Abstract
X-chromosome inactivation is a paradigmatic epigenetic phenomenon that results in the mitotically heritable transcriptional inactivation of one X-chromosome in female mammals, thereby equalizing X-linked gene dosage between the sexes. The epigenetic factors and mechanisms that execute X-inactivation overlap with those that regulate embryonic development and disease progression, thus offering a window into the epigenetic processes that regulate development and disease. Here I summarize some recent developments as well as open questions in X-inactivation research.
Cells retain their identity in part by inheriting gene expression profiles of their predecessors. Patterns of gene expression that are transmitted through cell division often propagate through epigenetic modifications of chromatin. Emerging evidence increasingly implicates this epigenetic mode of inheritance in a myriad of developmental processes as well as being a significant contributor to human diseases such as cancer. The epigenetic machinery offers a powerful mechanism to impact heritable changes in the expression of a wide variety of genes, both during normal development and in disease (Berdasco and Esteller, 2010; Sauvageau and Sauvageau, 2010; Surface et al., 2010).
Epigenetic regulation of gene expression is characterized by a number of defined covalent modifications of chromatin (Kouzarides, 2007). For example, methylation of cytosine residue of DNA and methylation of lysine 27 of histone H3 often mark transcriptionally silent genes; and, histone acetylation tags sites of active transcription. DNA and histone modifications are proposed to mediate the stable transmission of gene expression states through mitosis and cell division. Although many chromatin marks correlate with specific gene expression states, the factors and mechanisms that trigger epigenetic changes in gene expression are poorly defined (Bonasio et al., 2010).
X-chromosome inactivation offers a pliable model system to delineate factors and chromatin modifications that initiate epigenetic transcriptional changes (Payer and Lee, 2008). X-inactivation occurs in at least two distinct steps: initiation and maintenance. During the initiation phase, the prospective inactive X-chromosome undergoes epigenetic transcriptional inactivation. In the maintenance phase, replicated copies of the inactive X-chromosome are maintained inactive through multiple rounds of cell division. These two phases of X-inactivation occur during discrete stages of early mammalian embryogenesis and employ multiple and overlapping epigenetic mechanisms that can be molecularly dissected in the mouse (Heard and Disteche, 2006).
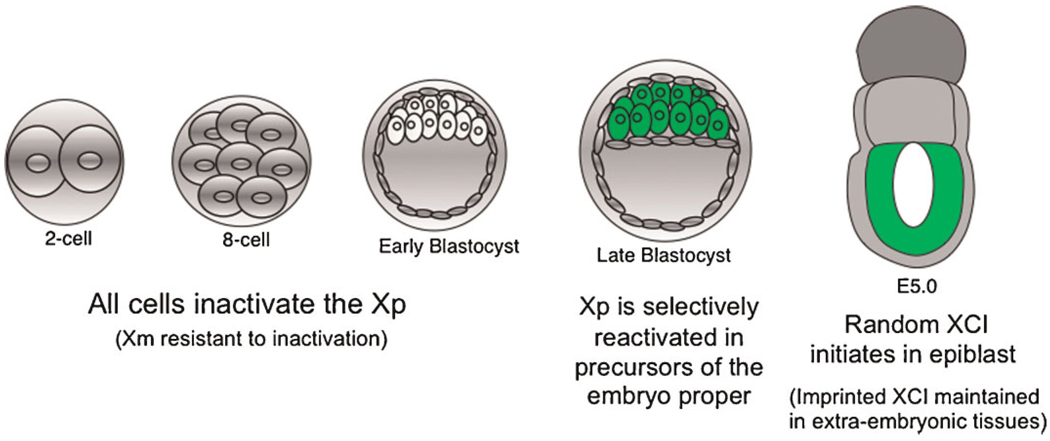
In the mouse, the pre-eminent X-inactivation model system, X-inactivation initiates early during embryogenesis when individual cells of the early female embryo undergo transcriptional silencing of genes along one of the two X-chromosomes (Kalantry et al., 2009; Patrat et al., 2009; Namekawa et al., 2010). Subsequently, with rare exceptions, the cellular epigenetic machinery ensures that replicated copies of the inactive and active X-chromosomes are maintained as such through mitosis and into descendant cells (Fig. 1). Initially, all cells of the pre-implantation-stage mouse embryo undergo exclusive inactivation of the paternal X-chromosome, in a process referred to as imprinted X-inactivation (Fig. 2; Kay et al., 1994). Later, at around the time of implantation, the inactive paternal-X selectively reactivates in the inner cell mass cells destined to form the fetus (Sheardown et al., 1997; Mak et al., 2004). These cells subsequently individually undergo random X-inactivation of either the maternal- or the paternal-X, thereby resulting in females being mosaic for X-linked gene expression (Monk and Harper, 1979; Rastan, 1982; McMahon et al., 1983). The remaining cells, all extra-embryonic in their fate (i.e., precursors of the placenta and the yolk-sac), maintain imprinted X-inactivation of the paternal-X (Takagi and Sasaki, 1975; West et al., 1977; Takagi, 1978).
