Abstract
Background
The Arabidopsis thaliana NPR1 gene encodes a transcription coactivator (NPR1) that plays a major role in the mechanisms regulating plant defense response. After pathogen infection and in response to salicylic acid (SA) accumulation, NPR1 translocates from the cytoplasm into the nucleus where it interacts with other transcription factors resulting in increased expression of over 2000 plant defense genes contributing to a pathogen resistance response.
Results
A putative Theobroma cacao NPR1 cDNA was isolated by RT-PCR using degenerate primers based on homologous sequences from Brassica, Arabidopsis and Carica papaya. The cDNA was used to isolate a genomic clone from Theobroma cacao containing a putative TcNPR1 gene. DNA sequencing revealed the presence of a 4.5 kb coding region containing three introns and encoding a polypeptide of 591 amino acids. The predicted TcNPR1 protein shares 55% identity and 78% similarity to Arabidopsis NPR1, and contains each of the highly conserved functional domains indicative of this class of transcription factors (BTB/POZ and ankyrin repeat protein-protein interaction domains and a nuclear localization sequence (NLS)). To functionally define the TcNPR1 gene, we transferred TcNPR1 into an Arabidopsis npr1 mutant that is highly susceptible to infection by the plant pathogen Pseudomonas syringae pv. tomato DC3000. Driven by the constitutive CaMV35S promoter, the cacao TcNPR1 gene partially complemented the npr1 mutation in transgenic Arabidopsis plants, resulting in 100 fold less bacterial growth in a leaf infection assay. Upon induction with SA, TcNPR1 was shown to translocate into the nucleus of leaf and root cells in a manner identical to Arabidopsis NPR1. Cacao NPR1 was also capable of participating in SA-JA signaling crosstalk, as evidenced by the suppression of JA responsive gene expression in TcNPR1 overexpressing transgenic plants.
Conclusion
Our data indicate that the TcNPR1 is a functional ortholog of Arabidopsis NPR1, and is likely to play a major role in defense response in cacao. This fundamental knowledge can contribute to breeding of disease resistant cacao varieties through the application of molecular markers or the use of transgenic strategies.
Background
Plants have evolved a complex network of defense responses, often associated with a response local to the site of infection [1-4]. In addition, defenses are also systemically induced in remote parts of the plant in a process known as systemic acquired resistance (SAR) [2,5,6]. Induction of the SAR pathway leads to heightened broad-spectrum resistance to secondary pathogen attacks by a variety of pathogens. Multiple studies in both monocots and dicots have shown that salicylic acid (SA) plays a central role as a signaling molecule in SAR [7-14]. Following pathogen attack, SA levels increase both locally and systemically in infected plants. In addition, SA is required for the induced expression of a set of pathogenesis-related (PR) genes [7,15-17].
NPR1 was originally identified by screening for mutants that were insensitive to SA (or its chemical analogs, 2,6-dichloroisonicotic acid (INA) or benzothiadiazole (BTH)) in Arabidopsis [7,18-20]. These screens identified a mutation designated as Non-Expressor of PR1 (NPR1). Studies that followed further documented that npr1 mutants displayed reduced expression of PR genes upon SA treatment and were more susceptible to pathogens [7,18,20,21]. Conversely, when NPR1 was overexpressed, the resulting transgenic plants displayed increased resistance to pathogens, and were able to induce increased levels of PR genes in a dose-dependent fashion [22].
NPR1 encodes a protein containing ankyrin repeats and a BTB/POZ domain, both of which mediate protein-protein interactions in animals [23]. NPR1 shares homology with IκBα transcription inhibitors, which regulate the innate immunity response [21,24]. Recent work has shed light onto the mechanisms of NPR1 function [5,6,10,17,25-27]. NPR1 is constitutively expressed, and NPR1 protein is present as inactive oligomers in the cytoplasm of the cell. Upon SAR induction, the redox state of the cell is altered, resulting in the reduction of NPR1 to its active monomeric form. Monomeric NPR1 moves into the nucleus where it can affect the induction of PR genes. Although NPR1 itself has no DNA binding domains, it participates in the regulation of defense gene transcription via interactions with TGA transcription factors [16,28-33]. In Arabidopsis, two conserved cysteine residues (C82 and C216) have been shown to be essential to the oligomerization and cytoplasmic localization of AtNPR1 [25]. Mutation of these residues results in constitutive monomerization and nuclear localization of NPR1.
It is believed that NPR1 also plays a role in the jasmonic acid (JA) signaling pathway and mediates the crosstalk between SA-JA defense pathways to fine-tune defense responses [27,30,34-36]. SA-mediated defenses are mainly effective against biotrophic pathogens, whereas JA-mediated defenses are predominantly efficient against necrotrophic pathogens and herbivorous insects. NPR1 mediates the antagonistic effect of SA on JA signaling by suppressing the expression of JA-responsive genes upon combined treatment of SA and methyl jasmonate (MeJA) [34].
A growing body of evidence has revealed that the salicylic acid dependent, NPR1-mediated defense pathway is also conserved in other plant species across wide phylogenetic distances. Two NPR1-like genes have been characterized from Vitis vinifera (grapevine) [14]. When translational fusions of the proteins encoded by the two genes with GFP were transiently expressed in Nicotiana benthamiana leaves, the proteins were localized predominantly to the nucleus and triggered the accumulation of pathogenesis-related proteins PR1 and PR2. In addition, the silencing of a tomato NPR1-like gene leads to increased bacterial growth upon Ralstonia solanacearum infection in tomato [12]. In tobacco, the suppression of NPR1-like gene leads to increased susceptibility to tobacco mosaic virus [8]. Similarly, overexpression of the apple MpNPR1 gene in transgenic apple plants resulted in the up-regulation of PR genes and enhanced resistance to bacterial and fungal pathogens [37]. In wheat, the expression of Arabidopsis NPR1 confers resistance to Fusarium head blight in susceptible cultivar Bobwhite [13]. Major efforts have been made to study the SA and NPR1-dependent pathway in rice, the model monocot plant. Treatment of rice plants with the salicylic acid analog probenazole results in enhanced resistance against rice blast fungus [38]. In addition, rice plants expressing bacterial salicylate hydrolase (nahG) are unable to accumulate salicylic acid and display increased susceptibility to rice blast [39]. Overexpression of the Arabidopsis NPR1 gene in rice leads to enhanced resistance to the bacterial pathogen Xanthomonas oryzae pv. oryzae [9]. An orthologue of NPR1 has been isolated from rice (OsNPR1/NH1), and the overexpression of OsNPR1 in rice leads to enhanced resistance to both bacterial and oomycete pathogens [40]. Moreover, OsNPR1 is able to complement the Arabidopsis npr1-1 mutant [11]. Like AtNPR1, OsNPR1 is also constitutively expressed and localizes to the cytoplasm. Treatment of rice cells with a reducing agent resulted in the movement of OsNPR1 into the nucleus. Similar to Arabidopsis NPR1, mutation of the corresponding cysteines (C82 and C216) in OsNPR1 also resulted in constitutive nuclear localization [11]. Thus, it appears that the mechanisms of SA-dependent, NPR1-mediated defense response likely evolved very early in the emergence of the plant kingdom.
