Abstract
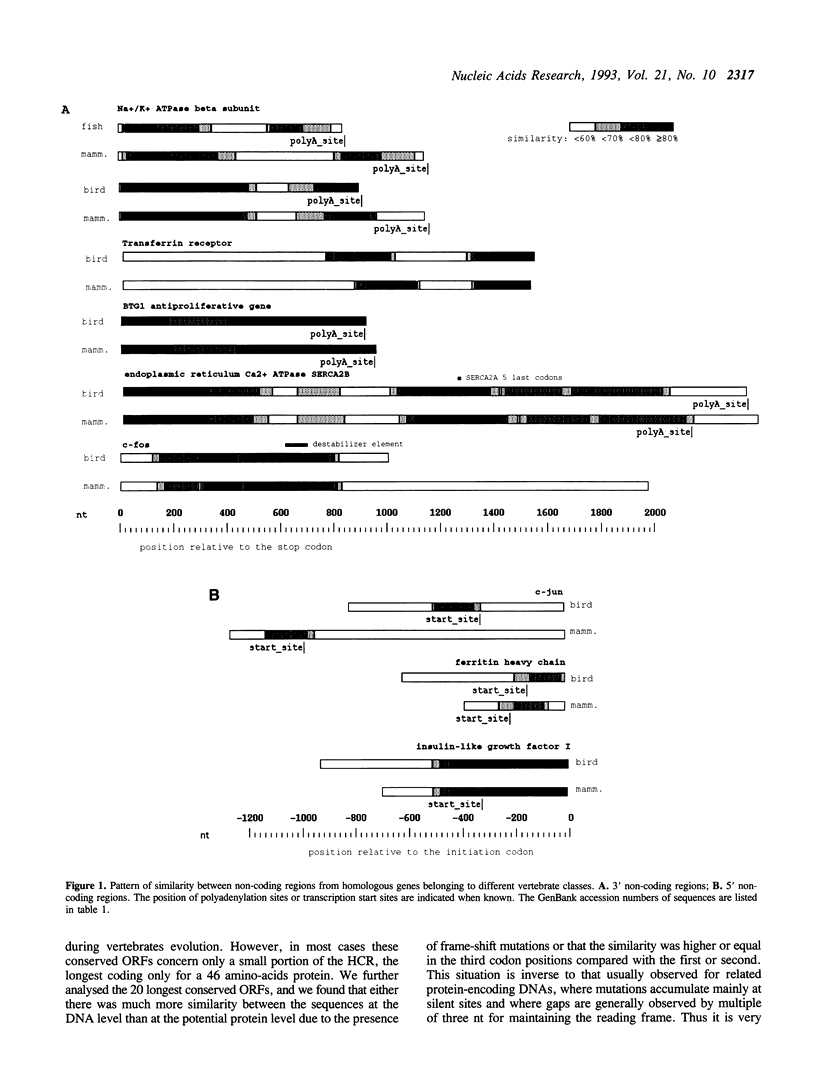
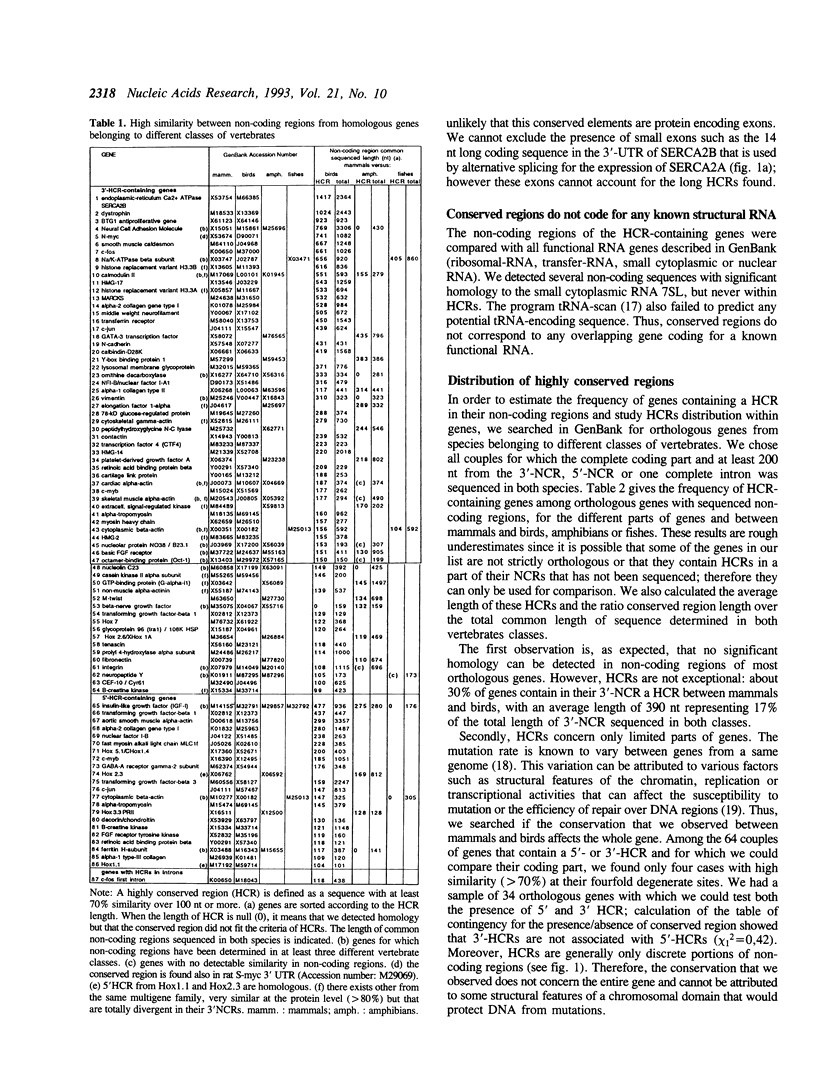
Comparison of nucleotide sequences from different classes of vertebrates that diverged more than 300 million years ago, revealed the existence of highly conserved regions (HCRs) with more than 70% similarity over 100 to 1450 nt in non-coding parts of genes. Such a conservation is unexpected because it is much longer and stronger than what is necessary for specifying the binding of a regulatory protein. HCRs are relatively frequent, particularly in genes that are essential to cell life. In multigene families, conserved regions are specific of each isotype and are probably involved in the control of their specific pattern of expression. Studying HCRs distribution within genes showed that functional constraints are generally much stronger in 3'-non-coding regions than in promoters or introns. The 3'-HCRs are particularly A + T-rich and are always located in the transcribed untranslated regions of genes, which suggests that they are involved in post-transcriptional processes. However, current knowledge of mechanisms that regulate mRNA export, localisation, translation, or degradation is not sufficient to explain the strong functional constraints that we have characterised.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Arrick B. A., Lee A. L., Grendell R. L., Derynck R. Inhibition of translation of transforming growth factor-beta 3 mRNA by its 5' untranslated region. Mol Cell Biol. 1991 Sep;11(9):4306–4313. doi: 10.1128/mcb.11.9.4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiss L. E., Friedman J. M. Regulation of N-myc gene expression: use of an adenovirus vector to demonstrate posttranscriptional control. Mol Cell Biol. 1990 Dec;10(12):6700–6708. doi: 10.1128/mcb.10.12.6700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckman J. S., Weber J. L. Survey of human and rat microsatellites. Genomics. 1992 Apr;12(4):627–631. doi: 10.1016/0888-7543(92)90285-z. [DOI] [PubMed] [Google Scholar]
- Benton M. J. Phylogeny of the major tetrapod groups: morphological data and divergence dates. J Mol Evol. 1990 May;30(5):409–424. doi: 10.1007/BF02101113. [DOI] [PubMed] [Google Scholar]
- Bernardi G. The isochore organization of the human genome. Annu Rev Genet. 1989;23:637–661. doi: 10.1146/annurev.ge.23.120189.003225. [DOI] [PubMed] [Google Scholar]