Fig. 1.
The mosaic coat color of the calico cat exemplifies X-chromosome inactivation (Lyon, 1961). Calico cats are almost exclusively female and comprise of two colors of fur, black and orange, on an otherwise white background. The black and orange patches are due to mutually exclusive expression of two different alleles of the same X chromosome-linked gene. In fact, in the alternating patches, genes along only one of the two Xs are active while most genes on the other X-chromosome are inactive. The discreteness of the patches is due to clonal expansion of melanocytes whose precursors had inactivated one or the other X-chromosome during embryogenesis; the patches therefore reflect the epigenetic stability of X-inactivation.
Fig. 2.
Two forms of X-inactivation, imprinted and random, occur in the mouse embryo. During early pre-implantation development of the embryo, all cells undergo imprinted inactivation of the paternal X-chromosome. During the late blastocyststage, however, the cells of the inner cell mass (ICM; in green) reactivate the Xp. These cells contain the precursors of the embryo proper or the fetus. The cells in gray comprise either to the trophectoderm or the yolk-sac, which give rise to the placenta or the yolk-sac, respectively. After the embryo implants, the ICM descendants randomly inactivate either the maternal- or the paternal-X. The remaining cells, all extra-embryonic, maintain imprinted X-inactivation of the paternal X-chromosome.
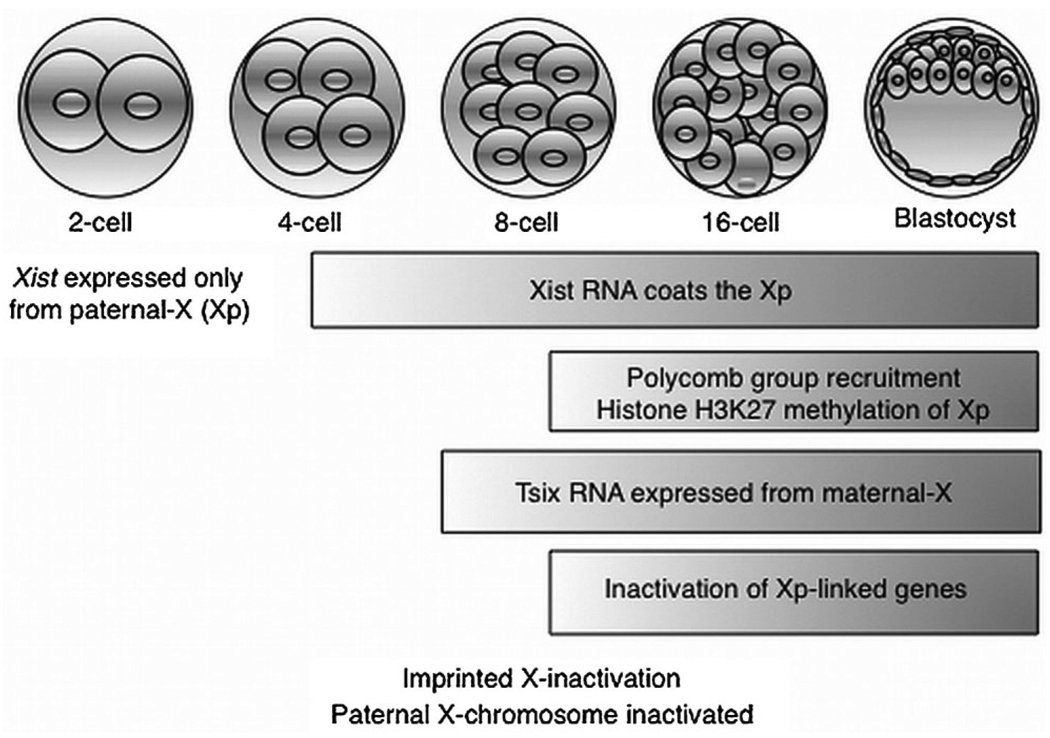
X-inactivation occurs gradually over several rounds of cleavage divisions of the zygote and correlates with an ordered series of epigenetic events, thus providing a window to explore the mechanisms that initiate and propagate gene silencing (Fig. 3). The two Xs in females are initially distinguished by the expression exclusively from the inactive-X of the long non-coding RNA Xist (X-inactive specific transcript) (Brown et al., 1991). Xist upregulation, in fact, presages transcriptional silencing of genes along the prospective inactive X-chromosome. Quite unusually and coincident with silencing of genes along the inactive X-chromosome, Xist RNA physically coats the X-chromosome from which its expression is induced (Brown et al., 1992; Clemson et al., 1996). Xist RNA accumulation in cis on the inactive X-elect is thought to result in recruitment of protein complexes, such as the Polycomb group, whose actions are posited to alter the structure of the chromosome, packaging the DNA into a transcriptionally inert heterochromatin (Fig. 3). While the inactive X-chromosome is distinguished by Xist RNA expression, the active-X is characterized by the expression of the Xist anti-sense long non-coding transcript Tsix (Lee et al., 1999). Tsix expression is thought to forestall expression of Xist RNA, thereby preventing inactivation of the active-X (Stavropoulos et al., 2001; Navarro et al., 2005; Ohhata et al., 2008).
