Abstract
Background
Cytochrome P450 monooxygenases (P450s) catalyze oxidation of various substrates using oxygen and NAD(P)H. Plant P450s are involved in the biosynthesis of primary and secondary metabolites performing diverse biological functions. The recent availability of the soybean genome sequence allows us to identify and analyze soybean putative P450s at a genome scale. Co-expression analysis using an available soybean microarray and Illumina sequencing data provides clues for functional annotation of these enzymes. This approach is based on the assumption that genes that have similar expression patterns across a set of conditions may have a functional relationship.
Results
We have identified a total number of 332 full-length P450 genes and 378 pseudogenes from the soybean genome. From the full-length sequences, 195 genes belong to A-type, which could be further divided into 20 families. The remaining 137 genes belong to non-A type P450s and are classified into 28 families. A total of 178 probe sets were found to correspond to P450 genes on the Affymetrix soybean array. Out of these probe sets, 108 represented single genes. Using the 28 publicly available microarray libraries that contain organ-specific information, some tissue-specific P450s were identified. Similarly, stress responsive soybean P450s were retrieved from 99 microarray soybean libraries. We also utilized Illumina transcriptome sequencing technology to analyze the expressions of all 332 soybean P450 genes. This dataset contains total RNAs isolated from nodules, roots, root tips, leaves, flowers, green pods, apical meristem, mock-inoculated and Bradyrhizobium japonicum-infected root hair cells. The tissue-specific expression patterns of these P450 genes were analyzed and the expression of a representative set of genes were confirmed by qRT-PCR. We performed the co-expression analysis on many of the 108 P450 genes on the Affymetrix arrays. First we confirmed that CYP93C5 (an isoflavone synthase gene) is co-expressed with several genes encoding isoflavonoid-related metabolic enzymes. We then focused on nodulation-induced P450s and found that CYP728H1 was co-expressed with the genes involved in phenylpropanoid metabolism. Similarly, CYP736A34 was highly co-expressed with lipoxygenase, lectin and CYP83D1, all of which are involved in root and nodule development.
Conclusions
The genome scale analysis of P450s in soybean reveals many unique features of these important enzymes in this crop although the functions of most of them are largely unknown. Gene co-expression analysis proves to be a useful tool to infer the function of uncharacterized genes. Our work presented here could provide important leads toward functional genomics studies of soybean P450s and their regulatory network through the integration of reverse genetics, biochemistry, and metabolic profiling tools. The identification of nodule-specific P450s and their further exploitation may help us to better understand the intriguing process of soybean and rhizobium interaction.
Background
Cytochrome P450 monooxygenases (P450s) are enzymes found in most organisms from bacteria, to plants and human [1]. They catalyze the oxidation of various substrates using oxygen and NAD(P)H. In plants, large numbers of P450 genes form complex super-families and play important roles in many plant metabolic processes. They are involved in biosynthesis of pigments (anthocyanins), accessory pigments (carotenoids), defense-related compounds (some phytoalexins), UV protectants (flavonoids and sinapoyl esters), structural polymers (lignins), and fatty acids. P450s also contribute to the homeostasis of signalling molecules such as plant hormones because they are frequently the rate-limiting enzymes of hormone biosyntheses [2]. Similar to their functions in animals, P450s are also responsible for degradation of endogenous as well as exogenous compounds such as herbicides, insecticides and pollutants [3].
Structurally, the bacterial P450s are soluble proteins. In contrast, all plant P450s studied so far are membrane-localized. Most of them are anchored on the cytoplasmic surface of the endoplasmic reticulum (ER) by a hydrophobic peptide present at the N-terminus, possibly forming a trans-membrane segment [4]. Analysis of the Arabidopsis and other plant P450 sequences predicts potential signal peptides that should target some of the P450s to the plastids or to the mitochondria [2], though no mitochondrial P450s are known in plants. The number of P450 genes in plants is much higher than in other organisms, correlating to the fact that plants produce a huge repertoire of primary and secondary metabolites. For nomenclature and classification of diverse P450 genes, a universal system has been set up based on the protein sequence identity and phylogeny [5]. Briefly, P450 proteins that share at least 40% identity are assigned to the same family. They are further grouped into sub-families that share at least 55% identity. A few exceptions to this nomenclature system do occur, especially in plants where gene duplication events make it more complex. In such cases, phylogeny and gene organization are taken as criteria for family assignment. P450 genes for all organisms are named and classified by a P450 nomenclature committee in chronological order of sequence submission (David Nelson: dnelson@uthsc.edu). To distinguish from other organisms, plant P450s are classified into families from CYP71A1 to CYP99XY, and then from CYP701A1 and above [2]. P450s in plants are traditionally classified in two types: the A-type and the non-A type [6,7]. Recently, plant P450s have been re-classified into 11 clans. The A-type is now grouped as the CYP71 clan and the non-A type has 10 clans including the CYP51, CYP72, CYP74, CYP85, CYP86, CYP97, CYP710, CYP711, CYP727, and CYP746 clan [7].
Here, we carried out a systematic analysis of the soybean genome for P450 gene families. Soybean (Glycine max) is one of the most important legume species and a leading oilseed crop in the world. Processed soybeans are the largest source of vegetable oil and protein feed. According to a USDA report, soybean represented 56% of world oilseed production in 2008 http://www.soystats.com/2009/. The soybean genome has recently been sequenced and various large-scale expression analyses have been established in soybean, providing unique resources for genomic analysis of this important gene family [8]. Importantly, the soybean P450 information can be compared to other fully sequenced plant genomes. As of September 2010, from the fully sequenced plant genomes, there are 71 full-length P450 genes in moss Physcomitrella patens, 245 in Arabidopsis, 332 in rice, and 310 in poplar [9].
Since membrane-bound enzymes are traditionally more difficult to study, few P450s have been functionally characterized. In the Arabidopsis genome, only 41 of the 245 coding sequences have been associated with a specific biochemical function [10]. Recently, a large-scale P450 co-expression analysis with functional annotation of the 245 Arabidopsis P450 genes was performed to predict the function of unknown P450 genes [11]. This co-expression analysis revealed expression patterns of the majority of Arabidopsis P450s, and provided novel clues on individual P450 functions, pathways, and their regulatory networks. A novel phenolic pathway in pollen development was identified based on this co-expression analysis [12].
In this study, using in silico resources and bioinformatics tools, we identified and annotated putative functional P450 encoding sequences in the soybean genome. Phylogenetic analysis using amino acid sequences allowed us to identify gene orthologs and clusters of orthologous groups for further characterizations. By analyzing large-scale microarray and Illumina sequencing data, we also analyzed the co-expression of P450 genes in soybean, which could provide important clues to their function. We were able to identify tissue-specific P450 genes that may play roles in biological process like nodulation, floral development, and seed maturation. The large-scale expression analyses were confirmed for selected P450 genes using quantitative real-time reverse-transcription polymerase chain reaction (qRT-PCR).
Results and Discussions
Soybean cytochrome P450 genes
A large number of soybean P450 genes have been deposited into the cytochrome P450 database http://drnelson.uthsc.edu/CytochromeP450.html following the completion of the soybean genome sequencing. A total of 332 full-length soybean P450 genes and 378 pseudogenes with Glyma location markers were retrieved from the database. To screen for additional P450 genes, we performed BLAST search using standard P450 domains against Phytozome 4.0 and confirmed all 332 putative P450 genes in the genome. No new full-length P450 gene was discovered by three sets of domain search algorithms, including Pfam, Panther, and KOG. Additionally, we were able to identify 13 new pseudogenes from the domain searches, which were later confirmed and classified by the P450 nomenclature committee and updated in the cytochrome P450 database (as of September, 2010). Currently, the cytochrome P450 database lists all the soybean P450 genes, pseudogenes, and their corresponding Glyma location markers. These genes are classified into A-type and Non-A type P450s. There are 20 families of the A-type and 28 families of the non-A type P450s in soybean, consisting of 195 and 137 full-length sequences, respectively (Tables 1 and 2).
Table 1.
Comparison of A-type P450 families among soybean, Medicago, Arabidopsis, rice, poplar, grape and moss.
| Family | Soybean | Medicago | Arabidopsis | Rice | Popular | Grape | Moss |
|---|---|---|---|---|---|---|---|
| A-Type | |||||||
| CYP71 Clan | |||||||
| CYP71 | 55 | 37 | 52 | 84 | 25 | 24 | 0 |
| CYP73 | 3 | 1 | 1 | 3 | 3 | 3 | 4 |
| CYP75 | 7 | 0 | 1 | 3 | 3 | 11 | 0 |
| CYP76 | 14 | 6 | 8 | 29 | 13 | 24 | 0 |
| CYP77 | 4 | 2 | 5 | 2 | 3 | 2 | 0 |
| CYP78 | 11 | 1 | 6 | 8 | 10 | 7 | 3 |
| CYP79 | 5 | 3 | 7 | 4 | 4 | 9 | 0 |
| CYP80 | 0 | 0 | 0 | 0 | 6 | 6 | 0 |
| CYP81 | 12 | 5 | 18 | 12 | 28 | 21 | 0 |
| CYP82 | 24 | 10 | 5 | 0 | 10 | 34 | 0 |
| CYP83 | 12 | 9 | 1 | 0 | 5 | 0 | 0 |
| CYP84 | 3 | 3 | 2 | 3 | 3 | 3 | 0 |
| CYP89 | 8 | 9 | 7 | 14 | 10 | 14 | 0 |
| CYP92 | 2 | 1 | 0 | 9 | 8 | 6 | 0 |
| CYP93 | 13 | 8 | 1 | 3 | 4 | 4 | 0 |
| CYP98 | 2 | 1 | 3 | 2 | 5 | 1 | 1 |
| CYP99 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| CYP701 | 2 | 1 | 1 | 5 | 1 | 1 | 1 |
| CYP703 | 1 | 1 | 1 | 1 | 1 | 1 | 3 |
| CYP705 | 0 | 0 | 26 | 0 | 0 | 0 | 0 |
| CYP706 | 3 | 1 | 7 | 4 | 5 | 9 | 0 |
| CYP712 | 2 | 1 | 2 | 0 | 9 | 2 | 0 |
| CYP723 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| CYP736 | 12 | 1 | 0 | 0 | 6 | 8 | 0 |
| Others | 0 | 0 | 0 | 0 | 0 | 0 | 29 |
| Total | 195 | 101 | 154 | 190 | 162 | 166 | 41 |
The families highlighted in bold have unique distributions among these species as described in the text.
Table 2.
