Abstract
The development of substance dependence requires the initiation of substance use, the conversion from experimental use to established use, and finally the development of dependence. Numerous large twin studies have indicated a significant genetic contribution to this process. Genetic studies to date have been most successful at identifying genetic factors that influence the transition from regular use to dependence. The availability of large cohort samples for nicotine and alcohol dependence has resulted in significant progress being made in understanding at least some of the genetic contributions to these addictions. Fewer studies have replicated specific genetic contributions to illicit drug use, though it is clear that there is a strong genetic component involved here as well. Substance dependence can be thought of as a pharmacogenetic illness, and most likely hundreds and more probably thousands of genetic variants will be required to fully explain the genetic input to this disease.
Keywords: nicotine dependence, alcohol dependence, substance dependence, genetics, nicotinic receptor, metabolism, pharmacogenetics, addiction
Introduction
Large segments of our population use tobacco, alcohol, and other drugs. Cigarette smoking is common in both industrialized and developing countries. In the United States, over 43 million people use tobacco, and worldwide, over one billion people are tobacco users (Centers for Disease Control, 2010; World Health Organization, 2010). In the U.S., over 400,000 people die every year from tobacco related illnesses, and smoking remains the greatest contributor to preventable death (Mokdad et al., 2004). With increasing tobacco use in developing countries, it is predicted that the worldwide death toll will rise to eight million people per year by 2030. Alcohol is the most commonly used and abused substance in the population, and 12.5% of adults in the U.S. develop alcohol dependence during their lifetime (Hasin et al., 2007). In 2004, the World Health Organization estimated that alcohol use disorders affected 76.3 million people globally (World Health Organization, 2004). In the U.S., almost 80,000 people die per year from the consequences of alcohol consumption, which includes alcohol related illnesses and accidents (Mokdad et al., 2004). Our society pays a high price for substance use, primarily through increased health care costs and judicial system expenditures. It is estimated that over 11% of federal and state government budgets ($374 billion in 2005) are spent dealing with the consequences of tobacco, alcohol, and other substance use, abuse, and dependence (The National Center on Addiction and Substance Abuse at Columbia University, 2009).
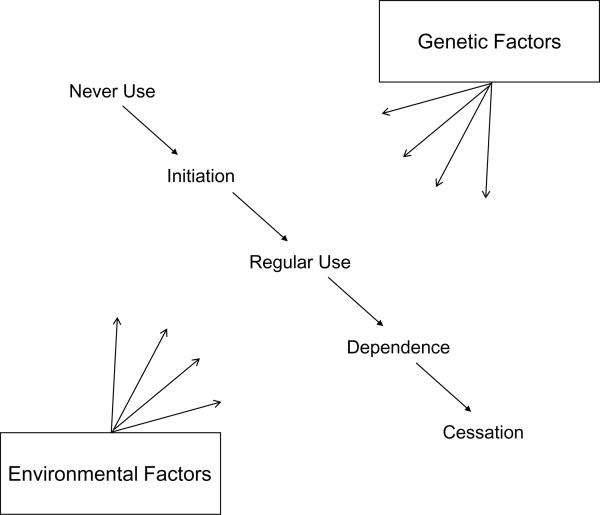
The development of addiction requires the use of a substance and a subsequent chain of behavioral events that leads to addiction. The key steps in the development of addiction include the initiation of substance use, the conversion from experimental use to established use, and finally the development of addiction (see Figure 1). Each step is influenced by environmental and genetic factors, some of which are common to all steps, and others that are specific. For example, environmental factors, such as the availability of nicotine, alcohol, and drugs, play a role in each stage in the development of addiction, but accessibility of a substance is relatively more important in the initiation of substance use. Similarly, high cost of a substance through taxation can reduce initiation, use, and addiction; however taxation has a stronger influence on teenagers who have less money, thus limiting initial use. Family, twin, and adoption studies also convincingly demonstrate a substantial genetic contribution to the development of addiction to nicotine, alcohol, and illicit drugs. Heritability estimates for nicotine, alcohol, and drug addiction are in the range of 50% to 60% (Heath et al., 1997; Tsuang et al., 1998; Kendler et al., 2003; Li, 2006). In general, it appears that environmental factors have a stronger effect on initiation, whereas genetic factors play a larger role in the transition from regular use to the development of addiction (Vink et al., 2005). Given the robust behavioral evidence for the role of genetic influence in addiction, genetic studies are warranted.
Figure 1.
Steps in the development of dependence.
Initial inroads into understanding the genetic influences of addiction in humans relied on both genetic linkage mapping and candidate gene association studies, resulting in the identification of hundreds of potential genes contributing to the addiction process. Yet, few of these associations have been replicated in independent studies, potentially reflecting a number of false positives and/or genetic heterogeneity in which multiple genes contribute modest effects. The last decade, however, has seen a revolution in genetic technologies so that hundreds of thousands of genetic variants (or single nucleotide polymorphisms; SNPs) can be queried in thousands of individuals in a cost effective manner. This technology facilitates genome wide association studies (GWAS) that test for an association of genetic variants with an illness in order to discover genetic contributions to complex diseases. Complex diseases are caused by many genetic and environmental factors working together, and GWAS has permitted the discovery of hundreds of genetic variants that alter the risk of developing multiple complex diseases, including type 2 diabetes, Crohn's disease, and Parkinson's disease (Hindorff et al., 2010). More recently, the genetic tools of GWAS have been applied to the study of addiction to identify genetic variations that contribute to this illness. The success of this approach has been in part due to the creation of genetic research consortia for the study of nicotine and illicit drugs (NIDA Genetics Consortium http://www.nida.nih.gov/about/organization/genetics/consortium/index.html) and alcohol (e.g. NIAAA's Collaborative Study on the Genetics of Alcoholism; COGA; http://www.niaaa.nih.gov/ResearchInformation/ExtramuralResearch/SharedResources/projcoga.htm) permitting the collection of the massive numbers of comprehensively assessed subjects and DNA samples required for large scale studies. These resources are also shared with the scientific community though the database of Genotypes and Phenotypes (dbGaP http://www.ncbi.nlm.nih.gov/gap) so that scientists around the world can test new hypotheses about the genetic underpinnings of addiction.
This review will give a synopsis of the current understanding of genetic contributions to the vulnerability of substance dependence. There have been extensive discussions about the terminology used to define substance use disorder – “dependence” versus “addiction.” Substance dependence is the official diagnostic nomenclature used in the 4th edition of the Diagnostic and Statistical Manual of Mental Disorders (DSM-IV; American Psychiatric Association, 2000) to represent the syndrome of substance misuse that leads to adverse consequences and includes a cluster of symptoms such as tolerance, withdrawal, and inability to stop using (See DSM-IV substance dependence for the complete diagnostic criteria). The creators of DSM-IV criteria selected the term “dependence” because of the concern of stigmatization associated with “addiction.” At this time, revisions to the 5th edition of the Diagnostic and Statistical Manual of Mental Disorders (DSM-5) are underway for release in 2013. In this revision, issues have again been raised about the term used to define this clinical syndrome. In order to differentiate from the normal physiologic development of tolerance and withdrawal that develops with substance use from the compulsive drug use with loss of control, DSM-5 proposes the use of the word “addiction” to define substance use disorder. The words “dependence“ and “addiction” are used interchangeably in this review to represent the same underlying concept of substance use disorder.
