Abstract
Background
Because of their known role as transcriptional regulators of key plant developmental processes, the diversification of MADS-box gene function is thought to be a major driving force in the developmental evolution of plants. Yet the function of some MADS-box gene subfamilies has remained elusive thus far. One such lineage, AGL6, has now been functionally characterized in three angiosperm species, but a phylogenetic framework for comparison of AGL6 gene function is currently missing.
Results
Based on phylogenetic analyses of newly isolated and EST-based sequences, we describe the duplication history of the AGL6 subfamily in angiosperms. Our analyses provide support for four ancient duplications in the evolution of the AGL6 lineage: one at the base of core eudicots resulting in euAGL6 and AGL6-like gene clades, one during basal angiosperm diversification and two in monocot evolution. To investigate whether the spatial domains in which AGL6 genes function have diverged after duplication, we use quantitative Real Time PCR. We show that the core eudicot AGL6-like clade acquired expression in vegetative tissues, while its paralog euAGL6 remains predominantly confined to reproductive tissues.
Conclusions
These and previous data lead us to propose that the AGL6 lineage in core eudicots, in addition to functions related to the expression in reproductive structures, may have acquired a function in developmental transitions of vegetative shoots.
Background
The AGAMOUS LIKE 6 lineage of MIKC-type MADS-box transcription factors is rooted in a superclade with both SEPALLATA-like genes and APETALA1/FRUITFUL-like genes, though the exact relationships between these three lineages are somewhat unclear [1-6]. The function of SEP- and AP1-like genes is known to contribute to floral meristem specification and floral organ identity in angiosperm reproductive development [5,7-11]. Yet the function of AGL6 genes, the third lineage in this superclade, had not been functionally characterized until recently.
The genome of the genetic plant model Arabidopsis thaliana harbors two AGL6 genes: AGL6 after which the subfamily was named and its paralog AGL13. No knockout phenotype has been described for either one of these two paralogs, possibly due to genetic redundancy with each other and other factors [12-15]. Just recently, knockout phenotypes have been described in three other angiosperm species: the grasses rice (Oryza) and maize (Zea) and in Petunia, an asterid species [16-18]. In Oryza, Ohmori et al. [16] characterized the function of MADS6 where it regulates floral organ identity and floral meristem determinacy and was renamed MOSAIC FLORAL ORGANS1 (MFO1). Mfo1 florets develop abnormal paleas and lodicules, mixed organs and extra floral organs [16]. In Zea, bearded ear, a loss-of-function mutant for AGL6 demonstrates a similar role in floral organ development and floral meristem identity [18]. Finally in Petunia, the flower specific function of PhAGL6 in petal and anther development was revealed in double and triple mutants with SEPALLATA-like genes, indicating that PhAGL6 functions redundantly with closely related genes [17]. In agreement with this role confined to reproductive development, PhAGL6 expression was not observed during vegetative developmental stages [17]. So far, the knockout phenotypes studied suggest that AGL6 plays a redundant role in establishing the flower and its organs.
Members of the AGL6 lineage have been isolated from gymnosperms [19-21] and all major angiosperm clades [1]. Comparing expression patterns of these genes throughout these plant lineages suggests that expression of AGL6 in floral meristems has been conserved since angiosperm origin [14,21,22]. Yet in Arabidopsis and gymnosperms, additional expression in vegetative tissues has been observed [12,13,19,20,22,23]. A detailed reporter analysis of the regulatory elements of Arabidopsis AGL13 and AGL6 observed expression of AGL13 in the vasculature underlying the shoot apical meristem [14]. In addition, expression of AGL6 was recently detected by in situ hybridization in cauline leaf primordia and in cryptic bract regions in response to floral induction [15]. Similarly, gymnosperm homologs have shown to be expressed in vegetative tissues [19,20]. For example, Mouradov et al. [20] detected expression of an AGL6-homolog (PrMADS3) in the group of cells initiating needle primordia in vegetative buds, suggestive of non-reproductive functions of the pre-angiosperm AGL6 lineage.
Though often frought with pleotropic effects unrelated to the original function, heterologous overexpression of AGL6 genes from Hyacinthus orientalis [24], Oncidium Gower Ramsey [25], Dendrocalamus latiflorus [26] and Picea abies [2] in Arabidopsis or Nicotiana suggested several other possible functions for AGL6 genes: a role in the juvenile to adult transition in Norway spruce [2] or a role as regulator of floral transition in the studied monocot species [24-26]. The latter function was corroborated by the constitutive expression in Arabidopsis of AGL6 or of AGL6- EAR, a repressor domain [15]. However, this early flowering phenotype was not obtained when an activated form (AGL6-VP16) was expressed under the AGL6 promoter, leaving a putative function for the AGL6 gene in the regulation of Arabidopsis flowering time to be confirmed.
The long-awaited functional characterization of AGL6 now highlights the need for a phylogenetic framework to understand the origin and diversification of AGL6 gene functions. The evolutionary history of the AGL6 gene lineage has been addressed in detail for grasses indicating that paralogs of AGL6 in Oryza sativa originate from an ancient duplication before the origin of the grass clade [23]. Here we present an analysis of the phylogenetic history of AGL6 genes in angiosperms by extending the available AGL6 sequence sampling with newly isolated asterid and EST-based eudicot sequences. Our inferences indicate four previously unidentified ancient duplications: one at the base of core eudicots resulting in what we have named AGL6-like and euAGL6 gene clades, two in monocots and a fourth one in basal angiosperms. Tracing expression patterns along the phylogeny of the AGL6 lineage indicates that expression of paralogs after the major core eudicot duplication has diverged to include expression in vegetative tissues.
Methods
Cloning of AGL6 MADS-box genes
Floral buds from Philadelphus pubescens (Hydrangeaceae), Alangium platinifolium (Cornaceae), Galax urceolata (Diapensiaceae), Diospyros digyna (Ebenaceae), Gustavia brasiliensis (Lecythidaceae), Roridula gorgonias (Roridulaceae), Saurauia zahlbruckneri, Actinidia chinensis (Actinidiaceae), Asarum europaeum (Aristolochiaceae), Papaver somniferum (Papaveraceae), Berberis julianae (Berberidaceae), Anemone nemorosa (Ranunculaceae) and Citrus sinensis (Rutaceae) were frozen in liquid nitrogen and stored at -80°C. Total RNA was isolated using the Invisorb Spin Plant RNA kit (Invitek, Berlin, DE) or Trizol (Invitrogen, Carlsbad, US). The mRNA was reverse transcribed into cDNA using AMV reverse transcriptase (Promega, Madison, US) and an oligo-dT primer. Our initial strategy was applying 3'-RACE [27] using primers (AP1MDS3: 5'-GTICARYTIARRMGIATIGARAAYAAGAT-3', RQVT: 5'-CGRCARGTGACSTTCTSCAARCG-3', oligodT: 5'-CCGGATCCTCTAGAGCGGCCGC(T)17-3') and according PCR-programs taken from the literature [21,28]. All PCR amplifications were carried out using Taq DNA Polymerase (Invitrogen, Carlsbad, US). PCR products were gel-purified with a Nucleospin extract 2 kit (Macherey-Nagel, Düren, DE) and cloned into the pGEM-T vector (Promega, Madison, US). After transformation, between 50 and 100 white clones were checked for inserts in a PCR reaction using the same primers and program. Plasmid DNA for selected clones was extracted with the Nucleospin Plasmid kit (Macherey-Nagel, Düren, DE). The plasmid inserts were sequenced using T7 and SP6 universal primers using the BigDye Terminator 1.1 kit (Applied Biosystems, Forster City, US) on an Applied Biosystems 310 sequencer or the plasmids were sent for sequencing (MacroGen Inc., Seoul, KP). AGL6-homologs from Asarum europaeum (AeAGL6 - 293 bp), Anemone nemorosa (AnAGL6 - 199 bp), Berberis julianae (BjAGL6 - 199 bp), Papaver somniferum (PasAGL6 - 199 bp) and Citrus sinensis (CsAGL62 - 98 bp) were cloned using a specific primer combination based on sequences of closely related species. In total, sixteen new AGL6 sequences were deposited in Genbank [Accession numbers HM121967 - HM121982]. In addition, SEPALLATA3-homologs were amplified from Asarum europaeum (AeSEP3 - 209 bp), Papaver somniferum (PsSEP3 - 420 bp), Berberis juliana (BSEP3 - 420 bp) and Anemone nemorosa (AnSEP3 - 420 bp) using specific primers from closely related species. New SEP3-sequences were deposited in Genbank [Accession numbers HM121963 - HM121966]. For all used primers see additional File 1: List of primer sequences used.