Comparison of non-A type P450 families among soybean, Medicago, Arabidopsis, rice, poplar, grape and moss.
| Family | Soybean | Medicago | Arabidopsis | Rice | Popular | Grape | Moss |
|---|---|---|---|---|---|---|---|
| Non A-Type | |||||||
| CYP51 clan | |||||||
| CYP51 | 2 | 1 | 1 | 10 | 2 | 2 | 1 |
| CYP72 clan | |||||||
| CYP72 | 12 | 7 | 9 | 13 | 6 | 22 | 0 |
| CYP709 | 0 | 1 | 3 | 9 | 1 | 1 | 0 |
| CYP714 | 6 | 3 | 2 | 5 | 6 | 6 | 0 |
| CYP715 | 6 | 1 | 1 | 1 | 2 | 1 | 0 |
| CYP721 | 2 | 1 | 1 | 2 | 6 | 5 | 0 |
| CYP734 | 3 | 1 | 1 | 4 | 2 | 2 | 0 |
| CYP735 | 3 | 1 | 1 | 2 | 2 | 1 | 0 |
| CYP749 | 0 | 0 | 0 | 0 | 9 | 0 | 0 |
| CYP74 clan | |||||||
| CYP74 | 6 | 4 | 2 | 4 | 6 | 7 | 3 |
| CYP85 clan | |||||||
| CYP85 | 5 | 1 | 2 | 1 | 3 | 2 | 0 |
| CYP87 | 2 | 2 | 1 | 11 | 12 | 7 | 0 |
| CYP88 | 3 | 3 | 2 | 1 | 2 | 2 | 0 |
| CYP90 | 12 | 4 | 4 | 5 | 7 | 4 | 0 |
| CYP702 | 0 | 0 | 6 | 0 | 0 | 0 | 0 |
| CYP707 | 10 | 3 | 4 | 3 | 7 | 5 | 0 |
| CYP708 | 0 | 0 | 4 | 0 | 0 | 0 | 0 |
| CYP716 | 7 | 3 | 2 | 0 | 17 | 15 | 1 |
| CYP718 | 1 | 0 | 1 | 0 | 17 | 15 | 1 |
| CYP720 | 2 | 1 | 1 | 0 | 1 | 1 | 0 |
| CYP722 | 2 | 1 | 1 | 1 | 1 | 1 | 0 |
| CYP724 | 1 | 0 | 1 | 1 | 2 | 2 | 0 |
| CYP728 | 2 | 0 | 0 | 11 | 7 | 6 | 0 |
| CYP729 | 0 | 1 | 0 | 2 | 1 | 0 | 0 |
| CYP733 | 3 | 0 | 0 | 1 | 1 | 1 | 0 |
| CYP86 clan | |||||||
| CYP86 | 9 | 3 | 11 | 5 | 8 | 6 | 2 |
| CYP94 | 14 | 4 | 6 | 18 | 13 | 9 | 2 |
| CYP96 | 7 | 5 | 13 | 12 | 9 | 5 | 0 |
| CYP704 | 5 | 14 | 3 | 7 | 6 | 6 | 6 |
| CYP97 clan | |||||||
| CYP97 | 5 | 4 | 3 | 3 | 3 | 3 | 3 |
| CYP710 clan | |||||||
| CYP710 | 2 | 1 | 4 | 4 | 1 | 1 | 2 |
| CYP711 clan | |||||||
| CYP711 | 4 | 2 | 1 | 5 | 2 | 1 | 0 |
| CYP727 clan | |||||||
| CYP727 | 1 | 0 | 0 | 1 | 2 | 0 | 0 |
| CYP746 clan | |||||||
| CYP746 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Others | 0 | 0 | 0 | 0 | 0 | 0 | 9 |
| Total | 137 | 72 | 91 | 142 | 148 | 124 | 30 |
Non-A type P450 families are organized into 10 clans. The families highlighted in bold have unique distributions among these species as described in the text.
Among these families, CYP71 is the largest A-type family, with 55 members (Table 1); while CYP94 is the largest non-A type family (Table 2), with 14 members. Only four families; CYP703, 718, 724, and 727 consist of a single gene. The remaining 44 families are all multi-gene families most likely due to two genome duplication events in soybean [8,13]. The number of pseudogenes matched well with the number of full-length genes in the A-type families (Additional File 1, Table S1A). For example, CYP71, the largest family also has 65 pseudogenes, the most in all families. In contrast, pseudugene numbers did not match the numbers of full-length genes in non-A type families (Additional File 1, Table S1B). For example, CYP710 has only two full-length genes but 17 pseudogenes. We speculate that non-A type P450s are evolutionally more ancient than A-type families, allowing more time for gene duplication and rearrangement, resulting in more diverse compositions than A-type genes.
The soybean P450 genes were compared to a few selected plant species including Medicago, Arabidopsis, rice, poplar, grape and moss (Tables 1 and 2). A comparison of P450s among soybean, Medicago, Arabidopsis and rice, where comprehensive analyses of the P450 genes have been published, revealed that the CYP92 family is present in most of the higher plants except Arabidopsis. CYP92A6 is proposed to be involved in brassinosteroid (BR) biosynthesis during etiolated hypocotyl growth of pea [14,15]. Since this family is absent in Arabidopsis that has a robust BR biosynthesis and signaling system, CYP92A6 might have additional functions that need to be evaluated. Similarly, CYP727, CYP728 and CYP733 families are present in rice and soybean and absent in Arabidopsis and Medicago. For example, CYP727A1 is represented by a single gene in rice and CYP727B5 is a single gene in soybean. These are probably solo-function genes similar to the CYP51G family [9]. CYP736 family is present in soybean and Medicago [16] but absent from - Arabidopsis and rice. However, like several other families, the CYP736 family genes are also found in grape and poplar (Table 1). We did not identify any unique "legume-specific" A-type P450s. In general, moss has the most distinct P450 family distribution, representing a significant evolutionary distance.
The CYP702 and CYP708 of non-A type families are present in Arabidopsis but absent in soybean, Medicago or rice (Table 2). The P450 genes from these two families are all unique to Arabidopsis and its closest relatives, such as Brassica napus, making these the only known Brassicaceae-specific CYP proteins thus far. Among them, CYP708A2 is a thalianol hydroxylase, and CYP702A2 and CYP702A3 are triterpene synthases [17]. In contrast, if grape and poplar P450s are included, we cannot find any P450 families that are unique to soybean, or unique to legumes (soybean and Medicago combined). Even though legumes P450 are involved in making unique compounds, such as isoflavones (CYP93C), these P450 genes are not distant enough to form their own families.
Phylogenetic analysis of soybean and Arabidopsis P450 genes
A neighbor-joining (N-J) phylogenetic tree for P450 protein sequences from soybean and Arabidopsis was constructed to determine the orthologous genes and cluster groups between these two species using P-distance in the MEGA4 package [18]. The soybean P450 genes were first classified into two major classes, A-type and non-A-type (Additional File 2, Figure S1A and Figure S1B). The A-type genes contain 59% of the soybean sequences (195 of the 332 sequences) and represent many of the plant-specific enzymes for the synthesis of secondary products (such as phenylpropanoids, etc.). The proteins encoded by the non-A-type sequences contain 137 of the 332 sequences and include enzymes involved in the synthesis of primary metabolic compounds (such as sterols, fatty acids, etc.) hormones and other signaling molecules. Based on the phylogenetic tree, CYP736 family that is present in soybean and Medicago closely resembles to the CYP83 and CYP81 gene families in Arabidopsis (Additional File 2, Figure S1A). The CYP83 family genes are involved in glucosinolate metabolism [19]. As shown in Additional File 2, Figure S1A, the CYP733A family in soybean and CYP81K in Arabidopsis belong to the same cluster, but no function has been assigned to either of these two gene families.
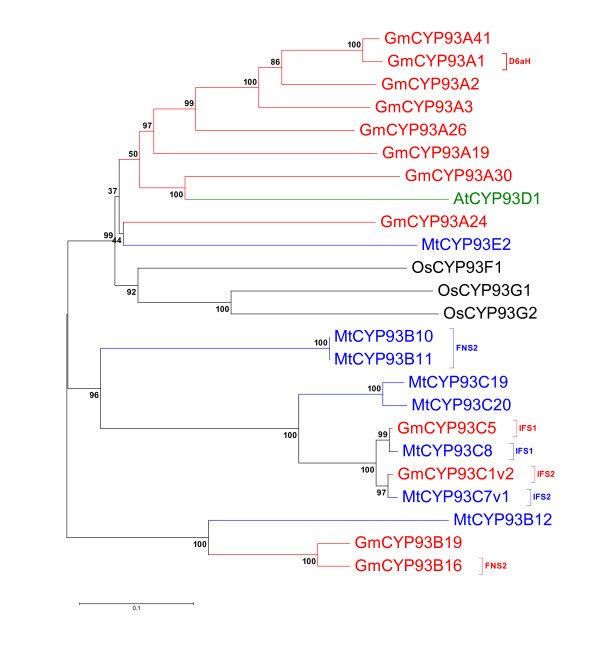
Flavones are widely distributed in higher plants and isoflavonoids are mainly produced in the leguminous plants. They play significant eco-physiological functions in the adaptation of plants in their biological environments [20]. Flavonoids are synthesized from the phenylpropanoid pathways that utilize several cytochrome P450s. One of the P450 families involved in flavonoid biosynthesis in soybean is the CYP93 family that contains 13 genes compared with 8 genes in Medicago, 1 gene in Arabidopsis and 3 genes in rice (Figure 1). The CYP93 family includes 9 subfamilies, among which CYP93A encodes dihydroxy-pterocarpan 6α-hydroxylase (D6aH) involved in legume phytoalexin biosynthesis [21]; CYP93B is flavone synthase (FNSII) involved in flavone biosynthesis [22,23]; CYP93C is isoflavone synthase (IFS) involved in isoflavone biosynthesis [24]. Arabidopsis CYP93D1 is the only member in the 93D subfamily and it is closely related to soybean CYP93A30, suggesting it can be orthologous to the soybean gene (Figure 1). There are a few sequences in the CYP93F and the CYP93G families, whose functions are still unclear. Sequence similarity indicated that the CYP93 family could have evolved from the more ancient CYP75 by duplication and divergence. The CYP93C family is mostly confined to legumes, whereas the CYP93B sequences have a wider distribution and are found in many species. It is possible that CYP93C might have evolved from the CYP93B subfamily which appears to be a more ancient subfamily [9].
Figure 1.
A phylogenetic tree of CYP93 family proteins. An unrooted phylogenetic tree of CYP93 family created with amino acid sequences of Arabidopsis (Green), soybean (Red), Medicago (Blue) and rice (Black). Isoflaovne synthase and flavonoid metabolism known genes were shown in parenthesis. Phylogentic tree was developed using MEGA4.
