeutics. Handb. Exp. Pharmacol. 2010;197:143–170. doi: 10.1007/978-3-642-00477-3_5. [DOI] [PubMed] [Google Scholar]
- 30.Grimm D., Streetz K.L., Jopling C.L., Storm T.A., Pandey K., Davis C.R., Marion P., Salazar F., Kay M.A. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 31.Castanotto D., Sakurai K., Lingeman R., Li H., Shively L., Aagaard L., Soifer H., Gatignol A., Riggs A., Rossi J.J. Combinatorial delivery of small interfering RNAs reduces RNAi efficacy by selective incorporation into RISC. Nucleic Acids Res. 2007;35:5154–5164. doi: 10.1093/nar/gkm543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boudreau R.L., Martins I., Davidson B.L. Artificial microRNAs as siRNA shuttles: improved safety as compared to shRNAs in vitro and in vivo. Mol. Ther. 2009;17:169–175. doi: 10.1038/mt.2008.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jackson A.L., Bartz S.R., Schelter J., Kobayashi S.V., Burchard J., Mao M., Li B., Cavet G., Linsley P.S. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 34.Birmingham A., Anderson E.M., Reynolds A., Ilsley-Tyree D., Leake D., Fedorov Y., Baskerville S., Maksimova E., Robinson K., Karpilow J., et al. 3' UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat. Methods. 2006;3:199–204. doi: 10.1038/nmeth854. [DOI] [PubMed] [Google Scholar]
- 35.McBride J.L., Boudreau R.L., Harper S.Q., Staber P.D., Monteys A.M., Martins I., Gilmore B.L., Burstein H., Peluso R.W., Polisky B., et al. Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. Proc. Natl Acad. Sci. USA. 2008;105:5868–5873. doi: 10.1073/pnas.0801775105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aguzzi A., O'Connor T. Protein aggregation diseases: pathogenicity and therapeutic perspectives. Nat. Rev. Drug Discov. 2010;9:237–248. doi: 10.1038/nrd3050. [DOI] [PubMed] [Google Scholar]
- 37.Bertram L., Lill C.M., Tanzi R.E. The genetics of Alzheimer disease: back to the future. Neuron. 2010;68:270–281. doi: 10.1016/j.neuron.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 38.Singer O., Marr R.A., Rockenstein E., Crews L., Coufal N.G., Gage F.H., Verma I.M., Masliah E. Targeting BACE1 with siRNAs ameliorates Alzheimer disease neuropathology in a transgenic model. Nat. Neurosci. 2005;8:1343–1349. doi: 10.1038/nn1531. [DOI] [PubMed] [Google Scholar]
- 39.Rodriguez-Lebron E., Gouvion C.M., Moore S.A., Davidson B.L., Paulson H.L. Allele-specific RNAi mitigates phenotypic progression in a transgenic model of Alzheimer's disease. Mol. Th. J. Am. Soc. Gene Ther. 2009;17:1563–1573. doi: 10.1038/mt.2009.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.LaFerla F.M., Oddo S. Alzheimer's disease: Abeta, tau and synaptic dysfunction. Trends Mol. Med. 2005;11:170–176. doi: 10.1016/j.molmed.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 41.Piedrahita D., Hernandez I., Lopez-Tobon A., Fedorov D., Obara B., Manjunath B.S., Boudreau R.L., Davidson B., Laferla F., Gallego-Gomez J.C., et al. Silencing of CDK5 reduces neurofibrillary tangles in transgenic alzheimer's mice. J. Neurosci. 2010;30:13966–13976. doi: 10.1523/JNEUROSCI.3637-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brunden K.R., Trojanowski J.Q., Lee V.M. Advances in tau-focused drug discovery for Alzheimer's disease and related tauopathies. Nat. Rev. Drug Discov. 2009;8:783–793. doi: 10.1038/nrd2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roberson E.D., Scearce-Levie K., Palop J.J., Yan F., Cheng I.H., Wu T., Gerstein H., Yu G.Q., Mucke L. Reducing endogenous tau ameliorates amyloid beta-induced deficits in an Alzheimer's disease mouse model. Science. 2007;316:750–754. doi: 10.1126/science.1141736. [DOI] [PubMed] [Google Scholar]
