Abstract
In medicine, mRNA transcripts are being developed as molecular biomarkers for the diagnosis and treatment of a number of diseases. These biomarkers offer early and more accurate prediction and diagnosis of disease and disease progression, and ability to identify individuals at risk. Use of microarrays also offers opportunity to identify orthogonal (uncorrelated) biomarkers not known to be linked with conventional biomarkers. Investigators are increasingly using blood as a surrogate tissue for biopsy and analysis; total RNA isolated from whole blood is predominantly from erythroid cells, and whole blood mRNA share more than 80% of the transcriptome with major tissues. Thus blood mRNA biomarkers for individualized disease prediction and diagnosis are an exciting area in medicine; mRNA biomarkers in nutrition have potential application that parallel these opportunities. Assessment of selenium (Se) status and requirements is one area where tissue mRNA levels have been used successfully. Selenoprotein-H and selenoprotein-W as well as glutathione peroxidase-1 (Gpx1) mRNAs are highly down-regulated in Se deficiency in rat liver, and the minimum dietary Se requirement is 0.06–0.07 μg Se/g based on these biomarkers, similar to requirements determined using conventional biomarkers. Blood Gpx1 mRNA can also be used to determine Se requirements in rats, showing that blood mRNA has potential for assessment of nutrient status. Future research is needed to develop mRNA biomarker panels for all nutrients that will discriminate between deficient, marginal, adequate, and supernutritional individuals and populations, and differentiate between individuals that will benefit versus be adversely affected by nutrient supplementation.
Keywords: Individualized nutrition, mRNA, Quantitative real-time-PCR, Requirements, Selenium, Selenoprotein
1. Introduction
A biomarker, as defined in current medical dictionaries, is “a distinctive biological or biologically-derived indicator (as a biochemical metabolite in the body) of a process, event, or condition (as aging, disease, or exposure to a toxic substance), such as age-related biomarkers of disease or degenerative change [1].” The Biomarkers Definitions Working Group, as part of the NIH Director’s Initiative on Biomarkers and Surrogate Endpoints, defined biomarker as “a characteristic that is objectively measured and evaluated as an indicator of normal biologic processes, pathogenic processes, or pharmacologic responses to a therapeutic intervention [2].” For the purposes of this review, a molecular biomarker is defined as “an mRNA transcript that indicates the (nutrient) status of an organism or tissue, as distinguished from a biochemical biomarker (eg. enzyme activity), or chemical biomarker (eg. plasma element concentration).”
The discoveries of reverse transcriptase and heat-stable DNA polymerase, the development of high-throughput qRT-PCR and microarray technologies, the sequencing and annotation of multiple genomes, and development of the parallel sophisticated bioinformatics, collectively have provided the ability to assess relative levels of individual gene transcripts at a quantity and a pace that was unimaginable just a decade ago. This review will discuss the increasing development of these tools to provide molecular biomarkers for application in medicine, including the potential for individualized medicine, and the efficacy of using blood as a source of RNA for these measurements. Lastly, the review proposes that molecular biomarkers have just as much potential in nutrition for assessment of nutrient status, using selenium as an example.
2. Molecular Biomarkers in Medicine
In his essay in Nature on “Predicting disease using genomics [3],” Bell suggests that genetic (genes) and genomic (RNA and protein) information will allow early and more accurate prediction and diagnosis of disease and disease progression, and that medicine will become oriented towards disease prevention rather than efforts to cure people at late stages of illness. He points out that common genetic risk factors are being identified for a number of diseases including leprosy, asthma, cancer, diabetes, and cardiovascular disease, and that these new genomic tools will create a new approach to clinical practice [3]. Similar reviews point out the promise of molecular biomarkers in cancer [4], cardiovascular disease [5], treatment of CNS disorders [6], monitoring of infectious disease [7], assessment of environmental health in children [8], and even chronic fatigue syndrome [9] and psychiatry [10]. Clearly there is excitement and promise in the use of molecular biomarkers for diagnosis and treatment of disease.
2.1 Biomarkers and individualized medicine
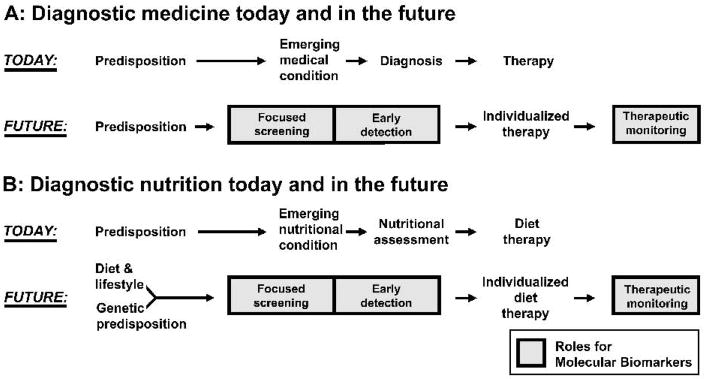
Bell [3] further points out that these new genomic tools will improve our ability to identify individuals at risk or in presymptomatic phases of disease, and will more precisely define disease subtypes on the basis of their individual pathophysiology and their responsiveness to therapy, including predictive diagnosis and risk profiling. Bell suggests (Fig. 1A) that treatment in medical practice today doesn’t begin until the patient visits the doctor with symptoms, often relatively late in the disease, and treatment often only alleviates symptoms and slows disease progression [3]. In contrast in the future, Bell foresees (Fig. 1A) that screening via molecular biomarkers for genetic predisposition will facilitate early detection, define individual disease subtypes based precisely on individual pathophysiology, and will inform treatment and responsiveness to therapy [3]. Treatment of HIV or hepatitis C are examples of this individualized treatment paradigm already in practice; chip-based diagnostics for predicting cytochrome P450 metabolism are also examples of biomarkers for monitoring therapy. In describing the search for new cardiovascular biomarkers, Gerszten & Wang describe successful new biomarkers “as improv(ing) the prediction of risk in an individual and hence contribut(ion) to personalized medicine [5].” Proposed future roles for molecular biomarkers in personalized medicine are even being extended to CNS disorders, where the traditional approach is described as focused on average patients and not on the outliers who are most likely to benefit from personalized prescription [6]. Clearly, the application of molecular biomarkers to individualized disease prediction and diagnosis is seen as an exciting area in medicine with high potential for success.