For the non-A type P450s, we selected the CYP74 family for more detailed comparisons. Members of CYP74 family are involved in the formation of plant oxylipins, a large family of metabolites derived from polyunsaturated fatty acids [25]. In Arabidopsis genome, only two CYP74s have been identified: CYP74A1 and CYP74B2. Arabidopsis CYP74A1 encodes an allene oxide synthase (AOS), which commits 13-hydroperoxy linolenic acid (13-HPOT) to the formation of plant defense hormone, jasmonic acid [26] whereas CYP74B2 is a hydroperoxide lyase (HPL), which converts 13-HPOT to 6-carbon aldehydes and 12-carbon ω-keto-fatty acids [27]. The volatile products from HPL pathway are collectively called green leaf volatiles (GLVs), which can attract the natural enemies of insect herbivores and play an important role in tritrophic interactions. As shown in Figure S1B, in the soybean genome, out of 6 CYP74s, there are 3 CYP74As: CYP74A1, CYP74A21 and CYP74A22. We can assume that these three CYP74As are bona fide AOSs. It has been reported that tomato and several other plant species have at least two AOSs leading to the formation of jasmonic acid [25]. Similar to Arabidopsis, soybean has only one member of CYP74B; CYP74B15, which is probably a functional HPL. The remaining two soybean CYP74s: CYP74C12 and CYP74C13 do not have homologs in Arabidopsis. CYP74Cs from cucumber, melon, almond and rice were shown to be HPLs that have a preference, but not absolute specificity, for 9-hydroperoxides of linoleic and linolenic acids [28-31]. A third AOS gene from tomato was identified, LeAOS3, which is a 9-AOS and a member of CYP74C. So far, 9-AOS has been identified in a few plants including tomato, barley and potato http://metacyc.org/META/new-image?type=PATHWAY&object=PWY-5407[32]. It is quite possible that CYP74C12 and CYP74C13 have activities against 9-HPOT and it would be interesting to identify the products of enzyme reaction to see if there are 9-HPL or 9-AOS.
P450 gene expression profiling using Affymetrix arrays showed organ-specific and stress-induced expression
To obtain expression profiles of soybean P450 genes, we first utilized the extensive Affymetrix array data publically available at the NCBI database. To identify P450 probe sets on the Affymetrix soybean array (Part #900526), we performed BLASTN searches using the sequences of each probe set against the predicted soybean cDNAs at the TIGR gene index and Phytozome 4.0 genomic sequences. A total of 178 probe sets were found to correspond to P450 genes. Out of these probe sets, 108 represented single genes (at least 100 bp in matching length); the rest matched with more than one gene (55 of them matched with more than two genes). Since multiple targets complicate interpretation of the expression profiles, we focused on the 108 single-target genes for further analysis.
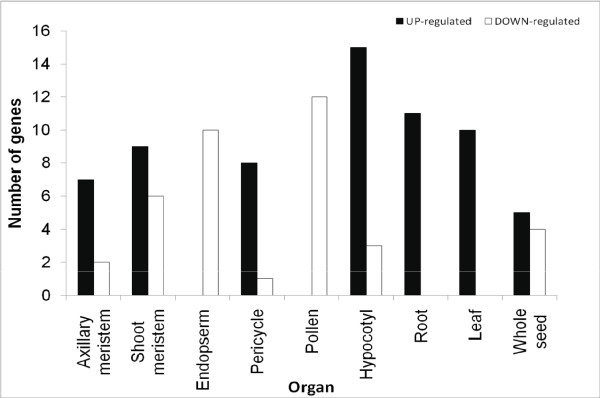
There are 28 microarray libraries that contain organ-specific information in the public database. Following standard microarray data analysis, we identified highly tissue-specific soybean P450 genes (with a cut-off > 3 for up-regulated genes and < −3 for down regulated genes). The data are summarized in Figures 2, 3, 4 and Additional File 3, Table S2. The P450 genes highly induced in leaves, roots, hypocotyl, seeds and axillary meristem are apparent (Figures 2 and 3). For example, out of 108 P450 genes, seven are highly induced and one significantly repressed in the axillary meristem tissues when all genes expressed in all tissues were averaged as control. Each tissue has its own unique set of differentially expressed P450s. Interestingly, CYP93C1 (IFS1) and CYP93C5 (IFS2) were highly expressed in roots and seeds. More specifically, IFS1 is mainly expressed in the root and seed coat; while IFS2 is mainly expressed in embryos and pods. These results agree with previous publications using Northern blot analysis, qRT-PCR, and promoter:GUS transgene assays [33].
Figure 2.
Numbers of up- and down-regulated soybean P450 genes in different organs extracted from Affymetrix soybean microarray dataset.
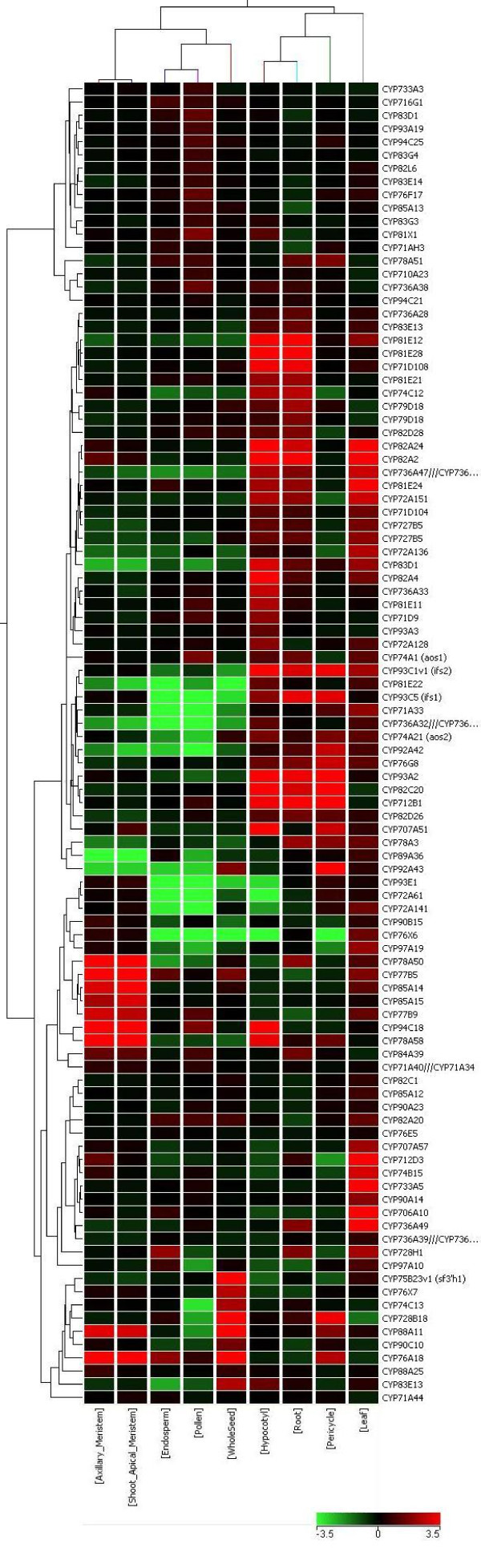
Figure 3.
Expression pattern of soybean P450 genes in different organs according to the analysis of Affymetrix soybean microarray dataset. The color scale indicates the degree of expression (green: low expression; red: high expression). Heat map was created using GeneSpring 10X.
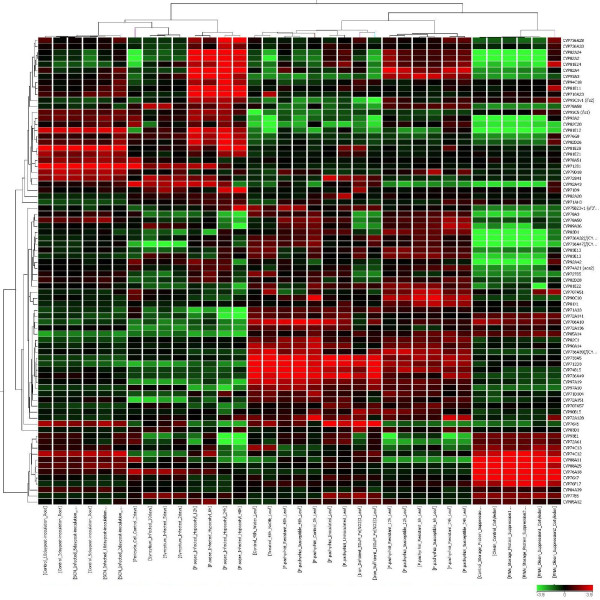
Figure 4.
Expression pattern of soybean P450 genes under stress treatments according to the analysis of Affymetrix soybean microarray dataset. The color scale indicates the degree of expression (green: low expression; red: high expression). Heat map was created using GeneSpring 10X.
CYP93A2, a member of the 93A subfamily, also showed high expression in roots. CYP81E12, CYP81E28 and CYP81E21 are also highly expressed in roots. The CYP81E family encodes isoflavone 2'- and 3'-hydroxylases which are involved in isoflavonoid phytoalexin synthesis. Not surprisingly, in Medicago CYP81E7 was reported to be highly expressed in roots as well [34]. Other up-regulated and down-regulated P450 genes in each tissue are listed in Additional File 4, Table S3. The functions of these genes are not known, however their strong tissue-specific expression suggests their involvement in those tissue types.
There are 99 microarray libraries in the public database from various stress treatments of soybean. As summarized in Figure 4, the genes CYP736A28, CYP93E1, CYP82A4, CYP94C18 and CYP81E11 are highly induced in soybean cyst nematode infection. Similarly, CYP71D9, CYP736A33, CYP72A128, CYP81E28, and CYP81E21 showed strong induction in hypocotyls infected with Phytophthora Sojae. Interestingly, CYP707A51 and CYP710A23 are highly expressed in soybean leaves treated with Nod factor from B. japonicum compared to untreated leaves, suggesting they may be involved in the auto-regulation process in which shoots synthesize a mobile signal to suppress additional nodulation in the root after initial rhizobial infection. Under abiotic stress conditions, CYP71D9 and CYP83D1 were highly expressed under iron deficiency compared to plants grown under sufficient iron. The exact functions of these two genes have not been reported before.
P450 gene expression profiling using Illumina transcriptome analysis identified organ-specific and rhizobium-induced expression
Given the limitations of the current soybean Affymetrix DNA microarray platform, we also utilized Illumina transcriptome sequencing technology to analyze all 332 soybean P450 gene expressions, using published data from Libault et al.[35]. These data sets were generated from total RNAs isolated from nodules, roots, root tips, leaves, flowers, green pods, apical meristem, mock-inoculated and B. japonicum-infected root hair cells harvested at 12, 24 and 48 h after inoculation [35]. For comparison, total RNA extracted from stripped roots (i.e. roots devoid of root hairs) were also harvested and sequenced at 48 h after inoculation with B. japonicum. Illumina transcriptome analysis allowed a more thorough assessment of all of the 332 soybean P450 genes. Additionally, since Affymetrix array analysis is considered to be less sensitive when comparing different experimental set ups, especially for low abundant RNAs, this deep transcriptome sequence analysis allowed a clear distinction between low-abundance and undetectable transcripts.