- 44.Roberson E.D., Halabisky B., Yoo J.W., Yao J., Chin J., Yan F., Wu T., Hamto P., Devidze N., Yu G.Q., et al. Amyloid-beta/Fyn-induced synaptic, network, and cognitive impairments depend on tau levels in multiple mouse models of Alzheimer's disease. J. Neurosci. 2011;31:700–711. doi: 10.1523/JNEUROSCI.4152-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Halliday G.M., McCann H. The progression of pathology in Parkinson's disease. Ann. N Y Acad. Sci. 2010;1184:188–195. doi: 10.1111/j.1749-6632.2009.05118.x. [DOI] [PubMed] [Google Scholar]
- 46.Spillantini M.G., Schmidt M.L., Lee V.M., Trojanowski J.Q., Jakes R., Goedert M. Alpha-synuclein in Lewy bodies. Nature. 1997;388:839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 47.Hardy J. Genetic analysis of pathways to Parkinson disease. Neuron. 2010;68:201–206. doi: 10.1016/j.neuron.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sapru M.K., Yates J.W., Hogan S., Jiang L., Halter J., Bohn M.C. Silencing of human alpha-synuclein in vitro and in rat brain using lentiviral-mediated RNAi. Exp. Neurol. 2006;198:382–390. doi: 10.1016/j.expneurol.2005.12.024. [DOI] [PubMed] [Google Scholar]
- 49.Fountaine T.M., Wade-Martins R. RNA interference-mediated knockdown of alpha-synuclein protects human dopaminergic neuroblastoma cells from MPP(+) toxicity and reduces dopamine transport. J. Neurosci. Res. 2007;85:351–363. doi: 10.1002/jnr.21125. [DOI] [PubMed] [Google Scholar]
- 50.Liu D.M., Jin L., Wang H., Zhao H.Y., Zhao C.L., Yang H. RNA interference mediated silencing of alpha-synuclein in MN9D cells and its effects on cell viability. Neurosci. Bull. 2008;24:96–104. doi: 10.1007/s12264-008-0096-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gorbatyuk O.S., Li S., Nash K., Gorbatyuk M., Lewin A.S., Sullivan L.F., Mandel R.J., Chen W., Meyers C., Manfredsson F.P., et al. In vivo RNAi-mediated alpha-synuclein silencing induces nigrostriatal degeneration. Mol. Ther. 2010;18:1450–1457. doi: 10.1038/mt.2010.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ross C.A., Tabrizi S.J. Huntington's disease: from molecular pathogenesis to clinical treatment. Lancet Neurol. 2011;10:83–98. doi: 10.1016/S1474-4422(10)70245-3. [DOI] [PubMed] [Google Scholar]
- 53.Durr A. Autosomal dominant cerebellar ataxias: polyglutamine expansions and beyond. Lancet Neurol. 2010;9:885–894. doi: 10.1016/S1474-4422(10)70183-6. [DOI] [PubMed] [Google Scholar]
- 54.Finsterer J. Perspectives of Kennedy's disease. J. Neurol. Sci. 2010;298:1–10. doi: 10.1016/j.jns.2010.08.025. [DOI] [PubMed] [Google Scholar]
- 55.Yamada M., Shimohata M., Sato T., Tsuji S., Takahashi H. Polyglutamine disease: recent advances in the neuropathology of dentatorubral-pallidoluysian atrophy. Neuropathology. 2006;26:346–351. doi: 10.1111/j.1440-1789.2006.00670.x. [DOI] [PubMed] [Google Scholar]
- 56.Scholefield J., Wood M.J. Therapeutic gene silencing strategies for polyglutamine disorders. Trends Genet. 2010;26:29–38. doi: 10.1016/j.tig.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 57.Boudreau R.L., Davidson B.L. RNAi therapeutics for CNS disorders. Brain Res. 2010;1338:112–121. doi: 10.1016/j.brainres.2010.03.038. [DOI] [PubMed] [Google Scholar]
- 58.Xia H., Mao Q., Eliason S.L., Harper S.Q., Martins I.H., Orr H.T., Paulson H.L., Yang L., Kotin R.M., Davidson B.L. RNAi suppresses polyglutamine-induced neurodegeneration in a mouse model of SCA1. Nat. Med. 2004;10:816–820. doi: 10.1038/nm1076. [DOI] [PubMed] [Google Scholar]
- 59.Harper S.Q., Staber P.D., He X., Eliason S.L., Martins I., Mao Q., Yang L., Kotin R.M., Paulson H.L., Davidson B.L. RNA interference improves motor and neuropathological abnormalities in a Huntington's disease mouse model. Proc. Natl Acad. Sci. USA. 2005;102:5820–5825. doi: 10.1073/pnas.0501507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rodriguez-Lebron E., Denovan-Wright E.M., Nash K., Lewin A.S., Mandel R.J. Intrastriatal rAAV-mediated delivery of anti-huntingtin shRNAs induces partial reversal of disease progression in R6/1 Huntington's disease transgenic mice. Mol. Ther. 2005;12:618–633. doi: 10.1016/j.ymthe.2005.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang Y.L., Liu W., Wada E., Murata M., Wada K., Kanazawa I. Clinico-pathological rescue of a model mouse of Huntington's disease by siRNA. Neurosci. Res. 2005;53:241–249. doi: 10.1016/j.neures.2005.06.021. [DOI] [PubMed] [Google Scholar]