Genome Wide Association Studies of Nicotine Dependence
The strongest genetic contribution to nicotine dependence comes from variation in the nicotinic receptor subunits, and the most compelling genetic evidence is provided by several large-scale GWAS meta-analyses of smoking behavior (Liu et al., 2010; Thorgeirsson et al., 2010; Tobacco and Genetics Consortium, 2010). Because smoking is a major contributor to many illnesses, cigarettes smoked per day (CPD), a proxy phenotype for nicotine dependence, has been measured in many genetic studies, and this has allowed meta-analyses of over 80,000 individuals of European ancestry. These genetic meta-analyses of cigarettes smoked per day confirm that two chromosomal regions containing nicotinic receptor subunit gene clusters influence smoking behavior.
The most robust genetic finding that alters the risk of developing heavy smoking is in the chromosome 15q25 region, which contains the α5, α3, and β4 nicotinic receptor subunit gene cluster (CHRNA5, CHRNA3, CHRNB4). The SNP rs16969968 is unequivocally associated with smoking behavior (p=5.57 × 10−72) (Tobacco and Genetics Consortium, 2010). Further examination of the chromosome 15 region demonstrates that there are at least two distinct genetic risk variants that contribute to heavy smoking behavior (Saccone et al., 2010a; Tobacco and Genetics Consortium, 2010).
Variation in an independent group of nicotinic receptors is also associated with the development of heavy smoking and nicotine dependence. The nicotinic receptor gene cluster on chromosome 8 that includes the α6 and β3 nicotinic receptor subunit gene cluster (CHRNA6, CHRNB3) is correlated with smoking behavior. This region generated genome-wide significant association with nicotine dependence, though the strength of this association is much less (rs6474412 p=1.4 × 10−8) (Thorgeirsson et al., 2010).
In addition to genetic variants in the nicotinic receptors contributing to the development of nicotine dependence, genetic variation in nicotine metabolism plays an important role in cigarette consumption (Schoedel et al., 2004; Minematsu et al., 2006) and nicotine dependence (Audrain-McGovern et al., 2007). Conversion of nicotine to cotinine accounts for 70% of initial nicotine metabolism and is performed by the CYP2a6 enzyme (Yamazaki et al., 1999; Su et al., 2000; Malaiyandi et al., 2006). Important functional polymorphisms of CYP2a6 include large deletions and gene recombinations that involve neighboring genes (Oscarson et al., 1999, 2002). The importance of nicotine metabolism and variation in the CYP2a6 region on chromosome 19 was recently reinforced by the GWAS meta-analysis studies in which variants in this region were associated with number of cigarettes smoked per day (Thorgeirsson et al., 2010; Tobacco and Genetics Consortium, 2010). The most significant SNP reported in this region, the intergenic variant rs41405144, lies within two large deletions (defined as CYP2a6*4 and CYP2a6*12). This variant, rs41405144, is correlated with rs1801272, a non-synonymous SNP that defines the CYP2a6*2 loss of function allele. These findings confirm that variation in nicotine metabolism contributes to the number of cigarettes smoked daily and the development of nicotine dependence.
Genetics of Alcohol Dependence
Alcohol dependence was one of the first behavioral disorders shown to have validated genetic contributions. Polymorphisms in the alcohol metabolizing enzymes are the most strongly associated genetic variants that influence alcohol consumption and alcohol dependence. In 1972, individuals of Asian descent were reported to have facial flushing and decreased tolerance when exposed to alcohol (Wolff, 1972). The flushing reaction after ingesting alcohol is secondary to a deficiency of aldehyde dehydrogenase (specifically ALDH2), an enzyme involved in the metabolism of ethanol (Goedde et al., 1980). The ALDH2 deficiency was found to be present in a large part of the general Japanese population, but uncommon in alcohol dependent individuals, implying a protective role for the deficiency of ALDH2 in alcohol dependence (Harada et al., 1982).
Since these initial discoveries, much has been learned about alcohol metabolism. Ethanol metabolism occurs predominantly in the liver in two steps: the oxidation of ethanol to acetaldehyde catalyzed by alcohol dehydrogenases (ADHs), and the oxidation of acetaldehyde to acetate by acetaldhyde dehydrogenases (ALDHs). Several known genetic variants cause amino acid changes in these proteins and alter enzymatic activity. For instance, ADH1B*2, or rs1229984, diminishes ADH1b enzymatic activity several fold, and ALDH2*2, or rs671, results in a nearly inactive enzyme (Edenberg, 2007). These genetic variants reduce the probability of heavy alcohol consumption and the development of alcohol dependence (Edenberg, 2007; Macgregor et al., 2009; Sherva et al., 2009). The mechanism by which variants of these enzymes influence the risk of developing alcohol dependence is hypothesized to be through an elevation of acetaldehyde levels after drinking, leading to facial flushing, nausea, and other adverse reactions.
In terms of GWAS assessments, in contrast to the GWAS of smoking behaviors to date, GWAS of alcohol dependence have been less consistent in identifying genetic variants associated with alcoholism (Treutlein et al., 2009; Bierut et al., 2010; Edenberg et al., 2010). One main reason for the differences in results is that these initial studies of alcohol dependence are of modest size by GWAS standards, with only a few thousand subjects compared to the tens of thousands of subjects in the GWAS of smoking behaviors. Each study identified novel regions that have suggestive evidence of association with alcohol dependence, including PECR (Treutlein et al., 2009), an enzyme involved in fatty acid metabolism, PKNOX2 (Bierut et al., 2010), which plays a role in cell proliferation, differentiation, and death, and SLC22A18 (Edenberg et al., 2010), a solute carrier. However, there is not consistent replication across studies. In addition, the alcohol metabolizing genes previously found to be associated with alcohol dependence are not well queried on the genetic platforms used by these studies, so remain to be validated by GWAS. For instance, rs1229984 and rs671 are not genotyped on many of the initial GWAS chips (www.broadinstitute.org/mpg/snap; Johnson et al., 2008). Overall, this variation in results suggests that individual genetic contributions to alcohol dependence will be of modest effect. Larger-scale meta-analyses are underway, and hopefully these studies will discover unique genetic associations with alcoholism.
While alcoholism GWAS studies await further validation, some of the candidates coming out of these earlier human genetic approaches have support from work in animal model systems, and therefore seem like potentially stronger alcoholism risk candidate genes. Animal models support the human genetic studies implicating the γ-aminobutyric acid (GABA) system as fundamentally involved in alcohol intoxication, withdrawal, and other behavioral aspects of alcoholism (Krystal et al., 2006). Ethanol enhances GABAA receptor function (Bowen et al., 1998) and electrophysiologic studies implicate GABAA receptors as targets for the effect of ethanol in the central nervous system (Suzdak et al, 1986). Multiple candidate gene reports show an association between variants in GABRA2 and alcohol dependence (Enoch, 2008). In a hypothesis driven approach to test this association as part of a GWAS study, we find a modest association with GABRA2 (OR=1.1 and p value ~0.01), further supporting the role of this candidate gene in the development of alcoholism.
Genetic influences for other Drug Addictions
Though less common in the general population, illicit addictions such as cocaine and opiate dependence can be more devastating socially, cause more physical illnesses, and represent an extreme of addiction. Because illicit drug addiction is less common, large scale GWAS have not been undertaken as yet. Instead, the approach to studying the genetics of drug addiction has been through candidate genes. Hundreds of candidate gene association studies have been performed in the pre-GWAS era for drug addiction as well as for nicotine and alcohol dependence. Thousands more candidate gene studies have been undertaken for other medical illnesses. However, a disconnect between these reported candidate gene associations and the findings from GWAS studies exist in both the addiction field and across all of medicine. If these candidate gene studies are valid, then the GWAS studies should identify thousands of genetic variants that play a role in disease. Though hundreds of genetic variants have been conclusively confirmed by GWAS as contributors to complex diseases, the number of confirmed genetic variants is more modest than what is expected from the candidate gene studies. Overall, only a modest percentage of the numerous genetic associations proposed in the candidate gene era have been subsequently replicated in genome wide association studies (Siontis et al., 2010), which suggests that many of the candidate gene studies were false positive reports. Interestingly, those that conclusively replicate have strong genetic effects.