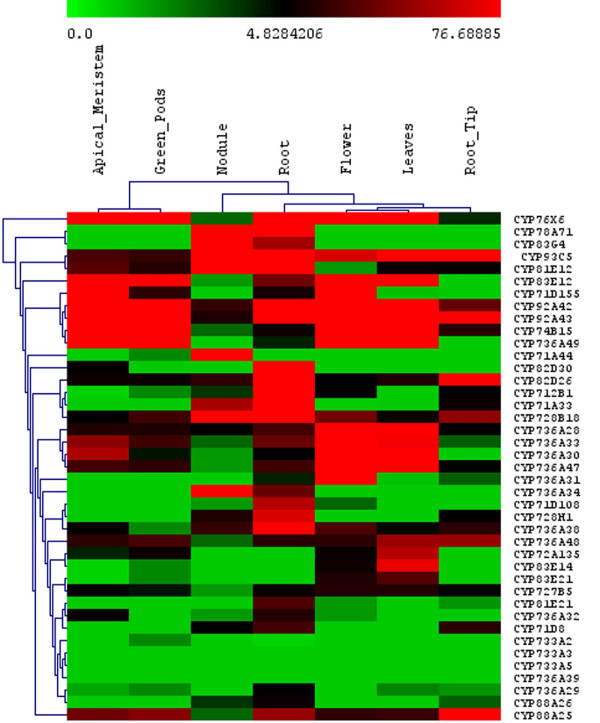
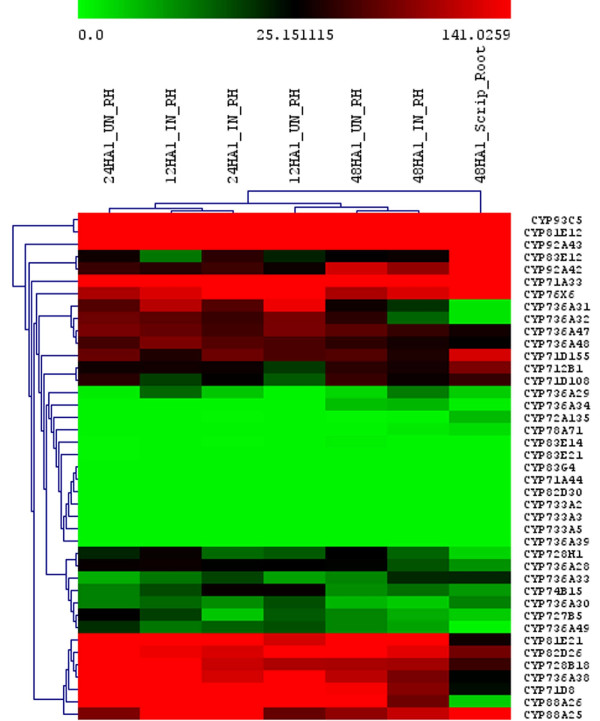
From these Illumina data, the P450 genes highly expressed in different tissue were hierarchically clustered using the MeV v4.5.1 software. The entire expression profile in all tissues and treatments is shown in Additional File 5, Figure S2. To simplify the figure, the expression of P450s involved in isoflavone biosynthesis, CYP93 family genes and P450 families present only in soybean and not in Arabidopsis, including CYP92, CYP733, CYP735 and CYP736 have been highlighted (Figures 5 and 6). In Figure 5, very different expression profiles in apical meristem, green pods, nodules, roots, flowers, leaves, and root tips can be seen among selected P450s. For example, CYP733A members were not detected in any of the tissues, suggesting a more narrow distribution beyond our collection of cDNA libraries. In Figure 6, root hairs and stripped roots were profiled with or without rhizobia induction. Isoflavone biosynthetic enzymes were expressed in all tissues and treatments, in contrast with CYP733A and CYP83 family where no expression could be detected in any tissues. Interestingly, CYP88A3 appeared to be expressed only in the root hairs, not in the roots stripped of root hairs. Similarly, CYP736A31 and CYP736A32 exhibit much higher expression in the root hairs than in the stripped roots. In contrast, the same family CYP736A47 and CYP736A48 had an opposite expression pattern, with significantly higher expression in roots than in root hairs.
Figure 5.
Expression pattern of soybean P450 genes in different organs according to the analysis of Illumina transcriptome dataset. The color scale indicates the degree of expression (green: low expression; red: high expression). Heat map was created using GeneSpring 10X.
Figure 6.
Expression of soybean P450 genes in root hair infection process according to the analysis of Illumina transcriptome dataset. The labels are 24HA1_UN_RH, root hair sample 24 hours after mock treatment; 12HA1_IN_RH, root hair sample 12 hours after infection; 24HA1_IN_RH, root hair sample 24 hours after infection; 12HA1_UN_RH, root hair sample 12 hours after mock treatment; 48HA1_UN_RH, root hair sample 48 hours after mock treatment 48HA1_IN_RH, root hair sample 48 hours after infection; 48HA1_Script_Root, stripped root sample 48 hours after infection.
Of all P450 genes analyzed, the ones highly expressed in nodules, roots, leaves and flowers are summarized in Table 3. The most abundant P450 genes in each tissue, as shown by the calculated tag sequence numbers, are CYP83G4 (nodules), CYP76X6 (apical meristem, green pods and leaves), CYP83E12 (flowers), and CYP93C5 (roots and root tips). Clearly, CYP76X6 is the most abundant P450 in green tissues (function unknown); while the isoflavone synthetic enzyme IFS is the major P450 in roots.
Table 3.
Organ specific expression of P450 genes using Illumina transcriptome.
| Nodule | Apical Meristem | Flower | Green Pods | Leaf | Root | Root Tips | |
| CYP78A71 | 7763.1 | 0 | 0 | 0 | 0 | 102.7 | 0 |
| CYP83G4 | 4737.8 | 0 | 0 | 0 | 0 | 50.1 | 0 |
| CYP71A44 | 524.1 | 0 | 0 | 2.2 | 0 | 0 | 0 |
| CYP736A34 | 95.2 | 0 | 0 | 0 | 0 | 34.0 | 0 |
| CYP93C5 | 889.5 | 25.0 | 65.7 | 20.1 | 159.2 | 3334.8 | 2596.7 |
| CYP81E12 | 404.8 | 30.0 | 1.9 | 15.6 | 8.1 | 2962.0 | 7.1 |
| CYP82D26 | 18.3 | 9.2 | 4.8 | 8.9 | 13.9 | 310.7 | 139.4 |
| CYP712B1 | 3.9 | 0.8 | 4.8 | 2.2 | 0 | 231.8 | 6.1 |
| CYP71A33 | 51.9 | 0 | 1.0 | 0 | 0 | 172.3 | 9.1 |
| CYP71D108 | 1.9 | 0 | 2.9 | 0 | 0 | 56.0 | 0 |
| CYP72A135 | 0 | 4.2 | 7.7 | 8.9 | 55.8 | 0 | 0 |
| CYP83E21 | 0 | 0.8 | 13.5 | 2.2 | 27.9 | 0 | 0 |
| CYP83E12 | 1.9 | 512.4 | 1403.1 | 967.1 | 145.2 | 36.5 | 0 |
| CYP74B15 | 2.9 | 136.6 | 468.4 | 113.9 | 560.1 | 9.3 | 16.2 |
| CYP71D155 | 0 | 691.6 | 89.8 | 17.9 | 1.1 | 11.0 | 0 |
| CYP76X6 | 2.9 | 8328.7 | 1044.9 | 12422.1 | 1119.0 | 188.5 | 4.0 |
List of P450 genes and their expression intensity values in different organs. Value zero indicates no expression.
We were interested in the rhizobium-induced P450 genes. Out of 12 genes in the CYP736 family, CYP736A34 showed high expression in nodules and roots (Table 3 and Figure 6). CYP78A71, CYP83G4 and CYP71A44 were highly expressed in nodules. Almost no expression was detected in other tissues. Considering there are only two members of the CYP83 family in Arabidopsis, but five members in M. truncatula and six members in Lotus japonicus, the CYP83 family may play important roles in legumes, although its function remains unknown [16].
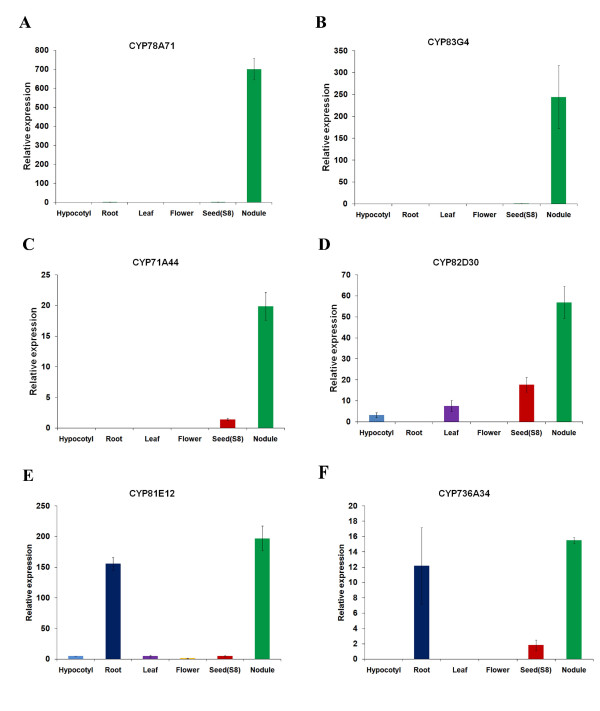
To confirm these findings from Illumina sequencing, we carried out qRT-PCR analysis on the four nodule-specific genes and a set of selected other P450 genes. Total RNAs were isolated from hypocotyls, roots, leaves, flowers, seed (stage R8), and dissected nodules. The expression levels of the above-mentioned CYP78A71, CYP83G4, CYP71A44, and CYP82D30 were measured. All four genes showed dominant nodule-specific expression, consistent with the Illumina data. Two additional genes, CYP81E12 and CYP736A34, showed strong root and nodule expression, also fitting the Illumina data very well. All six genes on Figure 7 had lower expression in other tissues.
Figure 7.
qRT-PCR confirmation of soybean P450 genes highly expressed in root and nodule as revealed by the analysis of Microarray and Solexa transcriptome datasets. Error bars represent SE (Standard error) of three replications.
