Abstract
TMEM106B has recently been identified as a genetic risk factor for frontotemporal lobar degeneration with TDP-43 inclusions (FTLD-TDP). Amyotrophic lateral sclerosis (ALS), like FTLD-TDP, is characterized by pathological TDP-43 inclusions. We therefore investigated whether FTLD-TDP-associated risk genotypes at TMEM106B (1) contribute to risk of developing ALS or (2) modify the clinical presentation in ALS. Detailed clinical and pathological information from 61 postmortem ALS patients was collected by database query, retrospective chart review, and histopathological slide review. DNA from these patients, as well as 24 additional ALS patients, was genotyped for three TMEM106B single nucleotide polymorphisms known to confer increased risk of FTLDTDP. Associations between TMEM106B genotype and ALS were investigated by comparing TMEM106B genotypes in ALS patients (n = 85) and normal controls (n = 553), and associations between TMEM106B genotype and clinical and pathologic features were explored using linear regression. Multivariate linear models were used to evaluate the contributions of TMEM106B genotype and TDP-43 pathology to cognitive performance in ALS as measured by a phonemic verbal fluency test. We found that TMEM106B genotypes did not differ between ALS patients and normal controls. However, protective alleles at TMEM106B were significantly associated with preserved cognition in ALS patients, with the strongest association seen under a major-allele-dominant genetic model. While lower TDP-43 pathology scores and protective alleles at TMEM106B both correlated with better cognitive scores, these factors were not correlated with each other and demonstrated independent effects. These findings implicate the FTLD-TDP risk gene TMEM106B in the development of cognitive impairment in ALS.
Keywords: TMEM106B, Frontotemporal lobar, degeneration, Amyotrophic lateral sclerosis, Cognitive impairment, Frontotemporal dementia, FTLD-TDP, ALS, TDP-43
Introduction
Amyotrophic lateral sclerosis (ALS) is the most common motor neuron disease, causing progressive weakness and leading to death in an average of 3 years [28]. Historically, the disease was defined by motor signs and symptoms characteristic of upper and lower motor neuron dysfunction, with cognitive impairment (CI) considered an exclusionary criterion. Over the past few decades, however, cognitive impairment has been increasingly recognized as a common feature in ALS (reviewed in [24]), with ~50% of ALS patients demonstrating some degree of CI [18]. In addition, the pattern of cognitive deficits in ALS resembles that of frontotemporal lobar degeneration (FTLD) [20, 26], with ~20% of ALS patients meeting Neary criteria [21] for FTLD [18, 20]. It is worth noting, however, that not all ALS patients develop cognitive deficits. While bulbar site of onset [18], specific radiological patterns of atrophy [20], and older age at onset [20] have been reported to associate with poorer cognitive performance in ALS, it is still difficult to predict which patients will develop CI or frank FTLD.
A mechanistic link between FTLD and ALS has emerged in the last few years. In 2006, the protein TAR DNA binding protein of 43 kD (TDP-43) was identified as the major component of the ubiquitinated inclusions of both ALS and the largest neuropathological subset of FTLD, accordingly named FTLD-TDP [1, 19, 22]. Since then, mutations in the gene encoding TDP-43 (TARDBP) have been linked to both ALS [13, 15, 25, 30] (reviewed in [23]) and FTLD [2], implicating TDP-43 in the pathogenesis of these two clinical syndromes. Mirroring the fact that many ALS patients develop FTLD, it has long been recognized that ~15% of patients with FTLD-TDP develop motor neuron disease (often referred to as FTLD-MND) reminiscent of ALS [4, 14]. Thus, we and others have argued that ALS and FTLDTDP may be different manifestations of a common pathogenesis affecting areas of the neuraxis extending from the spinal cord to the prefrontal cortex [7]. Corroborating this argument is the fact that clinical manifestations along the ALS-FTLD spectrum appear to correlate with the distribution and burden of TDP-43 pathology [12].