- 62.DiFiglia M., Sena-Esteves M., Chase K., Sapp E., Pfister E., Sass M., Yoder J., Reeves P., Pandey R.K., Rajeev K.G., et al. Therapeutic silencing of mutant huntingtin with siRNA attenuates striatal and cortical neuropathology and behavioral deficits. Proc. Natl Acad. Sci. USA. 2007;104:17204–17209. doi: 10.1073/pnas.0708285104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Franich N.R., Fitzsimons H.L., Fong D.M., Klugmann M., During M.J., Young D. AAV vector-mediated RNAi of mutant huntingtin expression is neuroprotective in a novel genetic rat model of Huntington's disease. Mol. Ther. 2008;16:947–956. doi: 10.1038/mt.2008.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Boudreau R.L., McBride J.L., Martins I., Shen S., Xing Y., Carter B.J., Davidson B.L. Nonallele-specific silencing of mutant and wild-type huntingtin demonstrates therapeutic efficacy in Huntington's disease mice. Mol. Ther. 2009;17:1053–1063. doi: 10.1038/mt.2009.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Drouet V., Perrin V., Hassig R., Dufour N., Auregan G., Alves S., Bonvento G., Brouillet E., Luthi-Carter R., Hantraye P., et al. Sustained effects of nonallele-specific Huntingtin silencing. Ann. Neurol. 2009;65:276–285. doi: 10.1002/ana.21569. [DOI] [PubMed] [Google Scholar]
- 66.Duyao M.P., Auerbach A.B., Ryan A., Persichetti F., Barnes G.T., McNeil S.M., Ge P., Vonsattel J.P., Gusella J.F., Joyner A.L., et al. Inactivation of the mouse Huntington's disease gene homolog Hdh. Science. 1995;269:407–410. doi: 10.1126/science.7618107. [DOI] [PubMed] [Google Scholar]
- 67.Nasir J., Floresco S.B., O'Kusky J.R., Diewert V.M., Richman J.M., Zeisler J., Borowski A., Marth J.D., Phillips A.G., Hayden M.R. Targeted disruption of the Huntington's disease gene results in embryonic lethality and behavioral and morphological changes in heterozygotes. Cell. 1995;81:811–823. doi: 10.1016/0092-8674(95)90542-1. [DOI] [PubMed] [Google Scholar]
- 68.Dragatsis I., Levine M.S., Zeitlin S. Inactivation of Hdh in the brain and testis results in progressive neurodegeneration and sterility in mice. Nat. Genet. 2000;26:300–306. doi: 10.1038/81593. [DOI] [PubMed] [Google Scholar]
- 69.Alves S., Nascimento-Ferreira I., Auregan G., Hassig R., Dufour N., Brouillet E., Pedroso de Lima M.C., Hantraye P., Pereira de Almeida L., Deglon N. Allele-specific RNA silencing of mutant ataxin-3 mediates neuroprotection in a rat model of Machado-Joseph disease. PLoS ONE. 2008;3:e3341. doi: 10.1371/journal.pone.0003341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Scholefield J., Greenberg L.J., Weinberg M.S., Arbuthnot P.B., Abdelgany A., Wood M.J. Design of RNAi hairpins for mutation-specific silencing of ataxin-7 and correction of a SCA7 phenotype. PLoS ONE. 2009;4:e7232. doi: 10.1371/journal.pone.0007232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pfister E.L., Kennington L., Straubhaar J., Wagh S., Liu W., DiFiglia M., Landwehrmeyer B., Vonsattel J.P., Zamore P.D., Aronin N. Five siRNAs targeting three SNPs may provide therapy for three-quarters of Huntington's disease patients. Curr. Biol. 2009;19:774–778. doi: 10.1016/j.cub.2009.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lombardi M.S., Jaspers L., Spronkmans C., Gellera C., Taroni F., Di Maria E., Donato S.D., Kaemmerer W.F. A majority of Huntington's disease patients may be treatable by individualized allele-specific RNA interference. Exp. Neurol. 2009;217:312–319. doi: 10.1016/j.expneurol.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 73.Hu J., Liu J., Corey D.R. Allele-selective inhibition of huntingtin expression by switching to an miRNA-like RNAi mechanism. Chem. Biol. 2010;17:1183–1188. doi: 10.1016/j.chembiol.2010.