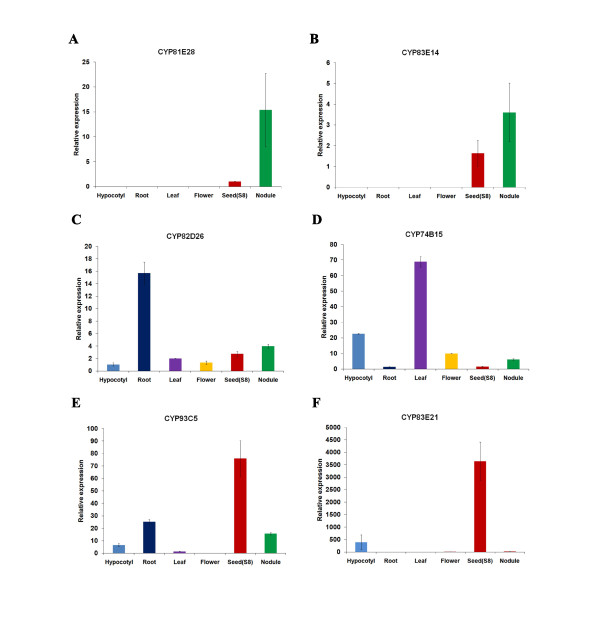
As controls, CYP82D26, CYP74B15, CYP93C5, and CYP83E21 genes were also analyzed by qRT-PCR (Figure 8). All four genes showed exactly the same expression pattern as suggested by Illumina analysis. CYP93C5 (IFS1) has been characterized by other expression analysis methods previously and all data are consistent with our qRT-PCR analysis. Taken together, we have discovered four major nodulation-specific P450 genes, which will be functionally characterized in the future.
Figure 8.
qRT-PCR confirmation of soybean P450 gene expression in different organs as revealed by the analysis of Microarray and Solexa transcriptome datasets. Error bars represent SE of three replications.
We also analyzed another set of Illumina data recently published by Severin et al. [36]. Since the normalization methods were different in these two data sets, we were unable to combine these two transcriptome data. However, the expression profiles of highlighted genes in the comparable tissue types were similar in these two data sets (see Additional File 6, Table S4). For example, the above-mentioned CYP78A71, CYP83G4, and CYP71A44 all showed nodule-specific expression while CYP82D30 expressed in roots and nodules.
Co-expression analysis revealed co-ordinately expressed metabolic enzymes of some P450s
Co-expression analysis assumes that some genes of the same biochemical pathway are co-ordinately regulated at the transcriptional level. In general, P450s catalyze slow and irreversible steps in many branches of the plant metabolic pathways. They have been shown to be co-expressed and regulated with non-P450 genes with known functions in the same pathway [37]. For example, the functions of several uncharacterized P450 genes in Arabidopsis were predicted using co-expression analysis and confirmed experimentally later [11,38]. In this study, we performed co-expression analysis comparing the expression of each P450 with that of genes present in the Affymetrix Soybean gene chip.
The annotation information of the current Affymetrix soybean gene chip was combined with expression data to identify genes and pathways co-expressed with the P450 genes. For each P450 gene, we calculated Pearson correlation coefficients (r-value) with genes present on the Affymetrix soybean array. Only the genes with significant coefficients (r > 0.7) were selected for pathway analysis. We first investigated a known soybean P450 gene to test our co-expression analysis approach. As shown in Table 4, CYP93C5 isoflavone synthase (IFS1) gene co-expresses strongly with several isoflavonoid-related metabolic enzymes, including chalcone isomerase 1B1 (r = 0.91), and chalcone isomerase 1B2 (r = 0.87). Interestingly, the few transcription factors, such as Myb76 and bZIP42 that are co-expressed with these genes could be the missing transcription factors that regulate isoflavone biosynthesis under stress conditions. Functional annotation of Arabidopsis and soybean are shown in Table 5.
Table 4.
Co-expression analysis of selected soybean P450 genes.
| Soybean Gene | Co-expressed genes | r value |
|---|---|---|
| CYP93C5-IFS1 | ||
| Isoflavone synthase 1 | 1 | |
| Chalcone isomerase 1B2 | 0.91 | |
| Chalcone isomerase 1B1 | 0.87 | |
| Peroxidase, pathogen-induced | 0.85 | |
| Copper amino oxidase | 0.82 | |
| NAC domain protein | 0.81 | |
| Peroxidase (PC7) | 0.80 | |
| Peroxidase precursor (GMIPER1) | 0.79 | |
| MYB transcription factor MYB67 | 0.79 | |
| Chalcone isomerase 2 | 0.79 | |
| NORK protein | 0.79 | |
| KNT1 | 0.78 | |
| Transcription factor bZIP42 | 0.78 | |
| Plastid glucose-6-phosphate | 0.77 | |
| Kunitz trypsin inhibitor p20-1-like protein | 0.77 | |
| Dof3 | 0.76 | |
| Chalcone isomerase | 0.76 | |
| Glutathione S-transferase GST 17 | 0.75 | |
| CYP728H1 | ||
| CYP728H1 | 1 | |
| 2'-hydroxy isoflavone/dihydroflavonol reductase homolog | 0.92 | |
| 4-coumarate:CoA ligase isoenzyme 2 | 0.90 | |
| Cytochrome P450 (CYP98A2) | 0.86 | |
| RecA/Rad51/DMC1-like protein | 0.83 | |
| Peroxidase | 0.81 | |
| Endo-xyloglucan transferase | 0.80 | |
| bZIP transcription factor bZIP17 | 0.79 | |
| Essex desiccation protectant protein Lea14 homolog | 0.79 | |
| NAC domain protein (NAC18) | 0.79 | |
| WD-repeat cell cycle regulatory protein | 0.79 | |
| 4-coumarate:coenzyme A ligase | 0.79 | |
| 31 kDa protein | 0.78 | |
| Transcription factor bZIP18 | 0.77 | |
| Hydroxyproline-rich glycoprotein (sbHRGP2) mRNA, 3' end | 0.76 | |
| Early nodulin | 0.76 | |
| Protease inhibitor | 0.76 | |
| Wee1 | 0.76 | |
| CYP736A32/34 | ||
| CYP736A32//CYP736A34 | 1 | |
| Lectin | 0.93 | |
| SOS2-like protein kinase | 0.92 | |
| Cultivar Wenfeng7 purple acid phosphatase-like protein (Pap3) | 0.91 | |
| Lipoxygenase L-5 | 0.88 | |
| Cysteine proteinase | 0.88 | |
| Peroxisomal ascorbate peroxidase | 0.88 | |
| Thiol protease isoform B | 0.86 | |
| Phosphoglycerate mutase-like protein | 0.86 | |
| Coronatine-insensitive 1 | 0.84 | |
| Catalase (cat4) | 0.83 | |
| Sorbitol-like transporter | 0.83 | |
| MYB transcription factor MYB48 | 0.82 | |
| Phosphoenolpyruvate carboxylase | 0.82 | |
| Indole-3-acetic acid induced protein ARG-2 homolog | 0.81 | |
| Lipoxygenase (lox7) | 0.79 | |
| WRKY39 protein | 0.77 | |
| Glutathione S-transferase GST 8 | 0.77 | |
| Mitogen-activated protein kinase 1 (MAPK1) | 0.76 | |
| Ribulose-1,5-bisphosphate carboxylase small subunit | 0.76 | |
| Clone pSXET1a xyloglucan endotransglycosylase precursor (XET1) | 0.76 | |
| SOC1 | 0.76 | |
| OAS-TL3 cysteine synthase | 0.76 | |
| WRKY23 (WRKY23) | 0.76 | |
| Nitrate reductase (NiR) | 0.75 | |
| NAC domain protein | 0.75 | |
| bZIP transcription factor bZIP105 | 0.74 | |
| Allene oxide synthase (AOS2) | 0.74 | |
| Chalcone synthase 1 | 0.72 | |
| CYP75B23v1 (sf3'h1) | 0.70 |
Selected P450 genes and their co-expressed genes are listed with Pearson correlation coefficient (r value >0.7)
Table 5.
List of Arabidopsis and soybean P450 genes and orthologues with known functions.