Fig. 3.
Dynamics of X-chromosome inactivation in the early mouse embryo. All cells initially undergo imprinted X-inactivation resulting in silencing of genes exclusively along the paternalX-chromosome. A hallmark of X-inactivation is the expression of Xist RNA, which is only expressed from the paternal-X at the stages shown. Soon after Xist is transcribed, it starts to physically coat the paternal-X, followed closely by accumulation of proteins such as the Polycomb group and associated histone modifications that are posited to bring about silencing of paternal X-linked genes. The active maternal X-chromosome expresses the Xist anti-sense RNA T six, which is proposed to counter inactivation of the maternal-X by suppressing Xist expression.
Both imprinted and random X-inactivation share many of the features that characterize the inactive X-chromosome. Most notably, during both random and imprinted X-inactivation, Xist RNA and the Polycomb group proteins decorate the inactive-X early during X-inactivation and Tsix transcription is an early hallmark of the active-X. The importance of these factors is further underscored by their absolute requirement during X-inactivation as evidenced by loss-of-function mutations. A defect in Xist RNA and components of the Polycomb repressive complex 2 (PRC2) renders the inactive-X prone to being active (Marahrens et al., 1997; Wang et al., 2001; Kalantry et al., 2006; Kalantry et al., 2009); and, vice versa, Tsix-deficiency results in inactivation of the normally active X-chromosome (Lee, 2000; Sado et al., 2001).
Combined with the phenotypes in mouse and cell culture models, the early localization of Xist RNA and the Polycomb group on the inactive-X have led to the widely held belief that these factors must function during the initiation phase of X-inactivation. In contrast to this widely accepted role for Xist and the Polycomb group, however, several lines of evidence suggest that both gene products are dispensable for chromosome-wide silencing of X-linked genes during the initiation phase of X-inactivation. For example, cells of female embryos inheriting a null mutation in Xist on the paternal X-chromosome nevertheless transiently initiate silencing of many paternal X-linked genes during imprinted X-inactivation (Kalantry et al., 2009). These data highlight a locus-by-locus variability in Xist RNA-dependent initiation of imprinted X-linked gene silencing and imply the existence of as-yet-undiscovered cis-limited triggers of X-linked gene silencing. While imprinted X-linked gene silencing can initiate in the absence of Xist RNA, it is not stable. Xist is absolutely required to solidify or stabilize the inactive state during imprinted X-inactivation (Marahrens et al., 1997; Kalantry et al., 2009).
The primacy of Xist in mediating chromosome-wide transcriptional inactivation is also inferred by the effects of ectopically expressed Xist transgenes. While multi-copy Xist transgenes can result in a variable degree of silencing of neighboring sequences, single-copy Xist expressing transgenes do not induce silencing in their vicinity (Heard et al., 1999). Clearly, Xist influences the probability of a sequence to be transcriptionally silenced; but, this effect appears to be dependent on local chromatin context (Chow et al., 2010; Tang et al., 2010).
Xist-independent imprinted X-inactivation appears to be the ancestral form of X-inactivation. Marsupial mammals, which shared a common ancestor with placental mammals about 150 Mya, display imprinted X-inactivation in all their cell types and also lack Xist (Davidow et al., 2007; Hore et al., 2007; Shevchenko et al., 2007). Cis-acting elements other than Xist must therefore mediate imprinted X-inactivation in marsupials. Evolutionarily, the appearance of Xist coincides with the divergence of eutherians and marsupial mammals and the origins of random X-inactivation that is unique to eutherians (Wallis et al., 2007). X-linked gene silencing during random X-inactivation is preceded by a choice step whereby the cellular machinery targets one of the two Xs to undergo inactivation. Xist presence on an X-chromosome is absolutely necessary for that X to be chosen to be inactivated. If an X-chromosome lacks Xist, the other X-chromosome is invariably chosen for inactivation (Marahrens et al., 1998). Formally, this result does not necessarily support (or oppose, for that matter) a role for Xist in initiating X-linked gene silencing: the mutant X is preempted from attempting to initiate silencing because it fails to first be chosen to be inactivated. Whether Xist triggers X-linked gene silencing during random X-inactivation, therefore, requires the generation of homozygous Xist-mutant embryos, where both Xs now have an equal chance of being chosen for inactivation.
That X-linked gene silencing is subject to regional regulation, as opposed strictly to a central element such as Xist, is also consistent with the notion that X-linked genes became subject to dosage compensation gradually. X-inactivation is thought to be a by-product of sexual differentiation of a pair of homologous chromosomes into an X- and a Y-chromosome (Jegalian and Page, 1998). Due to a declining ability of the Y to recombine with the X, it has progressively degenerated. The gradual loss of genes from the proto-Y during X–Y differentiation is believed to have driven silencing of homologous X-linked genes in females in a piecemeal fashion (Jegalian and Page, 1998; Lahn and Page, 1999). If Xist RNA is the sole trigger of imprinted X-inactivation, the expectation would be that X-chromosomal genes would have to await its arrival before undergoing dosage compensation. This idea is clearly inconsistent with evidence suggesting that Xist has originated subsequent to the loss of Y-linked genes and hence the proposed advent of dosage compensation resulting in silencing of X-linked genes (Lahn and Page, 1999).