Sampling, blast search and phylogenetic analysis
In order to reconstruct the timing of duplication that resulted in paralogous copies in Actinidia chinensis, Roridula gorgonias and Saurauia zahlbruckneri, we performed a BLAST search to look for all available AGL6 genes in the EST-database. The obtained sequences were combined with previously characterized AGL6-like sequences in a matrix (See additional File 2 - List of species used in the phylogenetic analysis with abbrevations and accession numbers). The short AGL6-sequences from Asarum, Berberis, Anemone, Papaver, Citrus and Eschscholzia were not included in this first matrix because they can be expected to artificially lower support values in resampling methods such as bootstrapping. Obtained nucleotide sequences were manually aligned using MacClade 4 [29], according to the reading frame of the conceptually translated amino acid sequences. The alignment starts at the RQVT-site in the MADS-domain and ends just before the stop codon. A multifasta alignment file can be found in the supplementary data (see additional File 3). After alignment, nucleotide sequences were analyzed with PAUP* 4b10 [30], Mr Bayes 3.1.2 [31], GARLI [32] and PHYML [33]. Paup* 4b10 [30] was used for parsimony bootstrap analysis. A Maximum Parsimony (MP) heuristic search was conducted using 1000 random addition sequences with TBR branch swapping and saving of multiple parsimonous trees (MulTrees on). Branch support values were obtained by nonparametric bootstrap analysis on 1000 pseudo-replicate data sets [34]. Parameters for the Bayesian analysis and maximum likelihood analysis were estimated using Modeltest 3.06 [35]. Modeltest selected the GTR+I+G substitution model using the Akaike Information criterion. MrBayes was run for 5 million generations where every 100 generations one tree was saved. The search reached stationarity around 50 000 generations. This number was considered the 'burnin period' and was excluded when the consensus phylogeny was constructed. PHYML [33] and GARLI [32] were used for maximum likelihood inference of the matrix. Confidence in the clades was estimated by the approximate likelihood ratio test method from Anisimova & Gascuel [36] and bootstrap analysis with 100 replicates. For the aLRT-tests, we used both the Chi2 probability and the more conservative SH-like test as branch support measures. The most likely tree was used and ML bootstrap values (>70) and BPP values (>90) were plotted on the three. In a second analysis, to identify the AGL6-sequences from Asarum europaeum, Anemone nemorosa, Berberis julianae, Papaver somniferum, Eschscholzia californica and Citrus sinensis as bona fide AGL6 sequences, we included these short sequences in the previous matrix and performed a second analysis using a similar strategy (See additional File 4 for the phylogenetic tree with bootstrap values from the likelihood analysis). To identify the newly identified SEPALLATA3 sequences as SEP3 homologs, we constructed a matrix with selected representatives from all SEPALLATA-subfamilies. We used AGL6-sequences as outgroup and constructed a neighbour Joining tree and performed parsimony bootstrap analyses with PAUP* 4b10 [30]. The resulting tree can be found in additional File 5. All included genes in this tree are listed in additional File 2.
qRT-PCR quantification of gene expression
To examine the expression patterns of selected AGL6 and SEPALLATA3 genes using qRT-PCR, vegetative parts and floral organs of Houttuynia cordata and Asarum europaeum (Piperales, magnoliids), Eschscholzia californica, Papaver somniferum and Berberis julianae (Ranunculales, eudicots), Vitis vinifera (Vitales, core eudicots), Citrus sinensis (Sapindales, rosids) and Actinidia chinensis (Ericales, asterids) were collected and frozen in liquid nitrogen. Floral material for Asarum, Papaver, Eschscholzia, Berberis, Vitis, Citrus and Actinidia were in the floral bud stage, except for Houttuynia where only mature flower material was available. The floral parts of Eschscholzia, Papaver and Actinidia were dissected from young mature flowers. RNA was extracted from each organ type separately using the Invisorb Spin Plant RNA kit (Invitek, Berlin, DE) or Trizol (Invitrogen, Carlsbad, US) and each RNA sample was DNase treated using TURBO DNA-free (Ambion, Austin, US). To verify the absence of gDNA in the total RNA, we used a PCR-reaction (40 cycles) with actin primers (data not shown). Based on this, we repeated the DNAse treatment for few samples. Total RNA was reverse transcribed into cDNA using AMV reverse transcriptase (Promega, Madison, US) and the included oligo-dT primer and random primers. Real-time PCR was performed on a StepOne Plus apparatus (Applied Biosystems, Forster City, US) using Fast SYBR Green Master Mix (Applied Biosystems, Forster City, US). Primers were constructed using the Primer Express software (Applied Biosystems, Forster City, US). The data presented here are the average of three technical replicates with standard error of the mean and two biological replicates are shown. All samples are normalized against ACTIN expression. Data analysis used the delta CT-method. For Houttuynia cordata, Asarum europaeum and Berberis julianae, ACTIN was cloned using a specific primer pair. New ACTIN sequences were deposited in Genbank [HM121983-HM121985]. The SEPALLATA3 sequences used are VvMADS4 for Vitis vinifera (XM_002275669), CitMADS3 for Citrus sinensis (AB218611), an EST-sequence from Actinidia chinensis (FG527965) identified as SEP3-like (see additional File 5). EScaAGL9 from Eschscholzia californica (AY850180) and HcSEP3 from Houttuynia cordata (AB089159). For all used primers see additional File 1: List of primer sequences used.
Results
Ancient duplications in the AGL6 lineage occurred at the base of the core eudicots and during monocot and magnoliid evolution
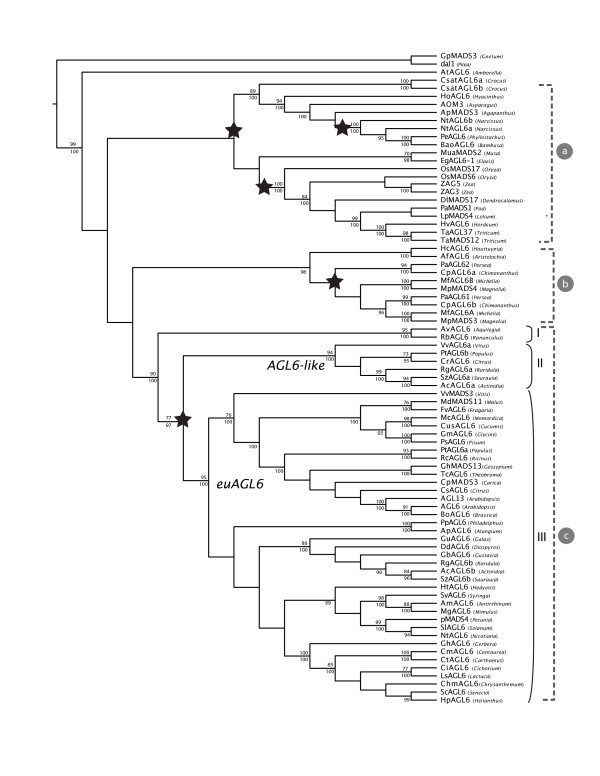
For the reconstruction of the evolution of the AGL6 lineage, we combined previously identified AGL6-homologs with newly isolated and EST-derived AGL6 genes. The obtained data matrix contains two gymnosperm sequences and 79 angiosperm accessions and we analyzed this under maximum parsimony (MP) and maximum likelihood (ML) optimality criteria and in addition we used Bayesian posterior probability distribution estimation (BPP). We rooted the phylogeny by selecting gymnosperm AGL6 genes as outgroup. Because the maximum likelihood estimated topology and the Bayesian phylogram were identical, we present this tree as single most optimal phylogenetic estimate (Figure 1). Bayesian posterior probabilities and bootstrap values from the likelihood analysis are plotted on the tree as support measures for clades (Figure 1). MP bootstrap values, which were almost identical to ML bootstrap values, and branch support values from the aLRT-test, which again confirm the proposed hypothesis, are not shown.