Fig. 1.
Impact of molecular biomarkers in medicine and in nutrition, leading to individualized therapy. (A) Diagram of diagnostic medicine today and in the future (adapted from [3] by permission from Macmillan Publishers Ltd (Nature)). (B) Diagram of diagnostic nutrition today and in the future. Boxes show key potential roles for molecular biomarkers to provide early detection of aberrant nutritional status due to genetics or diet/lifestyle, thus shaping individualized therapy and subsequent monitoring.
2.2 Discovery of orthogonal biomarkers
A second potentially important aspect of molecular biomarkers is the application of microarrays in discovery science to identify “orthogonal” (uncorrelated) biomarkers [5]. Conventional biomarkers have usually been identified as an extension of known pathways, whereas new emerging technologies can uncover and allow unbiased characterization of variations in genes and mRNA associated with disease conditions [5]. The identification of orthogonal biomarkers is likely to overcome limitations of current biomarkers which are often associated with the same pathways, such as in inflammation or cardiovascular disease. As a consequence, current biomarkers often only provide information that is correlated with what is already known about the disease. Furthermore, use of multiple conventional biomarkers provides diminishing diagnostic strength because they are all basically assessing the same pathways, whereas use of a much smaller number of orthogonal biomarkers, not associated with the same pathways or downstream conditions, would have considerably more diagnostic power [5]. Winegarden [4] in his Lancet essay on microarrays in cancer, states: “there are two major advantages to the use of microarrays to identify such molecular classifiers of cancer. First, microarrays allow a researcher to screen thousands of genes without a-priori knowledge about which genes might be involved. Second, with microarrays it is possible to identify a panel of genes, rather than a single gene, that when used together may be a more accurate and robust indicator of patients’ outcome [4]. Discovery studies using microarrays to examine the full set of transcripts thus are powerful tools to identify new biomarkers that may signal the status of distinct previously-unknown and/or previously un-linked pathways associated with the condition under study.
3. Molecular Biomarkers in Blood
One of the challenges of biomarker use is obtaining samples from the appropriate tissue. The Biomarkers Definitions Working Group [2] indicated that clinical endpoints or biomarkers are distinct characteristics of disease that reflect how a patient feels, functions or survives, whereas surrogate endpoints or biomarkers are a subset of biomarkers that substitute for a clinical endpoint, and that are expected to predict clinical benefit or harm. Blood is one of the obvious surrogate tissues that has been used since the onset of medicine.
3.1 Total whole blood RNA
Conventional wisdom a decade or more ago assumed that mammalian erythroid cells, lacking a nucleus, were devoid of mRNA and incapable of protein synthesis. Recent microarray and qRT-PCR studies on mRNA isolated from whole blood, however, indicate that as much as 70% of total RNA isolated from whole blood is hemoglobin mRNA in humans or rodents [11], demonstrating that erythroid cells are clearly the predominant source of RNA isolated from whole blood. The total RNA from human RBCs resembles typical eukaryotic RNA, with a 5S–80S sedimentation distribution similar to sedimentation distributions in avian (nucleated) erythrocytes; this RNA contains the standard 18S- and 28S-rRNA bands indicative of protein synthesis [12]. Furthermore, genes identified in total RNA include transcripts of genes encoding initiation, activation and regulation of transcription and translation, and include RNA-stabilizing factors and anti-apoptotic proteins, supporting nucleus-independent protein synthesis in erythrocytes [12]. Clearly total RNA isolated from whole blood is predominantly from erythroid cells and not the leukocyte fraction.