Theobroma cacao L, (cacao) is a small tropical tree species endemic to the Amazon rainforest of South America. Cacao seeds are harvested and processed into cocoa beans and chocolate, providing an income for millions of small-holder farmers in West Africa, Central and South America, the Caribbean, Malaysia, Indonesia and other tropical areas. Pathogens are a major problem for cacao production, causing annual crop losses estimated at 30-40% [41]. In its center of diversity, the Amazon basin, cacao is susceptible to several potentially devastating pathogens, such as Moniliophthora perniciosa, the causal agent of witches' broom disease, Moniliophthora roreri, the causal agent of frosty pod rot [41-45] and several Phytophthora spp., the causal agent of black pod disease [46,47]. Outside this region, cacao is susceptible to a number of opportunistic pathogens [48-50].
Several defense-related genes in Theobroma cacao have been identified through gene expression analyses after hormone treatments [46,47,51]. An endo-1,4-β-glucanase is induced by the application of ethylene, and a type III peroxidase and a class VII chitinase are induced by methyl jasmonate treatment in mature cacao leaves. Those genes are responsible for induced resistance to pests in cacao, though the responses to hormone induction are different depending on developmental stages. In addition, transgenic overexpression of a class I chitinase gene in cacao enhances foliar resistance against the fungal pathogen, Colletotrichum gloeosporioides [52]. Moreover, ESTs sharing sequence homology to known PR genes have been isolated from cacao [53-55]. Several of these genes have been shown to be up-regulated by treatment of plants with benzothiadiazole (BTH), the salicylic acid analog [53]. All together, recent evidence suggests that cacao may utilize SAR pathway during the defense response; however, the extent of conservation of the pathway in cacao is presently unknown.
In this paper, we report the isolation and characterization of an NPR1 homologue from the tropical tree, Theobroma cacao. We show that Theobroma cacao NPR1 (TcNPR1) shares similar functions as Arabidopsis NPR1. It is able to partially complement the Arabidopsis npr1-2 mutation in transgenic Arabidopsis plants in a leaf infection assay and translocate into nucleus upon SA induction in the same manner as the endogenous Arabidopsis NPR1 protein.
Results
Isolation of a putative TcNPR1 gene
Degenerate PCR was utilized to clone the full length cDNA of Theobroma cacao NPR1 (TcNPR1). The degenerate primers were designed based on the alignment of NPR1 homologs from Arabidopsis, Brassica and Carica papaya and cDNA from cacao genotype Scavina6 (SCA6) leaf was used as template. A fragment of 1776 bp was isolated, cloned into pGEM sequencing vector and sequenced to reveal an intact coding sequence of the expected length and with high homology to the Arabidopsis NPR1 gene.
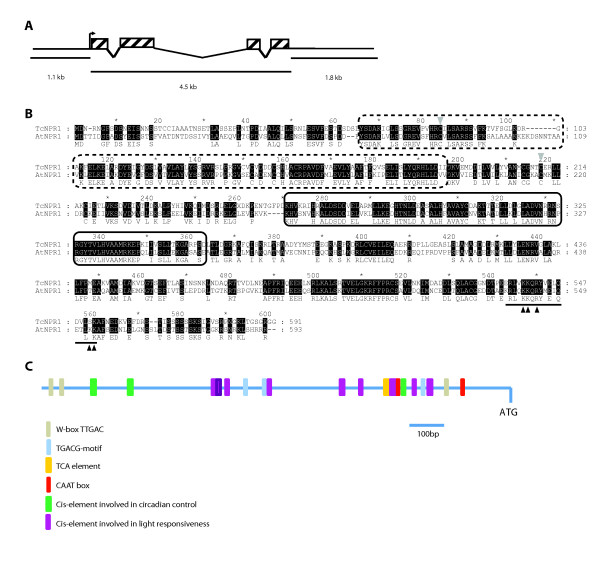
A genomic fragment containing a putative TcNPR1 gene was obtained by screening Clemson University Genomics Institute (CUGI) cacao BAC library using the putative cacao TcNPR1 cDNA clone as probe. Two BAC clones were found to contain the TcNPR1 gene: 2K13 and 11K17. The genomic sequence of TcNPR1 was isolated by primer walking sequencing from known sequence using clone 2K13. A similar strategy was performed to sequence a region of 1.1 kb containing the promoter sequence upstream of ATG start codon. The full sequence consisted of a 4.5 kb genomic region of TcNPR1 containing 1.1 kb promoter, four exons and three introns (depicted in Figure 1A), which is similar to the genomic structure of AtNPR1.
Figure 1.
Gene and protein structures of Theobroma cacao NPR1. A. Diagram of TcNPR1 gene structure. Boxes with diagonal stripes represent exons. Diagonal lines represent introns. The arrow represents the start site of transcription. The sizes of the promoter region, coding and the 3'-untranslated (UTR) regions of TcNPR1 are indicated. B. Alignment of AtNPR1 and TcNPR1 proteins. Protein alignment was carried out by ClustalW. Residues blocked in black are identical in both sequences. Numbers refer to the amino acid position in AtNPR1 protein. BTB/POZ and ankyrin repeats domains are highlighted by dashed line box and solid line box, respectively. Two of the conserved cysteines (C82 and C216 in AtNPR1) are shown with grey triangles. The potential nuclear localization signal identified in Arabidopsis is underlined. Amino acids demonstrated to be critical for AtNPR1 nuclear translocation are indicated with black triangles. C. Schematic representation of predicted cis-acting regulatory DNA element in cacao TcNPR1 promoter region. A 1.1 kb DNA fragment upstream of start codon was analyzed by querying the PLACE and PlantCare databases. The colored blocks represent different cis-elements as indicated.