Figure 1.
Phylogeny of AGL6 genes in angiosperms, as inferred from maximum likelihood and Bayesian analysis. Bayesian posterior probabilities (below) and bootstrap values from the likelihood analysis (above) are plotted on the tree. Black star indicates inferred duplication events. Included species, abbreviations and accession numbers are listed in additional file 2: List of species used in the phylogenetic analysis
While in previous phylogenetic analyses of the AGL6 lineage, eudicot sequences were sparsely sampled or the focus was on monocot representatives [16,17,23,37], the focus of this study is on the eudicot clade. Three major groups of AGL6 genes can be recognized in our estimate of phylogenetic relationships (Figure 1): a clade grouping monocot sequences (a); a clade grouping magnoliid sequences (b) and finally a large eudicot clade (c).
Within monocots (Figure 1a), gene relationships are not congruent with species relationships. Two Poales representatives (PeAGL6 from Phyllostachys and BoAGL6 from Bambusa) are nested strongly (ML 89 and BPP 100) in a clade grouping Liliales and Asparagales sequences. This is in turn sister to a clade containing all other Poales sequences (ML 100 and BP 100). The phylogeny thus indicates that during the divergence of monocots a first duplication event has occurred at least before the origin of Liliales [38]. However, no duplicated AGL6 genes were retrieved for any of the species in these groups, suggesting that paralogous copies may have been asymmetrically retained after duplication or remain to be identified. The fact that the Asparagales Narcissus sequence NtAGL6a groups more closely with Poales sequences PeAGL6 and BoAGL6 than with NtAGL6b, indicates a second ancient duplication tracing back to a time before the Asparagales branched off in monocot phylogeny. We would like to note here however, that sampling is particularly sparse to support the latter duplication and that although our analyses provide support, increased taxon sampling would improve accuracy for this inferred node [e.g. [39]]. In addition, our analyses show two duplicate AGL6 sequences for Crocus in Liliales (CsatAGL6a, CsatAGL6b) that we are unable to date more precisely. Finally we retrieve the ancient duplications in Poales shown by Reinheimer & Kellogg [23], leading to two copies in Oryza sativa (OsMADS6 and OsMADS17) and a second one leading to multiple copies in Zea mays (ZAG3 and ZAG5).
Within the magnoliid clade (Figure 1b), which is supported by a BPP-value of 96, both the representatives of Laurales (Persea and Chimonanthus) and Magnoliales (Magnolia and Michelia) possess multiple copies of AGL6. Although our analyses do not provide enough support for the timing of the duplication event, it probably occurred before the evolutionary origin leading to Magnoliales and Laurales [38]. However, another scenario involving two subsequent duplications can not be excluded.
Within the strongly supported eudicot group (Figure 1c, ML 90 and BPP 100), we recognize three supported clades: a basal eudicot clade with the Ranunculales representatives from Aquilegia (AvAGL6) and Ranunculus (RbAGL6) (Figure 1, I, ML 95 and BPP 100), a small core eudicot (Figure 1, II, ML 94 and BPP 100) and a large core eudicot clade (Figure 1, III, ML 95 and BPP 100). We propose to rename the large core eudicot clade the euAGL6 clade, similar to the eudicot duplications in the AGAMOUS, APETALA3 and APETALA1 lineage [5,28,40]. The small core eudicot clade will be referred to as the AGL6-like clade, as no functional characterization of any representative has been performed. In both euAGL6 and AGL6-like clades, rosid AGL6 sequences from Vitis (VvAGL6a and VvMADS3), Populus (PtAGL6a and PtAGL6b), Citrus (CrAGL6 and CsAGL6) and asterid AGL6 sequences from Roridula (RgAGL6a and RgAGL6b), Saurauia (SzAGL6a and SzAGL6b) and Actinidia (AcAGL6a and AcAGL6b) can be identified. Our data indicate that the duplication leading to these two paralogous clades occurred after the divergence of the basal eudicot Ranunculales clade and before the divergence of core eudicots [38]. This scenario is supported by a BPP-value of 97 and a bootstrap value of 77. Although duplications frequently occur in almost all MADS-box gene lineages, this ancient duplication event was not identified in previous analyses of the AGL6 lineage. The AGL6-like clade (Figure 1, I) groups only few species, despite our targeted efforts to identify more sequences belonging to this clade in sequence databases. Within the euAGL6 clade (Figure 1, II), no disruptions of gene to species relationships indicate later additional duplications. As expected from our knowledge of evolutionary relationships within angiosperms, a rosid (ML 76 and BPP 100) and an asterid clade are recognized.
Expression patterns of duplicated eudicot AGL6 genes have diverged and differ from SEP3 genes
Information on expression patterns of AGL6 genes for eudicot representatives is even more limited as compared to monocots [23] and magnoliids [4]. Until now, one rosid species (Arabidopsis thaliana) [13,14], one basal eudicot species (Vitis vinifera) [41] and one asterid species (Petunia hybrida) [17,42] have been investigated for their AGL6 expression patterns. Because our analyses indicate an ancient duplication at the base of the core eudicots with resulting paralogous copies, we studied the expression patterns of these duplicated AGL6 genes in Vitis, Citrus and Actinidia using qRT-PCR (Figure 2) in comparison to expression of SEP3 mRNA's [17].
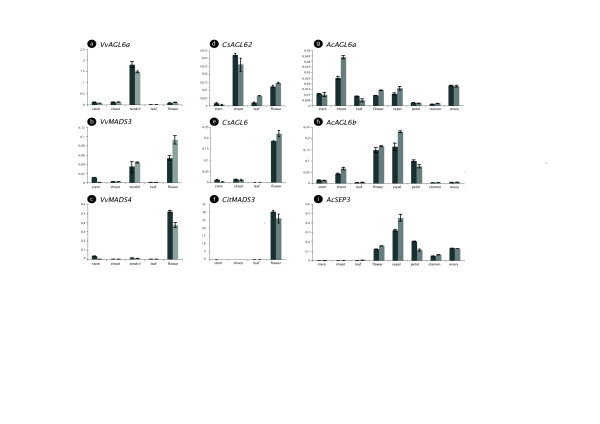
Figure 2.
Expression of duplicated AGL6- and SEPALLATA3-genes in core eudicot species using qRT-PCR. AGL6 and SEP3 qRT-PCR products from Vitis vinifera (a-c), Citrus sinensis (d-f) and Actinidia chinensis (g-i) are shown. Relative expression to ACTIN is shown using the delta Ct method. Floral material for Vitis, Citrus and Actinidia were in the floral bud stage. The floral parts of Actinidia were dissected from young mature flowers.