- Boulikas T. Evolutionary consequences of nonrandom damage and repair of chromatin domains. J Mol Evol. 1992 Aug;35(2):156–180. doi: 10.1007/BF00183227. [DOI] [PubMed] [Google Scholar]
- Burks C., Cassidy M., Cinkosky M. J., Cumella K. E., Gilna P., Hayden J. E., Keen G. M., Kelley T. A., Kelly M., Kristofferson D. GenBank. Nucleic Acids Res. 1991 Apr 25;19 (Suppl):2221–2225. doi: 10.1093/nar/19.suppl.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan L. N., Grammatikakis N., Banks J. M., Gerhardt E. M. Chicken transferrin receptor gene: conservation 3' noncoding sequences and expression in erythroid cells. Nucleic Acids Res. 1989 May 25;17(10):3763–3771. doi: 10.1093/nar/17.10.3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichant G. A., Burks C. Identifying potential tRNA genes in genomic DNA sequences. J Mol Biol. 1991 Aug 5;220(3):659–671. doi: 10.1016/0022-2836(91)90108-i. [DOI] [PubMed] [Google Scholar]
- Goodman M., Czelusniak J., Koop B. F., Tagle D. A., Slightom J. L. Globins: a case study in molecular phylogeny. Cold Spring Harb Symp Quant Biol. 1987;52:875–890. doi: 10.1101/sqb.1987.052.01.096. [DOI] [PubMed] [Google Scholar]
- Gouy M., Gautier C., Attimonelli M., Lanave C., di Paola G. ACNUC--a portable retrieval system for nucleic acid sequence databases: logical and physical designs and usage. Comput Appl Biosci. 1985 Sep;1(3):167–172. doi: 10.1093/bioinformatics/1.3.167. [DOI] [PubMed] [Google Scholar]
- Grens A., Scheffler I. E. The 5'- and 3'-untranslated regions of ornithine decarboxylase mRNA affect the translational efficiency. J Biol Chem. 1990 Jul 15;265(20):11810–11816. [PubMed] [Google Scholar]
- Hentze M. W. Determinants and regulation of cytoplasmic mRNA stability in eukaryotic cells. Biochim Biophys Acta. 1991 Nov 11;1090(3):281–292. doi: 10.1016/0167-4781(91)90191-n. [DOI] [PubMed] [Google Scholar]
- Higgins D. G., Bleasby A. J., Fuchs R. CLUSTAL V: improved software for multiple sequence alignment. Comput Appl Biosci. 1992 Apr;8(2):189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- Hraba-Renevey S., Kress M. Expression of a mouse replacement histone H3.3 gene with a highly conserved 3' noncoding region during SV40- and polyoma-induced Go to S-phase transition. Nucleic Acids Res. 1989 Apr 11;17(7):2449–2461. doi: 10.1093/nar/17.7.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M. T., Gorman C. M. Intervening sequences increase efficiency of RNA 3' processing and accumulation of cytoplasmic RNA. Nucleic Acids Res. 1990 Feb 25;18(4):937–947. doi: 10.1093/nar/18.4.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajimoto Y., Rotwein P. Structure of the chicken insulin-like growth factor I gene reveals conserved promoter elements. J Biol Chem. 1991 May 25;266(15):9724–9731. [PubMed] [Google Scholar]