Recently, polymorphisms at locus 7p21, within the gene TMEM106B, were identified by a genomewide association study (GWAS) as conferring risk for the development of FTLD-TDP [29]. Little is known about TMEM106B, and ALS GWAS have not yielded this locus or gene as conferring increased risk of ALS [8, 10]. However, it still remains possible that TMEM106B genotype may be a risk factor for ALS, or act as a genetic modifier of phenotype within ALS patients. In this paper, we examine the role of TMEM106B genotype on risk of developing ALS, on antemortem clinical features such as development of CI in ALS, and on postmortem TDP-43 neuropathological burden within ALS patients. We show that while TMEM106B may not be a risk factor for the development of ALS, risk genotypes at TMEM106B may confer added risk of CI in these patients.
Materials and methods
Cohort
Sixty-one patients with a clinical diagnosis of ALS in accordance with the modified El Escorial Criteria [5] and a neuropathological diagnosis of motor neuron disease (FTLD-MND or ALS) were identified within an autopsy database of patients with neurodegenerative diseases at the University of Pennsylvania as described [31]. These 61 patients, as well as an additional 24 patients with clinical ALS by the same criteria, were used in this study. Patients were included without bias toward the presence or absence of cognitive impairment although individuals with neuropathological evidence of Alzheimer's disease (Braak Stage V–VI) [3] at autopsy were excluded. For the 61 autopsy patients, detailed clinical characteristics (age at onset, age at death, site of onset, disease duration, ALS global disease severity as captured in a functional rating score (ALS-FRSR) [6], gender, performance on cognitive tests) were ascertained by retrospective chart review of clinic visits from 1980 through 2009 within the University of Pennsylvania Health System; the vast majority of patients were seen by one provider (L.M.), and chart reviewers (R.V. and E.A., under supervision by A.C.P.) were blind to TMEM106B genotype and autopsy neuropathology findings. Unless otherwise specified, results of testing used in this study were those from the visit most proximate to death, occurring within 12 months of death.
Cognitive testing
Antemortem cognitive testing was performed at 3–6-month intervals during routine clinic visits. Data for two cognitive tests were available for the majority of the autopsy cohort (n = 61), within 12 months of death. These included a test of letter-guided verbal fluency (FAS test, n = 32 patients) [16] and a test of frontal executive function (Trail-making test, n = 28 patients) [17]. For the FAS test, patients without significant dysarthria were asked to say as many unique words (places and names not permitted) beginning with a specified target letter (i.e. “F” or “D”) as possible in 1 min. Patients with significant dysarthria but preserved limb and hand function were asked to write the words; 90 s was allotted to these patients. The total number of unique words produced was recorded. For the Trail-making test, patients without significant dysarthria were asked to sequentially say letters of the alphabet, beginning with A, alternating with numbers, beginning with 1 (i.e. A-1, B-2) and ending at Z-26. Each error was subtracted from the maximum score of 52. Patients with significant dysarthria but relatively preserved hand and limb ability were asked to perform the same task in writing.
We have previously shown that both tests perform well compared to the Frontal Behavioral Inventory (FBI), a battery of neuropsychological tests used in assessing cognitive impairment in FTLD. Specifically, on receiver operating curve analysis compared to the FBI, the FAS test has an area under the curve (AUC) of 0.88, and the Trail-making test has an AUC of 0.80 (manuscript in preparation) in differentiating cognitively intact and cognitively impaired individuals.
Neuropathological characterization
In the autopsy cohort (n = 61), TDP-43 inclusions and tau pathology were examined in five regions of the brain: the middle frontal gyrus, cingulate gyrus, hippocampus (CA1/subiculum), amygdala, and the superior/middle temporal gyrus. Sections were fixed and cut into 6–10 lm sections, stained with hematoxylin and eosin and Thioflavin S, and immunohistochemistry was performed with antibodies to tau, α-synuclein, ubiquitin, TDP-43, and β-amyloid, as previously described [22]. Sections were then reviewed by a neuropathologist (F.G. or J.Q.T.), blind to TMEM106B genotype, and the extent of TDP-43 and tau pathology were scored for each of the five brain regions on a 4-point ordinal (0–3) scale, with 0 representing no observed pathology and 3 representing severe pathology. The pathology scores for all five regions were then summed to create a scale from 0 to 15, with 15 representing widespread cortical pathology and 0 representing no pathology observed. Patients were thus assigned a score reflecting both the extent and severity of meso-, allo- and neocortical TDP-43 or tau pathology on a scale from 0 to 15.
