Abstract
Chronic and excessive alcohol consumption has been related to an increased risk of several cancers, including that of the liver; however, studies in animal models have yet to conclusively determine whether ethanol acts as a tumor promoter in hepatic tumorigenesis. We examined whether prolonged alcohol consumption could act as a hepatic tumor promoter after initiation by diethylnitrosamine (DEN) in a rat model. Male Sprague-Dawley rats were injected with 20 mg DEN/kg body weight 1 wk before introduction of either an ethanol liquid diet or an isoenergic control liquid diet. Hepatic pathological lesions, hepatocyte proliferation, apoptosis, PPARα and PPARγ, and plasma insulin-like growth factor 1 (IGF-1) levels were assessed after 6 and 10 mo. Mean body and liver weights, plasma IGF-1 concentration, hepatic expressions of proliferating cellular nuclear antigen and Ki-67, and cyclin D1 in ethanol-fed rats were all significantly lower after 10 mo of treatment compared with control rats. In addition, levels of hepatic PPARγ protein, not PPARα, were significantly higher in the ethanol-fed rats after prolonged treatment. Although ethanol feeding also resulted in significantly fewer altered hepatic foci, hepatocellular adenoma was detected in ethanol-fed rats at 10 mo, but not in control rats given the same dose of DEN. Together, these results indicate that chronic, excessive ethanol consumption impairs normal hepatocyte proliferation, which is associated with reduced IGF-1 levels, but promotes hepatic carcinogenesis.
Introduction
Primary liver cancer is responsible for over 1 million deaths worldwide. Its poor prognosis and high fatality rate make it the 3rd leading cause of cancer mortality. Long-term, excessive alcohol consumption is a significant independent risk factor for liver cancer (1). When combined with other factors, such as hepatitis viral infection, the risk of developing liver cancer increases even further. In the United States, alcohol abuse is 5 times more prevalent than the incidence of hepatitis infection, and chronic alcohol intake accounts for 32–45% of all liver cancer cases (2). Furthermore, the incidence of liver cancer has been dramatically increased (3).
Ethanol is not a carcinogen per se; however, there is accumulating evidence in humans for the carcinogenicity of acetaldehyde, the first metabolite of ethanol in the body (1). Acetaldehyde has been shown to induce DNA strand breaks in vitro and to act as a mutagenic and carcinogenic agent in vivo (1). Numerous studies have provided evidence of a linkage between ethanol and hepatocellular carcinoma; however, whether alcohol can promote carcinogenesis by its own action or it does so by acting as a cofactor in the presence of other risk factors, such as hepatitis infection, dietary carcinogens, or nutritional deficiencies, has not been well defined.
The multistage, chemically induced rat hepatocarcinogenesis model has been widely reported in the literature, with diethylnitrosamine (DEN)7 often used as the carcinogen of choice. Because a normal liver has a low rate of proliferation and hepatocytes are mainly found in the G0 phase of the cell cycle (4), the induction of a highly proliferative state in the liver is an important aspect of any model of hepatocellular carcinoma (5). Strategies used to increase hepatocyte proliferation include partial hepatectomy (PH; surgical removal of two-thirds of the liver tissue) and exposure to hepatotoxins like carbon tetrachloride (6). Several studies have investigated the effect of feeding ethanol, either in drinking water or in a liquid diet, on the development of DEN-induced hepatocarcinogenesis in rats (7–10). However, these studies provide conflicting evidence, with some reporting a promotional effect (7), some reporting inhibition (9, 10), and others reporting no effects of ethanol on hepatocarcinogenesis in rats (8). Although the exact mechanisms are unclear, these differing observations in animal studies may be related to the timing of ethanol feeding in relation to PH or carcinogen injection. Among the studies where PH was used to induce a regenerative-proliferative state, only Takada et al. (7) fed ethanol after PH and were able to see a promoting effect of ethanol on tumorigenesis. Studies that performed PH at the same time as ethanol feeding reported either no effect (8) or inhibition of promotion (9, 10). Furthermore, previous studies that did not use PH either reported a co-carcinogenic effect (11, 12), a promoting effect (13, 14), or an inhibitory effect of ethanol on hepatocellular carcinoma (15). The data from former studies may be confounded by other factors that also influence chemically induced hepatic carcinogenesis, such as diets deficient in labile methyl donors or low in carbohydrates, or coadministration of carbon tetrachloride. In previous reports of ethanol inhibiting carcinogen-induced hepatocellular carcinoma, coadministration of ethanol with the carcinogen may have competitively inhibited activation of the carcinogen by cytochrome P450 enzymes (1).
In this study, we investigated the effects of long-term ethanol feeding for 6 and 10 mo with a nutritionally adequate Lieber-DeCarli liquid diet in a chemicallyvinitiated carcinogenic rat model without PH. We injected rats with a very low dose of DEN carcinogen prior to ethanol administration to eliminate the possible interaction between alcohol-induced CYP enzyme activation and carcinogen metabolism and to focus on the ability of ethanol to act on the promotion stage of carcinogenesis. We also investigated the effect of chronic ethanol consumption on the normal hepatocyte proliferation by examining cell proliferation and cyclin D1 and apoptosis, preneoplastic placental form of glutathione S-transferase (pGST) positive altered hepatic foci (AHF), and tumor formation in this study. Further, we examined plasma insulin-like growth factor 1 (IGF-1), an essential anabolic agent in the regulation of cellular proliferation and apoptosis in a wide variety of cells and tissues. Decreased IGF-1 levels in alcoholics have been attributed to both liver damage and metabolic dysfunction due to their prolonged and excessive alcohol consumption (16). In addition, we examined hepatic expressions of PPARα and PPARγ that have been shown to be involved in the development of alcoholic liver disease (17, 18).