In Vitis vinifera, expression of the two AGL6 genes differs markedly: while the AGL6-homolog from the AGL6-like clade (VvAGL6a, Figure 2a) is predominantly expressed in tendrils, expression of its paralog (VvMADS3, Figure 2b) is both in tendrils and flowers. Previously, it was shown that the expression of VvMADS3 was restricted to the floral parts of the plant and the expression in tendrils we show here was not detected [41]. This could be explained by the difference of specific sampling of tissues between both studies. The SEP3 gene (VvMADS4, Figure 2c) is most strongly expressed in the flower and confirms the observations of a previous study in Vitis [43]. Thus, both VvMADS3 and VvMADS4 are expressed in the flower, while VvAGL6a is most strongly expressed in tendrils. In contrast to VvMADS4, both AGL6 genes are also expressed in tendrils. Although tendrils would seem vegetative structures in nature, a putative homology of tendrils and reproductive shoots has been proposed, partly on the basis of AP1 and FUL expression [44].
For the rosid genus Citrus, we also found one paralog belonging to the AGL6-like lineage (CrAGL6 and CsAGL62) and one paralog grouping with the euAGL6 clade (CsAGL6). The expression patterns of both genes were examined in vegetative and floral parts of Citrus sinensis. Again, the expression patterns between the duplicated AGL6 genes differ significantly. While the AGL6-like gene (CsAGL62, Figure 2d) is expressed both in the vegetative shoot and the flower of C. sinensis, the euAGL6 gene (CsAGL6, Figure 2e) is predominantly expressed in the flower. We compared this to the expression pattern of a SEP3-homolog in Citrus, CitMADS3 (Figure 2f) [45]. Similar to the results of Endo et al. [45], the SEP3-homolog is strongly expressed in floral tissues. So both CitMADS3 and CsAGL6 are expressed in the flower, while CsAGL62 is expressed predominantly in the vegetative shoot.
Also, in Actinidia chinensis, the two AGL6-paralogs have acquired different expression patterns. AcAGL6a, which belongs to the AGL6-like clade is expressed both in floral parts and in all vegetative tissue samples (Figure 2g), but predominantly in the vegetative shoot. The euAGL6 gene in Actinidia, AcAGL6b is also expressed in both vegetative and floral tissues (sepals and petals), but predominantly in floral tissue (Figure 2h). The expression pattern in floral tissues is rather similar to the expression of the ortholog of AcAGL6a in Petunia, pMADS4, of which the expression is confined to floral organs [17,42]. According to expectations, the SEP3 gene in Actinidia is only expresssed in floral parts (AcSEP3, Figure 2i). The expression of pMADS4 in the floral organs of Petunia, which is mostly in the petal and the ovary, is different from the expression of the AGL6 genes in Actinidia. While the AGL6-like homolog is expressed in sepals and ovaries (AcAGL6a, Figure 2g), the euAGL6 copy is mainly expressed in sepals and petals (AcAGL6b, Figure 2h).
It is obvious from these results that the duplicated AGL6 genes in Vitis, Citrus and Actinidia have acquired different expression patterns after duplication. While in Vitis and Actinidia, the paralog from the AGL6-like clade is strongly expressed in tendrils (Vitis) or in the vegetative shoot (Actinidia), the euAGL6 paralog is expressed in tendrils and floral tissues for Vitis and shoot and floral tissues for Actinidia. Therefore, AGL6 expression in Vitis and Actinidia is different from SEP3 genes, which are predominantly expressed in floral tissues. In the rosid species Citrus, the situation is different; the gene belonging to the AGL6-like clade is expressed in both vegetative and floral parts while the euAGL6 gene has an expression restricted to floral tissues, similar to expression of the SEP3 gene. Thus, while SEP3-expression is always limited to floral tissues, euAGL6 genes are also predominantly expressed in floral tissues with some vegetative expression. Contrary, expression levels of AGL6-like genes are highest in the vegetative shoots and/or tendrils.
Vegetative expression was lost in angiosperms and regained in core eudicots
Our phylogenetic studies reveal that several plant lineages can be expected to harbor more than one AGL6 representative in their genome and that these paralogous sequences can be of ancient origin. Because gene duplicates in MIKC type MADS-box genes have been shown repeatedly to sub- or neofunctionalize, with gene expression divergence as an observable outcome [46-48], we attempted to infer ancestral AGL6 expression patterns in flowering plants. This should help to phylogenetically root and trace the evolution of gene function in this subfamily of MADS-box transcription factors. Previous expression patterns of AGL6 representatives in magnoliids [4,49], Nymphaeles and Proteales [22], non-grass monocots [24,25,37,50] and Poaceae [23] did not find expression in vegetative tissues. However, we showed that some duplicated eudicot AGL6 genes are expressed in vegetative tissues (cfr supra) and in gymnosperms an AGL6-homolog, PrMADS3, was detected in the group of cells initiating needle primordia in vegetative buds from Pinus radiata [20]. To investigate when expression in vegetative tissues may have originated, we quantitatively compared the expression level of AGL6 in vegetative and reproductive tissue samples of several species belonging to the magnoliids and Ranunculales, two clades that originated before the divergence of the core eudicot clade. Again here, we compared the expression of AGL6 to that of SEP3 mRNA's in both vegetative and reproductive tissue samples (Figure 3).
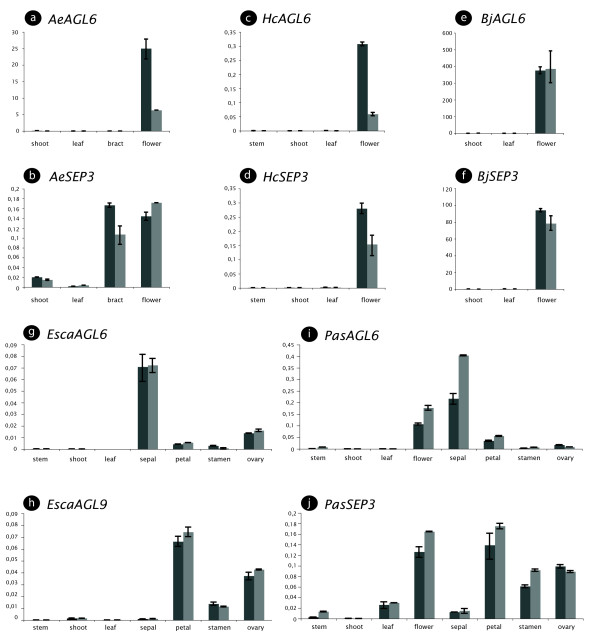
Figure 3.
Expression of AGL6- and SEPALLATA3-representatives in selected magnoliid and Ranunculales species using qRT-PCR. AGL6-like and SEP3-like qRT-PCR products from Asarum europaeum (a-b), Houttuynia cordata (c-d), Berberis julianae (e-f), Eschscholzia californica (g-h) and Papaver somniferum (i-j) are shown. Relative expression to ACTIN is shown using the delta Ct method. Floral material for Asarum, Berberis, Papaver and Eschscholzia were in the floral bud stage, except for Houttuynia where only mature flower material was available. The floral parts of Papaver and Eschscholzia were dissected from young mature flowers.
We selected two representatives from the Piperales (magnoliid clade), Asarum europaeum and Houttuynia cordata (Figure 3a-d). In both species, expression of the AGL6-representatives, AeAGL6 and HcAGL6, is restricted to floral tissue (Figure 3a,c). In contrast to AeAGL6, the SEP3-homolog from Asarum europaeum (AeSEP3) is not only expressed in floral tissue, but also in the bracts enveloping the flower (Figure 3b). In Houttuynia cordata, expression of a SEP3 gene (HcSEP3) is similar to that of a AGL6 gene (Fig 3d). This is in agreement with the observations for Persea americana and Magnolia grandiflora, for which no vegetative expression of AGL6 representatives was detected [4,49].