| AGI locus | Arabidopsis Gene | Soybean orthologue | Percentage Identity | Function in Arabidopsis | Reference -Arabidopsis function |
|---|---|---|---|---|---|
| At1g11680 | CYP51A2 | CYP51G1 | 80.9 | Obtusifoliol 14α-demethylase | (Kushiro et al. 2001); (Kim et al. 2005b) |
| At2g30770 | CYP71A13 | CYP71A9 | 37.9 | Conversion of indole-3-acetaldoxime, camalexin biosynthesis | (Nafisi et al. 2007) |
| At3g26830 | CYP71B15 | CYP83E8 | Conversion of s-dihydrocamalexic acid to camalexin | (Bottcher et al. 2009); (Zhou et al. 1999) | |
| At1g17060 | CYP72C1 | CYP72A69 | 44.3 | Exact substrate not identified | (Nakamura et al. 2005); (Takahashi et al. 2005) |
| At2g30490 | CYP73A5 | CYP73A11 | 84.4 | Cinnamic acid 4-hydroxylase (t-CAH) | (Mizutani et al. 1997) |
| At5g42650 | CYP74A | CYP74A1 | 56.4 | Allene oxide synthase (AOS)JA | (Laudert et al. 1996); (Park et al. 2002) |
| At4g15440 | CYP74B2 | CYP74B15 | 60.2 | Hydroperoxide lyase (HPL) JA | (Bate et al. 1998) |
| At5g07990 | CYP75B1 | CYP75B43 | 52.6 | 3′-hydroxylase for narigenin, dihydrokaempferol (F3′H) | (Schoenbohm et al. 2000) |
| At5g04630 | CYP77A4 | CYP77A3/A12 | 68.1 | Catalyze the formation of three mono-epoxides of alpha-linolenic acid | (Sauveplane et al. 2009) |
| At3g10570 | CYP77A6 | CYP77A12/A12 | 65 | Chain hydroxylase - for cutin synthesis for morphology of flower | (Li-Beisson et al. 2009) |
| At1g13710 | CYP78A5 | CYP78A72 | 62.8 | KLU control organ size, control leaf growth | (Wang et al. 2008); (Anastasiou et al. 2007) |
| At5g05260 | CYP79A2 | CYP79D17 | 49.1 | Conversion of phenylalanine to oxime | (Whittstock and Halkier 2000) |
| At4g39950 | CYP79B2 | CYP79D21 | 51.1 | Conversion of tryptophan, tryptophan analogs to oxime | (Hull et al. 2000); (Mikkelsen et al. 2000) |
| At2g22330 | CYP79B3 | CYP79D21 | 50.9 | Conversion of tryptophan to oxime | (Hull et al. 2000) |
| At1g16410 | CYP79F1 | CYP79D17/21 | 40.7 | Mono to hexahomomethionine in synthesis of aliphatic glucosinolates | (Hansen et al. 2001); (Reintanz et al. 2001); (Chen et al. 2003) |
| At1g16400 | CYP79F2 | CYP79D17/22 | 40.2 | Long chain penta and hexahomomethionine in synthesis of long chain aliphatic glucosinolates | (Reintanz et al. 2001); (Chen et al. 2003) |
| At5g57220 | CYP81F2 | CYP82A3 | 47.6 | Conversion of indole-3-yl-methyl to 4-hydroxy-indole-3-yl-methyl glucosinolate, | (Bednarek et al. 2009); (Pfalz et al. 2009) |
| At4g13770 | CYP83A1 | CYP83E8/CYP736A29 | 37 | Oxidation of methionine-derived oximes; | (Bak and Feyereisen 2001); (Naur et al. 2003) |
| At4g31500 | CYP83B1 | CYP83E8/CYP736A30 | 45 | Oxidation of indole-3-acetaldoxime | (Bak et al. 2001); (Naur et al. 2003) |
| At4g36220 | CYP84A1 | CYP8438/39/21 | 73.4 | 5-hydroxylase for coniferaldehyde, coniferyl alcohol and ferulic acid (F5H) | (Ruegger et al. 1999); (Humphreys et al. 1999) |
| At5g38970 | CYP85A1 | CYP85A12 | 68.9 | C6-oxidase for 6-deoxycastasterone, other steroids | (Shimada et al. 2001); (Shimada et al. 2003) |
| At3g30180 | CYP85A2 | CYP85A13 | 70.3 | C6-oxidase for 6-deoxycastasterone, other steroids;Conversion of castasterone to brassinolide | (Shimada et al. 2003); (Nomura et al. 2005) |
| At5g58860 | CYP86A1 | CYP86A37 | 72.8 | ω-hydroxylase for satur. and unsat. C12 to C18 fatty acids | (Benveniste et al. 1998) |
| At4g00360 | CYP86A2 | CYP86A66 | 71.2 | ω-hydroxylase for satur. and unsat. C12 to C18 fatty acids | (Duan and Schuler 2005); (Xiao et al. 2004) |
| At2g45970 | CYP86A8 | CYP86A66/67 | 76.4 | ω-hydroxylase for satur. and unsatur. C12 to C18 fatty acids | (Wellesen et al. 2001) |
| At5g23190 | CYP86B1 | CYP86B9/10/11 | 69.5 | C22 and C24 fatty acids, accumulated in the suberin polyester. | (Compagnon et al. 2009) |
| At1g05160 | CYP88A3 | CYP88A11/25 | 63.5 | Multifunctional ent-kaurenoic acid oxidase | (Helliwell et al. 2001) |
| At2g32440 | CYP88A4 | CYP88A26 | 53.2 | Multifunctional ent-kaurenoic acid oxidase | (Helliwell et al. 2001) |
| At5g05690 | CYP90A1 | CYP90A14/23/24 | 75.8 | 23α-hydroxylase for 6-oxo-cathasterone | (Szekeres et al. 1996) |
| At3g50660 | CYP90B1 | CYP90B15/18/17 | 71.3 | 22α-hydroxylase for 6-oxo-campestanol, campesterol and cholesterol | (Choe et al. 1998); (Fujita et al. 2006) |
| At4g36380 | CYP90C1 | CYP90C8/9/10 | 55.3 | Conversion of typhasterol to castasterone, C-23 hydroxylation | (Ohinishi et al.); (Kim et al. 2005a) |
| At3g13730 | CYP90D1 | CYP90D12/13 | 61.9 | Exact substrate in downstream BR synthesis not identified | (Kim et al. 2005a); (Greer et al. 2007); (Ohinishi et al. 2006) |
| At1g57750 | CYP96A15 | CYP94B13 | 42.4 | Formation of secondary alcohols and ketones in cuticular wax of stem, acyl CoA reductase | (Greer et al. 2007) |
| At1g31800 | CYP97A3 | CYP97A10/19 | 70.7 | β −ring hydroxylase on carotenes | (Kim and DellaPenna 2006) |
| At3g53130 | CYP97C1 | CYP97A16/17 | 79 | ε −ring hydroxylase on carotenes | (Tian et al. 2004) |
| At2g40890 | CYP98A3 | CYP98A2/47 | 80 | 3′-hydroxylase for p-coumaryl shikimic/quinic acids (C3′H) | (Schoch et al. 2001); (Kai et al. 2006) |
| At5g25900 | CYP701A3 | CYP701A25/16 | 61.2 | Multifunctional ent-kaurene oxidase | (Helliwell et al. 1998) |
| At1g01280 | CYP703A2 | CYP703A8 | 74.9 | Sporopollenin synthesis, pollen development | (Morant et al. 2007) |
| At1g69500 | CYP704B1 | CYP704B28 | 75.5 | Fatty acid -sporopollenin biosynthesis -pollen | (Dobritsa et al. 2009) |
| At4g19230 | CYP707A1 | CYP707A16 | 68.4 | 8′-hydroxylase for ABA inactivation | (Saito et al. 2004); (Kushiro et al. 2004) |
| At2g29090 | CYP707A2 | CYP707A45 | 61.9 | 8′-hydroxylase for ABA inactivation, Enhancement of ABA catabolism | (Saito et al. 2004); (Kushiro et al. 2004) |
| At5g45340 | CYP707A3 | CYP707A16/56 | 71.6 | 8′-hydroxylase for ABA inactivation | (Saito et al. 2004); (Kushiro et al. 2004) |
| At3g19270 | CYP707A4 | CYP707A53/59 | 65.1 | 8′-hydroxylase for ABA inactivation | (Saito et al. 2004); (Kushiro et al. 2004) |
| At2g34500 | CYP710A1 | CYP710A22/23 | 66.3 | C-22 desaturase for β-sitosterol | (Morikawa et al. 2006) |
| At2g34490 | CYP710A2 | CYP710A22/24 | 62 | C-22 desaturase on 24-epi-campesterol and β-sitosterol | (Morikawa et al. 2006) |
| At2g26170 | CYP711A1 | CYP711A23/24/25/26 | 70.6 | Caretnoid,core phenylpropanoid metabolism | (Booker et al. 2005) |
| At2g26710 | CYP734A1 | CYP734A17/20/21 | 76.9 | 26-hydroxylase for brassinolide and castasterone | (Neff et al. 1999) |
| Soybean gene | Arabidopsis orthologue | Percentage identity | Function in soybean | Reference-Soybean function | |
| CYP93C1 | CYP93A41 | 40.9 | Isoflavone synthase (IFS1) | (Jung et al. 2000) | |
| CYP93C5 | CYP93A41 | 40.3 | Isoflavone synthase (IFS2) | (Jung et al. 2000) | |
| CYP71D09 | CYP71B34 | 39.1 | Flavonoid 6-hydroxylase | (Latunde-Dada et al. 2001) | |
| CYP73A11 | CYP73A5 | 84.4 | Cinnamate 4-hydroxylase | (Schopfer et al. 1998) | |
| CYP93A1 | Dihydroxypterocarpan 6a-hydroxylase (D6aH) | (Schopfer and Ebel 1998); (Schopfer et al. 1998) | |||
| CYP71A10 | Metabolism of phenylurea herbicides | (Siminszky et al. 1999) | |||
We then performed the co-expression analysis on many of the 108 P450 genes on the Affymetrix arrays (Table 4 and Additional File 6, Table S4). We selected three examples and highlight them in Table 4. The CYP728H1 showed high expression in roots and nodules, and was co-expressed with 4-coumarate:CoA ligase isoenzyme 2, cytochrome P450 98A2 and 4- coumarate:coenzyme A ligase genes which are involved in phenylpropanoid metabolism. CYP728H1 is also co-expressed with endo-xyloglucan transferase, NAC transcription factor, 2'-hydroxy isoflavone and early nodulin genes. Gene expression and co-expression analysis suggests that CYP728H1 may play an important role in isoflavone metabolism, as well as during root and nodule development.
The CYP736 family is present in soybean but not in Arabidopsis. CYP736A34 showed high expression in roots and nodules (Figure 7). Co-expression analysis showed that the expression of this gene is highly correlated with lipoxygenase, lectin and CYP83D1, all of which are involved in root and nodule development (Table 4). Some of the co-expressed genes are defense related genes such as cysteine proteinase. Since defense response is one of the early nodulation events, CYP736A34 may be functioning at the early stages of this symbiotic process.
These analyses show that co-expression analysis combined with pathway mapping of co-expressed genes is a powerful tool to identify genes encoding enzymes acting in the same biochemical pathway or biological process. Unfortunately, annotation of soybean genes in the Affymetrix gene chip is not as comprehensive as other model species. Many of the co-expressed genes have unknown or un-confirmed functions. A majority of the P450s cannot be mapped to specific pathways. However, this approach still provides important leads to large sets of uncharacterized soybean P450s, and with improved annotation of soybean genes in the near future, we should be able to extract more functional information of soybean P450s.
Conclusions
Soybean is an important leguminous crop. With the advent of soybean genome sequencing, it is possible to study large gene families of soybean. We targeted one of the most challenging families in plants, the cytochrome P450 genes, and investigated their genetic make-up, gene distributions, expression profiles, and co-expressed associates. Cytochrome P450s are indispensable for soybean growth, development, and defense against pathogens. They may play important roles for soybean symbiotic interaction with rhizobacteria. Overall, we identified 332 full-length P450 genes and 378 pseudogenes in the genome. We used publicly available microarray libraries and identified few tissue-specific and stress responsive soybean P450s. The tissue-specific expression patterns of some P450 genes were confirmed by qRT-PCR. In addition, the expressions patterns of all 332 soybean P450 genes were obtained through the analysis of Illumina transcriptome datasets. The co-expression analysis on some of the P450 genes was performed using the Affymetrix array datasets. We demonstrate that gene co-expression analysis is a useful tool to guide our further study on the function of uncharacterized genes. Importantly, the identification of nodule-specific P450s and their further exploitation may help us to uncover the intriguing process of soybean and rhizobium interaction.
Methods
Identification of soybean cytochrome P450 genes
Existing soybean P450 genes and corresponding Glyma numbers (location markers in soybean genome) were retrieved from the Cytochrome P450 Homepage website http://drnelson.uthsc.edu/CytochromeP450.html. Additional full-length soybean genes were identified based on the cytochrome P450 domain predictions from Pfam [39], Panther [40], and KOG [41] separately. The search results were consolidated with existing soybean P450s. From these sequences, the pseudogenes were identified using existing criteria listed on the P450 homepage website.
Computational phylogenetic analysis
Protein sequences of soybean P450 genes were obtained based on Phytozome 4.0. For comparison, a collection of P450s from Arabidopsis thaliana (245 genes) and the corresponding CYP names were retrieved from Arabidopsis cytochrome P450 web-based resource http://www.p450.kvl.dk/p450.shtml[42]. In addition, CYP93 family proteins from Medicago truncatula and rice were selected from Genbank for relationship analysis across species. Multiple sequence alignment were performed using the BLOSUM matrix (Gap opening and extension penalties of 25 and 1, respectively), using the ClustalW algorithm-based AlignX module from the Mega4 software [18]. The phylogenetic tree was constructed using the Neighbour-Joining Tree method by P-distance in MEGA4. The significance level of the neighbor-joining analysis was examined by bootstrap testing with 1000 repeats.