Arabidopsis and cacao NPR1 protein sequences are highly similar
Conceptual translation of the cacao NPR1 protein revealed that it consists of 591 amino acid residues, only two amino acids shorter than AtNPR1. Alignment of the AtNPR1 and TcNPR1 protein sequences revealed that they are highly similar to each other (55% identity, 74% similarity). Both the Arabidopsis and cacao NPR1 genes encode predicted proteins that share a number of structural features (Figure 1B). Each has a BTB/POZ domain near its N-terminal end (dashed line box) which shares 65% identity. Similarly, an ankyrin repeat region (solid line box) is present in both proteins which shares about 72% identity. In other ankyrin containing proteins, these domains have been shown to play roles in protein-protein interactions [16,23,56,57]. In the AtNPR1 protein, the BTB/POZ domain has been shown to function in homo-dimerization of NPR1, and the ankyrin repeat region mediates interactions with TGA transcription factors [58]. In addition, two cysteine residues (C82 and C216 in AtNPR1), which have been shown to play a role in the redox regulated activation and nuclear localization [25], are also conserved in TcNPR1 (Figure 1B. grey triangles). In fact, the AtNPR1 and TcNPR1 proteins share eleven conserved cysteine residues, suggesting that they share a similar structural conformation. The C-terminal region of AtNPR1 has been shown to contain a nuclear localization signal (NLS) that directs NPR1 monomers into the nucleus upon induction [59]. Five basic amino acids in this region function directly in this role (Figure 1B, black arrows). Four out of five of these basic amino acids are identical in TcNPR1, suggesting that TcNPR1 may also contain functional nuclear localization sequences. These similarities in protein structure suggest that TcNPR1 gene may also share the same function as AtNPR1 during plant defense response.
Cacao NPR1 gene promoter contains putative SA regulatory elements
We analyzed the 1.1 kb promoter region of the TcNPR1 gene (Figure 1C) using plant cis-acting regulatory elements databases PLACE http://www.dna.affrc.go.jp/PLACE/[60] and PlantCare http://bioinformatics.psb.ugent.be/webtools/plantcare/html/[61,62]. Although a potential CAAT box was found 290 bp and 140 bp upstream of the ATG start codon, we did not observe an element resembling a TATA box. This is not surprising, as recent studies of core promoter regions in both plants and animals suggest that only 24%-29% of genes contain TATA-like elements [63,64]. A variety of other regulatory elements were also found. Several elements known to regulate inducibility by salicylic acid were found, such as the AS-1 element (TGACG). TGACG motifs were found involved in transcription activation by SA and this element was previously shown to be required for the SA-induced expression of PR1 [65]. In addition, there were multiple copies of the W-box (TTGAC), an element similar to the AS-1 element, which was also found in promoter of AtNPR1. W-box was shown to be the binding site for SA-induced WRKY DNA binding proteins [66], and was required for the SA induction of the tobacco (Nicotiana tabacum) class I chitinase gene [67]. All of the information suggests that the TcNPR1 gene might be regulated by SA in a manner similar to AtNPR1. Interestingly, several cis-elements involved in light responsiveness and circadian control are also presented in the TcNPR1 promoter, suggesting that TcNPR1 might be also regulated by light.
Basal and induced expression of TcNPR1 in cacao tissues
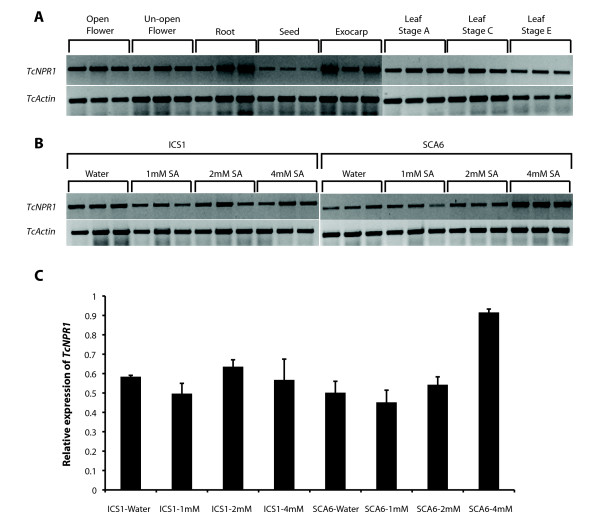
Semi-quantitative RT-PCR was performed to illustrate the basal expression level of TcNPR1 in various cacao tissues of Scavina6, including leaves from stage A (young/expanding), C (expanded/soft), E (mature/hardened), open flowers, unopened flowers, roots, seeds and fruit exocarps. TcNPR1 transcript was detected in all tissues tested (Figure 2A), an expression pattern similar to the Arabidopsis gene, however, the basal level of expression varies among different tissues. The expression of TcNPR1 was relatively high in the younger leaves (stage A and C) and lower in the later stages of development (stage E). The lowest expression of TcNPR1 in all tested tissue was observed in seeds whereas the expression was relatively high in fruit exocarps. In flowers, expression of TcNPR1 was higher in open flowers than in unopened ones. The expression of TcNPR1 in roots was at a moderate level, comparable to that in flowers and younger leaves.
Figure 2.
Gene expression analysis of TcNPR1 in cacao. A. Expression of TcNPR1 in various cacao tissues. Total RNA samples were collected from open flowers, unopened flowers, roots, seeds, exocarp and three different leaf developmental stages from youngest to oldest (A, C and E) from cacao genotype Scavina6 (SCA6). Semi- quantitative RT-PCR was performed and cacao actin (TcActin) was used as cDNA loading control. B. Expression of TcNPR1 in cacao leaf tissue after salicylic acid (SA) treatment. Semi-quantitative RT-PCR was performed with cDNA from stage C leaves of two different cacao genotypes ICS1 (left panel) and SCA6 (right panel), sampled 24 hrs after SA treatment in three different concentrations (1 mM, 2 mM and 4 mM). Water-treated samples served as a control and TcActin was used as cDNA normalization control. C. Calculated average relative gene expression levels from B. Gel images were quantified by ImageQuant and expression of TcNPR1 was normalized to TcActin. Expression levels are presented as the means ± standard errors of three biological replicates.