Of note, all of our autopsy-confirmed ALS cases had been evaluated from a diagnostic standpoint as previously described [11] for evidence of concomitant AD, PD, and other neuropathologies in addition to TDP-43 neuropathology. Briefly, of a starting total of 69 autopsy-confirmed ALS cases, 7 had concomitant AD, and 1 had concomitant PD. For simplicity, these eight cases were excluded from the current study of the effect of TMEM106B alleles on CI in ALS.
Genotyping
DNA was extracted from brain samples as previously described [29], and genotyping was performed using Taq-Man chemistry-based allelic discrimination assays [Applied Biosystems (ABI), Foster City, CA, USA] on the ABI 7500 Fast Real-Time System followed by analysis with SDS 7500 software v2.0.1. Two TMEM106B SNPs were genotyped: rs1020004 (ABI C_7604953_10) and rs1990622 (ABI C_11171598_10); these were the top SNPs previously described to confer increased risk of FTLD-TDP by GWAS [29]. In addition to the autopsy cohort of 61 patients for whom detailed clinical and neuropathological data were available, an additional 24 ALS patients underwent genotyping for TMEM106B. Thus, a total of 85 ALS patient samples were genotyped at rs1020004 and rs1990622 to assess whether TMEM106B genotype may be a risk factor for the development of ALS. Genotype frequencies in cases were compared to a set of 553 neurologically normal controls previously genotyped in the same way by our center [29].
All individuals were of self-described Caucasian ancestry with the exception of 3/85 ALS patients and 9/553 normal controls. Among the ALS patients, two were African-American, and one was multi-racial; among the controls, five were African-American, one was multiracial, and three had unknown ethnicities.
Statistical analyses
Cochran-Armitage trend tests were used to evaluate the association between ALS and TMEM106B genotype under a codominant model using 85 ALS patients and 553 neurologically normal controls. Linear regression analyses were then used to evaluate the association of TMEM106B genotype to endophenotypes within ALS, as well as demographic characteristics, using the autopsy cohort of 61 ALS patients. Specifically, the association of TMEM106B genotype with gender, age at onset of disease, disease duration, ALS-FRS-R score, TDP-43 pathology score, and tau pathology score were evaluated.
To evaluate factors associated with cognitive performance in ALS, univariate linear regressions were first used to assess for association between FAS test score and age at testing, gender, disease duration, ALS-FRS-R score, TDP-43 pathology score, tau pathology score, and TMEM106B genotype. Factors that were significant on univariate analyses were then incorporated into a multivariate model predicting cognitive performance on FAS testing, using a forward stepwise approach with additional variables added if they contributed significantly to the model (F-statistic p < 0.01).
Results
Patient cohort
After excluding ALS patients with concomitant AD or PD, 61 ALS patients were used in the clinico-pathologico-genetic part of the study, with an additional 24 ALS patients included in the genetic analysis only. Demographically, our autopsy cohort was fairly typical for ALS, with 64% men and a median age at symptom onset of 57 years (Table 1). Median disease duration was 31 months, with a median age at death of 61 years. 32.8% of patients had a bulbar site of onset, and the median ALS-FRS-R score on the last ante visit was 19 (0 most impaired, 40 least impaired). The 24 additional ALS patients who were included in the genetic analysis only were composed of individuals in whom the diagnosis was certain but histopathological or clinical data were incomplete.
Table 1.
Demographic characteristics of cohort
| Autopsy cohort (n = 61) | Additional genotyped patients (n = 24) | |
|---|---|---|
| Gender | 39 males (63.9%) | 12 males (50%) |
| 22 females (36.1%) | 12 females (50%) | |
| Site of onseta | 37 spinal (60.7%) | n/a |
| 20 bulbar (32.8%) | ||
| Age at onset, median years (IQR) | 57 (41–73) | 62 (55.5–71.5) |
| ALS-FRS-Rb, median (IQR) | 19 (15–22) | n/a |
| Duration, median months (IQR) | 31 (3–59) | n/a |
The additional cohort of 24 individuals was composed of individuals in whom the ALS diagnosis was certain, but histopathological or clinical data were less complete
n/a not available
For four patients in the autopsy cohort and most patients in the additional cohort, site of onset was not clear from chart review
Most proximate score to death
Cognitive performance ranged from normal to significantly impaired. For FAS testing, the median score was 11.5, with an interquartile range of 8–14. Trail-making testing showed a median score of 51, with a relatively small interquartile range of 50–52.