3.2 Blood molecular biomarkers in medicine
Genomics and transcriptomics investigators thus have begun to use blood as a “surrogate” tissue for biopsy and analysis. Studies using microarray analysis have shown that blood cells share more than 80% of the transcriptome with each of nine tissues studied (brain, colon, heart, kidney, liver, lung, prostate, spleen and stomach), and estimates are that the blood transcriptome contains 16,000–20,000 transcripts [13]. Other researchers have found, when looking at isolated total RNA from whole blood, liver, and spleen, that the total number of expressed transcripts in blood RNA was intermediate between liver (lowest) and spleen (highest), and that the concordance of gene expression between human and monkey was 73% for blood RNA [14]. A recent review cites more than 40 human disease conditions or risks that have been studied using gene-expression profiling on human blood cells [15], in specific conditions in the areas of health status, cancer, neurological disease, cardiovascular disease and autoimmune disease. The 2009 publication of a volume in Methods of Molecular Biology, entitled DNA and RNA Profiling in Human Blood [16], further illustrates the emerging focus on RNA from blood as a molecular biomarker.
For instance, total RNA from blood was used to define a gene panel (9 genes) to be used to detect pathological responses in chronic fatigue syndrome patients and for differential diagnosis of this syndrome [9]. Microarray analysis of whole blood RNA was used to develop a panel of molecular biomarkers for acetaminophen exposure in rats that predicted exposure 89–96% based on microarray bioinformatics analysis (molecular biomarkers) versus blinded analysis by board certified veterinary pathologists of 62% based on clinical chemistry, 78% using hematology, and 67–75% based on histopathology [17]. Collectively, these reports and reviews suggest that molecular biomarkers perhaps offer more potential than biochemical biomarkers to identify presymptomatic phases of disease, predict patient outcome, identify clinically-relevant patient subgroups, and increase understanding of disease mechanisms. This especially includes increased opportunities for individualized medicine [3–6].
3.3 Molecular biomarkers in population studies as well as clinical studies
The above discussion clearly indicates that biomarkers based on mRNA levels, especially in blood, are an emerging and exciting area of research in medicine. Initial uses are obviously going to be in clinical research as well as in basic research, and in diagnosis and treatment of patients. The advent of high-throughput technologies associated with these techniques, however, suggests that there will be far wider application, and that as these technologies evolve there will be increasing application for population studies and epidemiology studies. Ability to screen populations for presymptomatic phases of disease or to identify clinically-relevant patient subgroups [15] are but two obvious applications in population studies. The commercial availability of vacutainers already containing RNA-stabilizing solutions and associated kits for ready RNA isolation from whole blood (PAXgene, Qiagen) clearly illustrates this potential. Application to nutrition studies of all types is certainly in the near future as well.
3.4 Costs for molecular biomarker analyses
Currently, commercial full genome microarrays for humans or rodents cost around $500, and preparation of isolated RNA, hybridization, scanning, and analysis typically add about $400, resulting in a research cost of just under $1000 per sample. As these arrays typically now report the expression of over 47,000 transcripts for over 38,000 genes, the potential cost today is actually less than 3 cents per gene. Costs for qRT-PCR analyses in a research laboratory are about $3 per gene for reagents and supplies. For comparison, routine blood analysis for hemoglobin C-reactive protein, cell count and reticulocyte count in a clinical lab typically total $50–$100 or higher. Just as with advent of continuous-flow autoanalyzers for routine repetitive medical laboratory analyses forty years ago, mainstream clinical use of microarray analysis or qRT-PCR in the future will likely increase efficiency, reduce cost, and focus on predictive panels of biomarkers that will make these techniques a centerpiece of clinical laboratory practice in the future.
4. Blood Molecular Biomarkers in Nutrition
Development of molecular biomarkers for nutritional status based on blood, too, has potential application that parallels the opportunities in medicine, including providing solid data for individualized nutrition. One potential advantage of using total blood mRNA for nutritional assessment, due to the long lifespan of red cells (120 days for humans; 60 days for rodents), is that blood-based molecular biomarkers may effectively integrate nutrient status over the lifespan of red cells rather than reflecting recent nutrient intake. Few studies so far, however, have used molecular biology methods to determine nutrient requirements, especially for humans. Changes in white cell metallothionein mRNA were found to correlate with changes in zinc intake in human subjects [18], and microarray analysis found decreased levels of mRNA for a zinc influx transporter in women supplemented with zinc [19]. These studies included use of blood spots as the biopsy tissue [18], clearly illustrating the potential application in population studies as well as clinical nutrition research. To identify biomarkers for the health risk assessment of chronic low level exposure to cadmium, the gene expression profile of blood was determined on residents in a cadmium-polluted area and in a control population where whole blood was collected in Paxgene tubes for RNA isolation. Cadmium exposure significantly up-regulated 137 genes and down-regulated 80 genes, compared with the control group, leading to identification of 7 cadmium-responsive genes that were suggested as a biomarker panel for risk assessment [20]. These are but a few of the emerging studies that illustrate the potential use of molecular biomarkers in nutrition.