Induction by SA
Since it is well-characterized that NPR1 transcript accumulation can be increased by SA treatment of Arabidopsis leaves, we tested if TcNPR1 can respond to exogenous SA in the same manner. We applied various concentrations of SA to stage C leaves of two genotypes, Scavina6 and ICS1, which differ in their resistance to witches' broom disease (Scavina6 is more resistant) [68]. In Arabidopsis, the NPR1 gene is induced approximately 2-3 fold 24 hrs after treatment of leaves with 1 mM SA [2,69]. Semi-quantitative RT-PCR was employed to demonstrate the induced level of TcNPR1 24 hours after SA application (Figure 2B). To quantify the expression of TcNPR1 after SA treatment, we measured the fluorescence intensity of ethidium bromide stained DNA fragments irradiated with UV light using a high-sensitivity camera and ImageQuant software. Data were normalized to the expression level of an actin control. The results presented in Figure 2B showed that there was no significant change of TcNPR1 expression upon 1 mM, 2 mM and 4 mM SA treatment in ICS1 compared to water control. However, in the Scavina6, there was a statistically-significant 2-fold increase of TcNPR1 at 4 mM SA induction, though there was no change upon 1 mM and 2 mM SA treatment.
Complementation of Arabidopsis npr1-2 mutant
To assess the function of TcNPR1, we placed the cacao TcNPR1 gene under the control of the E-12 omega promoter and introduced it into the Arabidopsis npr1-2 mutant to test if it can restore the mutant phenotype. One of the well characterized phenotypes of this mutant is the lack of SA-dependent activation of the PR1 gene [18,21]. The PR1 gene is thought to encode a protein active in defense response and has been used as a marker of SA pathway activation in many studies and in different plant species.
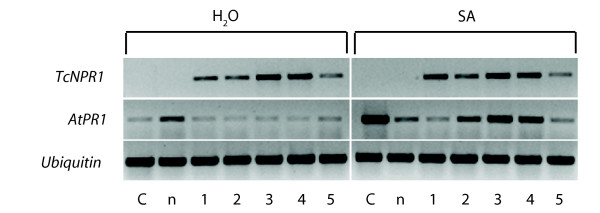
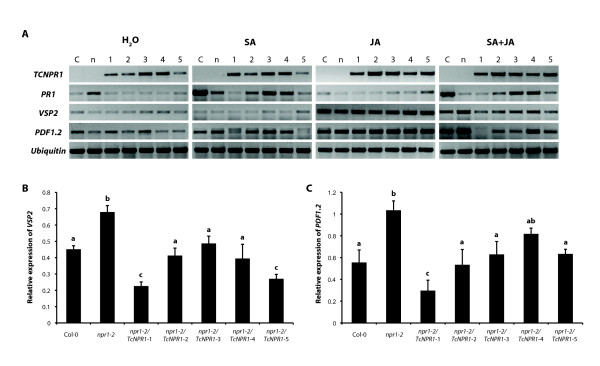
Five independent TcNPR1 transgenic lines, wild type Arabidopsis Col-0 along with the npr1-2 mutant were sprayed with 1 mM SA, and the expression of TcNPR1 and AtPR1 was determined by semi-quantitative RT-PCR 24 hr after induction. Five transgenic lines all showed heterologous TcNPR1 expression with varied expression levels (Figure 3). As expected, there was no significant up-regulation of the transgene after SA treatment because TcNPR1 was expressed constitutively from the E12-Ω promoter. The Arabidopsis PR1 gene showed a very large induction after SA treatment in wild type Arabidopsis Col-0 (Figure 3), but there was no up regulation in the npr1-2 mutant, which is consistent with previous report [69]. There was a small increase in PR1 expression in the mutant treated with water, which could be expected from plant to plant biological variation. However, the PR1 gene expression level did not change after SA treatment, as expected for the npr1-2 mutant. We observed a moderate induction of the PR1 gene in 3 out of 5 transgenic lines (Line 2, 3 and 4), though the level of induction was not as high as in wild type Col-0. No PR1 gene induction was observed for transgenic lines 1 and 5. These results suggest that the TcNPR1 gene can at least partially complement the Arabidopsis npr1mutant and act to mediate SA dependent PR1 gene expression in Arabidopsis leaves but it may not act as efficiently as the endogenous NPR1 itself.
Figure 3.
Gene expression of TcNPR1 and AtPR1 in transgenic Arabidopsis npr1-2 lines. Semi-quantitative RT-PCR was performed with cDNA prepared from the leaves of 4-week-old plants of wild type (C), npr1-2 (n) and 5 independent transgenic npr1-2 mutant lines overexpressing TcNPR1 (1-5). TcNPR1 and AtPR1 expression were evaluated 24 hrs after 1 mM SA treatment. Water-treated control leaves (left panel) from each genotype were also analyzed. Arabidopsis Ubiquitin (AtUbiquitin) expression was assayed as a non SA-induced, cDNA loading control.
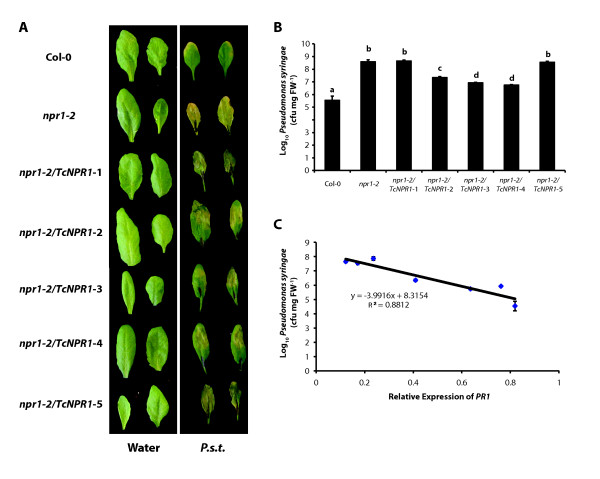
Another phenotype of the Arabidopsis npr1 mutation is increased pathogen growth after bacterial infection of leaves [18,21,69]. To test if TcNPR1 overexpression in npr1-2 mutant can complement the mutant disease susceptible phenotype, we infected leaves from 5 transgenic lines with Pseudomonas syringae pv. tomato DC3000 (P.s.t.) by syringe infiltration. The results indicated that the npr1-2 mutant was more susceptible than Col-0 (Figure 4A) three days after inoculation, exhibiting yellow necrosis similar to previous results [69]. Three transgenic lines overexpressing the TcNPR1 gene and exhibiting SA dependent PR1 up-regulation partially restored induced resistance compared to the control npr1-2 mutant (Figure 4A). Although several yellow necrotic spots were displayed on leaves of the transgenic plants, they did not exhibit severe necrosis or senescence. However, the other two transgenic lines, line 1 and 5, showed necrosis all over the leaves and the tissues were wilted. Water infiltration served as a control to demonstrate that the injection of water alone did not damage the tissues.