Materials and Methods
Rats and diets.
Male Sprague-Dawley rats (Charles River Laboratories) were maintained and fed as described in a previous report (19). To avoid interactions between ethanol and carcinogen activation, DEN (Sigma) was given as an i.p. injection of 20 mg/kg body weight 1 wk before the ethanol feeding began. This low dose has been shown to initiate cells that can develop into AHF without producing hepatic necrosis (13). The rats were assigned to weight-matched groups and fed either an ethanol liquid diet or a control liquid diet for a 6- or 10-mo period (n = 7 rats/group).
All ethanol-fed groups were given a Lieber-DeCarli liquid diet (Dyets) containing 36% of the total energy as ethanol, yielding an alcohol concentration of 6.2% (v:v). This liquid diet achieves higher ethanol blood levels than ethanol administered in their drinking water and maintains an adequate nutritional intake, as thoroughly demonstrated by Lieber et al. (20). Ethanol was gradually introduced into experimental diets over a 3-wk period before providing rats with the final concentration. In the control diet, ethanol was replaced by an isoenergic amount of maltose dextrin (Dyets). Both diets contained 18% of their total energy as protein and 35% as fat; in the control diet, 47% of the total energy was from carbohydrates, whereas 11% of the total energy was from carbohydrates in the ethanol diet.
At the end of each experimental period, the rats were terminally exsanguinated under isoflurane anesthesia (Aerrane, Fort Dodge Animal Health). The body and liver weights were recorded at the time of death for all rats. Liver tissue was collected and slices from the right and left lobes were fixed in 10% buffered formalin for further embedding in paraffin wax and immunohistochemical analysis. Remaining tissue was snap-frozen in liquid nitrogen and stored at −80°C until further analysis. All animal protocols were approved by the Institutional Animal Care and Use Committee at the USDA Human Nutrition Research Center on Aging at Tufts University.
Histopathology.
Liver samples were fixed with 10% buffered formalin and embedded in paraffin wax. Five-microliter sections were stained with hematoxylin and eosin. The slides were microscopically evaluated for the presence of inflammation, hepatic steatosis, hepatic foci of cellular alteration, adenoma, and carcinoma according to the criteria proposed by the WHO/International Agency for Research on Cancer (21). The sections were photographed and examined by 3 independent investigators who were unaware of the treatment groups.
Immunohistochemistry.
Liver tissue sections were immunostained with rabbit polyclonal anti-pGST (Novocastra Laboratories) to visualize the AHF. Hepatocyte proliferation in the liver was assessed by immunostaining with mouse monoclonal anti-rat Ki-67 (clone MIB-5, Dako) and mouse monoclonal anti-proliferating cellular nuclear antigen (PCNA) (PC-10, Santa Cruz Biotechnology), using a previously described method (22). Hepatocytes with brown-stained nuclei and cytoplasm were counted as pGST-positive cells or foci and expressed per unit area (cm2). At the 10-mo time point, AHF was also quantified from a digital image of 2 sections. To do this, we used STEREO software developed by Dr. Yihua Xu and Dr. Henry C. Pitot (23) to assess formation and growth expressed as volume percent of the liver occupied by AHF. For Ki-67 and PCNA staining, the sections were quantified under light microscopy at ×400 by 2 independent investigators who were blinded to the treatment groups, as described (22).
Western-blot analysis.
Western blotting was performed with whole cell homogenates of liver tissues by using the previously described method (22). Antibodies against cyclin D1, PPARα, PPARγ, and GAPDH were purchased from Santa Cruz Biotechnology and anti-cleaved caspase-3 was purchased from Cell Signaling. Blots were developed using either ECL Western Blotting system (Amersham) or Lumiglo (Cell Signaling) depending on the antibody used and analyzed with a densitometer (GS-710 calibrated imaging densitometer, Bio-Rad). Equal protein loading was evaluated by staining membranes after transfer with Ponceau S (Sigma-Aldrich) and performing densitometry analysis for GAPDH.
Plasma IGF-1 concentration.
Plasma levels of IGF-1 were measured by enzyme immunoassay using a mouse IGF-1 DuoSet ELISA system (R&D Systems) according to the manufacturer’s instructions.
Statistical methods.
Mortality rates were compared by Fisher’s exact test. Comparisons of the effect of ethanol on each gene or protein at the 2 different time points in the presence or absence of ethanol were made using 2-way ANOVA. When there was a significant effect, t tests were performed. Correlations were analyzed by means of Pearson coefficient. Two-sided P-values < 0.05 were considered significant. All data are presented as means ± SE.
Results
Mortality, body, and liver weight.
Over the course of the study, 1 rat died in each of the 6- and 10-mo control groups and 2 rats died in the 10-mo ethanol-fed group. No rats were lost from the 6-mo ethanol-fed group. All deaths appeared unrelated to carcinogen exposure or ethanol feeding, because deaths and early mortality rates were not different between groups receiving ethanol and control. Body weight and liver weight did not differ between groups after 6 mo of treatment. Despite pair-feeding, ethanol-treated rats had lower body weight (P < 0.01) and liver weight (P < 0.05) at 10 mo compared with the control rats (Table 1).
TABLE 1.