- Kislauskis E. H., Singer R. H. Determinants of mRNA localization. Curr Opin Cell Biol. 1992 Dec;4(6):975–978. doi: 10.1016/0955-0674(92)90128-y. [DOI] [PubMed] [Google Scholar]
- Koeller D. M., Casey J. L., Hentze M. W., Gerhardt E. M., Chan L. N., Klausner R. D., Harford J. B. A cytosolic protein binds to structural elements within the iron regulatory region of the transferrin receptor mRNA. Proc Natl Acad Sci U S A. 1989 May;86(10):3574–3578. doi: 10.1073/pnas.86.10.3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruys V., Marinx O., Shaw G., Deschamps J., Huez G. Translational blockade imposed by cytokine-derived UA-rich sequences. Science. 1989 Aug 25;245(4920):852–855. doi: 10.1126/science.2672333. [DOI] [PubMed] [Google Scholar]
- Kühn L. C., Hentze M. W. Coordination of cellular iron metabolism by post-transcriptional gene regulation. J Inorg Biochem. 1992 Aug 15;47(3-4):183–195. doi: 10.1016/0162-0134(92)84064-t. [DOI] [PubMed] [Google Scholar]
- Lawrence J. B., Singer R. H. Intracellular localization of messenger RNAs for cytoskeletal proteins. Cell. 1986 May 9;45(3):407–415. doi: 10.1016/0092-8674(86)90326-0. [DOI] [PubMed] [Google Scholar]
- Lemaire C., Heilig R., Mandel J. L. The chicken dystrophin cDNA: striking conservation of the C-terminal coding and 3' untranslated regions between man and chicken. EMBO J. 1988 Dec 20;7(13):4157–4162. doi: 10.1002/j.1460-2075.1988.tb03311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff W. F., Pandey N. B. Multiple regulatory steps control histone mRNA concentrations. Trends Biochem Sci. 1988 Feb;13(2):49–52. doi: 10.1016/0968-0004(88)90027-8. [DOI] [PubMed] [Google Scholar]
- Matsuo K., Sato K., Ikeshima H., Shimoda K., Takano T. Four synonymous genes encode calmodulin in the teleost fish, medaka (Oryzias latipes): conservation of the multigene one-protein principle. Gene. 1992 Oct 1;119(2):279–281. doi: 10.1016/0378-1119(92)90283-u. [DOI] [PubMed] [Google Scholar]