Xist RNA expression and accumulation on the presumptive inactive-X precedes the enrichment of Polycomb group proteins on the inactive-X (Okamoto et al., 2004). As detailed in the review by Trask and Mager in this issue, Polycomb group genes encode an evolutionarily conserved set of proteins that mediate cellular memory in diverse settings, including during X-chromosome inactivation. Female embryos and cells lacking the essential Polycomb group protein EED, resulting in a complete absence of histone methylation mediated by the PRC2 Polycomb complex, harbor defects in imprinted X-inactivation (Wang et al., 2001; Kalantry et al., 2006). These Eed−/− embryos nevertheless initiate imprinted X-inactivation; the defect is restricted to the maintenance phase of imprinted X-inactivation (Kalantry et al., 2006). During random X-inactivation, PRC2 appears to be dispensable for both the initiation as well as the maintenance phases of X-inactivation (Wang et al., 2001; Kalantry and Magnuson, 2006). It remains to be determined whether in the absence of PRC2, the sister PRC1 complex can compensate and enact X-inactivation (Schoeftner et al., 2006). These data nevertheless underscore both the differences in the epigenetic machinery required to mediate imprinted versus random X-inactivation as well as the need to definitively delineate epigenetic factors that function in trans to initiate and maintain X-inactivation.
Xist RNA expression is countered by transcription of the Xist anti-sense RNA Tsix. In fact, the exclusive function of Tsix is to repress Xist (Payer and Lee, 2008). The act of Tsix transcription modulates Xist chromatin and thereby is thought to repress Xist expression (Stavropoulos et al., 2001; Navarro et al., 2005; Sado et al., 2005). During random X-inactivation, Tsix functions in a complementary manner to Xist to mediate the choice of an X-chromosome to be inactivated. A Tsix-mutant X-chromosome is invariably chosen for inactivation, coinciding with Xist RNA being more readily induced from that X-chromosome (Lee and Lu, 1999; Luikenhuis et al., 2001; Sado et al., 2001; Kalantry and Magnuson, 2006). During imprinted X-inactivation, Tsix is expressed from the active maternal-X (Lee and Lu, 1999; Sado et al., 2001); and, its absence on the maternal-X predisposes the maternal-X to becoming inactivated (Lee, 2000; Sado et al., 2001). Tsix has therefore been postulated to be a component of the germline mark established in the oocyte that prevents inactivation of the maternal-X in the resultant embryo (Lee, 2000). However, if Xist is not absolutely required to initiate chromosome-wide silencing of X-linked genes during imprinted X-inactivation, then the role of Tsix in preventing imprinted X-inactivation is brought into question. If Xist and Tsix function in a mutually exclusive manner to establish the epigenotype of the inactive paternal- and the active maternal-X chromosomes during imprinted X-inactivation, then it is reasonable to expect that a paternally inherited Xist and maternally inherited Tsix mutation would fully complement each other. Yet, this appears to not be the case (Sado et al., 2001). This finding therefore implies that a Tsix-independent mechanism may serve to establish the maternal epigenotype in the oocyte.
The mechanism by which a cell counts the number of X-chromosomes has long fascinated X-inactivation enthusiasts. Before a cell can commence the process of inactivating an X, it must first count the number of X-chromosomes it harbors. Only if the number of X-chromosomes exceeds one per diploid set of autosomes will stable X-inactivation occur (Avner and Heard, 2001). This equation is especially true during random X-inactivation and may also apply, albeit perhaps less stringently, during imprinted X-inactivation (Goto and Takagi, 2000; Okamoto et al., 2000). One tempting hypothesis proposed to explain counting is that the two Xs sense the existence of each other by physically pairing prior to the triggering of X-inactivation (Marahrens, 1999). Since such pairing would only occur in XX females, XY males would be immune from inactivation. The X-inactivation center—a region of the X-chromosome containing Xist, Tsix as well as several hundred kilobases of sequence on either side—is thought to be the segment that mediates counting (Clerc and Avner, 2003). Although this hypothesis is quite attractive and is supported by several recent reports describing the physical pairing of the two Xs via elements in the X-inactivation center, there are a number of observations to the contrary (Bacher et al., 2006; Augui et al., 2007; Xu et al., 2007; Monkhorst et al., 2008; Jonkers et al., 2009). The primary counterpoint is that deletions encompassing a large region of the X-inactivation center, including Xist and Tsix, nevertheless result in proper inactivation of the wild-type X-chromosome (Monkhorst et al., 2008). Moreover, evidence implicating X-inactivation center segments in X-chromosome pairing is not consistently replicated (Jonkers et al., 2009).