Body and liver weights and hepatocellular adenoma incidence of rats that consumed control or ethanol diet for 6 or 10 mo
| Time, mo | Treatment | n | Body weight,1g | Liver weight,1g | Rats with hepatocellular adenoma, n |
| 6 | Control | 6 | 623 ± 12.7 | 12.9 ± 1.40 | 0 |
| Ethanol | 7 | 582 ± 14.6 | 14.9 ± 0.60 | 0 | |
| 10 | Control | 6 | 762 ± 17.5# | 21.7 ± 0.80# | 0 |
| Ethanol | 5 | 624 ± 38.3* | 17.9 ± 0.90* | 4 |
Values are mean ± SE. *Different from control, < 0.05. #Different from 6 mo, P < 0.05.
AHF quantification.
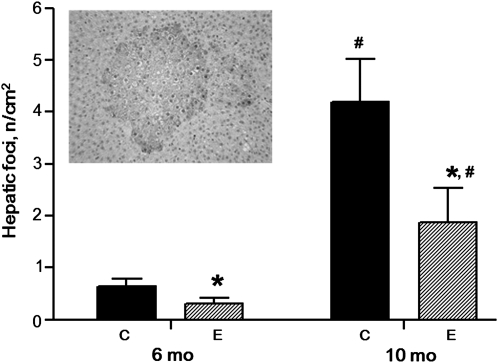
In the control rats, the number of pGST-positive AHF increased with time (Fig. 1). However, the number of pGST-positive AHF was lower in ethanol-fed rats than in the control groups at both 6 and 10 mo (P < 0.05). At 10 mo, the volume of the liver occupied by AHF in control rats was 0.42 ± 0.19%, whereas it was lower at only 0.05 ± 0.02% in the ethanol-fed group (P < 0.05).
FIGURE 1.
Numbers of AHF expressing pGST in livers of rats fed ethanol or control diets for 6 or 10 mo. Values are means ± SE, n = 5–7. *Different from control at that time, P < 0.05. #Different from 6 mo, P < 0.05. Inset: AHF from a 10-mo-old rat fed the control diet. C, control rats; E, ethanol-fed rats.
Pathology.
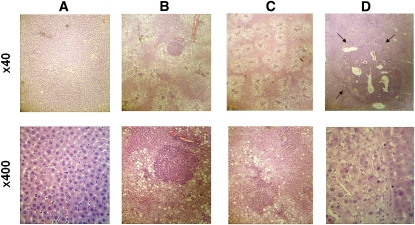
Massive hepatic steatosis and alcoholic foamy degeneration with a mixed infiltrate of inflammatory cells were detected in the all of the ethanol-fed rats after 6 and 10 mo of treatment, but not in control rats given the same dose of the DEN carcinogen (Fig. 2B,C). No tumors were detected on the surface of the liver. However, hepatocellular adenoma under microscopy (Fig. 2D) was detected in 4 of 5 ethanol-fed rats after 10 mo of treatment, but was not present in ethanol-fed rats after 6 mo of treatment nor in control rats given the same dose of DEN carcinogen at either time point (Table 1).
FIGURE 2.
Hepatic lesions under hematoxylin and eosin staining (upper, ×40; lower, ×400): normal (A); massive steatosis with tigroid basophilic focus (B); alcoholic foamy degeneration with a mixed infiltrate of inflammatory cells (C); and hepatocellular adenoma (D). The tumor (indicated by arrow) is composed of sheets of hepatocytes that are larger than those of the surrounding liver, which shows compression.
Proliferation and apoptosis.
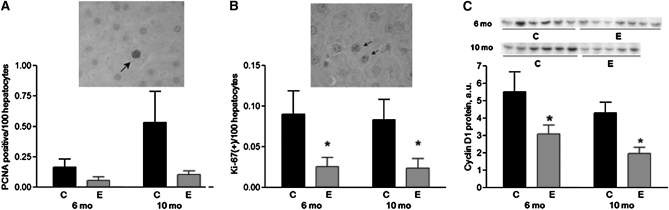
The effect of ethanol feeding on cell proliferation was assessed by both PCNA and Ki-67 immunohistochemistry. At both time points, PCNA labeling tended to be lower in the ethanol-fed groups (P = 0.08 at 6 mo and P = 0.06 at 10 mo) (Fig. 3A), and the ethanol-fed groups had fewer cells labeled with Ki-67 (P < 0.01) (Fig. 3B) compared with the control groups. At 10 mo, the number of Ki-67 was positively correlated with the number of AHF/cm2 area (r = 0.87; P < 0.001) and with the volume percent of liver occupied by AHF (r = 0.86; P < 0.01). Furthermore, the number of PCNA-positive cells was correlated with the liver weights at both time points (r = 0.46; P < 0.01) (data not shown).
FIGURE 3.
Number of hepatic PCNA and Ki-67 positively stained hepatocytes and hepatic cyclin D1 protein levels at in livers of rats fed ethanol or control diets for 6 or 10 mo. Values are means ± SE, n = 5–7. *Different from control at that time, P < 0.01. (A) PCNA-positive hepatocytes. Insert: PCNA-positive hepatocyte from a 10-mo-old rat fed the control diet. (B) Ki-67-positive hepatocytes. Insert: Ki-67-positive hepatocytes from a 10-mo-old rat fed the control diet. (C) Hepatic cyclin D1 protein levels. C, control rats; E, ethanol-fed rats.