We next selected three representatives from the Ranunculales, which is close to the ancient duplication event at the base of the core eudicots, to check for expression in vegetative tissue. Expression of AGL6 representatives was investigated in reproductive and vegetative tissue samples from Eschscholzia californica and Papaver somniferum (Papaveraceae) and Berberis julianae (Berberidaceae) using qRT-PCR (Figure 3e-j). In Berberis julianae, we found that AGL6 is solely expressed in the floral tissue (BjAGL6, Figure 3e), similar to SEP3 expression (BjSEP3, Figure 3f). Also for both representatives of the Papaveraceae, Eschscholzia californica (EscaAGL6, Figure 3g) and Papaver somniferum (PasAGL6, Figure 3i), AGL6 is restricted to floral parts. The SEP3 genes from these two species are also predominantly expressed in floral tissues (EscaAGL9 and PasSEP3, Figure 3h,j), yet low expression of PasSEP3 can be found in leaves and even stem (Figure 3j). Within the flower, expression of both EscaAGL6 and PasAGL6 is mainly maintained in the sepals with low expression in the ovary for EscaAGL6 and in the petals for PasAGL6 (Figure 3g,j). In contrast, the expression of EscaAGL9 and PasSEP3 (Figure 3h,j) is in all floral organs, except in sepals.
The detected vegetative expression pattern in gymnosperms and several core eudicots and the absence in monocots, magnoliids and Ranunculales suggests that expression in the shoot apical meristem or more broadly the meristem and young leaves (entire shoot) was lost around the origin of the angiosperm clade and regained after the duplication at the base of the core eudicots.
Discussion
The AGL6 gene phylogeny is not congruent with the corresponding species phylogeny
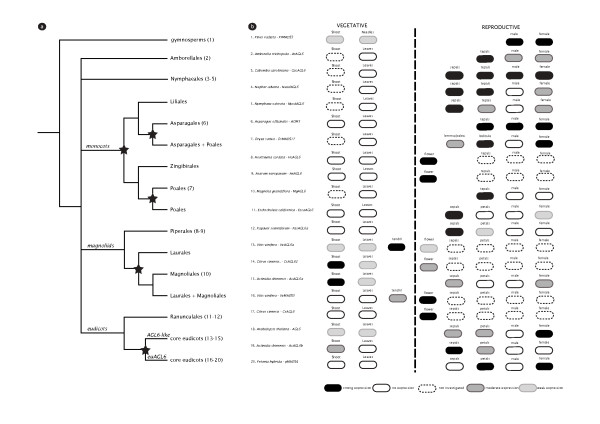
Our phylogenetic estimates indicate five ancient angiosperm duplications in the molecular evolution of the AGL6 lineage of which four were not detected in previous analyses [14,16,17,24,37] (summarized in Figure 4a). Two of these duplications occurred during monocot evolution: a first one before the Liliales branched off and a second one before Asparagales branched off. A third ancient duplication could be demonstrated during magnoliid diversification. Probably least expected, a fourth duplication event occurred before the origin of the core eudicot clade resulting in euAGL6 and AGL6-like genes. In contrast to previous phylogenetic analyses, it required the inclusion of sequences from basal asterids and EST-derived eudicot AGL6 sequences to generate support for the core eudicot duplication, illustrating that the pattern of gene loss or divergence in expression patterns of MADS-box genes requires elaborate sequence and taxon sampling to reveal the molecular evolutionary history of this gene family.
Figure 4.
Evolution of the AGL6 lineage and AGL6 expression patterns in gymno- and angiosperms. (a) Summary of the results of the phylogenetic analysis. Black stars indicate inferred duplication events. (b) Expression of selected representatives in both vegetative and reproductive structures is shown: strong expression (black), moderate expression (gray), weak expression (light gray), absence of expression (white) or not investigated expression (white with dashed line). Expression data were taken from Mouradov et al. [20], Kim et al. [4], Ohmori et al. [16], Reinheimer & Kellogg [23], Rijpkema et al. [17], Schauer et al. [14], Yoo et al. [22] and this study.
As is the case for the APETALA1, APETALA3, AGAMOUS and SEPALLATA gene lineages [5,28,40,51-53], the AGL6 lineage underwent a duplication that can be traced back to the origin of the core eudicot clade. It has been suggested that the resulting paralogous sequences could be the result of a whole genome duplication in an ancestor of core eudicots after which transcription factors have been preferentially retained [5,54]. Yet, an alternative scenario involving multiple independent duplication events has also been suggested as an explanation for the resulting duplicate lineages and both scenarios are not mutually exclusive [53]. Our results indicate that the euAGL6 and AGL6-like sequences were likely retained early after a genome duplication around the origin of the core eudicots. The phylogeny further shows that the euAGL6 lineage is represented by many more sequences than the AGL6-like lineage. One possible explanation for this observation is that the AGL6-like lineage was lost more frequently in later speciation events, resulting in a small core eudicot clade with few paralogous sequences. This is illustrated by the absence of the AGL6-like lineage in the genome of Arabidopsis thaliana as both AGL6 and AGL13 from Arabidopsis belong to the euAGL6 clade. Alternatively, the fact that one clade is much smaller than the other could equally well indicate that members of the smaller clade are expressed in a more restricted or different temporal or spatial domain not sampled by EST sequencing or missed in targeted cloning efforts. Although we are currently unable to attribute functions or diversification in function of the duplicated core eudicot AGL6-like sequences in this study, expression divergence between the paralogous copies is strong, suggesting significant evolution in the transcriptional regulation of these genes.
Maintenance in male organs appears ancestral in flowering plants, but was frequently lost and vegetative expression was gained in core eudicots
Ancestral expression patterns of AGL6 in reproductive structures were previously reconstructed in Schauer et al. [14] and Reinheimer & Kellogg [23]. It was suggested that expression of the AGL6 lineage in floral meristems has been conserved since angiosperm diversification [23]. In angiosperms, the inferred ancestral expression included both male and female reproductive tissues and male-specific expression was lost later in evolution [14]. Our updated summary of AGL6 expression patterns confirms this inference and adds to the complexity of the evolution of gene expression in the AGL6 lineage (Figure 4b). A recent study that included Nymphaeales AGL6 genes shows that expression of AGL6 in Nymphaea or Nuphar is absent from stamens, while out of three AGL6 paralogs from Cabomba, only CacaAGL6-2 is detected in male organs [22]. Also in grasses, expression in male organs was lost [23]. In basal eudicots, AGL6 expression in Nelumbo (Proteales) is weak in stamens [22] and in our study we find that in both Eschscholzia and Papaver, expression is not maintained in stamens relative to other organs. In core eudicots, for neither the euAGL6 or the AGL6-like lineages, we detected expression in stamens. Together these results indicate that although originally in gymnosperms and angiosperms expression was likely maintained in both male and female organs, from the early diversification of flowering plants, expression maintenance in male organs disappeared in most lineages. We have to mention that the expression levels summarized in Figure 4b may depend on the developmental stage of the (floral) tissues sampled, which in turn may influence a comprehensive summary of the data currently available.
We also investigated the expression in developing vegetative tissues. Consistent with previous observations, we find that the expression of AGL6 genes in angiosperms is confined to reproductive tissues before the origin of core eudicots (Figure 4b). After the origin of the core eudicot euAGL6 and AGL6-like lineages, these lineages gained expression in vegetative tissues. Yet in Actinidia (Ericales) and Citrus (Rutales), expression of AGL6-like is stronger in vegetative shoot tissue than in floral bud tissue. Also in Vitis (Vitales), we measure similar expression levels of the AGL6-like gene in vegetative shoots and flower tissue and intriguingly, expression is most pronounced in tendrils. In contrast, the euAGL6 lineage has overall retained its expression domain confined to floral tissues and moderate expression is also observed for vegetative tissues of Actinidia. For several other members of this lineage, expression in vegetative tissues is either confined to a restricted domain (e.g. Arabidopsis) or not detected at all (e.g. Petunia), suggesting that the euAGL6 lineage overall performs roles confined to reproductive development, though additional roles in vegetative development cannot be excluded for some species. Furthermore, functional diversification of the euAGL6 and AGL6-like lineages is not evident from characteristic motifs found at the C-terminal end of the proteins, as has been observed for several other MADS-box gene lineages that originated before the origin of core eudicots [5,28,40,51-53]. Rather, the phylogenetic signal that separates the two lineages is distributed throughout their sequences. Overall, our and previous data suggest that the expression in vegetative tissues of the AGL6 lineage is novel in core eudicots. More extensive studies are needed to determine when relative vegetative expression in core eudicots was acquired and how euAGL6 and AGL6-like lineages parsed an ancestral AGL6 function.