This lack of replication across methods reflects two distinct issues in these different studies designs: the low threshold for significance in candidate gene studies results in a high false positive rate, and the high threshold for significance in the GWAS design leads to a low sensitivity to detect true genetic contributions to disease. While candidate gene studies of addiction should therefore be interpreted with caution, they should not be dismissed because they may have captured unique phenotypes that allowed the detection of genetic variation that will not be seen in the large scale heterogeneous GWAS. Regardless, validation of human genetic mutations linked to illicit drug use awaits further study.
In the interim, animal models of addiction continue to provide insights into potential candidate genes that would benefit from more directed study in humans. A number of these studies have targeted the known neurobiological systems regulating the dopamine reward system and the endogenous opioid system. Dopamine plays a key role in reward behavior, yet the association with alcoholism and other drugs remains controversial. There are equally prominent association studies of DRD2 with alcoholism and other drug addictions and failures to replicate (Gelernter et al., 1991; Parsian et al., 1991, Le Foll et al, 2009). Similarly, the endogeneous opioid system clearly plays a role in addiction, and an amino acid change in the μ opioid receptor (OPRM1) displays functional changes with up to threefold variation in the affinity of the receptor to bind beta-endorphin, the endogenous opioid (Bond et al., 1998). However, a large scale meta-analysis does not demonstrate that this variant alters the risk of developing addiction (Arias et al., 2006).
We have the tools in hand now to directly test many of these candidate genes in large scale studies using uniform criteria for diagnosis and outcomes along with genotyping the specific variant needed, and so the contributions and controversy of these potential associations will be resolved in the coming years.
Common and Specific Factors in Addiction
The above sections have focused on candidate genes for specific addictions and several of the confirmed genetic findings support that specific genetic variants contribute to specific substance dependence risk. For instance, variants in the alcohol metabolizing genes specifically contribute to differences in alcohol consumption and alcohol dependence, but not to other addictive behaviors. Similarly, the variation in nicotine metabolizing genes contributes to smoking behavior and cigarettes smoked per day, but not alcoholism or other drug addiction. Yet data from family and twin analyses also support the idea that there is a strong contribution from common genetic factors to the development of dependence on various classes of drugs (Bierut et al., 1998; Merikangas et al., 1998; Tsuang et al., 1998; Kendler et al., 2003). In fact, twin studies have convincingly shown that most of the genetic variation to addiction is shared across the liability to develop nicotine, alcohol, and illicit drug addiction (Tsuang et al., 1998; Kendler et al., 2003). As a result, once an association is identified, the next step is to test whether this genetic variant influences multiple drug dependencies.
The chromosome 15 variant in the α5 nicotinic receptor, rs16969968, which influences the development of nicotine dependence, has also been independently shown to contribute to the occurrence of alcohol and cocaine dependence. The minor allele of rs16969968 that is correlated with an increased risk for nicotine dependence is associated with a decreased risk for alcohol and cocaine dependence (Grucza et al., 2008; Chen et al., 2009; Sherva et al., 2010). This bidirectional association is hypothesized to be due to the involvement of nicotinic receptors with both excitatory and inhibitory modulation of dopamine medicated reward pathways. These data reinforce the importance of variation in the CHRNA5-CHRNA3-CHRNB3 gene cluster for risk of dependence on multiple substances, although the direction of the effects varies across substances. In addition, variants in this region influence the initial responses to alcohol and nicotine in adolescents (Schlaepfer et al., 2008).
As we identify other genetic variants associated with addiction, it will therefore behoove us to test the potential contribution of each variant across the wide range of abused substances, as it is likely that some variants will be common risk factors of relevance to multiple addictive substances.
Where is the Unexplained Variance?
Though the GWAS based approach has been successful for nicotine dependence and other complex traits, a significant fraction of the genetic variance remains unexplained (Frazer et al., 2009). The heritability of addiction is approximately 50%, yet the confirmed genetic contributions to nicotine dependence (through the nicotinic receptors and nicotine metabolizing genes) and alcohol dependence (through alcohol metabolizing genes) explain only a small fraction of this heritability. There are two main explanations for the missing variance: rare variation not queried on the current GWAS chips, and many genes of small effect. It is likely that both of these reasons contribute to the missing genetic variance.
Clearly, part of this missing variance is related to coverage of the existing GWAS chips. By design, GWAS test for association with common variants (allele frequencies > 5%). The less common (or “rare” variants with allele frequencies < 5%) are not adequately represented on the existing arrays. For example, the well known genetic variants that alter alcohol metabolism, rs1229984 in ADH1b and rs671 in ALDH2, are not queried on most of the commercial GWAS chips. Although individually rare, these variants are collectively frequent, and their contribution to disease can be greater than those observed for common variants (Bodmer and Bonilla, 2008). Several other rare mechanisms can contribute to the modest explanation of variance to date. Structural variants, which include insertions and deletions, inversions, and translocations, can account for some of the unexplained heritability. Sequencing will be needed to allow us to definitively detect and test this class of variation.
Yet, there is also evidence that multiple common variants can begin to explain more of genetic variation in addiction. For smoking behavior, for example, we know that individual genetic variants contribute only a small effect to the development of nicotine dependence. Yet in combination, these genetic factors play a substantial role in the development of heavy smoking. For example, in our study from the Collaborative Genetic Study of Nicotine Dependence, approximately 9 variants in the nicotinic receptors explain 5% of the phenotypic variance in the sample (Saccone et al., 2010b). Though this explained variance estimate is likely higher than what will be seen in a general population study of smoking behavior, it demonstrates that collectively common genetic polymorphisms of small effect can begin to explain a larger proportion of genetic variation related to disease. Most likely hundreds, and more probably thousands, of genetic variants will be required to explain the genetic input to disease.
An additional potential drawback to GWAS studies is that there is heterogeneity of study design which may obscure true genetic contributors to disease, and careful consideration in the design of future addiction GWAS studies may help to alleviate this issue. An example is seen in the comparison of our study (Collaborative Genetic Study of Nicotine Dependence – COGEND) designed to examine genetic influences on smoking behavior and nicotine dependence, and the large scale GWAS of smoking (Saccone et al., 2009, 2010b; Thorgeirsson 2010). Our COGEND study compared very light smokers and current nicotine dependent smokers, thus focusing on differences between those who can smoke a little and not become addicted and individuals with addiction. In addition, our sample recruited subjects using a systematic strategy and in a relatively narrow age range (25–44) to avoid the confounding of secular trends in smoking. Conversely, the large scale GWAS of smoking were based on current and former smokers, and the entire range of smoking amount was included. The age range in these studies encompassed different generations in which we know smoking behavior has changed. In addition, some subjects were recruited for lung cancer, others for heart disease, and many other medical illnesses.
Recent meta-analyses have suggested that our more focused study design has paid off, with our ascertained sample that included a narrow age range and specific smoking behavior requirements increased power to detect genetic variation compared to a more heterogeneous GWAS. Two of the top genetic findings −rs16969968 in CHRNA5 and rs6474412 in CHRNB3—showed significance levels of 5.57 × 10−72 with a sample size of N = 73,853 (TAG, 2010) and 1.4 × 10−8 with a sample size of N = 84,956 (Thorgeirsson et al., 2010). In our COGEND sample of 2,062 subjects of European descent, we have a significance level of 4 × 10−7 for the CHRNA5 variant and 1.37 × 10−3 for the variant in CHRNB3 (Saccone et al., 2010a; Saccone et al., 2010b), representing a 3 and 10 fold increase in the power to detect genetic variation compared to a more heterogeneous GWAS. These comparisons demonstrate the amplified power of a study design through the systematic ascertainment, targeted age range, and phenotypic contrast of lifetime light smokers versus current heavy smokers.