1.3 Soybean Affymetrix array analysis
Publically available experiments and arrays were listed at the NCBI GEO and Array Express database http://www.ncbi.nlm.nih.gov/sites/entrez. For tissue-specific expression, six experiments (28 arrays) were analyzed using RNAs isolated from different tissues and developmental stages. For stress-induced experiments, eight experiments (99 arrays) were analyzed using RNAs isolated from various abiotic and biotic stress treatments. Expression analysis was carried out using GeneSpring 10GX (Agilent Technologies, Forster City, CA). First, all microarray data were normalized and summarized using the RMA algorithm. Signals from each probe set were then normalized to the median of their values across the entire dataset. Quality control of the dataset was performed using Principal Components Analysis (PCA) to confirm that there were no outlying replicate samples, and dye labeling had no associated bias. Data were combined from replicate samples and grouped in experiment interpretations. Gene list (CYP genes) were generated by expression with cut off P < 0.05. Statistical analysis was performed using One-Way ANOVA with Posthoc-TukeyHSD test to determine statistical significant difference between means. Signal intensities from organ and tissue samples were then compared to the average signal intensities during normalization. In stress experiments, signal intensities from treatment groups were compared to signal intensities from the corresponding control samples to generate fold changes. Genes that were significantly up or down regulated (>two fold) were selected for hierarchical clustering. Hierarchical clustering tree was created based on Pearson correlation coefficient under each experimental condition. Co-expression analysis and pathway mapping were performed in GeneSpring as well. The selected P450 genes were mean-centered and Pearson correlation coefficients (r-values) calculated between each P450 probe set. Co-expressed genes with r > 0.7 were retrieved and the corresponding available biochemical pathways were extracted from the Plant Metabolic Network http://www.plantcyc.org/[43].
1.4 Soybean Illumina expression data analysis
Solexa sequencing libraries for fourteen different conditions including nodules, roots, root tips, leaves, flowers, green pods, apical meristem, mock-inoculated and B. japonicum-infected root hair cells harvested at 12, 24 and 48 h after inoculation, generated and analyzed by Libault et al. [35,44] were utilized to quantify the transcriptomics expression of soybean genes (i.e. the number of sequence reads/million reads aligned). Read counts used in expression analyses were based on the subset of uniquely aligned reads that also overlapped the genomic spans of the Glyma1 gene predictions. Read counts for a given sample were normalized by using values for a gene's uniquely aligned read counts per million reads uniquely aligning within that sample. A total of 51,529 annotated soybean genes (74.5% of the 69,145 putative, annotated soybean genes) were found to be expressed in at least one condition.
1.5 Plant materials, growing conditions, and RNA extraction
Soybean (G. max L. cv. Jack) seeds were germinated in three-gallon pots containing Promix (Home Depot, Atlanta, GA). The seedlings were grown in Conviron growth chamber (26/20 °C day/night temperature, photoperiod of 14/10 h, 800 μmol m-2 s-1 light intensity and 60% humidity). When the seedlings developed four nodes and three fully opened trifoliate leaves, approximately 25 d after sowing, the roots, hypocotyls, leaves were collected. Flowers and early R8 stage seeds were harvested 55 and 100 d after sowing, respectively. Collected tissues were immediately frozen in liquid nitrogen. Soybean plants inoculated with B. japonicum USDA110 strain was used for nodulation as described previously [45].
Total RNA was isolated from plants using TRIZOL reagent (Invitrogen, Carlsbad, CA). For each sample, 10 μg of total RNA were digested with RNase-free DNaseI (Promega, Madison, WI) to remove any genomic DNA contamination. After DNaseI treatment, RNA concentration was determined again using a NanoDrop ND-1000 UV-Vis spectrophotometer (NanoDrop Technologies, Wilmington, DE). First-strand cDNA was synthesized from 2 μg total RNA using the Superscript III first strand synthesis system (Invitrogen). All cDNA samples were diluted 50-fold in sterile water for real time PCR reaction.
1.6 Quantitative RT-PCR analysis
Gene specific primers were designed using ProbeFinder Version 2.44 https://www.roche-applied-science.com. The list of genes and primers used for amplification are shown in Additional File 7, Table S5. Primer specificity was further confirmed by blasting each primer sequence against Phytozome http://www.phytozome.net/search.php?show=blast using the BLASTN algorithm. Soybean Actin and Ubiquitin genes were used as internal controls for gene expression studies [46]. Quantitative RT-PCR (qRT-PCR) reactions were performed in 96-well plates (StepOne Plus Real Time PCR System; Applied Biosystems, Foster City, CA) for all tissues tested. Clontech's SYBR Advantage qPCR Premix was used for the qRT-PCR reactions. Primer sets (0.2 μM final concentrations for each primer) were used in a final volume of 10 μL per well. The thermal profile of the qRT-PCR reactions was 95°C for 5 min, followed by 40 cycles of 95°C for 15 sec, and 60°C for 10 sec and 72°C for 20 sec. Melting curve of each PCR amplicon was obtained by adding the following cycling condition: 95°C for 15 sec followed by a constant increase of the temperature between 60 to 95°C at an increment of 0.3°C/cycle.
Authors' contributions
SKG, OY designed all the experiments. SKG performed Phylogentic tree construction, gene expression and co-expression analysis. JT carried out the computational and bioinformatics analysis. SKG, NB and SP designed qRT-PCR experiments and data analysis. NB performed all the qRT-PCR analysis. HC involved in design and co-expression analysis. OY, DX conceived of the project. YCA provided help on GeneSpring software. All authors read and approved the final manuscript.
Supplementary Material
Table S1 Comparison of P450 families among soybean, Medicago, Arabidopsis, rice, poplar, grape and moss. A. List of A-type P450 families. B. List of Non-Atype P450 families. For each family number of genes and pseudogenes were compared among different plant species.
Figure S1 Phylogenetic tree of all soybean P450s. S1A. A-type P450s of soybean and Arabidopsis. S1B. Non-A type P450s of soybean and Arabidopsis. Soybean P450s are shown in blue and Arabidopsis P450s are shown in green. Trees were constructed using MEGA4.
Table S2 Organ-specific expressions of soybean P450 genes based on Affymetrix arrays.
Table S3 Organ specific expression of P450 genes using Illumina transcriptome. List of P450 genes and their expression intensity values in different organs. Value zero indicates no expression.
Figure S2 Gene expression of 332 soybean P450 genes according to the analysis of Illumina transcriptome dataset. Gene expression in different organ and root hair inoculation process.
Table S4 Co-expression analysis of soybean P450 genes based on Affymetrix arrays.
Table S5 List of primer pairs used in qRT-PCR
Contributor Information
Satish K Guttikonda, Email: SKGuttikonda@dow.com.
Joshi Trupti, Email: joshitr@missouri.edu.
Naveen C Bisht, Email: NBisht@danforthcenter.org.
Hui Chen, Email: huchen@danforthcenter.org.
Yong-Qiang C An, Email: yong-qiang.an@ars.usda.gov.
Sona Pandey, Email: SPandey@danforthcenter.org.
Dong Xu, Email: xudong@missouri.edu.
Oliver Yu, Email: oyu@danforthcenter.org.
Acknowledgements
We thank Dr. Gary Stacey at University of Missouri-Columbia for providing Illumina data for the gene expression analysis. This project is support in part by grants from NSF (MCB0519634) and USDA (NRI2005-05190) to O.Y and Missouri Soybean Merchandising Council (#306) to D. X.
References
- Nelson DR. Cytochrome P450 and the individuality of species. Arch Biochem Biophys. 1999;369(1):1–10. doi: 10.1006/abbi.1999.1352. [DOI] [PubMed] [Google Scholar]
- Werck-Reichhart DBS, Paquette S. Cytochrome P450. Rockville, MD: American Society of Plant Biologists;; 2002. [Google Scholar]
- Werck-Reichhart D, Hehn A, Didierjean L. Cytochromes P450 for engineering herbicide tolerance. Trends Plant Sci. 2000;5(3):116–123. doi: 10.1016/S1360-1385(00)01567-3. [DOI] [PubMed] [Google Scholar]
- Williams PA, Cosme J, Sridhar V, Johnson EF, McRee DE. Mammalian microsomal cytochrome P450 monooxygenase: structural adaptations for membrane binding and functional diversity. Mol Cell. 2000;5(1):121–131. doi: 10.1016/S1097-2765(00)80408-6. [DOI] [PubMed] [Google Scholar]
- Nelson DR, Koymans L, Kamataki T, Stegeman JJ, Feyereisen R, Waxman DJ, Waterman MR, Gotoh O, Coon MJ, Estabrook RW. et al. P450 superfamily: update on new sequences, gene mapping, accession numbers and nomenclature. Pharmacogenetics. 1996;6(1):1–42. doi: 10.1097/00008571-199602000-00002. [DOI] [PubMed] [Google Scholar]
- Durst F, Nelson DR. Diversity and evolution of plant P450 and P450-reductases. Drug Metabol Drug Interact. 1995;12(3-4):189–206. doi: 10.1515/dmdi.1995.12.3-4.189. [DOI] [PubMed] [Google Scholar]
- Paquette SM, Bak S, Feyereisen R. Intron-exon organization and phylogeny in a large superfamily, the paralogous cytochrome P450 genes of Arabidopsis thaliana. DNA Cell Biol. 2000;19(5):307–317. doi: 10.1089/10445490050021221. [DOI] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J. et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463(7278):178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- Nelson DR. Cytochrome P450 nomenclature, 2004. Methods Mol Biol. 2006;320:1–10. doi: 10.1385/1-59259-998-2:1. [DOI] [PubMed] [Google Scholar]
- Schuler M, Duan H, Bilgin M, Ali S. Arabidopsis cytochrome P450s through the looking glass: a window on plant biochemistry. Phytochemistry Review. 2006;5:205–237. doi: 10.1007/s11101-006-9035-z. [DOI] [Google Scholar]