A putative role of AGL6 genes in vegetative development
In Arabidopsis, both AGL6 and AGL13 paralogs belong to the euAGL6 lineage, and no AGL6-like sequence has been annotated, suggesting that the AGL6-like lineage was lost in the evolution to Arabidopsis. Yet data from Arabidopsis may be indicative of multiple roles for the AGL6 subfamily. Several lines of evidence have now established a (redundant) function for the AGL6 lineage in reproductive development [16-18]. Recent two-hybrid and three-hybrid assays have indicated that the Arabidopsis AGL6 protein interacts with other MADS-domain proteins known to perform roles restricted to reproductive development such as AGAMOUS, APETALA1, SHATTERPROOF2 and SEPALLATA3 [11,55]. The function of PhAGL6 in Petunia was indeed only revealed in double mutant combinations with PhSEP3 [17] and similarly the moderate mfo1 phenotype in rice became severe in combination with a mutation in the SEP-like gene LHS1 [16]. In a third example, mutation of the maize AGL6 homolog bearded ear has a number of phenotypes shared with mutation in the maize AGAMOUS homolog ZEA AGAMOUS 3 and the combination of bde and zag3 results in the complete conversion of floral meristems into branch-like meristems [18].
In addition to the role in reproductive development, we have observed that members of the AGL6-like lineage acquired expression in vegetative shoot tissue. We speculate that this expression could be related to a function in flowering time. Several other observations can be interpreted to point in this direction. The protein interaction profile is again indicative: Arabidopsis AGL6 interacts with SHORT VEGETATIVE PHASE, SUPRESSOR OF OVEREXPRESSION OF CONSTANS1 and FRUITFUL [11]. These proteins have a well-described regulatory role in the transition to flowering, a process regulated in the leaf and shoot apical meristem. FUL and SOC1 are indeed expressed during vegetative development and their expression is apparent in the shoot apical meristem upon floral induction [56-58]. An additional combined function in the cambium has recently been described for these proteins [59]. Similar to FUL and SOC1, AGL13 appears to be expressed in the vasculature of Arabidopsis, leaving the possibility that AGL6 genes are regulating processes in the cambium together with FUL and SOC1. The idea that members of the AGL6 lineage, possibly in combination with FUL, may perform an additional role related to the phase transition of the adult shoot into the reproductive developmental program is further suggested by the fact that both Arabidopsis AGL6 and FUL are strongly upregulated in a microRNA172a mutant background [60]. MicroRNA172 targets members of the APETALA2 family of transcription factors such as SCHLAFMÜTZE, which in turn act as repressors of flowering [61]. Such a regulatory role for AGL6 or AGL13 might have gone unnoticed thus far as a result of redundancy with other factors. A recent study similarly suggests a function related to flowering time for Arabidopsis AGL6 [15]. Constitutive expression or fusion to the EAR repressor domain resulted in early flowering and in such lines, expression levels of known regulators of flowering time were modified. Earlier ectopic expression experiments of AGL6 genes from several monocot species [24-26] similarly resulted in early flowering Arabidopsis transformants. However, in another experiment, the effect on flowering time was absent when AGL6 was expressed under its native promoter, which is unexpected if AGL6 would play a role in controlling Arabidopsis flowering time [15]. A similar series of experiments should be performed for its paralog, AGL13, to be able to extend conclusions to the entire euAGL6 lineage in Arabidopsis. The shoot expression observed in this study contributes to the idea that the AGL6 lineage may have regained a function in regulating aspects of the floral transition in core eudicots. This should be functionally investigated in a species that retained both euAGL6 and AGL6-like gene lineages.
Conclusions
Phylogenetic inference of the AGL6 subfamily of MADS-box transcription factors indicates that four ancient duplications occurred during the evolution of this lineage. As is the case for other MADS-box gene lineages, one of these duplications occurred at the base of the core eudicots and resulted in euAGL6 and AGL6-like gene clades of which the representatives show strong expression divergence. Thus far, AGL6 gene expression was observed only in reproductive structures, but our analyses indicate additional expression in vegetative shoots after the core eudicot duplication. Though speculative, this may indicate that AGL6 genes perform a function in the developmental transitions of shoots, in addition to their function in the reproductive structures.
Authors' contributions
TV and KG layed out the work performed. TV performed experiments and TV and KG drafted the manuscript. DV provided a SEP3-alignment. AB provided material of Eschscholzia californica and critically revised the manuscript. SM provided additional ideas for the discussion of data in the manuscript. All authors read and approved the final manuscript.
Supplementary Material
List of primer sequences used in the expression analysis.
List of species used in the phylogenetic analysis of AGL6 and SEP3 with abbrevations and accession numbers (black stars indicate EST-data).
Multifasta alignment file of the AGL6-matrix.
Identification of new AGL6 representatives from Asarum europaeum, Anemone nemorosa, Berberis julianae, Papaver somniferum, Eschscholzia californica, Anemone nemorosa and Citrus sinensis (highlighted in gray). Bootstrap values from the likelihood analysis are plotted on the most likely tree as support meausures.
Identification of new SEP3-representatives from Asarum europaeum, Berberis julianae, Anemone nemorosa, Papaver somniferum and Actinidia chinensis. Neighbour-Joining tree with bootstrap values (above branches) and bootstrap values from parsimony analysis (below branches). Newly identified SEP3-sequences are highlighted in gray.
Contributor Information
Tom Viaene, Email: tom.viaene@bio.kuleuven.be.
Dries Vekemans, Email: dries.vekemans@bio.kuleuven.Be.
Annette Becker, Email: annette.becker@flower-evodevo.com.
Siegbert Melzer, Email: siegbert.melzer@bio.kuleuven.be.
Koen Geuten, Email: koen.geuten@bio.kuleuven.be.
Acknowledgements
We would like to thank Nand Van Belle and Viviane Leyman from the National Botanical Garden of Belgium, Paul Uyttebroeck from the Kruidtuin in Leuven, Art Vogel from the Hortus Botanicus in Leiden and the Berkeley Botanical Garden for the collection of fresh flower material. Thanks to Anja Vandeperre for technical assistance and support. Thanks to Sabrina Lange (Becker Lab) for providing Escholzia californica plant material. This research was supported by the Institute for the Promotion of Innovation through Science and Technology in Flanders (IWT Vlaanderen, no. 35040 to T.V.). General financial support was provided by K.U.Leuven (OT/05/35) and the Fund for Scientific Research-Flanders (Belgium) (FWO G.0418.08).