From Genetic Association to Function
The above sections have highlighted how human genetic tools have aided in the identification of genetic variants contributing to the addiction cycle. Yet it needs to be understood that a genetic association characterizes only the first stage in understanding the underlying biology that leads to disease. A genetic association represents not only an association with tested genetic variants, but also an association with untested, highly correlated SNPs that can span across many genes on the same chromosome. A challenge once a genetic association is confirmed is to then understand which of these variants contribute to the biological mechanism underlying the correlation with a disease.
In the chromosome 15 region, the most biologically credible variant associated with nicotine dependence is rs16969968, a polymorphism that causes an amino acid change from aspartic acid to asparagine (Asp398Asn) in the α5 nicotine receptor subunit. Several lines of evidence point to this variant as having functional importance. The specific region in the α5 protein that includes this polymorphism is highly conserved across different species, which implies biological importance (aspartic acid is conserved in chimpanzee, Bolivian squirrel monkey, domestic cow, mouse, chicken and African clawed frog) (Bierut et al., 2008). An in vitro functional study found that α4α5β2 receptors that only differed by the asparagine amino acid substitution exhibited altered response to a nicotine agonist compared with receptors containing the aspartic acid amino acid (Bierut et al., 2008). Further studies of the nicotinic receptors show that the α5 Asn 398 protein (high risk variant) in the nicotinic acetylcholine receptor lowers Ca2+permeability and increases short-term desensitization in (α4β2)α5, but does not alter the receptor sensitivity to activation (Kuryatov et al., 2011). The high sensitivity to activation and desensitization of (α4β2)α5 nicotine acetylcholine receptors by nicotine results in a narrow concentration range in which activation and desensitization curves overlap at nicotine concentrations typically sustained in smokers. It is predicted that smokers would desensitize most of these receptors while permitting a smoldering activation of the remainder of the receptors. In addition, the α5 nicotinic receptor subunit is expressed in the brain regions that are important in the pathways relevant to the development of dependence. Finally, this key α5 gene variant is associated with a dorsal anterior cingulate-ventral striatum/extended amygdala circuit, and the “risk allele” decreases the intrinsic resting functional connectivity strength in this circuit (Hong et al., 2010). Importantly, this effect is observed in nonsmokers and it appears to represent a trait circuitry biomarker.
In the chromosome 15 region, the second independent genetic association with nicotine dependence is marked by rs880395 (Saccone et al., 2010a), and functional studies suggest a distinct biological mechanism: altered α5 nicotinic receptor mRNA expression (Wang et al., 2009; Falvella et al., 2010; Smith et al., 2011). Variants tagged by rs880395, which are more than 10kb upstream of CHRNA5, result in a 2.5 to 4 fold difference in α5 nicotinic receptor mRNA expression in the brain. High expression of CHRNA5 mRNA is correlated with an increased risk of heavy smoking and nicotine dependence (Wang et al., 2009). This change in expression is not seen in lymphocytes, which demonstrates that genetic variants can have tissue specific biologic effects (Smith et al., 2011).
These findings of the α5 nicotinic receptor in humans have motivated further animal studies of this receptor subunit, which show that the habenulo-interpeduncluar pathway is a key neurocircuit controlling nicotine consumption (Fowler et al., 2011). This circuit acts as a negative feedback response, opposite to the mesoaccumbens positive reward pathway. This animal work suggests that individuals with the α5 nicotinic receptor risk alleles for nicotine dependence are relatively insensitive to the inhibitory effects in the reward pathway. This type of work – spanning human, animal, cells and then back to humans - represents the power of genetic studies. We can identify associations, target new genes for study, and then test hypotheses in both animals and man.
These genetic associations with nicotine and alcohol dependence and proposed mechanisms of biologic action including neurotransmission and metabolism provide new insights to the underlying biology associated with addiction. Identifying how specific variants and genes associated with addictive behavior affect brain function will be key to understanding the development of dependence; yet numerous questions remain. For instance, will these mechanisms of action associated with genetic variation be expressed in all regions of the brain, or will the genetic effect be region specific? Will these variants have a similar influence throughout the lifespan, or will there be critical periods when these genetic variations alter risk of developing addiction? Though these biological mechanisms are proposed to lead to the altered risk for the development of addiction, they represent an initial understanding of the mechanisms of dependence, and it is likely that there will be more complex biologic functions underlying these genetic associations.
Convergence of Genetic Findings of Addiction and Cancer
There is an intriguing convergence of genetic findings for nicotine and alcohol dependence with medical disorders. Smoking is the strongest risk factor for the development of lung cancer and chronic obstructive pulmonary disease (COPD). Large-scale genetic studies demonstrate that the same variants on chromosome 15 that are associated with smoking behavior are also the strongest genetic risk factors for lung cancer and COPD (Amos et al., 2008, 2010; Hung et al., 2008; Liu et al., 2008; Thorgeirsson et al., 2008; Broderick et al., 2009; Pillai et al., 2009; Shiraishi et al., 2009; Lips et al., 2010). The convergence of these genetic findings associated with smoking behavior and smoking related illnesses raises the question of whether this locus has a direct biologic effect on the risk of developing lung cancer and COPD, or if the increased genetic risk of lung cancer and COPD can be explained solely through the genetic influences on smoking behavior.
The data remain mixed as to whether the genetic risk on chromosome 15 and lung cancer and COPD is related to heavier smoking (an indirect effect) or whether a direct biological mechanism increases lung cancer and COPD risk independent of smoking (a direct effect). Evidence in favor of a direct biological effect are that this genetic risk for lung cancer and COPD association with these variants remains after statistically accounting for duration of smoking history and number of cigarettes smoked per day (Lips et al., 2010). The α5 nicotinic receptor subunit is expressed in lung tissue, and a 30-fold up-regulation of expression of CHRNA5 is seen in lung cancer tissue compared to normal lung tissue (Falvella et al., 2009).
On the other hand, this chromosomal region does not increase the risk of lung cancer among non-smokers (Lips et al., 2010). Furthermore, cigarettes per day may not fully account for the exposure to carcinogens in cigarette smoke. An intriguing study demonstrated that smokers with the risk variants in the chromosome 15 region ingested more toxins even after controlling for the number of cigarettes smoked (Le Marchand et al., 2008). This implies that the smokers with the risk variants are inhaling more intensely and increasing their exposure to nicotine and other carcinogens in cigarette smoke. Thus the measurement of cigarettes per day is an imprecise measure of the risk of smoking related to lung cancer and COPD.
A parallel finding is seen with genetic variants that influence alcohol consumption, alcohol dependence, and esophageal cancer. Large studies of esophageal cancer, a cancer related to alcohol use, identify two genetic variants in alcohol metabolizing genes that influence alcohol consumption and alcohol dependence (ADH1b variant, rs1229984, and ALDH2 variant, rs671) and also contribute to the risk of esophageal cancer (Hashibe et al., 2008; Tanaka et al., 2010). Even after controlling for alcohol consumption in the analyses, the protective effects of these variants for esophageal cancer remain strong. This implies that variants in alcohol metabolizing genes not only reduce alcohol consumption and decrease the risk for alcohol dependence, but also lower the susceptibility to esophageal cancer, perhaps by reducing the carcinogenic effects of alcohol, its metabolites and other toxins.