- Ehlting J, Sauveplane V, Olry A, Ginglinger JF, Provart NJ, Werck-Reichhart D. An extensive (co-)expression analysis tool for the cytochrome P450 superfamily in Arabidopsis thaliana. BMC Plant Biol. 2008;8:47. doi: 10.1186/1471-2229-8-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuno M, Compagnon V, Schoch GA, Schmitt M, Debayle D, Bassard JE, Pollet B, Hehn A, Heintz D, Ullmann P. et al. Evolution of a novel phenolic pathway for pollen development. Science. 2009;325(5948):1688–1692. doi: 10.1126/science.1174095. [DOI] [PubMed] [Google Scholar]
- Gill N, Findley S, Walling JG, Hans C, Ma J, Doyle J, Stacey G, Jackson SA. Molecular and chromosomal evidence for allopolyploidy in soybean. Plant Physiol. 2009;151(3):1167–1174. doi: 10.1104/pp.109.137935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura TBGJ. Cytochrome P450s in plant steroid hormone synthesis and metabolism. Phytochemistry Review. 2006;5:421–432. doi: 10.1007/s11101-006-9024-2. [DOI] [Google Scholar]
- Kang JG, Yun J, Kim DH, Chung KS, Fujioka S, Kim JI, Dae HW, Yoshida S, Takatsuto S, Song PS. et al. Light and brassinosteroid signals are integrated via a dark-induced small G protein in etiolated seedling growth. Cell. 2001;105(5):625–636. doi: 10.1016/S0092-8674(01)00370-1. [DOI] [PubMed] [Google Scholar]
- Li L, Cheng H, Gai J, Yu D. Genome-wide identification and characterization of putative cytochrome P450 genes in the model legume Medicago truncatula. Planta. 2007;226(1):109–123. doi: 10.1007/s00425-006-0473-z. [DOI] [PubMed] [Google Scholar]
- Nelson DR, Ming R, Alam M, Schuler MA. Comparison of Cytochrome P450 Genes from SixPlant Genomes. Tropical Plant Biology. 2008;1:216–235. doi: 10.1007/s12042-008-9022-1. [DOI] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24(8):1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Naur P, Petersen BL, Mikkelsen MD, Bak S, Rasmussen H, Olsen CE, Halkier BA. CYP83A1 and CYP83B1, two nonredundant cytochrome P450 enzymes metabolizing oximes in the biosynthesis of glucosinolates in Arabidopsis. Plant Physiol. 2003;133(1):63–72. doi: 10.1104/pp.102.019240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon RA, Steele CL. Flavonoids and isoflavonoids - a gold mine for metabolic engineering. Trends Plant Sci. 1999;4(10):394–400. doi: 10.1016/S1360-1385(99)01471-5. [DOI] [PubMed] [Google Scholar]
- Schopfer CR, Ebel J. Identification of elicitor-induced cytochrome P450s of soybean (Glycine max L.) using differential display of mRNA. Mol Gen Genet. 1998;258(4):315–322. doi: 10.1007/s004380050736. [DOI] [PubMed] [Google Scholar]
- Otani K, Takahashi T, Furuya T, Ayabe S. Licodione Synthase, a Cytochrome P450 Monooxygenase Catalyzing 2-Hydroxylation of 5-Deoxyflavanone, in Cultured Glycyrrhiza echinata L. Cells. Plant Physiol. 1994;105(4):1427–1432. doi: 10.1104/pp.105.4.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi T, Aoki T, Ayabe S. Cloning and functional expression of a cytochrome P450 cDNA encoding 2- hydroxyisoflavanone synthase involved in biosynthesis of the isoflavonoid skeleton in licorice. Plant Physiol. 1999;121(3):821–828. doi: 10.1104/pp.121.3.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung W, Yu O, Lau SM, O'Keefe DP, Odell J, Fader G, McGonigle B. Identification and expression of isoflavone synthase, the key enzyme for biosynthesis of isoflavones in legumes. Nat Biotechnol. 2000;18(2):208–212. doi: 10.1038/72671. [DOI] [PubMed] [Google Scholar]
- Howe GA, Schilmiller AL. Oxylipin metabolism in response to stress. Current opinion in plant biology. 2002;5(3):230–236. doi: 10.1016/S1369-5266(02)00250-9. [DOI] [PubMed] [Google Scholar]
- Laudert D, Pfannschmidt U, Lottspeich F, Hollander-Czytko H, Weiler EW. Cloning, molecular and functional characterization of Arabidopsis thaliana allene oxide synthase (CYP 74), the first enzyme of the octadecanoid pathway to jasmonates. Plant Mol Biol. 1996;31(2):323–335. doi: 10.1007/BF00021793. [DOI] [PubMed] [Google Scholar]
- Bate NJ, Sivasankar S, Moxon C, Riley J, Thompson JE, Rothstein SJ. Molecular characterization of an Arabidopsis gene encoding hydroperoxide lyase, a cytochrome P-450 that is wound inducible. Plant Physiology. 1998;117(4):1393. doi: 10.1104/pp.117.4.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui K, Ujita C, Fujimoto S, Wilkinson J, Hiatt B, Knauf V, Kajiwara T, Feussner I. Fatty acid 9-and 13-hydroperoxide lyases from cucumber1. FEBS letters. 2000;481(2):183–188. doi: 10.1016/S0014-5793(00)01997-9. [DOI] [PubMed] [Google Scholar]
- Chehab EW, Raman G, Walley JW, Perea JV, Banu G, Theg S, Dehesh K. Rice HYDROPEROXIDE LYASES with unique expression patterns generate distinct aldehyde signatures in Arabidopsis. Plant Physiology. 2006;141(1):121. doi: 10.1104/pp.106.078592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe I, Oguro S, Utsumi Y, Sano Y, Noguchi H. Engineered biosynthesis of plant polyketides: chain length control in an octaketide-producing plant type III polyketide synthase. J Am Chem Soc. 2005;127(36):12709–12716. doi: 10.1021/ja053945v. [DOI] [PubMed] [Google Scholar]
- Tijet N, Schneider C, Muller BL, Brash AR. Biogenesis of volatile aldehydes from fatty acid hydroperoxides: molecular cloning of a hydroperoxide lyase (CYP74C) with specificity for both the 9-and 13-hydroperoxides of linoleic and linolenic acids. Archives of Biochemistry and Biophysics. 2001;386(2):281–289. doi: 10.1006/abbi.2000.2218. [DOI] [PubMed] [Google Scholar]
- Itoh A, Schilmiller AL, McCaig BC, Howe GA. Identification of a jasmonate-regulated allene oxide synthase that metabolizes 9-hydroperoxides of linoleic and linolenic acids. Journal of Biological Chemistry. 2002;277(48):46051. doi: 10.1074/jbc.M207234200. [DOI] [PubMed] [Google Scholar]
- Dhaubhadel S, McGarvey BD, Williams R, Gijzen M. Isoflavonoid biosynthesis and accumulation in developing soybean seeds. Plant Mol Biol. 2003;53(6):733–743. doi: 10.1023/B:PLAN.0000023666.30358.ae. [DOI] [PubMed] [Google Scholar]
- Hermansen K, Dinesen B, Hoie LH, Morgenstern E, Gruenwald J. Effects of soy and other natural products on LDL:HDL ratio and other lipid parameters: a literature review. Advances in Therapy. 2003;20(1):50–78. doi: 10.1007/BF02850119. [DOI] [PubMed] [Google Scholar]
- Libault M, Farmer A, Brechenmacher L, Drnevich J, Langley RJ, Bilgin DD, Radwan O, Neece DJ, Clough SJ, May GD. et al. Complete transcriptome of the soybean root hair cell, a single-cell model, and its alteration in response to Bradyrhizobium japonicum infection. Plant Physiol. 2010;152(2):541–552. doi: 10.1104/pp.109.148379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severin AJ, Woody JL, Bolon YT, Joseph B, Diers BW, Farmer AD, Muehlbauer GJ, Nelson RT, Grant D, Specht JE, RNA-Seq Atlas of Glycine max: a guide to the soybean transcriptome. BMC Plant Biol. p. 160. [DOI] [PMC free article] [PubMed]
- Aoki K, Ogata Y, Shibata D. Approaches for extracting practical information from gene co-expression networks in plant biology. Plant Cell Physiol. 2007;48(3):381–390. doi: 10.1093/pcp/pcm013. [DOI] [PubMed] [Google Scholar]
- Dobritsa AA, Shrestha J, Morant M, Pinot F, Matsuno M, Swanson R, Moller BL, Preuss D. CYP704B1 is a long-chain fatty acid omega-hydroxylase essential for sporopollenin synthesis in pollen of Arabidopsis. Plant Physiol. 2009;151(2):574–589. doi: 10.1104/pp.109.144469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, The Pfam protein families database. Nucleic Acids Res. pp. D211–222. [DOI] [PMC free article] [PubMed]
- Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13(9):2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Landweber LF. BLASTO: a tool for searching orthologous groups. Nucleic Acids Res. 2007. pp. W678–682. [DOI] [PMC free article] [PubMed]
- Paquette SM, Jensen K, Bak S. A web-based resource for the Arabidopsis P450, cytochromes b5, NADPH-cytochrome P450 reductases, and family 1 glycosyltransferases http://www.P450.kvl.dk. Phytochemistry. 2009;70(17-18):1940–1947. doi: 10.1016/j.phytochem.2009.08.024. [DOI] [PubMed] [Google Scholar]
- Rhee SY, Dickerson J, Xu D. Bioinformatics and its applications in plant biology. Annu Rev Plant Biol. 2006;57:335–360. doi: 10.1146/annurev.arplant.56.032604.144103. [DOI] [PubMed] [Google Scholar]
- Libault M, Farmer A, Joshi T, Takahashi K, Langley RJ, Franklin LD, He J, Xu D, May G, Stacey G. An integrated transcriptome atlas of the crop model Glycine max and its use in comparative analyses in plants. Plant J. 2010. [DOI] [PubMed]
- Subramanian S, Stacey G, Yu O. Endogenous isoflavones are essential for the establishment of symbiosis between soybean and Bradyrhizobium japonicum. Plant J. 2006;48(2):261–273. doi: 10.1111/j.1365-313X.2006.02874.x. [DOI] [PubMed] [Google Scholar]
- Subramanian S, Hu X, Lu G, Odelland JT, Yu O. The promoters of two isoflavone synthase genes respond differentially to nodulation and defense signals in transgenic soybean roots. Plant Mol Biol. 2004;54(5):623–639. doi: 10.1023/B:PLAN.0000040814.28507.35. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Comparison of P450 families among soybean, Medicago, Arabidopsis, rice, poplar, grape and moss. A. List of A-type P450 families. B. List of Non-Atype P450 families. For each family number of genes and pseudogenes were compared among different plant species.
Figure S1 Phylogenetic tree of all soybean P450s. S1A. A-type P450s of soybean and Arabidopsis. S1B. Non-A type P450s of soybean and Arabidopsis. Soybean P450s are shown in blue and Arabidopsis P450s are shown in green. Trees were constructed using MEGA4.
Table S2 Organ-specific expressions of soybean P450 genes based on Affymetrix arrays.
Table S3 Organ specific expression of P450 genes using Illumina transcriptome. List of P450 genes and their expression intensity values in different organs. Value zero indicates no expression.
Figure S2 Gene expression of 332 soybean P450 genes according to the analysis of Illumina transcriptome dataset. Gene expression in different organ and root hair inoculation process.
Table S4 Co-expression analysis of soybean P450 genes based on Affymetrix arrays.
Table S5 List of primer pairs used in qRT-PCR