Although all autopsy-confirmed ALS cases had evidence of brainstem and spinal cord TDP-43 inclusions, 41 patients had no TDP-43 pathology in any of the 5 brain regions sampled [middle frontal gyrus, cingulate gyrus, hippocampus (CA1/subiculum), amygdala, and the superior/middle temporal gyrus], 6 had TDP-43 pathology in 1 region only, 3 had TDP-43 pathology in 2 regions, 7 in 3 regions, 2 in 4 regions, and 2 in all 5 regions. Concomitant tau pathology was seen less frequently in the ALS cohort after excluding the seven ALS cases with concomitant AD having a Braak and Braak neurofibrillary tangle stage of V/VI [3]. 48 of the 61 ALS patients studied here had no tau pathology in the 5 brain regions, 4 had tau pathology in only 1 region, 7 in 2 regions, and 2 in 3 regions.
TMEM106B genotype frequencies do not differ between ALS and normal controls
Given the observed association between TMEM106B genotype and disease for another TDP-43 proteinopathy, FTLD-TDP, we asked whether risk genotypes at TMEM106B were more common among ALS patients. However, for TMEM106B SNPs rs1990622 and rs1020004, allele and genotype frequencies did not differ significantly between ALS patients (n = 85) and normal controls (n = 553; Table 2).
Table 2.
TMEM106B SNP frequencies among ALS cases (n = 85) and neurologically normal controls (n = 553)
| SNP ID | Locus | Genotypes controls (n = 553) | Genotypes ALS (n = 85) | p value | MAF controls (n = 553) | MAF ALS (n = 85) | p value |
|---|---|---|---|---|---|---|---|
| rs1020004 | 12222303 | AA 258 (47) | AA 38 (45) | n/s | 0.307 | 0.312 | n/s |
| AG 249 (45) | AG 41 (48) | ||||||
| GG 45 (8) | GG 6 (7) | ||||||
| rs1990622 | 12250312 | TT 190 (34) | TT 27 (32) | n/s | 0.41 | 0.418 | n/s |
| TC 272 (49) | TC 45 (53) | ||||||
| CC 91 (16) | CC 13 (15) |
SNPs are listed in genomic order based on location on chromosome 7 (Locus within Genome Build 36). For genotypes, absolute number is shown, with percentage in parentheses. One genotyping reaction failed for the rs1020004 controls, so only 552 individuals were successfully genotyped
MAF minor allele frequency, n/s non-significant
TMEM106B genotype is correlated with FAS performance in ALS
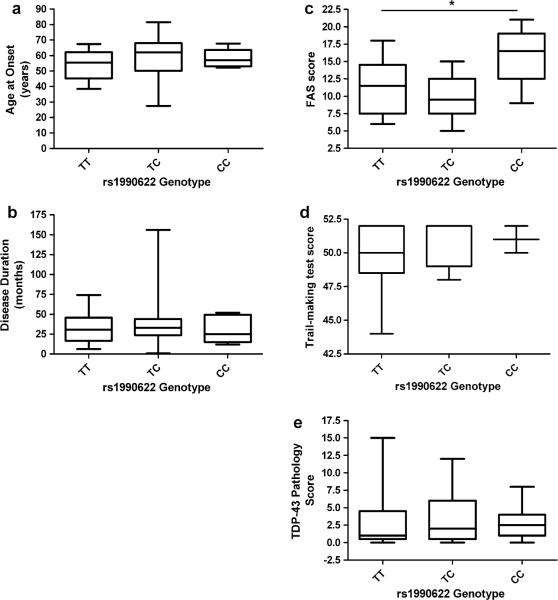
We next explored whether TMEM106B genotype could serve as a genetic modifier for phenotype in ALS. Specifically, we asked whether FTLD-TDP risk genotypes at TMEM106B SNP rs1990622 (the top SNP in the FTLD-TDP GWAS) [29] were associated with differences in age at onset, disease duration, cognitive performance, or degree of TDP-43 pathology among ALS patients (Fig. 1). Age at onset did not differ among carriers of different TMEM106B genotypes (p = 0.403; Fig. 1a), nor did disease duration (p = 0.705; Fig. 1b). Cognitive performance, as measured by FAS testing, did differ significantly among TMEM106B genotypes (p = 0.029 under codominant model, p = 0.018 under major allele dominant model), with homozygotes for the FTLD-TDP protective allele at SNP rs1990622 (CC) having higher FAS scores (Fig. 1c). CC homozygotes also performed better than the other TMEM106B genotype groups on the Trail-making test although differences were not statistically significant (Fig. 1d). Surprisingly, despite differences in performance on cognitive testing, TDP-43 pathology score did not differ significantly among TMEM106B genotypes (p = 0.777; Fig. 1e).