- Mohun T., Garrett N., Stutz F., Sophr G. A third striated muscle actin gene is expressed during early development in the amphibian Xenopus laevis. J Mol Biol. 1988 Jul 5;202(1):67–76. doi: 10.1016/0022-2836(88)90519-0. [DOI] [PubMed] [Google Scholar]
- Mouchiroud D., D'Onofrio G., Aïssani B., Macaya G., Gautier C., Bernardi G. The distribution of genes in the human genome. Gene. 1991 Apr;100:181–187. doi: 10.1016/0378-1119(91)90364-h. [DOI] [PubMed] [Google Scholar]
- Murti J. R., Bumbulis M., Schimenti J. C. High-frequency germ line gene conversion in transgenic mice. Mol Cell Biol. 1992 Jun;12(6):2545–2552. doi: 10.1128/mcb.12.6.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller W. E., Slor H., Pfeifer K., Hühn P., Bek A., Orsulic S., Ushijima H., Schröder H. C. Association of AUUUA-binding protein with A+U-rich mRNA during nucleo-cytoplasmic transport. J Mol Biol. 1992 Aug 5;226(3):721–733. doi: 10.1016/0022-2836(92)90628-w. [DOI] [PubMed] [Google Scholar]
- Müllner E. W., Kühn L. C. A stem-loop in the 3' untranslated region mediates iron-dependent regulation of transferrin receptor mRNA stability in the cytoplasm. Cell. 1988 Jun 3;53(5):815–825. doi: 10.1016/0092-8674(88)90098-0. [DOI] [PubMed] [Google Scholar]
- Nojima H. Structural organization of multiple rat calmodulin genes. J Mol Biol. 1989 Jul 20;208(2):269–282. doi: 10.1016/0022-2836(89)90388-4. [DOI] [PubMed] [Google Scholar]
- O'Brien T. P., Yang G. P., Sanders L., Lau L. F. Expression of cyr61, a growth factor-inducible immediate-early gene. Mol Cell Biol. 1990 Jul;10(7):3569–3577. doi: 10.1128/mcb.10.7.3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson P. L., Matheson N. W., Flescher D. C., Robbins R. J. The GDB Human Genome Data Base Anno 1992. Nucleic Acids Res. 1992 May 11;20 (Suppl):2201–2206. doi: 10.1093/nar/20.suppl.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltz S. W., Brewer G., Bernstein P., Hart P. A., Ross J. Regulation of mRNA turnover in eukaryotic cells. Crit Rev Eukaryot Gene Expr. 1991;1(2):99–126. [PubMed] [Google Scholar]
- Pomeroy M. E., Lawrence J. B., Singer R. H., Billings-Gagliardi S. Distribution of myosin heavy chain mRNA in embryonic muscle tissue visualized by ultrastructural in situ hybridization. Dev Biol. 1991 Jan;143(1):58–67. doi: 10.1016/0012-1606(91)90054-7. [DOI] [PubMed] [Google Scholar]
- Ryseck R. P., MacDonald-Bravo H., Zerial M., Bravo R. Coordinate induction of fibronectin, fibronectin receptor, tropomyosin, and actin genes in serum-stimulated fibroblasts. Exp Cell Res. 1989 Feb;180(2):537–545. doi: 10.1016/0014-4827(89)90080-3. [DOI] [PubMed] [Google Scholar]