5. Molecular Biomarkers for Selenium Status and Requirements
Selenium (Se) deficiency results in dramatic decreases in glutathione peroxidase-1 (Gpx1) mRNA as well as Gpx1 activity in rats [21] and other species. This down-regulation of Gpx1 mRNA in Se-deficient animals is specific for selenoprotein mRNAs and occurs due to nonsense mediated decay [22,23]. We have now used liver Gpx1 mRNA to assess Se requirements in growing rats [24–26], showing that mRNA levels can be used as specific biomarkers for nutrient status. These biomarkers were especially effective in determining Se requirements in pregnant and lactating rats as it explained that the drop in Gpx1 activity in pregnancy was caused by a natural drop in Gpx1 mRNA that was unrelated to changes in Se status [27], illustrating the usefulness of this molecular biomarker. Recently, we conducted a mouse microarray study to screen the complete selenoprotein proteome (selenoproteome) for selenoprotein mRNAs that are significantly decreased in Se deficiency, confirmed this regulation using qRT-PCR, and discovered that several additional selenoprotein mRNAs (Selh, Sepw1) are also highly down-regulated in Se deficiency [28]. We have more recently extended this study to rats to characterize Se regulation of mRNA levels of all 24 rat selenoproteins over the range from Se-deficiency to super-nutritional Se levels (8X the requirement), and to determine minimum dietary Se requirements using these molecular biomarkers [29]. Fig. 2 shows the Se response curves for these three highly-regulated selenoprotein mRNAs, Gpx1, Selh, and Sepw1, in rat liver, illustrating the efficacy of using molecular biomarkers for Se status. The result is that the minimum dietary Se requirement in the growing rat is 0.06–0.07 μg Se/g based on these selenoprotein mRNA biomarkers as compared to 0.06 μg Se/g based on plasma glutathione peroxidase-3 (Gpx3) activity, 0.08 μg Se/g based on liver Se concentration, and 0.09 μg Se/g based on liver Gpx1 activity in these same rats [29].
Fig. 2.
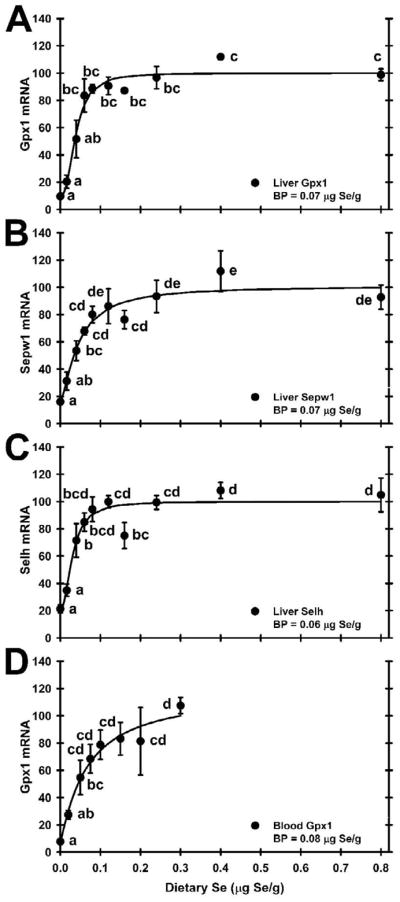
Relative levels of selenoprotein mRNA for liver Gpx1 (A), liver Sepw1 (B), liver Selh (C), and whole blood Gpx1 (D) in male weanling rats fed diets containing the indicated levels of Se for 28 days, as determined by qRT-PCR on total RNA isolated from rat liver (A–C, n=3/diet, as described in [29]), and as determined by ribonuclease protection analysis on total RNA isolated from rat blood (D, n=4/diet, as described in [31]). Values are means ± SEM. Data in each panel is significant (P < 0.0001) by ANOVA; values with a common letter are not significantly different (P < 0.05). The calculated plateau breakpoint (BP) for each response curve is also indicated.
5.1 Blood molecular biomarkers for selenium
We have now found that total RNA from rat whole blood can also be used to assess Se status [30], and that blood Gpx1 mRNA levels can be used to determine minimum dietary Se requirements in rats [31]. Fig. 2D shows that the minimum Se requirement based on Gpx1 mRNA in total RNA from rat blood is 0.08 μg Se/g diet. We have also begun to assess human Se status using blood selenoprotein mRNAs as biomarkers, but found that we could not distinguish between subjects consuming 48 μg Se/day verus 116 μg Se/day [32], presumably because both groups were on the plateau of the Se response curves for both biochemical biomarkers (plasma Gpx3 activity) and molecular biomarkers (blood selenoprotein mRNA levels) [32]. Studies in more Se-deficient populations will be necessary to evaluate the efficacy of using molecular biomarkers for assessment of human Se status.
5.2 Challenges in Se research where molecular biomarkers may help
Tissue levels of Se have long been used as chemical biomarkers to determine Se status and requirements in Se-deficient versus Se-adequate animals or humans [33]. Tissue levels with supernutritional Se supplementation, however, are far less indicative of level of Se exposure because organic (selenomethionine) and inorganic (selenite, selenate) forms are metabolized differently, incorporated differently into tissue selenoproteins and general-proteins [34], and even excreted in different proportions [35]. mRNAs for enzymes involved in these various pathways, such as synthesis of the urinary selenosugar (1-β-methylseleno-N-acetyl-D-galactosamine) under low Se conditions [36], or the methylation pathways for synthesis of methylated selenium forms with high Se intakes [37] might provide orthologous biomarkers for Se status. Microarrays are likely to detect additional unknown pathways that are also modulated by high Se.