Fig. 1.
Box and whiskers plots comparing a age at onset, b disease duration, c FAS score, d Trail-making test score, and e TDP-43 pathology score, among TMEM106B genotypes. While FAS score (p = 0.029) is significantly associated with genotype, the extent of TDP-43 pathology (p = 0.788) is not. Box indicates interquartile range, with median indicated by line; whiskers denote full range
TMEM106B genotype and TDP-43 pathology are independently correlated with FAS cognitive performance in ALS
Given the observed association between cognitive performance and TMEM106B genotype, we sought to further characterize this relationship. We first evaluated the effects of other demographic and clinical characteristics (age at testing, gender, disease duration, ALS-FRS score, and TDP-43 pathology) on cognitive performance. In univariate linear regressions, ALS-FRS-R scores (p = 0.033) and TDP-43 pathology scores (p = 0.020) both correlated significantly with FAS scores (Table 3).
Table 3.
Univariate and multivariate analyses of factors affecting cognitive performance
| Univariate analyses: FAS score (factor) | |||
|---|---|---|---|
| Factor | R2 | Direction | p value |
| Age at testing | 0.04 | − | 0.274 |
| Gender (male) | 0.055 | + | 0.199 |
| Disease duration | 0.007 | − | 0.651 |
| ALS-FRS-R | 0.143 | − | 0.033 |
| TDP-43 pathology score | 0.178 | − | 0.02 |
| Tau pathology score | 0.035 | − | 0.306 |
| TMEM106B genotype | |||
| Codominant (minor allele) | 0.216 | + | 0.029 |
| Major allele dominant | 0.172 | 0.018 | |
| Minor allele dominant | 0.004 | 0.747 | |
| Best-fit multivariate model: FAS score predicted by TMEM106B Genotype * TDP-43 pathology | |||
|---|---|---|---|
| R2 | Coefficient | p value | |
| Overall model | 0.388 | 0.005 | |
| TMEM106B genotype (minor allele) | 7.907 | 0.009 | |
| TDP-43 pathology score | −0.410 | 0.033 | |
| TMEM106B:TDP-43 pathology score | −1.690 | 0.124 | |
Top: Univariate linear regressions were used to evaluate associations between various factors and cognitive performance on FAS testing. Higher ALS-FRS-R score, higher TDP-43 pathology score, and risk genotype at TMEM106B were all significantly associated with poorer cognitive performance
Bottom: Best-fit multivariate model by a forward stepwise approach, with an R2 value of 0.388, included TMEM106B genotype, TDP-43 pathology and their interaction term as predictors of FAS test score. Within this model, risk genotypes at TMEM106B and higher TDP-43 pathology appear to contribute independently to poorer cognitive performance
TMEM106B genotype at rs1990622 effects shown. TMEM106B risk genotypes at the other SNP (rs1020004) performed similarly, as expected given the strong linkage between these two SNPs
Direction direction of association
Bold values are nominally significant (p < 0.05)
Multivariate linear regression using a forward stepwise approach arrived at a best-fit model in which FAS test score was predicted by the factors TMEM106B genotype, TDP-43 pathology score, and their interaction term (Table 3). Mirroring our prior finding that TDP-43 pathology score and TMEM106B genotype are not correlated with each other, these two factors appear to contribute independently to cognitive performance, with a p value for association of 0.033 for TDP-43 pathology score, a p value of 0.009 for TMEM106B genotype, and a non-significant p value of 0.124 for the interaction of these two factors. The best-fit multivariate model used TMEM106B genotype under a major allele dominant model, in accordance with our finding that minor allele homozygotes performed differently from major allele homozygotes and heterozygotes on cognitive testing.