Figure 4.
Pseudomonas syringae infection assay of transgenic Arabidopsis npr1-2 mutant lines. A. Disease symptoms on leaves of Col-0, npr1-2 and five independent lines of npr1-2 plants transformed with TcNPR1 (npr1-2/TcNPR1) inoculated with Pseudomonas syringae pv. tomato DC3000 (P.s.t.) (OD600 = 0.002) at three days post inoculation and on leaves of the same seven genotypes infiltrated with water as a control treatment. B. Growth of P.s.t. in leaves from Col-0, npr1-2 and five individual transgenic lines (npr1-2/TcNPR1). Three days after inoculation, leaf disks were collected and bacterial titers were measured. Data represents the means ± standard errors of three biological replicates, each containing three leaf disks from three individual plants. Letters above the histogram indicate statistically significant differences among genotypes (P < 0.01) using the single factor ANOVA. C. Correlation of bacterial growth with relative AtPR1 expression level. Average growth of Pseudomonas syringae pv. tomato DC3000 (Figure 4B) and average AtPR1 gene expression (Figure 3) were evaluated in leaf tissue of Col-0, npr1-2 mutant and five transgenic lines expressing TcNPR1. Data was plotted and analyzed by liner regression analysis.
To quantify the disease symptom, bacterial assays were carried out to measure the titer of bacterial on infected leaves. The levels of bacterial in infected npr1-2 mutant leaf disks increased more than 250 fold as compared to Col-0 controls (Figure 4B). The three transgenic lines overexpressing the TcNPR1 gene (Line 2, 3 and 4), which exhibited significant up-regulation of the PR1 after SA treatment, showed a 30 to 100 fold reduction of bacterial growth compared to the npr1-2 mutant. There was no significant change in bacterial growth rates in leaf disks of the other two transgenic lines tested (Line 1 and 5). To assess the relationship between the level of SA-dependent induction of PR1 and the degree of bacterial growth in the transgenic lines, we plotted the values as depicted in Figure 4C. A significant negative correlation between SA dependent gene induction and bacterial growth was observed (R2 = 0.88), suggesting that the resistance conferred by TcNPR1 is via the SA dependent resistance pathway and further supports our hypothesis that TcNPR1 plays a similar function to Arabidopsis NPR1 in plant defense response.
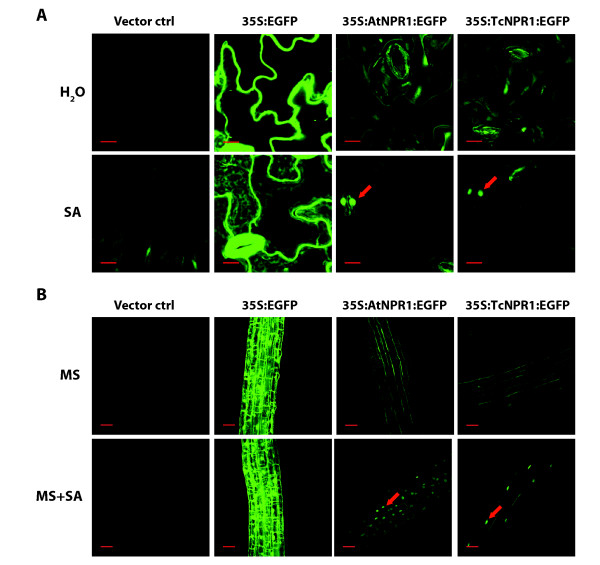
Nuclear translocation of TcNPR1 after SA induction
Another hallmark of AtNPR1 function is its nuclear localization in response to treatment with SA [2,25,59,70,71]. To determine if TcNPR1 can also translocate into the nucleus in response to SA in a manner similar to Arabidopsis NPR1, we created transgenic Arabidopsis plants containing a TcNPR1-EGFP translational fusion and observed the subcellular localization of the fusion protein using confocal microscopy (Figure 5). This construct (35S:TcNPR1:EGFP) was stably transformed into the npr1-2 mutant and we observed the localization of EGFP fusion protein before and 24 hrs after SA treatment in both leaf and root cells of four independent transgenic lines. We observed no EGFP fluorescence in negative control plants transformed with the identical vector lacking the TcNPR1-EGFP fusion gene (Figure 5A and 5B). As an additional control, transgenic plants overexpressing EGFP without a fusion to TcNPR1 were imaged, and we observed strong fluorescence in both cytoplasm and nucleus with no localization changes after SA treatment. A final control consisted of a construct designed for the overexpression of the Arabidopsis NPR1 protein translationally fused to EGFP (35S:AtNPR1:EGFP). Consistent with the findings of others [25,59], we observed an extremely strong nuclear translocation of the fusion protein in leaf guard cells and in root cells 24 hrs after SA treatment.
Figure 5.
Nuclear localization of TcNPR1-EGFP in transgenic Arabidopsis plants in response to SA. A. Confocal images of EGFP fluorescence in Arabidopsis leaves of 4-week-old soil-grown plants 24 hrs after H2O (upper images) or 1 mM SA (lower images) treatment. All images were taken at the same magnification and exposure times. Arrows indicate the accumulation of green fluorescence in guard cell nuclei after SA treatment. Scale bar, 10 μm. B. Confocal images of EGFP fluorescence in Arabidopsis roots from 10-day-old seedlings grown on MS (upper images) or MS supplemented with 0.5 mM SA (lower images). All images were captured using the same exposure settings. Arrows indicate the accumulation of EGFP in nuclei of root cells after SA treatment. Scale bar, 30 μm. Samples from transgenic plants generated with pCAMBIA1300 (vector ctrl) was used as negative control and samples from transgenic plants expressing 35S:EGFP served as positive control in A and B.
The TcNPR1-EGFP fusion protein appeared to be evenly distributed in cytoplasm of leaf guard cells from water-treated 4-week-old soil grown plants, however, the protein accumulated moderately in guard cell nucleus 24 hours after SA application (Figure 5A, red arrow). Similarly, a modest level of nuclear translocation could also be observed in the root cells from 10-day-old seedlings grown on MS medium supplemented with 0.5 mM SA (Figure 5B). Although protein translocation of TcNPR1 is of lesser extent than observed with the Arabidopsis NPR1-EGFP protein based on reduced nuclear fluorescence observed in TcNPR1-EGFP transgenic plants, our results taken together indicate that TcNPR1, like Arabidopsis NPR1, can translocate into nucleus after SA induction and participate in the induction of defense related gene expression.