In light of the discord in the pairing data, very recently and quite excitingly the protein encoded by the X-linked Rnf12 gene has been put forth as a factor that senses the number of X-chromosomes and triggers Xist expression in a dose-dependent manner during random X-inactivation (Jonkers et al., 2009). This discovery would also potentially explain why X-inactivation occurs in XX female but not in XY male cells. Since Rnf12 is X-linked, the two active-Xs in female cells would express twice as much RNF12 protein prior to X-inactivation compared to XY male cells. The increased RNF12 levels would then induce X-inactivation only in female and not in male cells; RNF12 would therefore serve to “count” the number of X-chromosomes. Indeed, overexpression of RNF12 in male embryonic stem (ES) cells—an in vitro system to model random X-inactivation—leads to ectopic Xist expression from the single X-chromosome in a percentage of the transfected male cells (Jonkers et al., 2009). In the converse, loss-of-function experiment, the reduction in the level of RNF12 in female cells to that found in male cells by deleting one copy of Rnf12 the female ES cells, however, did not result in the expected absence of X-inactivation (Jonkers et al., 2009). Thus, while RNF12 appears to be sufficient to trigger X-inactivation in vitro, it may not be necessary.
In a separate study, Shin et al. (2010) tested the in vivo requirement of Rnf12 in X-inactivation by generating mice harboring a conditionally mutant Rnf12 allele. Consistent with the previous study, cells that would normally undergo random X-inactivation commenced X-inactivation when devoid of or deficient in Rnf12 (albeit with slight differences compared to wild-type samples). However, upon deletion of both alleles of Rnf12 in mouse oocytes the authors strikingly discovered defects in Xist RNA induction and X-linked gene silencing from the paternal X-chromosome in the resulting early female embryos; this defect coincided with lethality of the mutant embryos (Shin et al., 2010). Thus, while random X-inactivation can occur in the absence of RNF12, imprinted X-inactivation requires RNF12. Moreover, the authors suggest that this RNF12 function is oocyte-derived (Shin et al., 2010). However, animals generated from oocytes where one but not both Rnf12 alleles is subject to deletion argue against this conclusion. While female embryos derived from these oocytes that inherit a mutant Rnf12 allele perish in utero, presumably due to a defect in imprinted X-inactivation, females that inherit the wild-type Rnf12 allele are born at expected frequencies. If in fact maternally deposited RNF12 present in the oocyte drives imprinted X-inactivation in the early embryo, then its deficiency should impact embryos that inherit either the mutant or wild-type Rnf12 allele from heterozygous oocytes equally. One explanation for this seeming discrepancy may be that zygotically active maternal Rnf12 allele, perhaps in addition to oocyte-derived RNF12 protein is necessary for proper imprinted X-inactivation.
Formally, though, neither of the two aforementioned studies necessarily reveals a role for RNF12 in the cellular mechanism that counts the number of X-chromosomes. For example, parthenogenetic embryos harboring two maternal-Xs and no paternal X do not typically undergo X-inactivation (Matsui et al., 2001); presumably, these embryos harbor similar levels of RNF12 compared to wild-type female embryos yet neither X is inactivated. The Rnf12 findings by Shin et al. (2010) highlight that the maternal X-chromosome in the early embryo is immune from X-inactivation. The answer to how RNF12 selectively targets the paternal-X and avoids the maternal X-chromosome for Xist expression may impinge on the nature of the germline mark placed on the maternal-X during oogenesis that forestalls its inactivation in the early embryo (Tada et al., 2000).
Since the discovery of Xist, the field of X-inactivation has progressed rapidly. By harnessing the power of newer technologies, including (but not limited to) mouse mutagenesis, genome sequencing efforts, improved techniques to detect X-linked gene expression, and high-resolution microscopy, many groups have contributed to the molecular dissection of the epigenetic machinery that controls how a female cell stably inactivates one of the two X-chromosomes. These findings have broad implications, as the molecular themes that operate during X-inactivation apply to autosomal gene regulation during development and are increasingly being found to go awry during disease progression. Insights into the inactivation of an X-chromosome therefore promise to provide a broader understanding of normal and abnormal epigenetic regulation of gene expression. Despite the many advances in X-inactivation in the past two decades, however, some key questions remain to be answered. The coming years promise to provide a stream of exciting new findings in X-inactivation.
Acknowledgments
I thank the members of my laboratory, including Clair Harris, Emily Buttiegieg, & Miguel Pedraza, for constructive discussions. I apologize to the authors whose work may not have been referenced in this mini-review.