Discussion
In the present study, we evaluated the contribution of TMEM106B genotype to risk for developing ALS and to endophenotypes, particularly with respect to cognition, within ALS. While our data do not implicate TMEM106B genotype as a significant risk factor for ALS, among ALS patients, carriers of FTLD-TDP-associated TMEM106B genotypes (i.e. T allele for SNP rs1990622) have poorer cognitive performance. Moreover, the association between TMEM106B genotype and cognitive performance appears to be independent of the degree of cortical TDP-43 pathology.
Our findings support the notion that FTLD-TDP and ALS may be differing phenotypic manifestations of at least partly shared pathogenic mechanisms since the same gene (TMEM106B) appears to contribute to risk of CI/dementia in both diseases. In addition, the current study is most compatible with a model in which the TMEM106B risk allele (i.e. T allele at SNP rs1990622) is dominant over the protective allele (i.e. C allele at SNP rs1990622), i.e. the presence of either one or two T alleles is adequate to increase risk of CI in ALS. This possible dominant effect accords with preliminary findings from our previous FTLD-TDP GWAS showing homozygous carriers of the protective allele as having significantly longer disease durations than carriers of one or two risk alleles [29].
Interestingly, the extent of cortical TDP-43 pathology and TMEM106B genotype appear to contribute independently to risk of developing cognitive impairment in ALS. This is somewhat surprising since FTLD-TDP and ALS share pathological inclusions of TDP-43 as a common neuropathological feature, and one might expect shared mechanisms of pathogenesis to directly involve TDP-43. Such a situation is not wholly unprecedented, however; despite both being implicated in the pathogenesis of FTLD-TDP, TDP-43 and the protein progranulin (haploinsufficiency of which has been shown to cause FTLD-TDP) [9]do not appear by current evidence to be related mechanistically in either physiological or pathophysiological scenarios.
In addition, our findings do raise the question of what the underlying substrate of TMEM106B-mediated cognitive impairment in ALS might be. Possibilities include TDP-43 pathology in other areas of the brain (cortical or subcortical), or pathological inclusions of other proteins such as tau or a-synuclein. In this regard, it is notable that others have also reported ALS with significant hippocampal TDP-43 pathology and/or hippocampal degeneration in the absence of clinical dementia, as well as clinical dementia in ALS in the absence of significant hippocampal TDP-43 pathology or hippocampal degeneration [27]. Of note, we excluded cases with concomitant tau or α-synuclein pathology of severity great enough to warrant neuropathological diagnoses of Alzheimer's disease or Parkinson's disease. However, lesser degrees of mixed pathology might still be contributing to cortical dysfunction. Alternatively, TMEM106B may be acting via other pathogenic mechanisms entirely.
Several caveats should be kept in mind in interpreting our present findings. First, while our autopsy cohort size of 61 is reasonably large for a study with detailed clinical, pathological, and genetic data, there is some risk of both Type I and II errors. Second, our current findings apply to cognitive performance as captured on FAS testing. While we observed similar trends with a second cognitive test (Trail-making test), these did not reach statistical significance, possibly due to insufficient sample size and the relatively collapsed range of results for this test. In any case, further studies with greater sample sizes and a wider range of cognitive tests would be a valuable addition to the data presented here.
In summary, in the present clinico-pathologico-genetic study, we demonstrate an association between genotype at TMEM106B and cognitive performance in ALS. As such, the recently discovered gene TMEM106B appears to be a bona fide genetic risk factor for CI in at least two diseases characterized by TDP-43 proteinopathy. Further investigations of its function may lead to insights into the pathogenesis of, and possible therapeutic avenues for, these otherwise fatal diseases.