Cyclin D1 protein levels in the liver were 44% lower in the ethanol-fed groups after 6 mo of treatment and 54% lower in the ethanol-fed groups after 10 mo of treatment compared with control rats (P < 0.01 for both time points) (Fig. 3C). We also examined the effect of ethanol feeding on apoptosis by Western analysis of cleaved caspase-3. Hepatic cleaved caspase-3 levels were not significantly different between treatment groups at either time point (data not shown).
Plasma IGF-1 concentrations.
Plasma IGF-1 concentrations were lower in the ethanol-fed rats at 6 mo (267 ± 55.5 μg/L) compared with the control rats (631 ± 122 μg/L; P < 0.05). Similarly, plasma IGF-1 concentrations at 10 mo were also lower in the ethanol-fed rats (312 ± 66.0 μg/L) compared with control rats (499 ± 87.1 μg/L; P < 0.05).
PPAR expressions.
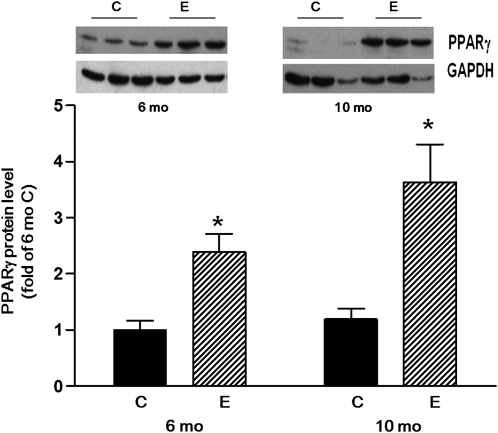
Hepatic PPARα levels in the ethanol-fed groups at 6 and 10 mo did not differ from their respective control groups (data not shown). However, hepatic PPARγ levels were 2-fold higher in the ethanol-fed group at 6 mo and 3-fold higher at 10 mo compared with the respective control groups (P < 0.05 for both time points) (Fig. 4).
FIGURE 4.
PPARγ protein levels in livers of rats fed ethanol or control diet for 6 or 10 mo. Values are means ± SE, n = 5–7. *Different from control at that time, P < 0.01. C, control rats; E, ethanol-fed rats. Insert: Hepatic PPARγ of 3 rats from each group.
Discussion
The present study clearly shows that alcohol can act as a promoter in hepatic carcinogenesis independently of any role in carcinogen activation or tumor initiation and uncomplicated by viral infection, inadequate diet, or PH of the liver. In addition, chronic ethanol consumption led to impaired proliferation of normal hepatocytes in this rat model, characterized by decreased PCNA and Ki-67, decreased cyclin D1, and lower plasma IGF-1 levels. Whereas we observed declines in pGST-positive AHF in rodent liver, we found that chronic ethanol consumption over a 10-mo period led to the development of hepatocellular adenoma in the DEN-initiated rats but not in control rats given the same dose of DEN. This suggests that traditional biomarkers associated with hepatic cancer risk such as pGST-positive AHF do not accurately reflect the later stage of carcinogenic changes in models of prolonged alcohol consumption due to alcohol-related changes in the ability of liver cells to proliferate and regenerate after insult. Instead, we see impaired compensatory liver cell proliferation alongside increased tumor formation.
To our knowledge, this is the first report outside the context of PH of ethanol-fed rats having a lower proliferation-labeling index in the liver. A decrease in proliferation by ethanol has been well documented in liver regeneration after PH (24, 25). Most of the studies documenting negative effects of alcohol on liver regeneration were carried out in a model involving a major surgery (removal of two-thirds of the liver), and proliferative changes were assessed only over a very short period of time (days). Also, there are conflicting reports regarding whether ethanol impairs the early proliferative response of hepatocytes and inhibits the regeneration of hepatocytes after PH (26), with some studies showing that ethanol feeding can induce a proliferative state in the livers of rats after a relatively short time (22, 26, 27). Our data complement previous research showing that 8 mo of ethanol exposure can promote hepatic carcinogenesis after PH (7). We detected only hepatocellular adenoma, not carcinoma, in alcohol-fed rats, which could be due to the low dose of DEN and the duration of study being <10 mo. The traditional hepatocellular carcinoma model in rats was developed by i.p. injecting 200 mg/kg body weight of DEN. In our preliminary study we observed that this dose of DEN resulted in strong hepatic carcinogenetic responses with necrosis in both alcohol diet-fed rats and control diet-fed rats. To investigate the ability of ethanol to act as a promoter of hepatic carcinogenesis, we injected rats with a very low dose of DEN (20 mg/kg i.p.) carcinogen. This low dose did not result in tumor formation in nonethanol-fed rats. In addition, given the absence of PH and other complicating factors, e.g. the high dose of carcinogen injection, our long-term alcohol-feeding model may offer a useful method by which to study the effects of prolonged alcohol consumption on liver malignant transformation and dietary intervention.