The recent unblinding and stopping of the Se and Vitamin E Cancer Prevention Trial (SELECT) in October, 2008 [38], illustrates an additional challenge in the Se area where molecular biomarkers may prove to be powerful tools. A previous human trial (using selenized yeast) had indicated that higher intakes of Se are associated with prevention of prostate cancer [39]. The SELECT trial (using selenomethionine, the major Se component in selenized yeast), however, was stopped after 5 years because an independent monitoring committee found that Se and vitamin E, taken alone or together, did not prevent prostate cancer, and because it was unlikely Se and vitamin E supplementation would produce a 25% reduction in prostate cancer (the study goal) even if the trial continued to its planned endpoint. In addition, while not significant, there were slightly more cases of diabetes in men taking only Se, as well as slightly more cases of prostate cancer in men taking only vitamin E [38], matching with other recent studies indicating increased risk of type-2 diabetes [40] as well as cancer [41,42] with Se supplementation. Differences in initial Se status of the two populations, and differences in form of Se supplementation are both likely to play some role in the differences between the two trials. The identification of new molecular biomarkers of Se status, especially orthogonal biomarkers, has the potential to further discriminate between individuals that will respond positively to high-Se supplementation versus individuals that will be adversely affected.
The are many examples for other nutrients where new orthogonal biomarkers, identified by discovery science, might be able to distinguish between subjects that would benefit from nutrient supplementation versus be adversely affected, or that could be used effectively to confirm nutrient requirements in less-studied groups such as children, the elderly, or even certain disease states.
6. Nutrition Assessment in the Future
What will the future hold in the area of nutrient assessment using molecular biomarkers? The underlying hypothesis is that homeostatic mechanisms exist for each essential nutrient – element, vitamin, amino acid, essential fatty acid – that detect and maintain the concentration and/or body burden of these nutrients at relatively constant levels in healthy individuals, thus adjusting for concentrations of dietary nutrients that can vary over a much wider range. Nutritional assessment of an individual using molecular biomarkers could be conducted for all nutrients in one assay by using a panel of biomarkers for each nutrient, which in the future might span the full range from deficient to toxic. As quality molecular biomarkers are identified and validated for each nutrient, inclusion of probe sets on RNA microarrays would allow rapid and full assessment of nutrient status for an individual. Such molecular analysis (Fig. 1B) could provide early detection of aberrant nutritional conditions due to individual genetics, or to poor nutritional and/or lifestyle choices, in parallel with what has been suggested for how molecular biomarkers will impact medical diagnosis. Just as in medicine, application of molecular biomarkers to nutrition will provide opportunities for early diagnosis, individualized treatment, and therapeutic monitoring of nutritional disorders.
The currently-available Chicken Genome Array from Affymetrix illustrates in one small way how molecular biomarkers are already providing powerful diagnostic tools [43]. This microarray covers over 32,000 transcripts corresponding to over 28,000 genes, but it also contains 689 probe sets for detecting 17 avian viruses. Use of this microarray thus empowers a veterinarian or producer to rapidly diagnose viral infection in a flock. Imagine how useful and rapid a complete nutrition assessment microarray could have been when the FDA recently was asked to diagnose the nature of over 40 cases of adverse reaction to a health food supplement which was ultimately found to contain up to >700 times the US RDA (recommended dietary allowance) for Se and contain up to >130 times the US AI (adequate intake) for chromium in females (19–50 yr) [44,45].
There are many pitfalls in this scenario. The range of individual variation in homeostatic set-points for these hypothetical biomarkers is unknown, and certainly will be wider in human populations than in studies using standardized diets and animals with homogeneous genetics. For some nutrients, a molecular biomarker revealing the level of nutrient stores (eg. iron or vitamin A) may not exist. For others, the reliable biomarker may be a metabolite or hormone signal rather than an mRNA biomarker in blood RNA. None-the-less, just as in medicine, discovery science and emerging RNA technologies offer tremendous potential for nutrition as well as medicine to provide powerful molecular biomarkers.
Just as for biomarkers in other areas, the ethics of the research in nutritional molecular biomarkers and of the translation to public practice will need to be considered. Application of biomarkers to combating misdescription of food contents on product labels [46], for instance, has little downside, but study and application of biomarkers in human subjects is a completely different matter, as illustrated by recent discussion about the challenges in research and use in studying biomarkers in the environmental health of children [8] and use of biomarkers to predict human behavior and psychiatric disorders [10]. Important social, legal, and ethical issues, such as confidentiality, risk profiling, privacy and children’s rights, informed consent, intervention, labeling, and discrimination [8,10], will apply in nutrition as well as in medicine and other areas. Biomarker research in nutrition is needed to show the efficacy of molecular biomarker use and to realize the potential of early identification and individualized treatment, but these studies and subsequent application will need to be conducted under the scrutiny of institutional review boards to carefully to protect the rights of individuals and society. Focused panels of biomarkers, as indicated in Fig 1, may be the mechanism to provide diagnostic strength in assessing nutrient status without compromising other ethical considerations.