Both of these examples challenge paradigms about the relationship between addiction and cancer. Epidemiologic data clearly support the association of addiction with cancer: smoking and nicotine dependence are associated with lung cancer; and alcohol consumption and alcohol dependence are associated with esophageal cancer. As a result, exposure to smoking and alcohol has been considered an environmental variable to be controlled in the study of cancer. However, the strongest genetic findings for the development of addiction are also the strongest genetic predictors for the correlated cancers. These findings blur the distinction between genetic and environmental risks with nicotine and alcohol addiction. It also remains unclear if the mechanism of these associations of cancer with the genetic variants can be completely explained through addictive behaviors, or if biologic mechanisms act in the brain to increase the risk of addiction while also acting in the lung and esophagus to increase the risk of cancer. Only through animal models will we be able to separate the genetic influence of these variants on the development of dependence from the genetic contribution to the development of cancer.
Genetic Implications for Different World Populations
Although dependence is common in all populations, to date all of large-scale GWAS have been performed in populations of European descent. Though the underlying biological mechanisms that lead to the development of substance dependence are most likely the same across populations, varying allele frequencies can alter the relative importance of specific genetic risk factors in different populations. For example, the variant rs16969968, which is relatively frequent in populations of European descent (37% allele frequency), is rare in populations of African or Asian descent (0% to 3% allele frequency) (Bierut et al., 2008). Similarly, the polymorphisms that cause amino acid changes in alcohol metabolizing genes, rs671 and rs1229984, are common in Asian populations, but are rare in populations of European and African descent (Edenberg et al., 2007). Thus, rs16969968 will play a larger role in the development of heavy smoking and nicotine dependence in populations of European ancestry compared to populations of African and Asian ancestry. Similarly, rs671 and rs1229984 will more strongly influence alcohol consumption and alcohol dependence in Asian populations compared to European and African populations. A new genetic frontier is to leverage these differences in genetic architecture across populations to refine association signals and narrow down associations to the most likely biologically causative variants. This strategy highlights the importance of recruiting, assessing, and studying diverse populations.
Future of Genetic Studies
As we are beginning to understand some of the genetic factors that alter our individual vulnerability to dependence, the future of genetic studies has the potential to personalize our treatment for addiction. We have unequivocal evidence of genetic variation that contributes to the development of this behavioral disorder. Addiction represents a great success in psychiatric genetics. The strongest specific genetic contributors to dependence are related to the pharmacologic responses to nicotine and alcohol and include variation in nicotinic receptor genes, nicotine metabolizing genes, and alcohol metabolizing genes.
Though some might say that GWAS have failed because we cannot account for the genetic variance associated with disease, this represents a very narrow view of the field. We have convincingly identified genetic variants that contribute to addiction. If the progress in other medical disorders can be used as an example, the “big science” consortia that include the study of tens of thousands and potentially hundreds of thousands of people will soon discover new variants that contribute to addiction. We now know that the genetic risk is modest (OR 1.3 or less) for variants that are common in the population, but rarer variants may have somewhat stronger effect. The genetic vulnerability to addiction represents the combination of hundreds or thousands of genes of modest effect.
We are at a stage where we can take several productive paths. First, we must integrate the results from candidate gene studies with the findings GWAS experiments. By synthesizing both approaches into a cohesive model, we will be able to balance the high false positive rate in candidate gene studies with this high false negative rate in GWAS. This will allow us to separate the wheat from the chaff in the candidate gene studies and aid in the discovery of more variants from the GWAS approach. Secondly, in genetic studies, ascertainment and phenotypes matter and size is not everything. Though we have thrown together GWAS studies from many different fields, it is time to go back and more carefully select studies for inclusion so that similar ascertainments can be used to reduce heterogeneity. An improvement of phenotypes should also aid in the discovery of genes. For example, cigarettes per day is an effective, but imprecise measurement of nicotine addiction. Though a large sample size can overcome a crude measure, there is a gain of power with more exact assessments. A balance must be reached between a smaller sample sized genetic studies with comprehensive assessments and a large samples with simpler phenotypes.
Tools are under development to aid scientist, physicians, and the public in the sythesis, interpretation, and dissemination of findings in human genetic variation in health and disease. One mechanism is the Human Genome Epidemiology Network (HuGENet™) (http://hugenavigator.net/HuGENavigator/home.do). Since 2001, HuGENet™ has maintained a searchable database of published, population-based epidemiologic studies of human genes extracted and curated from PubMed. This website allows the user to search by disease and gene with the goal of aiding in the translation and integration of genomics into public health research, policy, and practice.
Finally, we must remember that the major purpose for the study of the genetics of addiction is to ultimately improve our care for individuals with this disorder. Our current treatments for alcohol and nicotine dependence are related to the pharmacologic response of these substances. For example, we exploit the aversion to alcohol by administering disulfiram, a medication that interferes with ALDH, and thus increases acetaldehyde levels when alcohol is ingested. This build up of acetaldehyde causes symptoms of nausea, vomiting, flushing and headache, and is similar to the biologic response seen in individuals who carry an alcohol metabolizing gene deficiency. We may be able to utilize the variation in nicotinic receptors and nicotine metabolizing genes to improve our treatments for smoking. As we begin to understand more of the genetic diversity that influences an individual's specific risk to dependence, we will highlight new biologic pathways and neural circuitry that may be exploited pharmacologically. By identifying genetic risks that contribute to dependence, we can begin to dissect different contributions of genes and environments that lead to dependence, and in turn we can improve interventions to reduce dependence and improve cessation.
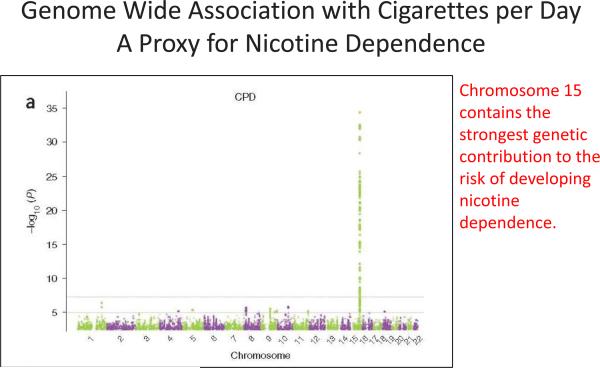
Figure 2.
Genome-Wide Association Results for cigarettes per day Manhattan plot, indicating significance of association of all SNPs in the TAG Consortium meta-analysis for cigarettes per day. Manhattan plot show SNPs plotted on the x axis according to their position on each chromosome, and plotting on the y axis (shown as negative log10 P value). Chromosome 15 contains the strongest genetic contribution to the risk of developing nicotine dependence. Figure courtesy of TAG Consortium (2010).
Table 1.
Genes associated with addiction.
| Nicotine Dependence |
| CHRNA5-CHRNA3-CHRNB4 |
| CHRNA6-CHRNB3 Region |
| Cyp2a6 Region |
| Alcohol Dependence |
| ADH1B |
| ALDH2 |
| Cocaine Dependence |
| CHRNA5-CHRNA3-CHRNB4 |
Acknowledgments
This work was supported by grants from the National Institute on Drug Abuse (K02 DA021237, R01 DA019963), the National Cancer Institute (P01 CA089392), and the National Institute on Alcohol Abuse and Alcoholism (U10 AA008401). The assistance of Sherri Fisher was key in the preparation of this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures Dr. Bierut is listed as an inventor on a patent, “Markers of Addiction,” covering the use of certain SNPs in diagnosing, prognosing and treating addiction. Dr. Bierut served as a consultant to Pfizer in 2008.