Literature Cited
- Augui S, Filion GJ, Huart S, Nora E, Guggiari M, Maresca M, Stewart AF, Heard E. Sensing X chromosome pairs before X inactivation via a novel X-pairing region of the Xic. Science. 2007;318:1632–1636. doi: 10.1126/science.1149420. [DOI] [PubMed] [Google Scholar]
- Avner P, Heard E. X-chromosome inactivation: Counting, choice and initiation. Nat Rev Genet. 2001;2:59–67. doi: 10.1038/35047580. [DOI] [PubMed] [Google Scholar]
- Bacher CP, Guggiari M, Brors B, Augui S, Clerc P, Avner P, Eils R, Heard E. Transient colocalization of X-inactivation centres accompanies the initiation of X inactivation. Nat Cell Biol. 2006;8:293–299. doi: 10.1038/ncb1365. [DOI] [PubMed] [Google Scholar]
- Berdasco M, Esteller M. Aberrant epigenetic landscape in cancer: How cellular identity goes awry. Dev Cell. 2010;19:698–711. doi: 10.1016/j.devcel.2010.10.005. [DOI] [PubMed] [Google Scholar]
- Bonasio R, Tu S, Reinberg D. Molecular signals of epigenetic states. Science. 2010;330:612–616. doi: 10.1126/science.1191078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, Willard HF. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Hendrich BD, Rupert JL, Lafreniere RG, Xing Y, Lawrence J, Willard HF. The human XIST gene: Analysis of a 17kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- Chow JC, Ciaudo C, Fazzari MJ, Mise N, Servant N, Glass JL, Attreed M, Avner P, Wutz A, Barillot E, Greally JM, Voinnet O, Heard E. LINE-1 activity in facultative heterochromatin formation during X chromosome inactivation. Cell. 2010;141:956–969. doi: 10.1016/j.cell.2010.04.042. [DOI] [PubMed] [Google Scholar]
- Clemson CM, McNeil JA, Willard HF, Lawrence JB. XIST RNA paints the inactive X chromosome at interphase: Evidence for a novel RNA involved in nuclear/chromosome structure. J Cell Biol. 1996;132:259–275. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerc P, Avner P. Multiple elements within the Xic regulate random X inactivation in mice. Semin Cell Dev Biol. 2003;14:85–92. doi: 10.1016/s1084-9521(02)00140-4. [DOI] [PubMed] [Google Scholar]
- Davidow LS, Breen M, Duke SE, Samollow PB, McCarrey JR, Lee JT. The search for a marsupial XIC reveals a break with vertebrate synteny. Chromosome Res. 2007;15:137–146. doi: 10.1007/s10577-007-1121-6. [DOI] [PubMed] [Google Scholar]
- Goto Y, Takagi N. Maternally inherited X chromosome is not inactivated in mouse blastocysts due to parental imprinting. Chromosome Res. 2000;8:101–109. doi: 10.1023/a:1009234217981. [DOI] [PubMed] [Google Scholar]
- Heard E, Disteche CM. Dosage compensation inmammals: Fine-tuning the expression of the X chromosome. Genes Dev. 2006;20:1848–1867. doi: 10.1101/gad.1422906. [DOI] [PubMed] [Google Scholar]
- Heard E, Mongelard F, Arnaud D, Avner P. Xist yeast artificial chromosome transgenes function asX-inactivation centers only in multicopy arrays and not as single copies. Mol Cell Biol. 1999;19:3156–3166. doi: 10.1128/mcb.19.4.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hore TA, Koina E, Wakefield MJ, Marshall Graves JA. The region homologous to the X-chromosome inactivation centre has been disrupted in marsupial and monotreme mammals. Chromosome Res. 2007;15:147–161. doi: 10.1007/s10577-007-1119-0. [DOI] [PubMed] [Google Scholar]
- Jegalian K, Page DC. A proposed path by which genes common to mammalian X and Y chromosomes evolve to become X inactivated. Nature. 1998;394:776–780. doi: 10.1038/29522. [DOI] [PubMed] [Google Scholar]
- Jonkers I, Barakat TS, Achame EM, Monkhorst K, Kenter A, Rentmeester E, Grosveld F, Grootegoed JA, Gribnau J. RNF12 is an X-encoded dose-dependent activator of X chromosome inactivation. Cell. 2009;139:999–1011. doi: 10.1016/j.cell.2009.10.034. [DOI] [PubMed] [Google Scholar]
- Kalantry S, Magnuson T. The Polycomb group protein EED is dispensable for the initiation of random X-chromosome inactivation. PLoS Genet. 2006;2:e66. doi: 10.1371/journal.pgen.0020066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalantry S, Mills KC, Yee D, Otte AP, Panning B, Magnuson T. The Polycomb group protein Eed protects the inactive X-chromosome from differentiation-induced reactivation. Nat Cell Biol. 2006;8:195–202. doi: 10.1038/ncb1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalantry S, Purushothaman S, Bowen RB, Starmer J, Magnuson T. Evidence of Xist RNA-independent initiation of mouse imprinted X-chromosome inactivation. Nature. 2009;460:647–651. doi: 10.1038/nature08161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Lahn BT, Page DC. Four evolutionary strata on the human X chromosome. Science. 1999;286:964–967. doi: 10.1126/science.286.5441.964. [DOI] [PubMed] [Google Scholar]