7. Conclusion
The 1998–2004 Institute of Medicine, while making recommendations for dietary nutrient intakes (both RDAs and tolerable upper intake levels(ULs)), consistently called for new biomarkers for both deficiency and toxicity for nearly all of the essential nutrients [47]. A recent review of the status of supplement use concluded that “present evidence is insufficient to recommend either for or against the use of MVMs [Multivitamins and Minerals] by the American public to prevent chronic disease” [48], but the panel further stated, “With the strong trends of increasing MVM and other dietary supplement consumption, and the increasing fortification of the U.S. diet, we are concerned that a growing proportion of the population maybe consuming levels considerably above the upper level, thus increasing the possibility of adverse effects.” Thus the MVM Supplements panel’s sixth suggestion was to capitalize “on the progressing state of biomedical science .. to identify important new biomarkers, early in the disease process… [48].” Clearly there is broad and high interest and need for biomarkers for both low and high nutrient levels. These biomarkers could have tremendous impact in both the US and world-wide.
Future discovery microarray studies have high potential to identify new molecular biomarkers, perhaps related to known pathways of metabolism but also related to orthogonal pathways completely dissociated from any known biological connection. Resulting panels of biomarkers may be able to discriminate between deficient, marginal, adequate, and supernutritional individuals and populations, and differentiate between individuals that will benefit versus be adversely affected by nutrient supplementation. Such mRNA biomarkers, including those in whole blood, will make powerful contributions to the study and practice of nutrition in the future.
Acknowledgments
Supported in part by the National Institutes of Health grant DK74184, and by the University of Wisconsin Agricultural Experiment Station grant WIS04909.
SUPPORT: Supported in part by the National Institutes of Health grant DK74184, and by the University of Wisconsin Agricultural Experiment Station grant WIS04909.
Abbreviations used
- AI
adequate intake
- Gpx1
glutathione peroxidase-1
- MVMs
multivitamins and minerals
- qRT-PCR
quantitative real time polymerase chain reaction
- RDA
recommended dietary allowance
- Se
selenium
- Selh
selenoprotein H
- Sepw1
selenoprotein W
- UL
tolerable upper intake level
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Merriam-Webster’s Medical Dictionary. [accessed 7/28/2009];[Internet] 2009 Available from: http://www2.merriam-webster.com/cgi-bin/mwmedsamp?book=Medical.
- 2.Biomarkers Definitions Working Group. Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther. 2001;69:89–95. doi: 10.1067/mcp.2001.113989. [DOI] [PubMed] [Google Scholar]
- 3.Bell J. Predicting disease using genomics. Nature. 2004;429:453–6. doi: 10.1038/nature02624. [DOI] [PubMed] [Google Scholar]
- 4.Winegarden N. Microarrays in cancer: moving from hype to clinical reality. Lancet. 2003;362:1428. doi: 10.1016/S0140-6736(03)14724-1. [DOI] [PubMed] [Google Scholar]
- 5.Gerszten RE, Wang TJ. The search for new cardiovascular biomarkers. Nature. 2008;451:949–52. doi: 10.1038/nature06802. [DOI] [PubMed] [Google Scholar]
- 6.de Leon J. Pharmacogenomics: the promise of personalized medicine for CNS disorders. Neuropsychopharmacology. 2009;34:159–72. doi: 10.1038/npp.2008.147. [DOI] [PubMed] [Google Scholar]
- 7.Mikhailovich V, Gryadunov D, Kolchinsky A, Makarov AA, Zasedatelev A. DNA microarrays in the clinic: infectious diseases. Bioessays. 2008;30:673–82. doi: 10.1002/bies.20781. [DOI] [PubMed] [Google Scholar]
- 8.Sly PD, Eskenazi B, Pronczuk J, Sram R, Diaz-Barriga F, Machin DG, Carpenter DO, Surdu S, Meslin EM. Ethical issues in measuring biomarkers in children’s environmental health. Environ Health Perspect. 2009;117:1185–90. doi: 10.1289/ehp.0800480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saiki T, Kawai T, Morita K, Ohta M, Saito T, Rokutan K, Ban N. Identification of marker genes for differential diagnosis of chronic fatigue syndrome. Mol Med. 2008;14:599–607. doi: 10.2119/2007-00059.Saiki. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh I, Rose N. Biomarkers in psychiatry. Nature. 2009;460:202–7. doi: 10.1038/460202a. [DOI] [PubMed] [Google Scholar]