References
- American Psychiatric Association . Diagnostic and Statistical Manual of Mental Disorders. Fourth Edition American Psychiatric Association; Washington DC: 1994. [Google Scholar]
- Amos CI, Gorlov IP, Dong Q, Wu X, Zhang H, Lu EY, Scheet P, Greisinger AJ, Mills GB, Spitz MR. Nicotinic acetylcholine receptor region on chromosome 15q25 and lung cancer risk among African Americans: a case-control study. J Natl Cancer Inst. 2010;102:1199–1205. doi: 10.1093/jnci/djq232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos CI, Wu X, Broderick P, Gorlov IP, Gu J, Eisen T, Dong Q, Zhang Q, Gu X, Vijayakrishnan J, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40:616–622. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias A, Feinn R, Kranzler HR. Association of an Asn40Asp (A118G) polymorphism in the mu-opioid receptor gene with substance dependence: a meta-analysis. Drug Alcohol Depend. 2006;83:262–268. doi: 10.1016/j.drugalcdep.2005.11.024. [DOI] [PubMed] [Google Scholar]
- Audrain-McGovern J, Al Koudsi N, Rodriguez D, Wileyto EP, Shields PG, Tyndale RF. The role of CYP2A6 in the emergence of nicotine dependence in adolescents. Pediatrics. 2007;119:e264–274. doi: 10.1542/peds.2006-1583. [DOI] [PubMed] [Google Scholar]
- Bierut LJ, Agrawal A, Bucholz KK, Doheny KF, Laurie C, Pugh E, Fisher S, Fox L, Howells W, Bertelsen S, et al. A genome-wide association study of alcohol dependence. Proc Natl Acad Sci U S A. 2010;107:5082–5087. doi: 10.1073/pnas.0911109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Dinwiddie SH, Begleiter H, Crowe RR, Hesselbrock V, Nurnberger JI, Jr., Porjesz B, Schuckit MA, Reich T. Familial transmission of substance dependence: alcohol, marijuana, cocaine, and habitual smoking: a report from the Collaborative Study on the Genetics of Alcoholism. Arch Gen Psychiatry. 1998;55:982–988. doi: 10.1001/archpsyc.55.11.982. [DOI] [PubMed] [Google Scholar]
- Bierut LJ, Stitzel JA, Wang JC, Hinrichs AL, Grucza RA, Xuei X, Saccone NL, Saccone SF, Bertelsen S, Fox L, et al. Variants in nicotinic receptors and risk for nicotine dependence. Am J Psychiatry. 2008;165:1163–1171. doi: 10.1176/appi.ajp.2008.07111711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodmer W, Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat Genet. 2008;40:695–701. doi: 10.1038/ng.f.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond C, LaForge KS, Tian M, Melia D, Zhang S, Borg L, Gong J, Schluger J, Strong JA, Leal SM, et al. Single-nucleotide polymorphism in the human mu opioid receptor gene alters beta-endorphin binding and activity: possible implications for opiate addiction. Proc Natl Acad Sci U S A. 1998;95:9608–9613. doi: 10.1073/pnas.95.16.9608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen CA, Grant KA. Pharmacological analysis of the heterogeneous discriminative stimulus effects of ethanol in rats using a three-choice ethanol-dizocilpine-water discrimination. Psychopharmacology (Berl) 1998;139:86–94. doi: 10.1007/s002130050693. [DOI] [PubMed] [Google Scholar]
- Broderick P, Wang Y, Vijayakrishnan J, Matakidou A, Spitz MR, Eisen T, Amos CI, Houlston RS. Deciphering the impact of common genetic variation on lung cancer risk: a genome-wide association study. Cancer Res. 2009;69:6633–6641. doi: 10.1158/0008-5472.CAN-09-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control Current cigarette smoking among adults aged ≥18 years – United States, 2009. Morbidity & Mortality Weekly Report. 2010;59:1135–1140. [PubMed] [Google Scholar]
- Chen X, Chen J, Williamson VS, An SS, Hettema JM, Aggen SH, Neale MC, Kendler KS. Variants in nicotinic acetylcholine receptors alpha5 and alpha3 increase risks to nicotine dependence. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:926–933. doi: 10.1002/ajmg.b.30919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ. The genetics of alcohol metabolism: role of alcohol dehydrogenase and aldehyde dehydrogenase variants. Alcohol Res Health. 2007;30:5–13. [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Dick DM, Xuei X, Tian H, Almasy L, Bauer LO, Crowe RR, Goate A, Hesselbrock V, Jones K, et al. Variations in GABRA2, encoding the alpha 2 subunit of the GABA(A) receptor, are associated with alcohol dependence and with brain oscillations. Am J Hum Genet. 2004;74:705–714. doi: 10.1086/383283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Koller DL, Xuei X, Wetherill L, McClintick JN, Almasy L, Bierut LJ, Bucholz KK, Goate A, Aliev F, et al. Genome-wide association study of alcohol dependence implicates a region on chromosome 11. Alcohol Clin Exp Res. 2010;34:840–852. doi: 10.1111/j.1530-0277.2010.01156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch MA. The role of GABA(A) receptors in the development of alcoholism. Pharmacol Biochem Behav. 2008;90:95–104. doi: 10.1016/j.pbb.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falvella FS, Galvan A, Colombo F, Frullanti E, Pastorino U, Dragani TA. Promoter polymorphisms and transcript levels of nicotinic receptor CHRNA5. J Natl Cancer Inst. 2010;102:1366–1370. doi: 10.1093/jnci/djq264. [DOI] [PubMed] [Google Scholar]
- Fowler CD, Lu Q, Johnson PM, Marks MJ, Kenny PJ. Habenular alpha5 nicotinic receptor subunit signalling controls nicotine intake. Nature. 2011 doi: 10.1038/nature09797. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer KA, Murray SS, Schork NJ, Topol EJ. Human genetic variation and its contribution to complex traits. Nat Rev Genet. 2009;10:241–251. doi: 10.1038/nrg2554. [DOI] [PubMed] [Google Scholar]
- Gelernter J, O'Malley S, Risch N, Kranzler HR, Krystal J, Merikangas K, Kennedy JL, Kidd KK. No association between an allele at the D2 dopamine receptor gene (DRD2) and alcoholism. JAMA. 1991;266:1801–1807. [PubMed] [Google Scholar]
- Goedde HW, Agarwal DP, Harada S. Genetic studies on alcohol-metabolizing enzymes: detection of isozymes in human hair roots. Enzyme. 1980;25:281–286. doi: 10.1159/000459265. [DOI] [PubMed] [Google Scholar]
- Grucza RA, Wang JC, Stitzel JA, Hinrichs AL, Saccone SF, Saccone NL, Bucholz KK, Cloninger CR, Neuman RJ, Budde JP, et al. A risk allele for nicotine dependence in CHRNA5 is a protective allele for cocaine dependence. Biol Psychiatry. 2008;64:922–929. doi: 10.1016/j.biopsych.2008.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada S, Agarwal DP, Goedde HW, Tagaki S, Ishikawa B. Possible protective role against alcoholism for aldehyde dehydrogenase isozyme deficiency in Japan. Lancet. 1982;2:827. doi: 10.1016/s0140-6736(82)92722-2. [DOI] [PubMed] [Google Scholar]
- Hashibe M, McKay JD, Curado MP, Oliveira JC, Koifman S, Koifman R, Zaridze D, Shangina O, Wunsch-Filho V, Eluf-Neto J, et al. Multiple ADH genes are associated with upper aerodigestive cancers. Nat Genet. 2008;40:707–709. doi: 10.1038/ng.151. [DOI] [PubMed] [Google Scholar]
- Hasin DS, Stinson FS, Ogburn E, Grant BF. Prevalence, correlates, disability, and comorbidity of DSM-IV alcohol abuse and dependence in the United States: results from the National Epidemiologic Survey on Alcohol and Related Conditions. Arch Gen Psychiatry. 2007;64:830–842. doi: 10.1001/archpsyc.64.7.830. [DOI] [PubMed] [Google Scholar]
- Heath AC, Bucholz KK, Madden PA, Dinwiddie SH, Slutske WS, Bierut LJ, Statham DJ, Dunne MP, Whitfield JB, Martin NG. Genetic and environmental contributions to alcohol dependence risk in a national twin sample: consistency of findings in women and men. Psychol Med. 1997;27:1381–1396. doi: 10.1017/s0033291797005643. [DOI] [PubMed] [Google Scholar]
- Hindorff LA, Junkins HA, Hall PN, Mehta JP, Manolio TA. [Accessed 12/15/10];A catalog of published genome-wide association studies. Available at: www.genome.gov/gwastudies.