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Watase K., Barrett C.F., Miyazaki T., Ishiguro T., Ishikawa K., Hu Y., Unno T., Sun Y., Kasai S., Watanabe M., et al. Spinocerebellar ataxia type 6 knockin mice develop a progressive neuronal dysfunction with age-dependent accumulation of mutant CaV2.1 channels. Proc. Natl Acad. Sci. USA. 2008;105:11987–11992. doi: 10.1073/pnas.0804350105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fonseca S.B., Pereira M.P., Kelley S.O. Recent advances in the use of cell-penetrating peptides for medical and biological applications. Adv. Drug Deliv. Rev. 2009;61:953–964. doi: 10.1016/j.addr.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 76.Bonoiu A.C., Mahajan S.D., Ding H., Roy I., Yong K.T., Kumar R., Hu R., Bergey E.J., Schwartz S.A., Prasad P.N. Nanotechnology approach for drug addiction therapy: gene silencing using delivery of gold nanorod-siRNA nanoplex in dopaminergic neurons. Proc. Natl Acad. Sci. USA. 2009;106:5546–5550. doi: 10.1073/pnas.0901715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cardoso A.L., Simoes S., de Almeida L.P., Plesnila N., Pedroso de Lima M.C., Wagner E., Culmsee C. Tf-lipoplexes for neuronal siRNA delivery: a promising system to mediate gene silencing in the CNS. J. Control Release. 2008;132:113–123. doi: 10.1016/j.jconrel.2008.08.014. [DOI] [PubMed] [Google Scholar]
- 78.Pulford B., Reim N., Bell A., Veatch J., Forster G., Bender H., Meyerett C., Hafeman S., Michel B., Johnson T., et al. Liposome-siRNA-peptide complexes cross the blood-brain barrier and significantly decrease PrP on neuronal cells and PrP in infected cell cultures. PLoS ONE. 2010;5:e11085. doi: 10.1371/journal.pone.0011085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Uno Y., Piao W., Miyata K., Nishina K., Mizusawa H., Yokota T. HDL facilitates in vivo delivery of -tocopherol-conjugated siRNA to the brain. Hum. Gene Ther. 2010 doi: 10.1089/hum.2010.083. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kumar P., Wu H., McBride J.L., Jung K.E., Kim M.H., Davidson B.L., Lee S.K., Shankar P., Manjunath N. Transvascular delivery of small interfering RNA to the central nervous system. Nature. 2007;448:39–43. doi: 10.1038/nature05901. [DOI] [PubMed] [Google Scholar]
- 81.Robbins M., Judge A., MacLachlan I. siRNA and innate immunity. Oligonucleotides. 2009;19:89–102. doi: 10.1089/oli.2009.0180. [DOI] [PubMed] [Google Scholar]
- 82.Iguchi H., Kosaka N., Ochiya T. Secretory microRNAs as a versatile communication tool. Commun. Integr. Biol. 2010;3:478–481. doi: 10.4161/cib.3.5.12693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Valadi H., Ekstrom K., Bossios A., Sjostrand M., Lee J.J., Lotvall J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 84.Chen X., Ba Y., Ma L., Cai X., Yin Y., Wang K., Guo J., Zhang Y., Chen J., Guo X., et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 85.Kosaka N., Iguchi H., Yoshioka Y., Takeshita F., Matsuki Y., Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J. Biol. Chem. 2010;285:17442–17452. doi: 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pegtel D.M., Cosmopoulos K., Thorley-Lawson D.A., van Eijndhoven M.A., Hopmans E.S., Lindenberg J.L., de Gruijl T.D., Wurdinger T., Middeldorp J.M. Functional delivery of viral miRNAs via exosomes. Proc. Natl Acad. Sci. USA. 2010;107:6328–6333. doi: 10.1073/pnas.0914843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang K., Zhang S., Weber J., Baxter D., Galas D.J. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 2010;38:7248–7259. doi: 10.1093/nar/gkq601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hong H., Zhang Y., Cai W. In vivo imaging of RNA interference. J. Nucl. Med. 2010;51:169–172. doi: 10.2967/jnumed.109.066878. [DOI] [PMC free article] [PubMed] [Google Scholar]