- 11.Tian Z, Palmer N, Schmid P, Yao H, Galdzicki M, Berger B, Wu E, Kohane IS. A practical platform for blood biomarker study by using global gene expression profiling of peripheral whole blood. PLoS ONE. 2009;4:e5157. doi: 10.1371/journal.pone.0005157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kabanova S, Kleinbongard P, Volkmer J, Andree B, Kelm M, Jax TW. Gene expression analysis of human red blood cells. Int J Med Sci. 2009;6:156–9. doi: 10.7150/ijms.6.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liew CC, Ma J, Tang HC, Zheng R, Dempsey AA. The peripheral blood transcriptome dynamically reflects system wide biology: a potential diagnostic tool. J Lab Clin Med. 2006;147:126–32. doi: 10.1016/j.lab.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 14.Lu YR, Wang LN, Jin X, Chen YN, Cong C, Yuan Y, Li YC, Tang WD, Li HX, et al. A preliminary study on the feasibility of gene expression profile of rhesus monkey detected with human microarray. Transplant Proc. 2008;40:598–602. doi: 10.1016/j.transproceed.2008.01.029. [DOI] [PubMed] [Google Scholar]
- 15.Mohr S, Liew CC. The peripheral-blood transcriptome: new insights into disease and risk assessment. Trends Mol Med. 2007;13:422–32. doi: 10.1016/j.molmed.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 16.DNA and RNA Profiling in Human Blood. New York NY: Humana Press; 2009. [Google Scholar]
- 17.Bushel PR, Heinloth AN, Li J, Huang L, Chou JW, Boorman GA, Malarkey DE, Houle CD, Ward SM, et al. Blood gene expression signatures predict exposure levels. Proc Natl Acad Sci U S A. 2007;104:18211–6. doi: 10.1073/pnas.0706987104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao J, Cousins RJ. Metallothionein mRNA in monocytes and peripheral blood mononuclear cells and in cells from dried blood spots increases after zinc supplementation of men. J Nutr. 2000;130:2180–7. doi: 10.1093/jn/130.9.2180. [DOI] [PubMed] [Google Scholar]
- 19.Andree KB, Kim J, Kirschke CP, Gregg JP, Paik HY, Joung H, Woodhouse L, King JC, Huang L. Investigation of lymphocyte gene expression for use as biomarkers for zinc status in humans. J Nutr. 2004;134:1716–23. doi: 10.1093/jn/134.7.1716. [DOI] [PubMed] [Google Scholar]
- 20.Dakeshita S, Kawai T, Uemura H, Hiyoshi M, Oguma E, Horiguchi H, Kayama F, Aoshima K, Shirahama S, et al. Gene expression signatures in peripheral blood cells from Japanese women exposed to environmental cadmium. Toxicology. 2009;257:25–32. doi: 10.1016/j.tox.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 21.Saedi MS, Smith CG, Frampton J, Chambers I, Harrison PR, Sunde RA. Effect of selenium status on mRNA levels for glutathione peroxidase in rat liver. Biochem Biophys Res Commun. 1988;153:855–61. doi: 10.1016/s0006-291x(88)81174-4. [DOI] [PubMed] [Google Scholar]
- 22.Weiss SL, Sunde RA. Cis-acting elements are required for selenium regulation of glutathione peroxidase-1 mRNA levels. RNA. 1998;4:816–27. doi: 10.1017/s1355838298971990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moriarty PM, Reddy CC, Maquat LE. Selenium deficiency reduces the abundance of mRNA for Se-dependent glutathione peroxidase 1 by a UGA-dependent mechanism likely to be nonsense codon-mediated decay of cytoplasmic mRNA. Mol Cell Biol. 1998;18:2932–9. doi: 10.1128/mcb.18.5.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weiss SL, Evenson JK, Thompson KM, Sunde RA. Dietary selenium regulation of glutathione peroxidase mRNA and other selenium-dependent parameters in male rats. J Nutr Biochem. 1997;8:85–91. doi: 10.1016/S0955-2863(96)00178-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weiss SL, Evenson JK, Thompson KM, Sunde RA. The selenium requirement for glutathione peroxidase mRNA level is half of the selenium requirement for glutathione peroxidase activity in female rats. J Nutr. 1996;126:2260–7. doi: 10.1093/jn/126.9.2260. [DOI] [PubMed] [Google Scholar]
- 26.Lei XG, Evenson JK, Thompson KM, Sunde RA. Glutathione peroxidase and phospholipid hydroperoxide glutathione peroxidase are differentially regulated in rats by dietary selenium. J Nutr. 1995;125:1438–46. doi: 10.1093/jn/125.6.1438. [DOI] [PubMed] [Google Scholar]
- 27.Sunde RA, Evenson JK, Thompson KM, Sachdev SW. Dietary selenium requirements based on glutathione peroxidase-1 activity and mRNA levels and other selenium parameters are not increased by pregnancy and lactation in rats. J Nutr. 2005;135:2144–50. doi: 10.1093/jn/135.9.2144. [DOI] [PubMed] [Google Scholar]
- 28.Sunde RA, Raines AM, Barnes KM, Evenson JK. Selenium status highly-regulates selenoprotein mRNA levels for only a subset of the selenoproteins in the selenoproteome. Biosci Rep. 2009;29:329–38. doi: 10.1042/BSR20080146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barnes KM, Evenson JK, Raines AM, Sunde RA. Transcript analysis of the selenoproteome indicates that dietary selenium requirements in rats based on selenium-regulated selenoprotein mRNA levels are uniformly less than those based on glutathione peroxidase activity. J Nutr. 2009;139:199–206. doi: 10.3945/jn.108.098624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Evenson JK, Wheeler AD, Blake SM, Sunde RA. Selenoprotein mRNA is expressed in blood at levels comparable to major tissues in rats. J Nutr. 2004;134:2640–5. doi: 10.1093/jn/134.10.2640. [DOI] [PubMed] [Google Scholar]