- Lee JT. Disruption of imprinted X inactivation by parent-of-origin effects at Tsix. Cell. 2000;103:17–27. doi: 10.1016/s0092-8674(00)00101-x. [DOI] [PubMed] [Google Scholar]
- Lee JT, Lu N. Targeted mutagenesis of Tsix leads to nonrandom X inactivation. Cell. 1999;99:47–57. doi: 10.1016/s0092-8674(00)80061-6. [DOI] [PubMed] [Google Scholar]
- Lee JT, Davidow LS, Warshawsky D. Tsix, a gene antisense to Xist at theX-inactivation centre. Nat Genet. 1999;21:400–404. doi: 10.1038/7734. [DOI] [PubMed] [Google Scholar]
- Luikenhuis S, Wutz A, Jaenisch R. Antisense transcription through the Xist locus mediates Tsix function in embryonic stem cells. Mol Cell Biol. 2001;21:8512–8520. doi: 10.1128/MCB.21.24.8512-8520.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L.) Naturwissenschaften. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- Mak W, Nesterova TB, de Napoles M, Appanah R, Yamanaka S, Otte AP, Brockdorff N. Reactivation of the paternal X chromosome in early mouse embryos. Science. 2004;303:666–669. doi: 10.1126/science.1092674. [DOI] [PubMed] [Google Scholar]
- Marahrens Y. X-inactivation by chromosomal pairing events. Genes Dev. 1999;13:2624–2632. doi: 10.1101/gad.13.20.2624. [DOI] [PubMed] [Google Scholar]
- Marahrens Y, Panning B, Dausman J, Strauss W, Jaenisch R. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 1997;11:156–166. doi: 10.1101/gad.11.2.156. [DOI] [PubMed] [Google Scholar]
- Marahrens Y, Loring J, Jaenisch R. Role of the Xist gene in X chromosome choosing. Cell. 1998;92:657–664. doi: 10.1016/s0092-8674(00)81133-2. [DOI] [PubMed] [Google Scholar]
- Matsui J, Goto Y, Takagi N. Control of Xist expression for imprinted and random X chromosome inactivation in mice. Hum Mol Genet. 2001;10:1393–1401. doi: 10.1093/hmg/10.13.1393. [DOI] [PubMed] [Google Scholar]
- McMahon A, Fosten M, Monk M. X-chromosome inactivation mosaicism in the three germ layers and the germ line of the mouse embryo. J Embryol Exp Morphol. 1983;74:207–220. [PubMed] [Google Scholar]
- Monk M, Harper MI. Sequential X chromosome inactivation coupled with cellular differentiation in early mouse embryos. Nature. 1979;281:311–313. doi: 10.1038/281311a0. [DOI] [PubMed] [Google Scholar]
- Monkhorst K, Jonkers I, Rentmeester E, Grosveld F, Gribnau J. X inactivation counting and choice is a stochastic process: Evidence for involvement of an X-linked activator. Cell. 2008;132:410–421. doi: 10.1016/j.cell.2007.12.036. [DOI] [PubMed] [Google Scholar]
- Namekawa SH, Payer B, Huynh KD, Jaenisch R, Lee JT. Two-step imprinted X inactivation: Repeat versus genic silencing in the mouse. Mol Cell Biol. 2010;30:3187–3205. doi: 10.1128/MCB.00227-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro P, Pichard S, Ciaudo C, Avner P, Rougeulle C. Tsix transcription across the Xist gene alters chromatin conformation without affecting Xist transcription: Implications for X-chromosome inactivation. Genes Dev. 2005;19:1474–1484. doi: 10.1101/gad.341105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohhata T, Hoki Y, Sasaki H, Sado T. Crucial role of antisense transcription across the Xist promoter in Tsix-mediated Xist chromatin modification. Development. 2008;135:227–235. doi: 10.1242/dev.008490. [DOI] [PubMed] [Google Scholar]
- Okamoto I, Tan S, Takagi N. X-chromosome inactivation in XX androgenetic mouse embryos surviving implantation. Development. 2000;127:4137–4145. doi: 10.1242/dev.127.19.4137. [DOI] [PubMed] [Google Scholar]
- Okamoto I, Otte AP, Allis CD, Reinberg D, Heard E. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science. 2004;303:644–649. doi: 10.1126/science.1092727. [DOI] [PubMed] [Google Scholar]
- Patrat C, Okamoto I, Diabangouaya P, Vialon V, Le Baccon P, Chow J, Heard E. Dynamic changes in paternal X-chromosome activity during imprinted X-chromosome inactivation in mice. Proc Natl Acad Sci USA. 2009;106:5198–5203. doi: 10.1073/pnas.0810683106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payer B, Lee JT. X chromosome dosage compensation: How mammals keep the balance. Annu Rev Genet. 2008;42:733–772. doi: 10.1146/annurev.genet.42.110807.091711. [DOI] [PubMed] [Google Scholar]
- Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, de la Cruz CC, Otte AP, Panning B, Zhang Y. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- Rastan S. Timing of X-chromosome inactivation in postimplantation mouse embryos. J Embryol Exp Morphol. 1982;71:11–24. [PubMed] [Google Scholar]