- Schröder H. C., Bachmann M., Diehl-Seifert B., Müller W. E. Transport of mRNA from nucleus to cytoplasm. Prog Nucleic Acid Res Mol Biol. 1987;34:89–142. doi: 10.1016/s0079-6603(08)60494-8. [DOI] [PubMed] [Google Scholar]
- Schwartz M. L., Shneidman P. S., Bruce J., Schlaepfer W. W. Actinomycin prevents the destabilization of neurofilament mRNA in primary sensory neurons. J Biol Chem. 1992 Dec 5;267(34):24596–24600. [PubMed] [Google Scholar]
- Shyu A. B., Greenberg M. E., Belasco J. G. The c-fos transcript is targeted for rapid decay by two distinct mRNA degradation pathways. Genes Dev. 1989 Jan;3(1):60–72. doi: 10.1101/gad.3.1.60. [DOI] [PubMed] [Google Scholar]
- Singer R. H. The cytoskeleton and mRNA localization. Curr Opin Cell Biol. 1992 Feb;4(1):15–19. doi: 10.1016/0955-0674(92)90053-f. [DOI] [PubMed] [Google Scholar]
- Steward O., Banker G. A. Getting the message from the gene to the synapse: sorting and intracellular transport of RNA in neurons. Trends Neurosci. 1992 May;15(5):180–186. doi: 10.1016/0166-2236(92)90170-d. [DOI] [PubMed] [Google Scholar]
- Thompson C. B., Challoner P. B., Neiman P. E., Groudine M. Expression of the c-myb proto-oncogene during cellular proliferation. 1986 Jan 30-Feb 5Nature. 319(6052):374–380. doi: 10.1038/319374a0. [DOI] [PubMed] [Google Scholar]
- Watson G., Paigen K. mRNA synthesis rates in vivo for androgen-inducible sequences in mouse kidney. Mol Cell Biol. 1988 May;8(5):2117–2124. doi: 10.1128/mcb.8.5.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells D., Hoffman D., Kedes L. Unusual structure, evolutionary conservation of non-coding sequences and numerous pseudogenes characterize the human H3.3 histone multigene family. Nucleic Acids Res. 1987 Apr 10;15(7):2871–2889. doi: 10.1093/nar/15.7.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe K. H., Sharp P. M., Li W. H. Mutation rates differ among regions of the mammalian genome. Nature. 1989 Jan 19;337(6204):283–285. doi: 10.1038/337283a0. [DOI] [PubMed] [Google Scholar]
- Yaffe D., Nudel U., Mayer Y., Neuman S. Highly conserved sequences in the 3' untranslated region of mRNAs coding for homologous proteins in distantly related species. Nucleic Acids Res. 1985 May 24;13(10):3723–3737. doi: 10.1093/nar/13.10.3723. [DOI] [PMC free article] [PubMed] [Google Scholar]