The observed change in liver weight correlated with the decreased levels of PCNA and cyclin D1 protein levels, suggesting these markers are reflective of changes in the entire liver. Hepatocyte proliferation, as assessed by Ki-67 immunohistochemistry, was lower after 6 and 10 mo of ethanol feeding. Although there were no significant changes in the PCNA levels in the ethanol-fed groups, this could be due to increased DNA repair, because PCNA measurements reflect change due to both increased cellular proliferation and DNA repair. Lower levels of hepatocyte proliferation in the ethanol-fed rats were associated with lower protein levels of cyclin D1 at 6 and 10 mo. Cyclin D1 is upregulated after PH and cyclin D1 induction facilitates the progress of hepatocytes through the G1 phase of the cell cycle (28). In addition, ethanol inhibition of PH-induced liver regeneration has previously been associated with decreased expression of cyclin D1 (24). In contrast, apoptosis, as assessed by Western analysis of cleaved caspase-3, did not differ between the ethanol-fed rats and control rats. These data indicate that a lower hepatocyte proliferation index found in the ethanol-fed groups could be leading to the lower numbers of AHF, independent of apoptosis. In the present study, ethanol feeding resulted in lower numbers of AHF at 6 and 10 mo and decreased the volume percentage of the liver occupied by the AHF at 10 mo compared with the control rats. Lower levels of hepatic proliferation can explain the lower numbers of AHF found in the ethanol groups. We therefore conclude that the dysfunction of either cell cycle control or differentiation machinery by chronic alcohol feeding is responsible for dysregulated growth and transformed phenotype. Previous studies, including ours, showed that ethanol feeding can induce a proliferative state in the livers of rats after 1 mo of treatment (22, 26, 27), which may reflect a regenerative, proliferative response as a result of the alcohol insult at a relatively short time. This proliferative response at an earlier stage may increase genomic instability or survival of carcinogen DEN-initiated hepatocytes, which could then develop into tumors through both genetic or epigenetic alterations. Indeed, certain dietary components, such as retinoic acid (22, 29), can inhibit acute or subacute, alcohol-induced cell proliferation and reduce the risk of tumorigenesis. However, the effects of long-term alcohol treatment are more dynamic, causing a number of biochemical and molecular alterations and pathological lesions (e.g. oxidative stress, steatohepatitis, reduced vitamin A levels, fibrogenesis, etc.). In the present study, alcohol significantly reduced levels of IGF-1 (stimulating factor for cell growth) but markedly increased levels of PPAR (inhibiting cell growth), after 6 and 10 mo of treatment, which may contribute to inhibiting the proliferative activity of mature hepatocytes. Because the transformed cells or tumor cells usually become resistant to growth inhibition and differentiation, they can therefore “escape” under certain conditions. Recently, we observed that treatment with chlormethiazole, an inhibitor of alcohol-induced cytochrome p4502E1 enzyme, for 10 mo can counter the tumor-promoting action of ethanol by restoring normal hepatic levels of retinoic acid, a strong anticancer agent (P.R.G. Chavez, Q. Ye, H.K. Seitz, and X.D. Wang, unpublished data). Therefore, we speculate that the alcohol-reduced levels of hepatic vitamin A and retinoic acid could be a potential mechanism for the promoting effect of long-term ethanol consumption on hepatic tumorigenesis. This notion was supported by our observation that the oval cells, as bipotent hepatic progenitor cells that are capable of differentiating into hepatocytes and are putative targets for transformation in liver cancer (30, 31), were detected in the ethanol-fed rats after 10 mo of treatment but not in the ethanol-fed rats given the cytochrome p4502E1 inhibitor (P.R.G. Chavez, C. Liu, and X.D. Wang, unpublished data). Clearly, further studies are needed to evaluate this notion.
In agreement with previous research that showed chronic ethanol feeding decreased plasma IGF-1 levels in rats (32, 33), we have shown that plasma IGF-1 concentrations in rats fed an alcohol-based diet for 6 and 10 mo were decreased by 58 and 37%, respectively, compared with rats fed a control diet. Experiments in liver-specific IGF-1 receptor knockout mice suggest that IGF-1 may substantially contribute to liver regeneration after PH through a signaling pathway that controls cyclins A and D, thereby controlling cell cycle progression (34). Disruptions in IGF-1 system components leading to alterations in cell cycle and survival signals have also been implicated in carcinogenesis in that significantly lower levels of both plasma IGF-1 and IGF-1 mRNA in tumor tissue have been shown in patients with hepatocellular cancer (35, 36). Correspondingly, we found that decreased plasma IGF-1 in rats fed ethanol for 10 mo was associated with increased incidence of liver tumor. This may be related to alcohol-reduced vitamin A status (37, 38), because low vitamin A levels have been associated with decreased plasma IGF-1 and mRNA gene expression in liver, lung, testis, and heart in Japanese quail and rats (39). We have previously demonstrated that retinoic acid supplementation can partially restore both the plasma IGF-1 and the hepatic IGF-1 expression in alcohol-fed rats (33), and evidence suggests that this may provide beneficial protection against certain alcohol-related injuries (22, 29). Further investigation into the role of the IGF axis in hepatic carcinogenesis is warranted.
The role of PPAR receptors in the development of alcohol liver disease has been intensively investigated both in cell culture systems and ethanol-fed rodents (40). In Sv/129 mice and rats, ethanol administration decreased PPARα protein levels (41, 42). Activation of PPARα by clofibrate in ethanol-fed rats ameliorates fatty liver and decreases necroinflammatory injury (43). A recent study showed that polyenephosphatidylcholine significantly ameliorated ethanol-induced hepatocyte damage and hepatitis in PPARα-null mice (44). Although we did not detect any changes in hepatic PPARα levels in the ethanol-fed groups at 6 and 10 mo in the present study, we cannot exclude alcohol-impaired PPARα/RXR binding to DNA and downregulated PPARα target genes, as was shown by a previous study in ethanol-fed C57BL/6J mice (45). PPARγ is normally expressed in both human and murine livers at only 10−30% of the levels found in adipose tissue (46). In contrast to a previous study in which PPARγ expressions remained unchanged in chronic alcohol-fed rats (47), we found that PPARγ expressions were upregulated in the livers of rats with alcohol feeding and were correlated with severe fatty liver at both 6 and 10 mo. Our data are in agreement with previous observations that PPARγ is expressed at markedly elevated levels in the fatty livers associated with a number of murine models of diabetes or obesity (48, 49). A recent study demonstrated that PPARγ is capable of activating the expression of genes involved in TG accumulation in hepatocytes and promoting the generation of fatty liver (50).