- Hong LE, Hodgkinson CA, Yang Y, Sampath H, Ross TJ, Buchholz B, Salmeron BJ, Srivastava V, Thaker GK, Goldman D, Stein EA. A genetically modulated, intrinsic cingulate circuit supports human nicotine addiction. Proc Natl Acad Sci U S A. 2010;107:13509–13514. doi: 10.1073/pnas.1004745107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung RJ, McKay JD, Gaborieau V, Boffetta P, Hashibe M, Zaridze D, Mukeria A, Szeszenia-Dabrowska N, Lissowska J, Rudnai P, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633–637. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O'Donnell CJ, de Bakker PI. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Prescott CA, Myers J, Neale MC. The structure of genetic and environmental risk factors for common psychiatric and substance use disorders in men and women. Arch Gen Psychiatry. 2003;60:929–937. doi: 10.1001/archpsyc.60.9.929. [DOI] [PubMed] [Google Scholar]
- Krystal JH, Staley J, Mason G, Petrakis IL, Kaufman J, Harris RA, Gelernter J, Lappalainen J. Gamma-aminobutyric acid type A receptors and alcoholism: intoxication, dependence, vulnerability, and treatment. Arch Gen Psychiatry. 2006;63:957–968. doi: 10.1001/archpsyc.63.9.957. [DOI] [PubMed] [Google Scholar]
- Kuryatov A, Berrettini W, Lindstrom J. Acetylcholine receptor (AChR) alpha5 subunit variant associated with risk for nicotine dependence and lung cancer reduces (alpha4beta2)alpha5 AChR function. Mol Pharmacol. 2011;79:119–125. doi: 10.1124/mol.110.066357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Foll B, Gallo A, Le Strat Y, Lu L, Gorwood P. Genetics of dopamine receptors and drug addiction: a comprehensive review. Behav Pharmacol. 2009;20:1–17. doi: 10.1097/FBP.0b013e3283242f05. [DOI] [PubMed] [Google Scholar]
- Le Marchand L, Derby KS, Murphy SE, Hecht SS, Hatsukami D, Carmella SG, Tiirikainen M, Wang H. Smokers with the CHRNA lung cancer-associated variants are exposed to higher levels of nicotine equivalents and a carcinogenic tobacco-specific nitrosamine. Cancer Res. 2008;68:9137–9140. doi: 10.1158/0008-5472.CAN-08-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MD. The genetics of nicotine dependence. Curr Psychiatry Rep. 2006;8:158–164. doi: 10.1007/s11920-006-0016-0. [DOI] [PubMed] [Google Scholar]
- Lips EH, Gaborieau V, McKay JD, Chabrier A, Hung RJ, Boffetta P, Hashibe M, Zaridze D, Szeszenia-Dabrowska N, Lissowska J, et al. Association between a 15q25 gene variant, smoking quantity and tobacco-related cancers among 17 000 individuals. Int J Epidemiol. 2010;39:563–577. doi: 10.1093/ije/dyp288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JZ, Tozzi F, Waterworth DM, Pillai SG, Muglia P, Middleton L, Berrettini W, Knouff CW, Yuan X, Waeber G, et al. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat Genet. 2010;42:436–440. doi: 10.1038/ng.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Vikis HG, Wang D, Lu Y, Wang Y, Schwartz AG, Pinney SM, Yang P, de Andrade M, Petersen GM, et al. Familial aggregation of common sequence variants on 15q24–25.1 in lung cancer. J Natl Cancer Inst. 2008;100:1326–1330. doi: 10.1093/jnci/djn268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macgregor S, Lind PA, Bucholz KK, Hansell NK, Madden PA, Richter MM, Montgomery GW, Martin NG, Heath AC, Whitfield JB. Associations of ADH and ALDH2 gene variation with self report alcohol reactions, consumption and dependence: an integrated analysis. Hum Mol Genet. 2009;18:580–593. doi: 10.1093/hmg/ddn372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malaiyandi V, Goodz SD, Sellers EM, Tyndale RF. CYP2A6 genotype, phenotype, and the use of nicotine metabolites as biomarkers during ad libitum smoking. Cancer Epidemiol Biomarkers Prev. 2006;15:1812–1819. doi: 10.1158/1055-9965.EPI-05-0723. [DOI] [PubMed] [Google Scholar]
- Merikangas KR, Stolar M, Stevens DE, Goulet J, Preisig MA, Fenton B, Zhang H, O'Malley SS, Rounsaville BJ. Familial transmission of substance use disorders. Arch Gen Psychiatry. 1998;55:973–979. doi: 10.1001/archpsyc.55.11.973. [DOI] [PubMed] [Google Scholar]
- Minematsu N, Nakamura H, Furuuchi M, Nakajima T, Takahashi S, Tateno H, Ishizaka A. Limitation of cigarette consumption by CYP2A6*4, *7 and *9 polymorphisms. Eur Respir J. 2006;27:289–292. doi: 10.1183/09031936.06.00056305. [DOI] [PubMed] [Google Scholar]
- Mokdad AH, Marks JS, Stroup DF, Gerberding JL. Actual causes of death in the United States, 2000. Journal of the American Medical Association. 2004;291:1238–1245. doi: 10.1001/jama.291.10.1238. [DOI] [PubMed] [Google Scholar]
- Oscarson M, McLellan RA, Asp V, Ledesma M, Bernal Ruiz ML, Sinues B, Rautio A, Ingelman-Sundberg M. Characterization of a novel CYP2A7/CYP2A6 hybrid allele (CYP2A6*12) that causes reduced CYP2A6 activity. Hum Mutat. 2002;20:275–283. doi: 10.1002/humu.10126. [DOI] [PubMed] [Google Scholar]
- Oscarson M, McLellan RA, Gullsten H, Agundez JA, Benitez J, Rautio A, Raunio H, Pelkonen O, Ingelman-Sundberg M. Identification and characterisation of novel polymorphisms in the CYP2A locus: implications for nicotine metabolism. FEBS Lett. 1999;460:321–327. doi: 10.1016/s0014-5793(99)01364-2. [DOI] [PubMed] [Google Scholar]
- Parsian A, Todd RD, Devor EJ, O'Malley KL, Suarez BK, Reich T, Cloninger CR. Alcoholism and alleles of the human D2 dopamine receptor locus. Studies of association and linkage. Arch Gen Psychiatry. 1991;48:655–663. doi: 10.1001/archpsyc.1991.01810310073013. [DOI] [PubMed] [Google Scholar]
- Paul SM. Alcohol-sensitive GABA receptors and alcohol antagonists. Proc Natl Acad Sci U S A. 2006;103:8307–8308. doi: 10.1073/pnas.0602862103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai SG, Ge D, Zhu G, Kong X, Shianna KV, Need AC, Feng S, Hersh CP, Bakke P, Gulsvik A, et al. A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet. 2009;5:e1000421. doi: 10.1371/journal.pgen.1000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone NL, Wang JC, Breslau N, Johnson EO, Hatsukami D, Saccone SF, Grucza RA, Sun L, Duan W, Budde J, et al. The CHRNA5-CHRNA3-CHRNB4 nicotinic receptor subunit gene cluster affects risk for nicotine dependence in African-Americans and in European-Americans. Cancer Res. 2009;69:6848–6856. doi: 10.1158/0008-5472.CAN-09-0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone NL, Culverhouse RC, Schwantes-An TH, Cannon DS, Chen X, Cichon S, Giegling I, Han S, Han Y, Keskitalo-Vuokko K, et al. Multiple independent loci at chromosome 15q25.1 affect smoking quantity: a meta-analysis and comparison with lung cancer and COPD. PLoS Genet. 