TcNPR1 and SA-JA crosstalk
It has been previously demonstrated that Arabidopsis NPR1 can mediate the antagonism between SA and jasmonic acid (JA) by suppressing JA-responsive genes [27,34,35], suggesting that it plays an important role in fine tuning the cross-talk between different regulatory pathways. To explore the role of TcNPR1 in cross-talk, we tested the effect of SA and JA treatments on defense gene expression in wild type Col-0, npr1-2 mutant and five independent 35S:TcNPR1 transgenic Arabidopsis lines. Semi-quantitative RT-PCR showed that all five lines carrying the cacao transgene expressed TcNPR1 at moderate levels, and these did not change much during hormone treatments (Figure 6A). Exogenous application of 1 mM SA activated PR1 in Col-0 and three transgenic lines, but not in npr1-2 mutant. Additionally, 48 hrs after treatment with 0.1 mM methyl jasmonate (MeJA) in 0.015% Silwet L-77, two well established MeJA inducible genes (VSP2 and PDF1.2) were up-regulated in wild-type plant and in npr1-2 mutant, consistent with previous reports [34,72]. Two DNA bands were detected in some of the PDF1.2 PCR products, and we determined that the smaller molecular weight band resulted from cDNA amplification and the large fragment resulted from amplification of genomic DNA (data not shown). As predicted, all five 35S:TcNPR1 transgenic lines exhibited levels of increased VSP2 and PDF1.2 that were similar to those seen in Col-0 plants. Upon treatment with a combination of 1 mM SA and 0.1 mM MeJA in 0.015% Silwet L-77, PR1 was expressed at a level similar to seen when plants were treated with SA alone, indicating that MeJA had no effects on SA-responsive PR1 expression. Both VSP2 and PDF1.2 expressed at significantly lower levels in Col-0 compared to that in npr1-2 mutant after SA and MeJA combined treatment, demonstrating the function of AtNPR1 in antagonistic repression of JA-responsive genes. All five transgenic lines containing TcNPR1 gene displayed reduced expression levels of JA-responsive gene expression upon SA and JA combined treatment compared to npr1-2 mutant, suggesting that TcNPR1 can also mediate SA-JA cross-talk in a manner similar to AtNPR1.
Figure 6.
Gene expression of SA- and JA-responsive genes in transgenic Arabidopsis npr1-2 mutants. A. Semi-quantitative RT-PCR was performed with cDNA prepared from leaves of 4-week-old plants of wild type(C), npr1-2 (n) and 5 independent transgenic npr1-2 mutant lines overexpressing TcNPR1 (1-5). The expression of TcNPR1, AtPR1, AtVSP2 and AtPDF1.2 was evaluated 48 hrs after treatment with water control, 1 mM SA water solution alone, 0.1 mM MeJA alone in 0.015% Silwet L-77 and the combination of 1 mM SA and 0.1 mM MeJA in 0.015% Silwet L-77. AtUbiquitin was used as a cDNA loading and normalization control. B. The intensity of AtVSP2 and AtPDF1.2 RT-PCR gel bands in Figure 6A were quantified by ImageQuant software for total pixel intensity and the expression levels were normalized by AtUbiquitin. The bar charts represent the means ± standard errors of relative expression value of AtVSP2 and AtPDF1.2 following 48 hrs treatment of SA-MeJA combination of three biological replicates. Letters above the bar chart indicate statistically significant differences among genotypes (P < 0.05) determined by single factor ANOVA.
To quantify the expression of VSP2 and PDF1.2 after the treatment of the combination of SA and MeJA, we measured the band intensity as above (Figure 6B). The data was normalized to an Ubiquitin control for loading effects. The relative expression levels of VSP2 and PDF1.2 were significantly decreased in TcNPR1 expressing transgenic lines compared to npr1-2 mutant (P < 0.05), a pattern similar to wild-type Col-0, suggesting that TcNPR1 restored the npr1 phenotype. These data support our hypothesis that TcNPR1 may play a role in mediating SA-JA cross talk as does Arabidopsis NPR1.
Discussion
We have isolated an NPR1 homologous gene from the tropical tree, Theobroma cacao, and have generated transgenic Arabidopsis npr1-2 mutant lines overexpressing TcNPR1. All of our results are consistent with the hypothesis that TcNPR1 is a functional orthologue of the well characterized Arabidopsis gene. TcNPR1 complemented each of the major Arabidopsis npr1-2 mutant phenotypes that were tested. Over-expression of TcNPR1 in the npr1-2 mutant conferred PR1 up-regulation after SA treatment and increased resistance to Pseudomonas syringae pv. tomato DC3000 (Figure 3 and 4A, B). TcNPR1 was shown to be translocated into the nucleus in response to SA and to participate in SA-JA cross talk regulation (Figure 5 and 6). In our data, we found that transgenic lines 1 Line exhibited reduced complementation in SA-induced PR1 expression and disease resistance (Figure 3 and 4), while at the same time same two lines efficiently mediated crosstalk between SA and JA (Figure 6). In previous studies, the activation of defense related genes was shown to involve the nuclear translocation of NPR1 [59] while the crosstalk between SA and JA signaling was shown to be mediated by cytosolic NPR1 [34], thus it appears that very different mechanisms exist for these two functions of NPR1. The differential efficiencies of complementation of TcNPR1 we observed may reflect these different mechanisms. It is well known that positional effects (differential transgene transcription levels due to different genomic insertion sites in individual transgenic events) can have a large effect on protein expression levels. As suggested by RNA expression levels of the different TcNPR1 expressing transgenic lines (Figure 3), lines 1 and 5 may have lower protein expression than lines 2-4. It seems plausible that the differential complementation of the two NPR1 functions resulted from the differences in expression levels, potentially as a result of different protein accumulation levels in the cytosol vs nuclear compartments. Consistent with this idea, only the higher levels of expression seen in lines 2-4 was sufficient to complement the nuclear gene induction function, but the levels of expression were high enough in all lines to complement the cytosolic SA/JA crosstalk function.
In all, our results demonstrate a high degree of evolutionary and functional conservation of NPR1 from the Brassicales to the Malvales. NPR1 is also conserved in species as diverse as grapevine [14], tomato [12], apple [37], banana [73], cotton [74], tobacco [8] and rice [11]. This high degree of functional conservation suggests that NPR1 function evolved very early in the development of higher plants and that it plays a very critical role in plant development and reproductive success.