- Sado T, Wang Z, Sasaki H, Li E. Regulation of imprinted X-chromosome inactivation in mice by Tsix. Development. 2001;128:1275–1286. doi: 10.1242/dev.128.8.1275. [DOI] [PubMed] [Google Scholar]
- Sado T, Hoki Y, Sasaki H. Tsix silences Xist through modification of chromatin structure. Dev Cell. 2005;9:159–165. doi: 10.1016/j.devcel.2005.05.015. [DOI] [PubMed] [Google Scholar]
- Sauvageau M, Sauvageau G. Polycomb group proteins: Multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell. 2010;7:299–313. doi: 10.1016/j.stem.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeftner S, Sengupta AK, Kubicek S, Mechtler K, Spahn L, Koseki H, Jenuwein T, Wutz A. Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. EMBO J. 2006;25:3110–3122. doi: 10.1038/sj.emboj.7601187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheardown SA, Duthie SM, Johnston CM, Newall AE, Formstone EJ, Arkell RM, Nesterova TB, Alghisi GC, Rastan S, Brockdorff N. Stabilization of Xist RNA mediates initiation of X chromosome inactivation. Cell. 1997;91:99–107. doi: 10.1016/s0092-8674(01)80012-x. [DOI] [PubMed] [Google Scholar]
- Shevchenko AI, Zakharova IS, Elisaphenko EA, Kolesnikov NN, Whitehead S, Bird C, Ross M, Weidman JR, Jirtle RL, Karamysheva TV, Rubtsov NB, VandeBerg JL, Mazurok NA, Nesterova TB, Brockdorff N, Zakian SM. Genes flanking Xist in mouse and human are separated on the X chromosome in American marsupials. Chromosome Res. 2007;15:127–136. doi: 10.1007/s10577-006-1115-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Bossenz M, Chung Y, Ma H, Byron M, Taniguchi-Ishigaki N, Zhu X, Jiao B, Hall LL, Green MR, Jones SN, Hermans-Borgmeyer I, Lawrence JB, Bach I. Maternal Rnf12/RLIM is required for imprinted X-chromosome inactivation in mice. Nature. 2010;467:977–981. doi: 10.1038/nature09457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J, Mak W, Zvetkova I, Appanah R, Nesterova TB, Webster Z, Peters AH, Jenuwein T, Otte AP, Brockdorff N. Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Dev Cell. 2003;4:481–495. doi: 10.1016/s1534-5807(03)00068-6. [DOI] [PubMed] [Google Scholar]
- Stavropoulos N, Lu N, Lee JT. A functional role for Tsix transcription in blocking Xist RNA accumulation but not in X-chromosome choice. Proc Natl Acad Sci USA. 2001;98:10232–10237. doi: 10.1073/pnas.171243598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surface LE, Thornton SR, Boyer LA. Polycomb group proteins set the stage for early lineage commitment. Cell Stem Cell. 2010;7:288–298. doi: 10.1016/j.stem.2010.08.004. [DOI] [PubMed] [Google Scholar]
- Tada T, Obata Y, Tada M, Goto Y, Nakatsuji N, Tan S, Kono T, Takagi N. Imprint switching for non-random X-chromosome inactivation during mouse oocyte growth. Development. 2000;127:3101–3105. doi: 10.1242/dev.127.14.3101. [DOI] [PubMed] [Google Scholar]
- Takagi N. Preferential inactivation of the paternally derived X chromosome in mice. Basic Life Sci. 1978;12:341–360. doi: 10.1007/978-1-4684-3390-6_24. [DOI] [PubMed] [Google Scholar]
- Takagi N, Sasaki M. Preferential inactivation of the paternally derived X chromosome in the extraembryonic membranes of the mouse. Nature. 1975;256:640–642. doi: 10.1038/256640a0. [DOI] [PubMed] [Google Scholar]
- Tang YA, Huntley D, Montana G, Cerase A, Nesterova TB, Brockdorff N. Efficiency of Xist-mediated silencing on autosomes is linked to chromosomal domain organisation. Epigenetics Chromatin. 2010;3:10. doi: 10.1186/1756-8935-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis MC, Waters PD, Delbridge ML, Kirby PJ, Pask AJ, Grutzner F, Rens W, Ferguson-Smith MA, Graves JA. Sex determination in platypus and echidna: Autosomal location of SOX3 confirms the absence of SRY from monotremes. Chromosome Res. 2007;15:949–959. doi: 10.1007/s10577-007-1185-3. [DOI] [PubMed] [Google Scholar]
- Wang J, Mager J, Chen Y, Schneider E, Cross JC, Nagy A, Magnuson T. Imprinted X inactivation maintained by a mouse Polycomb group gene. Nat Genet. 2001;28:371–375. doi: 10.1038/ng574. [DOI] [PubMed] [Google Scholar]
- West JD, Frels WI, Chapman VM, Papaioannou VE. Preferential expression of the maternally derived X chromosome in the mouse yolk sac. Cell. 1977;12:873–882. doi: 10.1016/0092-8674(77)90151-9. [DOI] [PubMed] [Google Scholar]
- Xu N, Donohoe ME, Silva SS, Lee JT. Evidence that homologous X-chromosome pairing requires transcription and Ctcf protein. Nat Genet. 2007;39:1390–1396. doi: 10.1038/ng.2007.5. [DOI] [PubMed] [Google Scholar]