References
- Becker A, Theissen G. The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol Phyl Evol. 2003;29:464–489. doi: 10.1016/S1055-7903(03)00207-0. [DOI] [PubMed] [Google Scholar]
- Carlsbecker A, Sundström J, Tandre K, Englund M, Kvarnhede A, Johanson U, Engström P. The DAL10 gene from Norway spruce (Picea abies) belongs to a potentially gymnosperm-specific subclass of MADS-box genes and is specifically active in seed cones and pollen cones. Evol Dev. 2003;5:551–561. doi: 10.1046/j.1525-142X.2003.03060.x. [DOI] [PubMed] [Google Scholar]
- Nam J, dePamphilis CW, Ma H, Nei M. Antiquity and evolution of the MADS-box gene family controlling flower development in plants. Mol Biol Evol. 2003;20:1435–1447. doi: 10.1093/molbev/msg152. [DOI] [PubMed] [Google Scholar]
- Kim S, Koh J, Yoo MJ, Kong H, Hu Y, Ma Y, Soltis P, Soltis E. Expression of floral MADS-box genes in basal angiosperms: implications for the evolution of floral regulators. Plant J. 2005;43:724–744. doi: 10.1111/j.1365-313X.2005.02487.x. [DOI] [PubMed] [Google Scholar]
- Zahn LM, Kong H, Leebens-Mack JH, Kim S, Soltis PS, Landherr LL, Soltis DE, dePamphilis CW, Ma H. The evolution of SEPALLATA subfamily of MADS-box genes: a preangiosperm origin with multiple duplications throughout angiosperm history. Genetics. 2005;169:2209–2223. doi: 10.1534/genetics.104.037770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theissen G, Melzer R. Molecular mechanisms underlying origin and diversification of the angiosperm flower. Ann Bot. 2007;100:603–619. doi: 10.1093/aob/mcm143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrandiz D, Gu Q, Martienssen R, Yanofsky MF. Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development. 2000;127:725–734. doi: 10.1242/dev.127.4.725. [DOI] [PubMed] [Google Scholar]
- Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature. 2000;405:200–203. doi: 10.1038/35012103. [DOI] [PubMed] [Google Scholar]
- Favaro R, Pinyopich A, Battaglia R, Kooiker M, Borghi L, Ditta G, Yanofsky MF, Kater MM, Colomba L. MADS-box proteins complexes control carpel and ovule development in Arabidopsis. Plant Cell. 2003. pp. 2603–2611. [DOI] [PMC free article] [PubMed]
- Ditta G, Pinyopich A, Robles P, Pelaz S, Yanofsky MF. The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr Biol. 2004;14:1935–1940. doi: 10.1016/j.cub.2004.10.028. [DOI] [PubMed] [Google Scholar]
- Immink GH, Tonaco IAN, de Folter S, Shchennikova A, van Dijk AJ, Busscher-Lange J, Borst JW, Angenent GC. SEPALLATA3: the glue for MADS box transcription factor complex formation. Genome Biology. 2009;10:R24. doi: 10.1186/gb-2009-10-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Yanofsky MF, Meyerowitz EM. AGL1-AGL6, an Arabidopsis gene family with similarity to floral homeotic and transcription factor genes. Genes Dev. 1991;5:484–495. doi: 10.1101/gad.5.3.484. [DOI] [PubMed] [Google Scholar]
- Rounsley SD, Ditta GS, Yanofsky MF. Diverse roles for MADS box genes in Arabidopsis development. Plant Cell. 1995;7:1259–1269. doi: 10.1105/tpc.7.8.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer SE, Schlüter PM, Baskar R, Gheyselinck J, Bolanos A, Curtis MD, Grossniklaus U. Intronic regulatory elements determine the divergent expression patterns of AGAMOUS-LIKE6 subfamily members in Arabidopsis. Plant J. 2009;59:987–1000. doi: 10.1111/j.1365-313X.2009.03928.x. [DOI] [PubMed] [Google Scholar]
- Koo SC, Bracko O, Park MS, Schwab R, Chun HJ, Park KM, Seo JS, Grbic V, Balasubramanian S, Schmid M, Godard F, Yun DJ, Lee SY, Cho MJ, Weigel D, Kim MC. Control of lateral organ development and flowering time by the Arabidopsis thaliana MADS-box gene AGAMOUS-LIKE6. Plant J. 2010. published online 2/03. [DOI] [PubMed]
- Ohmori S, Kimizu M, Sugita M, Miyao A, Hirochika H, Uchida E, Nagato Y, Yoshida H. MOSAIC FLORAL ORGANS1, an AGL6-like MADS box gene regulates floral organ identity and meristem fate in rice. Plant Cell. 2009;21:3008–3025. doi: 10.1105/tpc.109.068742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijpkema AS, Zethof J, Gerats T, Vandenbussche M. The petunia AGL6 gene has a SEPALLATA-like function in floral patterning. Plant J. 2009;60:1–9. doi: 10.1111/j.1365-313X.2009.03917.x. [DOI] [PubMed] [Google Scholar]
- Thompson BE, Bartling L, Whipple C, Hall DH, Sakai H, Schmidt R, Hake S. Bearded-ear encodes a MADS box transcription factor critical for maize floral development. Plant Cell. 2009;21:2578–2590. doi: 10.1105/tpc.109.067751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tandre K, Albert VA, Sundas A, Engström P. Conifer homologues to genes that control floral development in angiosperms. Plant Mol Biol. 1995;27:69–78. doi: 10.1007/BF00019179. [DOI] [PubMed] [Google Scholar]
- Mouradov A, Glassick TV, Hamdorf BA, Murphy LC, Maria SS, Yang Y, Teasdale RD. Family of MADS-box genes expressed early in male and female reproductive structures of Monterey pine. Plant Phys. 1998;117:55–61. doi: 10.1104/pp.117.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter KU, Becker A, Münster T, Kim JT, Saedler H, Theissen G. MADS-box genes reveal that gnetophytes are more closely related to conifers than to flowering plants. Proc Natl Acad Sci USA. 1999;96:7342–7347. doi: 10.1073/pnas.96.13.7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo MJ, Soltis PS, Soltis DE. Expression of floral MADS-box genes in two divergent water lilies: Nymphaeales and Nelumbo. Int J Plant Sci. 2010;171:121–146. doi: 10.1086/648986. [DOI] [Google Scholar]
- Reinheimer R, Kellogg EA. Evolution of AGL6-like MADS-box genes in grasses (Poaceae): ovule expression is ancient and palea expression is new. Plant Cell. 2009;21:2591–2605. doi: 10.1105/tpc.109.068239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan JH, Li WQ, Dong XC, Guo W, Shu HR. Ectopic expression of a hyacinth AGL6 homolog caused ealier flowering and homeotic conversion in Arabidopsis. Sci China Ser C-Life Sci. 2007;50:676–689. doi: 10.1007/s11427-007-0083-4. [DOI] [PubMed] [Google Scholar]
- Hsu HF, Huang CH, Chou LT, Yang CH. Ectopic expression of an orchid (Oncidium Gower Ramsey) AGL6-like gene promotes flowering by activating flowering time genes in Arabidopsis thaliana. Plant Cell Physiol. 2003;44:783–794. doi: 10.1093/pcp/pcg099. [DOI] [PubMed] [Google Scholar]
- Tian B, Chen Y, Yan Y, Li D. Isolation and ectopic expression of a bamboo MADS-box gene. Chin Sci Bull. 2005;50:217–224. [Google Scholar]
- Frohman MA, Dush MK, Martin GR. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litt A, Irish VF. Duplication and diversification in the APETALA1/FRUITFULL floral homeotic gene lineage: implactions for the evolution of floral development. Genetics. 2003;165:821–833. doi: 10.1093/genetics/165.2.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison RP, Maddison WP. MacClade 4: analysis of phylogeny and character evolution, version 4.06. Sunderland MA: Sinauer Associates; 2003. [Google Scholar]
- Swofford DL PAUP*Phylogenetic analysis using parsimony (and other methods) Version 4 2002Sunderland, MA: Sinauer Associates; 12504223 [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Zwickl DJ. PhD thesis. The University of Texas. Austin; 2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. [Google Scholar]
- Guindon S, Gascuel O. PhyML - A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- Posada P, Crandall KA. Selecting the best-fit model of nucleotide substitution. Syst Biol. 2001;50:580–601. doi: 10.1080/106351501750435121. [DOI] [PubMed] [Google Scholar]