Little is known about the mechanisms of defense signaling in cacao. Our data suggests that the central mechanisms operative in Arabidopsis are likely to be conserved in cacao. At a minimum, our data suggests that the mechanisms and molecules that interact with NPR1 during SA and JA signaling and nuclear translocation are also conserved in cacao. If this were not the case, we would not expect the cacao NPR1 protein to function normally in Arabidopsis. However, the cacao protein in some cases only partially restored function of the npr1 mutant, which is likely the result of transgene expression level differences compared to the endogenous gene and/or partial molecular incompatibility with its interacting protein partners. It is possible that the binding affinities between the cacao NPR1-interacting proteins are reduced as compared to the endogenous Arabidopsis coevolved partners. Partial complementation has commonly been observed in heterologous complementation analysis in many other systems [75-77].
Further investigation is needed to explore the entire defense response pathway in Theobroma cacao and to understand the similarities and differences with Arabidopsis overall. For example, our expression data shows that TcNPR1 can be up-regulated only at 4 mM SA treatment but not at lower concentrations, which is higher than the optimal level of 1 mM in Arabidopsis as previously indicated [69]. It would be interesting to test the endogenous SA level of cacao and to determine dose response dynamics in various tissues and during different stages of development. Another area of interest is to identify and characterize the downstream targets of TcNPR1 and to compare them to the approximately 2,248 genes that are regulated by NPR1 during systemic acquired resistance in Arabidopsis [78]. Surveying these genes in cacao could reveal interesting differences in the defense responses unique to this tropical tree relative to Arabidopsis. Furthermore, Arabidopsis NPR1 has been shown to interact with several different proteins such as the TGA transcription factors [16,28,33,70,79]. Thus another area of interest is to isolate TcNPR1 interacting cacao proteins, which will further enhance our knowledge of this pathway in cacao. We are also interested in studying other NPR1-like genes of cacao and the recent completion of a draft cacao genome sequence has led to the identification of three additional NPR1-like cacao genes [80].
Plant diseases, especially pathogenic fungi, are estimated to cause about 30-40% yield loss on cacao annually [41,81], and thus disease resistance is of substantial interest to cacao breeders. Our findings can be utilized in several approaches to help develop varieties of cacao with enhanced disease resistance. The sequence of the TcNPR1 gene could possibly be used to develop molecular markers and probes that can be employed to select disease resistant varieties with specific allelic variations. Interestingly, the major quantitative trait locus (QTLs) for witches' broom disease resistance is tightly linked to the TcNPR1 gene [80], thus the TcNPR1 gene serves as a key candidate gene for generation of molecular markers that can be used for marker assisted selection of new disease resistant varieties. In addition, TcNPR1 expression levels could be modified in transgenic cacao varieties to develop broad-spectrum disease resistance. This approach has already been successful in several species but to our knowledge, has not yet been deployed in commercial production for any species. However, consumer and industry reluctance to accept transgenic plant technology remains a formidable barrier to development of any transgenic cacao varieties for commercialization.
Conclusion
The isolation of the TcNPR1 gene and its heterologous complementation in Arabidopsis allowed us to rapidly characterize the function of this defense-related gene. The up-regulation of PR1 and increased bacterial resistance in transgenic Arabidopsis npr1-2 mutants strongly supported that TcNPR1 is a functional ortholog of Arabidopsis NPR1, and vital component in SA-dependent signaling pathway in Theobroma cacao. Our results provide potential opportunities to enhance disease resistance in this crop species through conventional breeding or biotechnological approaches. Further investigation is needed to identify the TcNPR1 interacting transcription factors and their downstream targets in cacao and to reveal further details of the molecular mechanisms of the role TcNPR1 plays as a central mediator of the plant defense response.
Methods
Full-length cDNA Cloning by Degenerate PCR
NPR1 cDNA sequences from Arabidopsis (U76707), Brassica napus (AF527176), and Carica papaya (AY550242) were aligned using the ClustalW program v1.8 [82]. Degenerate primers (TcNPR1dg-5', TATTGTCAARTCTRATGTAGAT; TcNPR1dg-3', GAARAAYCGTTTCCCKAGTTCCAC) were designed to regions highly conserved among all three sequences.
Total RNA was isolated from cacao leaves from variety Scavina6 as previously described [53]. Cacao leaf cDNA was synthesized using the SMART RACE cDNA Amplification Kit (Clontech Laboratories Inc., Mountain view, CA http://www.clontech.com/) according to the manufacturer's instructions.
PCR reactions were performed using cacao leaf 2.5 μl cDNA from first strand synthesis from SMART RACE cDNA Amplification Kit, 10 μl Redi-prime PCR mix (GeneChoice, Inc., Frederick, MD) and 5 μM of the above degenerate primers. Following denaturation (94° for 5 min.), PCR was performed for 32 cycles using the following condition (94° for 30 sec., 45° for 30 sec, 72° for 1 min.), followed by a 5 min. extension at 72°. PCR products were resolved on 1% agarose gels, purified with the GENECLEAN II Kit (Q-Biogene Inc., Solon OH) and cloned into the pGEM-T-Easy vector (Promega Corporation, Madison WI) according to the manufacturer's instructions. DNA sequencing was performed at the Penn State Genomics Core Facility using an ABI Hitachi 3730XL DNA Analyzer. The resulting clone was designated as pGEM-TcNPR1.
Genomic DNA cloning by BAC library screening
Theobroma cacao BAC filter arrays constructed using genomic DNA from genotype LCT-EEN 37 were purchased from the Clemson University Genomic Institute http://www.genome.clemson.edu/. Filter arrays were blocked for 4 hours at 60°C in a solution containing 1% BSA, 1 mM EDTA, 7% SDS, and 0.25 M sodium phosphate. PCR generated TcNPR1 cDNA fragment labeled with 32P dCTP using the MegaPrimer DNA Labeling System (GE Healthcare, Buckinghamshire, UK) according to the manufacturer's instructions was added and hybridized overnight at 60°C. The next day, the filter arrays were washed twice in 2× sodium chloride/sodium citrate (SSC), 0.5% sodium dodecyl sulfate (SDS) for 20 minutes at 60°C. Radiographic imaging was performed via storage phosphor imaging (Molecular Dynamics, http://www.mdyn.com/). After filter alignment and clone number identification, a BAC clone (2K13) containing a putative TcNPR1 fragment was obtained from a frozen stock. The sequence of TcNPR1 genomic fragment was acquired by series sequencing of the BAC clone from ATG start codon. Sequencing primer was designed based on the TcNPR1 cDNA at the first round and following series primers were designed based on the known sequence resulting from previous sequencing. Introns were identified by aligning the genomic sequence and full length cDNA using SPIDEY software http://www.ncbi.nlm.nih.gov/spidey/. The same strategy was applied to clone the 1.1 kb promoter region upstream of the ATG. Forward and reverse sequencing was also performed to validate the sequence.