In summary, these data indicate that the presence of decreased hepatocyte proliferation alongside increased tumor formation is an important facet of alcohol-induced hepatocarcinogenesis and should be examined in future investigations of the etiology and prevention of alcoholic liver cancer. Our model of alcohol-promoted hepatocarcinogenesis independent of dietary deficiency and PH may be useful for the future research of the effects of chronic ethanol consumption on the differentiation and proliferation of different populations of liver progenitor cells as well as for studying dietary interventions against alcohol-related hepatic carcinogenesis.
Acknowledgments
We thank Dr. Yihua Xu and Dr. Henry C. Pitot of the McArdle Laboratory for Cancer Research at the University of Wisconsin, Madison, and Dr. Lauren J. Richey of the Animal Pathology Core, Tufts Medical Center, Boston, MA for kindly providing assistance in the pathology of hepatic lesions. We also thank Kang-Quan Hu for her technical assistance and Stephanie-Jo McGehee for her assistance in the preparation of this manuscript. P.R.G.C., H.K.S. and X-D.W. designed research; P.R.G.C., F.L., C.L., and S.A.R.P. conducted research; P.R.G.C., F.L., J.C., and X-D.W. analyzed data; P.R.G.C., J.C., and X-D.W. wrote the paper; and X-D.W. had primary responsibility for final content. All authors read and approved the final manuscript.
Footnotes
Supported by NIH grants R01AA12682 and R01CA104932, USDA grant 1950-51000-064S, and the Dietmar-Hopp and Manfred-Lautenschlaeger Foundations. Any opinions, findings, conclusion, or recommendations expressed in this publication are those of the author(s) and do not necessarily reflect the views of NIH and the USDA.
Abbreviations used: AHF, altered hepatic foci; DEN, diethylnitrosamine; IGF-1, insulin-like growth factor 1; PCNA, proliferating cellular nuclear antigen; pGST, placental glutathione-S-transferase; PH, partial hepatectomy.
Literature Cited
- 1.Seitz HK, Stickel F. Molecular mechanisms of alcohol-mediated carcinogenesis. Nat Rev Cancer. 2007;7:599–612 [DOI] [PubMed] [Google Scholar]
- 2.Morgan TR, Mandayam S, Jamal MM. Alcohol and hepatocellular carcinoma. Gastroenterology. 2004;127:S87–96 [DOI] [PubMed] [Google Scholar]
- 3.El-Serag HB. Hepatocellular carcinoma: recent trends in the United States. Gastroenterology. 2004;127:S27–34 [DOI] [PubMed] [Google Scholar]
- 4.Zarnegar R, DeFrances MC, Michalopoulos GK. Hepatocyte growth factor: its role in hepatic growth and pathobiology. Arias IM, editor New York: Raven Press; 1994. p. 1047–58 [Google Scholar]
- 5.Columbano A, Rajalakshmi S, Sarma DS. Requirement of cell proliferation for the initiation of liver carcinogenesis as assayed by three different procedures. Cancer Res. 1981;41:2079–83 [PubMed] [Google Scholar]
- 6.Diehl AM. Recent events in alcoholic liver disease V. effects of ethanol on liver regeneration. Am J Physiol Gastrointest Liver Physiol. 2005;288:G1–6 [DOI] [PubMed] [Google Scholar]
- 7.Takada A, Nei J, Takase S, Matsuda Y. Effects of ethanol on experimental hepatocarcinogenesis. Hepatology. 1986;6:65–72 [DOI] [PubMed] [Google Scholar]
- 8.Puapairoj P, Cui L, Ogawa K, Akagi K, Hasegawa R, Ito N. Effect of ethanol on paraquat toxicity in F344 rats. Food Chem Toxicol. 1994;32:379–86 [DOI] [PubMed] [Google Scholar]
- 9.Ikawa E, Tsuda H, Sakata T, Masui T, Satoh K, Sato K, Ito N. Modification potentials of ethyl alcohol and acetaldehyde on development of preneoplastic glutathione S-transferase P-form-positive liver cell foci initiated by diethylnitrosamine in the rat. Cancer Lett. 1986;31:53–60 [DOI] [PubMed] [Google Scholar]
- 10.Mandl J, Banhegyi G, Garzo T, Lapis K, Antoni F, Schaff Z. Ethanol treatment inhibits the development of diethylnitrosamine-induced tumors in rats. J Exp Pathol. 1989;4:227–35 [PubMed] [Google Scholar]
- 11.Effects of tocopherol and deprenyl on the progression of disability in early Parkinson's disease The Parkinson Study Group. N Engl J Med. 1993;328:176–83 [DOI] [PubMed] [Google Scholar]
- 12.Holick CN, Michaud DS, Stolzenberg-Solomon R, Mayne ST, Pietinen P, Taylor PR, Virtamo J, Albanes D. Dietary carotenoids, serum {beta}-carotene, and retinol and risk of lung cancer in the Alpha-Tocopherol, Beta-Carotene Cohort Study. Am J Epidemiol. 2002;156:536–47 [DOI] [PubMed] [Google Scholar]