- Anisimova M, Gascuel O. Approximate likelihood ratio test for branches: a fast, accurate and powerful alternative. Syst Biol. 2006;55:539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- Kanno A, Hienuki H, Ito T, Nakamura T, Fukuda T, Yun PY, Song IJ, Kamimura T, Ochiai T, Yokoyama J, Maki M, Kameya T. The structure and expression of SEPALLATA-like genes in Asparagus species (Asparagaceae) Sex Plant Reprod. 2006;19:133–144. doi: 10.1007/s00497-006-0030-8. [DOI] [Google Scholar]
- APG III. Bot J Linean Soc. 2009;161:105–121. doi: 10.1111/j.1095-8339.2009.00996.x. [DOI] [Google Scholar]
- Geuten K, Massingham T, Darius P, Smets E, Goldman N. Experimental design criteria in phylogenetics: where to add taxa. Syst Biol. 2007;56:609–622. doi: 10.1080/10635150701499563. [DOI] [PubMed] [Google Scholar]
- Kramer EM, Dorit RL, Irish VF. Molecular evolution of genes controlling petal and stamen development: duplication and divergence within the APETALA 3 and PISTILLATA MADS-box gene lineages. Genetics. 1998;149:765–783. doi: 10.1093/genetics/149.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boss PK, Sensi E, Hua C, Davies C, Thomas MR. Cloning and characterization of grapevine (Vitis vinifera L.) MADS-box genes expressed during inflorescence and berry development. Plant Sci. 2002;162:887–895. doi: 10.1016/S0168-9452(02)00034-1. [DOI] [Google Scholar]
- Tsuchimoto S, Mayama T, van der Krol A, Ohtsubo E. The whorl-specific action of a petunia class B floral homeotic gene. Genes Cells. 2000;5:89–99. doi: 10.1046/j.1365-2443.2000.00308.x. [DOI] [PubMed] [Google Scholar]
- Joly D, Perrin M, Gertz C, Kronenberger J, Demangeat G, Masson JE. Expression analysis of flowering genes from seedling-stage to vineyard life of grapevine cv. Riesling. Plant Sci. 2004;166:1427–1436. doi: 10.1016/j.plantsci.2003.12.041. [DOI] [Google Scholar]
- Calonje M, Cubas P, Martinez-Zapatar JM, Carmona MJ. Floral meristem identity genes are expressed during tendril development in grapevine. Plant Phys. 2005;135:1491–1501. doi: 10.1104/pp.104.040832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo T, Shimada T, Fuji H, Omura M. Cloning and characterization of 5 MADS-box cDNA's isolated from citrus fruit tissue. Sci Hort. 2006;109:315–321. doi: 10.1016/j.scienta.2006.06.008. [DOI] [Google Scholar]
- Vandenbussche M, Zethof J, Royaert S, Weterings K, Gerats T. The duplicated B-class heterodimer model: whorl-specific effects and complex genetic interactions in Petunia hybrida flower development. Plant Cell. 2004;16:741–754. doi: 10.1105/tpc.019166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte JM, Cui L, Wall PK, Zhang Q, Zhang X, Leebens-Mack J, Ma H, Altman N, dePamphilis CW. Expression pattern shifts following duplication indicative of subfunctionalization and neofunctionalization in regulatory genes of Arabidopsis. Mol Biol Evol. 2006;23:469–478. doi: 10.1093/molbev/msj051. [DOI] [PubMed] [Google Scholar]
- Chapman M, Leebens-Mack J, Burke J. Positive selection and expression divergence following gene duplication in the sunflower CYCLOIDEA gene family. Mol Biol Evol. 2008;25:1260–1273. doi: 10.1093/molbev/msn001. [DOI] [PubMed] [Google Scholar]
- Chanderbali AS, Kim S, Buzgo M, Zheng Z, Oppenheimer DG, Soltis DE, Soltis PS. Genetic footprints of stamen ancestors guide perianth evolution in Persea (Lauraceae) Int J Plant Sci. 2006;167:1075–1089. doi: 10.1086/507586. [DOI] [Google Scholar]
- Losa A, Caporali E, Spada A, Martinelli S, Marziani G. AOM3 and AOM4: two MADS box genes expressed in reproductive structures of Asparagus officinalis. Sex Plant Reprod. 2004;16:215–221. doi: 10.1007/s00497-003-0199-z. [DOI] [Google Scholar]
- Kramer EM, Jaramillo MA, Di Stilio V. Patterns of gene duplication and functional evolution during the diversification of the AGAMOUS subfamily of MADS box genes in angiosperms. Genetics. 2004;166:1011–1023. doi: 10.1534/genetics.166.2.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer EM, Su HJ, Wu CC, Hu JM. A simplified explanation for the frameshift mutation that created a novel C-terminal motif in the APETALA 3 gene lineage. BMC Evol Biol. 2006;6:30. doi: 10.1186/1471-2148-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan H, Zhang N, Liu C, Xu G, Zhang J, Chen Z, Kong H. Patterns of gene duplication and functional diversification during the evolution of the AP1/SQUA subfamily of plant MADS-box genes. Mol Phyl Evol. 2007;44:26–41. doi: 10.1016/j.ympev.2007.02.016. [DOI] [PubMed] [Google Scholar]
- De Bodt S, Maere S, Van de Peer Y. Genome duplication and the origin of angiosperms. Trends Ecol Evolut. 2005;20:591–597. doi: 10.1016/j.tree.2005.07.008. [DOI] [PubMed] [Google Scholar]
- de Folter S, Immink R GH, Kieffer M, Parenicova L, Henz SR, Weigel D, Busscher M, Kooiker M, Colombo L, Kater MM. Comprehensive interaction map of the Arabidopsis MADS box transcription factors. Plant Cell. 2005;17:1424–1433. doi: 10.1105/tpc.105.031831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hempel FD, Weigel D, Mandel MA, Ditta G, Zambryski PC, Feldman LJ, Yanofsky MF. Floral determination and expression of floral regulatory genes in Arabidopsis. Development. 1997;124:3845–3853. doi: 10.1242/dev.124.19.3845. [DOI] [PubMed] [Google Scholar]
- Borner R, Kampmann G, Chandler J, Gleissner R, Wisman E, Apel K, Melzer S. A MADS domain gene involved in the transition to flowering in Arabidopsis. Plant J. 2000;24:591–599. doi: 10.1046/j.1365-313x.2000.00906.x. [DOI] [PubMed] [Google Scholar]
- Lee H, Suh SS, Park E, Cho E, Ahn J H, Kim SG, Lee J S, Kwon Y M, Lee I. The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev. 2000;14:2366–2376. doi: 10.1101/gad.813600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melzer S, Lens F, Gennen J, Vanneste S, Rohde A, Beeckman T. Flowering-time genes modulate meristem determinacy and growth form in Arabidopsis thaliana. Nat Genet. 2009;40:1489–1492. doi: 10.1038/ng.253. [DOI] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmerman P. Genevestigator V3: a reference expression database for the meta-analysis of transcriptomes. Adv in Bioinformatics. 2008. 420747. [DOI] [PMC free article] [PubMed]
- Mathieu J, Yant LJ, Mürdter F, Küttner F, Schmid M. Repression of flowering by the miR172 target SMZ. PloS Biol. 2009;7:e1000148. doi: 10.1371/journal.pbio.1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of primer sequences used in the expression analysis.
List of species used in the phylogenetic analysis of AGL6 and SEP3 with abbrevations and accession numbers (black stars indicate EST-data).
Multifasta alignment file of the AGL6-matrix.
Identification of new AGL6 representatives from Asarum europaeum, Anemone nemorosa, Berberis julianae, Papaver somniferum, Eschscholzia californica, Anemone nemorosa and Citrus sinensis (highlighted in gray). Bootstrap values from the likelihood analysis are plotted on the most likely tree as support meausures.
Identification of new SEP3-representatives from Asarum europaeum, Berberis julianae, Anemone nemorosa, Papaver somniferum and Actinidia chinensis. Neighbour-Joining tree with bootstrap values (above branches) and bootstrap values from parsimony analysis (below branches). Newly identified SEP3-sequences are highlighted in gray.