Acknowledgments
We thank the patients who contributed samples to this study. We thank Robert Greene and Young Baek for technical assistance. This work was supported by the NIH (AG033101, AG17586, AG10124, AG17586, AG32953, NS44266), as well as a Burroughs Wellcome Fund Career Award for Medical Scientists and the Benaroya Fund (to ACP), and the Koller Family Foundation. VMYL is the John H. Ware, 3rd, Professor of Alzheimer's Disease Research. JQT is the William Maul Measey-Truman G. Schnabel, Jr., Professor of Geriatric Medicine and Gerontology.
Footnotes
Conflict of interest The authors declare that they have no conflict of interest.
References
- 1.Arai T, Hasegawa M, Akiyama H, et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem Biophys Res Commun. 2006;351:602–611. doi: 10.1016/j.bbrc.2006.10.093. [DOI] [PubMed] [Google Scholar]
- 2.Benajiba L, Le Ber I, Camuzat A, et al. TARDBP mutations in motoneuron disease with frontotemporal lobar degeneration. Ann Neurol. 2009;65:470–473. doi: 10.1002/ana.21612. [DOI] [PubMed] [Google Scholar]
- 3.Braak H, Braak E. Staging of Alzheimer's disease-related neurofibrillary changes. Neurobiol Aging. 1995;16:271–278. doi: 10.1016/0197-4580(95)00021-6. discussion 278–284. [DOI] [PubMed] [Google Scholar]
- 4.Brandmeir NJ, Geser F, Kwong LK, et al. Severe subcortical TDP-43 pathology in sporadic frontotemporal lobar degeneration with motor neuron disease. Acta Neuropathol. 2008;115:123–131. doi: 10.1007/s00401-007-0315-5. [DOI] [PubMed] [Google Scholar]
- 5.Brooks BR, Miller RG, Swash M, Munsat TL. El Escorial revisited: revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph Lateral Scler Other Motor Neuron Disord. 2000;1:293–299. doi: 10.1080/146608200300079536. [DOI] [PubMed] [Google Scholar]
- 6.Cedarbaum JM, Stambler N, Malta E, et al. The ALSFRSR: a revised ALS functional rating scale that incorporates assessments of respiratory function. BDNF ALS Study Group (Phase III) J Neurol Sci. 1999;169:13–21. doi: 10.1016/s0022-510x(99)00210-5. [DOI] [PubMed] [Google Scholar]
- 7.Chen-Plotkin AS, Lee VM, Trojanowski JQ. TAR DNA-binding protein 43 in neurodegenerative disease. Nat Rev Neurol. 2010;6:211–220. doi: 10.1038/nrneurol.2010.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chio A, Schymick JC, Restagno G, et al. A two-stage genome-wide association study of sporadic amyotrophic lateral sclerosis. Hum Mol Genet. 2009;18:1524–1532. doi: 10.1093/hmg/ddp059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cruts M, Van Broeckhoven C. Loss of progranulin function in frontotemporal lobar degeneration. Trends Genet. 2008;24:186–194. doi: 10.1016/j.tig.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 10.Dunckley T, Huentelman MJ, Craig DW, et al. Whole-genome analysis of sporadic amyotrophic lateral sclerosis. N Engl J Med. 2007;357:775–788. doi: 10.1056/NEJMoa070174. [DOI] [PubMed] [Google Scholar]
- 11.Geser F, Brandmeir NJ, Kwong LK, et al. Evidence of multisystem disorder in whole-brain map of pathological TDP-43 in amyotrophic lateral sclerosis. Arch Neurol. 2008;65:636–641. doi: 10.1001/archneur.65.5.636. [DOI] [PubMed] [Google Scholar]
- 12.Geser F, Martinez-Lage M, Robinson J, et al. Clinical and pathological continuum of multisystem TDP-43 proteinopathies. Arch Neurol. 2009;66:180–189. doi: 10.1001/archneurol.2008.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gitcho MA, Baloh RH, Chakraverty S, et al. TDP-43 A315T mutation in familial motor neuron disease. Ann Neurol. 2008;63:535–538. doi: 10.1002/ana.21344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hodges JR, Davies RR, Xuereb JH, et al. Clinicopathological correlates in frontotemporal dementia. Ann Neurol. 2004;56:399–406. doi: 10.1002/ana.20203. [DOI] [PubMed] [Google Scholar]