2010a;6 doi: 10.1371/journal.pgen.1001053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone NL, Schwantes-An TH, Wang JC, Grucza RA, Breslau N, Hatsukami D, Johnson EO, Rice JP, Goate AM, Bierut LJ. Multiple cholinergic nicotinic receptor genes affect nicotine dependence risk in African and European Americans. Genes Brain Behav. 2010b;9:741–750. doi: 10.1111/j.1601-183X.2010.00608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaepfer IR, Hoft NR, Collins AC, Corley RP, Hewitt JK, Hopfer CJ, Lessem JM, McQueen MB, Rhee SH, Ehringer MA. The CHRNA5/A3/B4 gene cluster variability as an important determinant of early alcohol and tobacco initiation in young adults. Biol Psychiatry. 2008;63:1039–1046. doi: 10.1016/j.biopsych.2007.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoedel KA, Hoffmann EB, Rao Y, Sellers EM, Tyndale RF. Ethnic variation in CYP2A6 and association of genetically slow nicotine metabolism and smoking in adult Caucasians. Pharmacogenetics. 2004;14:615–626. doi: 10.1097/00008571-200409000-00006. [DOI] [PubMed] [Google Scholar]
- Sherva R, Kranzler HR, Yu Y, Logue MW, Poling J, Arias AJ, Anton RF, Oslin D, Farrer LA, Gelernter J. Variation in nicotinic acetylcholine receptor genes is associated with multiple substance dependence phenotypes. Neuropsychopharmacology. 2010;35:1921–1931. doi: 10.1038/npp.2010.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherva R, Rice JP, Neuman RJ, Rochberg N, Saccone NL, Bierut LJ. Associations and interactions between SNPs in the alcohol metabolizing genes and alcoholism phenotypes in European Americans. Alcohol Clin Exp Res. 2009;33:848–857. doi: 10.1111/j.1530-0277.2009.00904.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraishi K, Kohno T, Kunitoh H, Watanabe S, Goto K, Nishiwaki Y, Shimada Y, Hirose H, Saito I, Kuchiba A, et al. Contribution of nicotine acetylcholine receptor polymorphisms to lung cancer risk in a smoking-independent manner in the Japanese. Carcinogenesis. 2009;30:65–70. doi: 10.1093/carcin/bgn257. [DOI] [PubMed] [Google Scholar]
- Siontis KC, Patsopoulos NA, Ioannidis JP. Replication of past candidate loci for common diseases and phenotypes in 100 genome-wide association studies. Eur J Hum Genet. 2010;18:832–837. doi: 10.1038/ejhg.2010.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RM, Alachkar H, Papp AC, Wang D, Mash DC, Wang JC, Bierut LJ, Sadee W. Nicotinic alpha5 receptor subunit mRNA expression is associated with distant 5' upstream polymorphisms. Eur J Hum Genet. 2011;19:76–83. doi: 10.1038/ejhg.2010.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su T, Bao Z, Zhang QY, Smith TJ, Hong JY, Ding X. Human cytochrome P450 CYP2A13: predominant expression in the respiratory tract and its high efficiency metabolic activation of a tobacco-specific carcinogen, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Cancer Res. 2000;60:5074–5079. [PubMed] [Google Scholar]
- Suzdak PD, Schwartz RD, Skolnick P, Paul SM. Ethanol stimulates gamma-aminobutyric acid receptor-mediated chloride transport in rat brain synaptoneurosomes. Proc Natl Acad Sci U S A. 1986;83:4071–4075. doi: 10.1073/pnas.83.11.4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka F, Yamamoto K, Suzuki S, Inoue H, Tsurumaru M, Kajiyama Y, Kato H, Igaki H, Furuta K, Fujita H, et al. Strong interaction between the effects of alcohol consumption and smoking on oesophageal squamous cell carcinoma among individuals with ADH1B and/or ALDH2 risk alleles. Gut. 2010;59:1457–1464. doi: 10.1136/gut.2009.205724. [DOI] [PubMed] [Google Scholar]
- The National Center on Addiction and Substance Abuse at Columbia University . Shoveling up II: The impact of substance abuse on federal, state and local budgets. New York: 2009. [Google Scholar]
- Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP, Manolescu A, Thorleifsson G, Stefansson H, Ingason A, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorgeirsson TE, Gudbjartsson DF, Surakka I, Vink JM, Amin N, Geller F, Sulem P, Rafnar T, Esko T, Walter S, et al. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat Genet. 2010;42:448–453. doi: 10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobacco and Genetics Consortium Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet. 2010;42:441–447. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein J, Cichon S, Ridinger M, Wodarz N, Soyka M, Zill P, Maier W, Moessner R, Gaebel W, Dahmen N, et al. Genome-wide association study of alcohol dependence. Arch Gen Psychiatry. 2009;66:773–784. doi: 10.1001/archgenpsychiatry.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuang MT, Bar JL, Harley RM, Lyons MJ. The Harvard Twin Study of Substance Abuse: what we have learned. Harv Rev Psychiatry. 2001;9:267–279. [PubMed] [Google Scholar]
- Tsuang MT, Lyons MJ, Meyer JM, Doyle T, Eisen SA, Goldberg J, True W, Lin N, Toomey R, Eaves L. Co-occurrence of abuse of different drugs in men: the role of drug-specific and shared vulnerabilities. Arch Gen Psychiatry. 1998;55:967–972. doi: 10.1001/archpsyc.55.11.967. [DOI] [PubMed] [Google Scholar]
- Vink JM, Willemsen G, Boomsma DI. Heritability of smoking initiation and nicotine dependence. Behav Genet. 2005;35:397–406. doi: 10.1007/s10519-004-1327-8. [DOI] [PubMed] [Google Scholar]
- Wang JC, Cruchaga C, Saccone NL, Bertelsen S, Liu P, Budde JP, Duan W, Fox L, Grucza RA, Kern J, et al. Risk for nicotine dependence and lung cancer is conferred by mRNA expression levels and amino acid change in CHRNA5. Hum Mol Genet. 2009;18:3125–3135. doi: 10.1093/hmg/ddp231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff PH. Ethnic differences in alcohol sensitivity. Science. 1972;175:449–450. doi: 10.1126/science.175.4020.449. [DOI] [PubMed] [Google Scholar]
- World Health Organization . World Health Organization; Geneva: 2004. Global status report on alcohol 2004. [Google Scholar]
- World Health Organization Tobacco key facts. 2010 http://www.who.int/topics/tobacco/facts/en/index.html.
- Yamazaki H, Inoue K, Hashimoto M, Shimada T. Roles of CYP2A6 and CYP2B6 in nicotine C-oxidation by human liver microsomes. Arch Toxicol. 1999;73:65–70. doi: 10.1007/s002040050588. [DOI] [PubMed] [Google Scholar]
- Yu W, Gwinn M, Clyne M, Yesupriya A, Khoury MJ. A navigator for human genome epidemiology. Nat Genet. 2008;40:124–125. doi: 10.1038/ng0208-124. [DOI] [PubMed] [Google Scholar]