- 31.Sunde RA, Thompson KM, Evenson JK, Thompson BM. Blood glutathione peroxidase-1 mRNA levels can be used as molecular biomarkers to determine selenium requirements in rats. Exp Biol Med. 2009;234:1271–1279. doi: 10.3181/0906-RM-182. [DOI] [PubMed] [Google Scholar]
- 32.Sunde RA, Paterson E, Evenson JK, Barnes KM, Lovegrove JA, Gordon MH. Longitudinal selenium status in healthy British adults: assessment using biochemical and molecular biomarkers. Br J Nutr. 2008;99:S37–S47. doi: 10.1017/S0007114508006831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Food and Nutrition Board. Selenium. Dietary Reference Intakes for Vitamin C, Vitamin E, Selenium and Carotenoids. Washington, D.C: National Academy Press; 2000. pp. 284–324. [PubMed] [Google Scholar]
- 34.Sunde RA. Selenium. In: Bowman BA, Russell RM, editors. Present Knowledge in Nutrition. 9. Washington, D.C: ILSI Press; 2006. pp. 480–97. [Google Scholar]
- 35.Burk RF, Norsworthy BK, Hill KE, Motley AK, Byrne DW. Effects of chemical form of selenium on plasma biomarkers in a high-dose human supplementation trial. Cancer Epidemiol Biomarkers Prev. 2006;15:804–10. doi: 10.1158/1055-9965.EPI-05-0950. [DOI] [PubMed] [Google Scholar]
- 36.Kobayashi Y, Ogra Y, Ishiwata K, Takayama H, Aimi N, Suzuki KT. Selenosugars are key and urinary metabolites for selenium excretion within the required to low-toxic range. Proc Natl Acad Sci. 2002;99:15932–6. doi: 10.1073/pnas.252610699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsieh HS, Ganther HE. Biosynthesis of dimethyl selenide from sodium selenite in rat liver and kidney cell-free systems. Biochim Biophys Acta. 1977;497:205–17. doi: 10.1016/0304-4165(77)90153-2. [DOI] [PubMed] [Google Scholar]
- 38.Lippman SM, Klein EA, Goodman PJ, Lucia MS, Thompson IM, Ford LG, Parnes HL, Minasian LM, Gaziano JM, et al. Effect of selenium and vitamin E on risk of prostate cancer and other cancers: the Selenium and Vitamin E Cancer Prevention Trial (SELECT) JAMA. 2009;301:39–51. doi: 10.1001/jama.2008.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clark LC, Combs GF, Turnbull BW, Slate EH, Chalker DK, Chow J, Davis LS, Glover RA, Graham GF, et al. Effects of selenium supplementation for cancer prevention in patients with carcinoma of the skin. JAMA. 1996;276:1957–63. [PubMed] [Google Scholar]
- 40.Stranges S, Marshall JR, Natarajan R, Donahue RP, Trevisan M, Combs GF, Cappuccio FP, Ceriello A, Reid ME. Effects of long-term selenium supplementation on the incidence of type 2 diabetes: a randomized trial. Ann Intern Med. 2007;147:217–23. doi: 10.7326/0003-4819-147-4-200708210-00175. [DOI] [PubMed] [Google Scholar]
- 41.Duffield-Lillico AJ, Slate EH, Reid ME, Turnbull BW, Wilkins PA, Combs GF, Jr, Park HK, Gross EG, Graham GF, et al. Selenium supplementation and secondary prevention of nonmelanoma skin cancer in a randomized trial. J Natl Cancer Inst. 2003;95:1477–81. doi: 10.1093/jnci/djg061. [DOI] [PubMed] [Google Scholar]
- 42.Duffield-Lillico AJ, Reid ME, Turnbull BW, Combs GF, Jr, Slate EH, Fischbach LA, Marshall JR, Clark LC. Baseline characteristics and the effect of selenium supplementation on cancer incidence in a randomized clinical trial: a summary report of the Nutritional Prevention of Cancer Trial. Cancer Epidemiol Biomarkers Prev. 2002;11:630–9. [PubMed] [Google Scholar]
- 43.Affymetrix. GeneChip chicken array. [Internet] 2009 Available from: http://www.affymetrix.com/products_services/arrays/specific/chicken.affx.
- 44.FDA. FDA finds hazardous levels of selenium in samples of “Total Body Formula” and “Total Body Mega Formula”. [Internet] 2008 April 9; [cited 2008 May 5]; Available from: http://www.fda.gov/bbs/topics/news/2008/new01818.html.
- 45.FDA. FDA completes final analysis of “Total Body Formula” and “Total Body Mega Formula” products. [Internet] 2008 May 1; Available from: http://www.fda.gov/bbs/topics/news/2008/new01831.html.
- 46.Woolfe M, Primrose S. Food forensics: using DNA technology to combat misdescription and fraud. Trends Biotechnol. 2004;22:222–6. doi: 10.1016/j.tibtech.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 47.Food and Nutrition Board. Dietary Reference Intakes for Vitamin C, Vitamin E, Selenium and Carotenoids. Washington, D.C: National Academy Press; 2000. [PubMed] [Google Scholar]
- 48.NIH State-of-the-Science Panel. National Institutes of Health state-of-the-science conference statement: multivitamin/mineral supplements and chronic disease prevention. Am J Clin Nutr. 2007;85:257S–64S. doi: 10.1093/ajcn/85.1.257S. [DOI] [PubMed] [Google Scholar]