- 13.Driver HE, McLean AE. Dose-response relationships for initiation of rat liver tumours by diethylnitrosamine and promotion by phenobarbitone or alcohol. Food Chem Toxicol. 1986;24:241–5 [DOI] [PubMed] [Google Scholar]
- 14.Schwarz M, Buchmann A, Wiesbeck G, Kunz W. Effect of ethanol on early stages in nitrosamine carcinogenesis in rat liver. Cancer Lett. 1983;20:305–12 [DOI] [PubMed] [Google Scholar]
- 15.Habs M, Schmahl D. Inhibition of the hepatocarcinogenic activity of diethylnitrosamine (DENA) by ethanol in rats. Hepatogastroenterology. 1981;28:242–4 [PubMed] [Google Scholar]
- 16.Ronis MJ, Wands JR, Badger TM, de la Monte SM, Lang CH, Calissendorff J. Alcohol-induced disruption of endocrine signaling. Alcohol Clin Exp Res. 2007;31:1269–85 [DOI] [PubMed] [Google Scholar]
- 17.Purohit V, Gao B, Song BJ. Molecular mechanisms of alcoholic fatty liver. Alcohol Clin Exp Res. 2009;33:191–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mello T, Polvani S, Galli A. Peroxisome proliferator-activated receptor and retinoic x receptor in alcoholic liver disease. PPAR Res. 2009;2009:748174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chung J, Veeramachaneni S, Liu C, Mernitz H, Russell RM, Wang XD. Vitamin E supplementation does not prevent ethanol-reduced hepatic retinoic acid levels in rats. Nutr Res. 2009;29:664–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lieber CS, DeCarli LM, Sorrell MF. Experimental methods of ethanol administration. Hepatology. 1989;10:501–10 [DOI] [PubMed] [Google Scholar]
- 21.Mohr U. International classification of rodent tumors. Part I: The Rat 10. Digestive system. The International Agency for Research on Cancer, editor. New York: Springer; 1997 [Google Scholar]
- 22.Chung J, Liu C, Smith DE, Seitz HK, Russell RM, Wang XD. Restoration of retinoic acid concentration suppresses ethanol-enhanced c-Jun expression and hepatocyte proliferation in rat liver. Carcinogenesis. 2001;22:1213–9 [DOI] [PubMed] [Google Scholar]
- 23.Xu YH, Dragan YP, Campbell HA, Pitot HC. STEREO: a program on a PC-Windows 95 platform for recording and evaluating quantitative stereologic investigations of multistage hepatocarcinogenesis in rodents. Comput Methods Programs Biomed. 1998;56:49–63 [DOI] [PubMed] [Google Scholar]
- 24.Koteish A, Yang S, Lin H, Huang J, Diehl AM. Ethanol induces redox-sensitive cell-cycle inhibitors and inhibits liver regeneration after partial hepatectomy. Alcohol Clin Exp Res. 2002;26:1710–8 [DOI] [PubMed] [Google Scholar]
- 25.Diehl AM, Chacon M, Wagner P. The effect of chronic ethanol feeding on ornithine decarboxylase activity and liver regeneration. Hepatology. 1988;8:237–42 [DOI] [PubMed] [Google Scholar]
- 26.Baroni GS, Marucci L, Benedetti A, Mancini R, Jezequel AM, Orlandi F. Chronic ethanol feeding increases apoptosis and cell proliferation in rat liver. J Hepatol. 1994;20:508–13 [DOI] [PubMed] [Google Scholar]
- 27.Colantoni A, Idilman R, De Maria N, La Paglia N, Belmonte J, Wezeman F, Emanuele N, Van Thiel DH, Kovacs EJ, et al. Hepatic apoptosis and proliferation in male and female rats fed alcohol: role of cytokines. Alcohol Clin Exp Res. 2003;27:1184–9 [DOI] [PubMed] [Google Scholar]
- 28.Rickheim DG, Nelsen CJ, Fassett JT, Timchenko NA, Hansen LK, Albrecht JH. Differential regulation of cyclins D1 and D3 in hepatocyte proliferation. Hepatology. 2002;36:30–8 [DOI] [PubMed] [Google Scholar]
- 29.Chung J, Chavez PR, Russell RM, Wang XD. Retinoic acid inhibits hepatic Jun N-terminal kinase-dependent signaling pathway in ethanol-fed rats. Oncogene. 2002;21:1539–47 [DOI] [PubMed] [Google Scholar]
- 30.Lowes KN, Brennan BA, Yeoh GC, Olynyk JK. Oval cell numbers in human chronic liver diseases are directly related to disease severity. Am J Pathol. 1999;154:537–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuhlmann WD, Peschke P. Hepatic progenitor cells, stem cells, and AFP expression in models of liver injury. Int J Exp Pathol. 2006;87:343–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sonntag WE, Boyd RL. Chronic ethanol feeding inhibits plasma levels of insulin-like growth factor-1. Life Sci. 1988;43:1325–30 [DOI] [PubMed] [Google Scholar]
- 33.Lian F, Chung J, Russell RM, Wang XD. Alcohol-reduced plasma IGF-I levels and hepatic IGF-I expression can be partially restored by retinoic acid supplementation in rats. J Nutr. 2004;134:2953–6 [DOI] [PubMed] [Google Scholar]