- 15.Kabashi E, Valdmanis PN, Dion P, et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat Genet. 2008;40:572–574. doi: 10.1038/ng.132. [DOI] [PubMed] [Google Scholar]
- 16.Lezak M. Neuropsychological assessment. Oxford University Press; New York: 1983. [Google Scholar]
- 17.Libon DJ, Massimo L, Moore P, et al. Screening for frontotemporal dementias and Alzheimer's disease with the Philadelphia Brief Assessment of Cognition: a preliminary analysis. Dement Geriatr Cogn Disord. 2007;24:441–447. doi: 10.1159/000110577. [DOI] [PubMed] [Google Scholar]
- 18.Lomen-Hoerth C, Murphy J, Langmore S, et al. Are amyotrophic lateral sclerosis patients cognitively normal? Neurology. 2003;60:1094–1097. doi: 10.1212/01.wnl.0000055861.95202.8d. [DOI] [PubMed] [Google Scholar]
- 19.Mackenzie IR, Neumann M, Bigio EH, et al. Nomenclature for neuropathologic subtypes of frontotemporal lobar degeneration: consensus recommendations. Acta Neuropathol. 2009;117:15–18. doi: 10.1007/s00401-008-0460-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murphy JM, Henry RG, Langmore S, et al. Continuum of frontal lobe impairment in amyotrophic lateral sclerosis. Arch Neurol. 2007;64:530–534. doi: 10.1001/archneur.64.4.530. [DOI] [PubMed] [Google Scholar]
- 21.Neary D, Snowden JS, Gustafson L, et al. Frontotemporal lobar degeneration: a consensus on clinical diagnostic criteria. Neurology. 1998;51:1546–1554. doi: 10.1212/wnl.51.6.1546. [DOI] [PubMed] [Google Scholar]
- 22.Neumann M, Sampathu DM, Kwong LK, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 23.Pesiridis GS, Lee VM-Y, Trojanowski JQ. Mutations in TDP-43 link glycine-rich domain functions to amyotrophic lateral sclerosis. Hum Mol Genet. 2009;18(R2):R156–R162. doi: 10.1093/hmg/ddp303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Phukan J, Pender NP, Hardiman O. Cognitive impairment in amyotrophic lateral sclerosis. Lancet Neurol. 2007;6:994–1003. doi: 10.1016/S1474-4422(07)70265-X. [DOI] [PubMed] [Google Scholar]
- 25.Sreedharan J, Blair IP, Tripathi VB, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strong MJ, Grace GM, Freedman M, et al. Consensus criteria for the diagnosis of frontotemporal cognitive and behavioural syndromes in amyotrophic lateral sclerosis. Amyotroph Lateral Scler. 2009;10:131–146. doi: 10.1080/17482960802654364. [DOI] [PubMed] [Google Scholar]
- 27.Takeda T, Uchihara T, Arai N, Mizutani T, Iwata M. Progression of hippocampal degeneration in amyotrophic lateral sclerosis with or without memory impairment: distinction from Alzheimer disease. Acta Neuropathol. 2009;117:35–44. doi: 10.1007/s00401-008-0447-2. [DOI] [PubMed] [Google Scholar]
- 28.Testa D, Lovati R, Ferrarini M, Salmoiraghi F, Filippini G. Survival of 793 patients with amyotrophic lateral sclerosis diagnosed over a 28-year period. Amyotroph Lateral Scler Other Motor Neuron Disord. 2004;5:208–212. [PubMed] [Google Scholar]
- 29.Van Deerlin VM, Sleiman PM, Martinez-Lage M, et al. Common variants at 7p21 are associated with frontotemporal lobar degeneration with TDP-43 inclusions. Nat Genet. 2010;42:234–239. doi: 10.1038/ng.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van Deerlin VM, Leverenz JB, Bekris LM, et al. TARDBP mutations in amyotrophic lateral sclerosis with TDP-43 neuro-pathology: a genetic and histopathological analysis. Lancet Neurol. 2008;7:409–416. doi: 10.1016/S1474-4422(08)70071-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie SX, Baek Y, Grossman M, et al. Building an integrated neurodegenerative disease database at an academic health center. Alzheimers Dement. 2010 doi: 10.1016/j.jalz.2010.08.233. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]