- 34.Desbois-Mouthon C, Wendum D, Cadoret A, Rey C, Leneuve P, Blaise A, Housset C, Tronche F, Le Bouc Y, et al. Hepatocyte proliferation during liver regeneration is impaired in mice with liver-specific IGF-1R knockout. FASEB J. 2006;20:773–5 [DOI] [PubMed] [Google Scholar]
- 35.Su TS, Liu WY, Han SH, Jansen M, Yang-Fen TL. P'Eng FK, Chou CK. Transcripts of the insulin-like growth factors I and II in human hepatoma. Cancer Res. 1989;49:1773–7 [PubMed] [Google Scholar]
- 36.Stuver SO, Kuper H, Tzonou A, Lagiou P, Spanos E, Hsieh CC, Mantzoros C, Trichopoulos D. Insulin-like growth factor 1 in hepatocellular carcinoma and metastatic liver cancer in men. Int J Cancer. 2000;87:118–21 [DOI] [PubMed] [Google Scholar]
- 37.Wang XD, Liu C, Chung J, Stickel F, Seitz HK, Russell RM. Chronic alcohol intake reduces retinoic acid concentration and enhances AP-1 (c-Jun and c-Fos) expression in rat liver. Hepatology. 1998;28:744–50 [DOI] [PubMed] [Google Scholar]
- 38.Liu C, Russell RM, Seitz HK, Wang XD. Ethanol enhances retinoic acid metabolism into polar metabolites in rat liver via induction of cytochrome P4502E1. Gastroenterology. 2001;120:179–89 [DOI] [PubMed] [Google Scholar]
- 39.Fu ZW, Kubo T, Sugahara K, Noguchi T, Kato H. Retinoid nutritional status differently affects the expression of Japanese quail retinoic acid receptor-beta isoform transcripts in a tissue-specific manner. J Endocrinol. 2001;169:281–90 [DOI] [PubMed] [Google Scholar]
- 40.Mello T, Polvani S, Galli A. Peroxisome proliferator-activated receptor and retinoic x receptor in alcoholic liver disease. PPAR Res. 2009;2009:748–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wan YJ, Morimoto M, Thurman RG, Bojes HK, French SW. Expression of the peroxisome proliferator-activated receptor gene is decreased in experimental alcoholic liver disease. Life Sci. 1995;56:307–17 [DOI] [PubMed] [Google Scholar]
- 42.Nakajima T, Kamijo Y, Tanaka N, Sugiyama E, Tanaka E, Kiyosawa K, Fukushima Y, Peters JM, Gonzalez FJ, et al. Peroxisome proliferator-activated receptor alpha protects against alcohol-induced liver damage. Hepatology. 2004;40:972–80 [DOI] [PubMed] [Google Scholar]
- 43.Nanji AA, Dannenberg AJ, Jokelainen K, Bass NM. Alcoholic liver injury in the rat is associated with reduced expression of peroxisome proliferator-alpha (PPARalpha)-regulated genes and is ameliorated by PPARalpha activation. J Pharmacol Exp Ther. 2004;310:417–24 [DOI] [PubMed] [Google Scholar]
- 44.Okiyama W, Tanaka N, Nakajima T, Tanaka E, Kiyosawa K, Gonzalez FJ, Aoyama T. Polyenephosphatidylcholine prevents alcoholic liver disease in PPARalpha-null mice through attenuation of increases in oxidative stress. J Hepatol. 2009;50:1236–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fischer M, You M, Matsumoto M, Crabb DW. Peroxisome proliferator-activated receptor alpha (PPARalpha) agonist treatment reverses PPARalpha dysfunction and abnormalities in hepatic lipid metabolism in ethanol-fed mice. J Biol Chem. 2003;278:27997–8004 [DOI] [PubMed] [Google Scholar]
- 46.Tontonoz P, Graves RA, Budavari AI, Erdjument-Bromage H, Lui M, Hu E, Tempst P, Spiegelman BM. Adipocyte-specific transcription factor ARF6 is a heterodimeric complex of two nuclear hormone receptors, PPAR gamma and RXR alpha. Nucleic Acids Res. 1994;22:5628–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lieber CS, Leo MA, Wang X, Decarli LM. Effect of chronic alcohol consumption on Hepatic SIRT1 and PGC-1alpha in rats. Biochem Biophys Res Commun. 2008;370:44–8 [DOI] [PubMed] [Google Scholar]
- 48.Bedoucha M, Atzpodien E, Boelsterli UA. Diabetic KKAy mice exhibit increased hepatic PPARgamma1 gene expression and develop hepatic steatosis upon chronic treatment with antidiabetic thiazolidinediones. J Hepatol. 2001;35:17–23 [DOI] [PubMed] [Google Scholar]
- 49.Memon RA, Tecott LH, Nonogaki K, Beigneux A, Moser AH, Grunfeld C, Feingold KR. Up-regulation of peroxisome proliferator-activated receptors (PPAR-alpha) and PPAR-gamma messenger ribonucleic acid expression in the liver in murine obesity: troglitazone induces expression of PPAR-gamma-responsive adipose tissue-specific genes in the liver of obese diabetic mice. Endocrinology. 2000;141:4021–31 [DOI] [PubMed] [Google Scholar]
- 50.Matsusue K, Kusakabe T, Noguchi T, Takiguchi S, Suzuki T, Yamano S, Gonzalez FJ. Hepatic steatosis in leptin-deficient mice is promoted by the PPARgamma target gene Fsp27. Cell Metab. 2008;7:302–11 [DOI] [PMC free article] [PubMed] [Google Scholar]