For sequence verification the Arabidopsis NPR1 protein sequence (At1g64280) and putative cacao NPR1 protein sequences (genbank accession HM117159) were aligned using the ClustalW program v1.83 [82]. The TcNPR1 protein sequence was analyzed for potential functional sites by querying the Simple Modular Architecture Research Tool (SMART) database http://smart.embl-heidelberg.de/.
Semi-quantitative RT-PCR analysis of TcNPR1 expression in cacao tissues
Total RNA was isolated from Scavina6 leaves stages A, C and E (corresponding to stages YR, IG, MG respectively, as described in [83]), open flowers, unopened flowers, roots, exocarps and seeds as previously described [53]. For each tissue, three biological replicates were collected and analyzed. Cacao cDNA was synthesized in a final volume of 25 μl from 2 μg of total cacao RNA using M-MLV reverse transcriptase (New England Biolabs, Inc., Ipswich, MA). RNA and 0.5 μg oligo(dT) were added to sterile water to final volume of 18 μl. The mixture was then incubated at 70°C for 5 min, chilled on ice, which was followed by adding 10× reverse transcription buffer (New England Biolabs, Inc., Ipswich, MA), 0.1 M fresh made DTT and 10 mM dNTP. The mixture was further incubated at 42°C for 2 min, followed by incubation at 42°C for 1 hr with 10 units of reverse transcriptase MMLV (New England Biolabs, Inc., Ipswich, MA). The reaction was terminated at 70°C for 15 min.
Semi-quantitative RT-PCR was performed using intron-spanning primers for TcNPR1 (TcNPR1RT-5': ATGGATTCCCGTCTGGAACTTGGT; TcNPR1RT-3': TCTGGAGTGTCATTTCCTCCGCAT) and TcActin (CL33contig2 in Esttik Database http://esttik.cirad.fr/ used as an internal normalization and cDNA loading control (TcActinRT-5': AGCTGAGAGATTCCGTTGTCCAGA and TcActinRT-3': CCCACATCAACCAGACTTTGAGTTC). RT-PCR reactions were set up using 1 μl of 1/2 diluted cDNA and 5 μM of the TcNPR1 or TcActin primers. Titration of cycles was carried out and it was determined that the PCR amplification of TcNPR1 was within its linear range at 27 cycles using the following condition: 94°C for 30 sec., 56°C for 30 sec, 72°C for 1 min. Similarly, PCR of TcActin was performed under non-saturation conditions within the linear range (22 cycles at 94°C for 30 sec., 55°C for 30 sec, 72°C for 1 min). TcActin served as a cDNA loading control.
SA treatment of cacao seedlings
The leaves of two to three-month old cacao plants generated by rooted cuttings from two different genotypes (ICS1 and Scavina6) were sprayed with salicylic acid (SA) dissolved in water at three different concentrations, 1 mM, 2 mM and 4 mM. Control plants were treated with water. Plants were grown in a greenhouse under conditions previously described [53] and leaf tissue from fully expanded young leaves (developmental stage C, corresponding to stage IG in [46]) was harvested at 24 hrs after treatment and frozen in liquid nitrogen. Total RNA was isolated and cDNA was synthesized as described above. For each genotype and each treatment, three biological replicates were collected. Semi-quantitative RT-PCR and expression analysis were performed to assay the levels of TcNPR1 expression as described above. The PCR products were analyzed on 1% agarose gel, stained with ethidium bromide. The expression values of TcNPR1 and TcActin were quantified using ImageQuant software (Molecular Dynamics, Amersham Bioscience) as described in [84] and relative expression of TcNPR1 was calculated by comparing with the expression of TcActin.
Transgenic Arabidopsis mutant complementation assay
All binary plant transformation vectors were constructed by incorporating the genes of interest into pCAMBIA-1300 binary transformation vector containing plant selectable marker for hygromycin resistance [85].
Binary Vector p35S:TcNPR1 - The TcNPR1 coding sequence fragment was generated by PCR using pGEM-TcNPR1 as described above and included XmaI and NotI restriction sites at the 5'- and 3'-ends respectively (TcNPR1-5'-XmaI, CCCGGGATGGATAACAGAAATGGCTT; TcNPR1-3'-NotI, GCGGCCGCTTGCATTAGGCCTATGGTCTA). The fragment was cloned into pGEM T-Easy (Promega Corporation, Madison WI) according to the manufacturer's instructions and sequenced for integrity. The TcNPR1 coding sequence was then cloned into the XmaI and NotI sites of an intermediate cloning vector (pE2113) between E12-Ω promoter [86] and 35SCaMV terminator. A 3 kb restriction fragment containing TcNPR1 gene cassette was excised from pE2113 using PvuII and ligated into the SmaI site of pCAMBIA-1300.
Ligations were performed overnight at 16° with 3Units of T4 DNA ligase (Promega Corporation, Madison WI).
Binary vector p35S:AtNPR1 - The AtNPR1 coding sequence fragment was generated by PCR using the AtNPR1 cDNA clone U13446 from Arabidopsis Biological Resource Center http://www.biosci.ohio-state.edu/~plantbio/Facilities/abrc/abrchome.htm and included NcoI sites at the 5'- and 3'-ends (AtNPR1-5'-NcoI, CCATGGACACCACCATTGATGGATTC; AtNPR1-3'-NcoI, CCATGGTCCGACGACGATGAGAGAGTTTACG). The PCR fragment was cloned into pGEM T-Easy and sequenced. The resulting intermediate plasmid was designated pGEM-AtNPR1. The AtNPR1 coding sequence was then excised by NcoI from pGEM-AtNPR1, and blunt-end cloned into pE2113 between E12-Ω promoter [86] and 35SCaMV terminator as XmaI and NotI fragment. Contently 3.1 kb fragment containing the AtNPR1 gene cassette was obtained by digestion with PvuII, and blunt-end ligated into the SmaI site of pCAMBIA-1300.
Binary Vector p35S:TcNPR1:EGFP - The cassette of E12-Ω promoter and EGFP on the intermediate cloning vector pE2113 was cloned into EcoRI and HindIII sites of pCambia1300. The resulting binary vector was designated pXCGH. PCR generated TcNPR1fragment, including SmaI and KpnI sites at the 5'- and 3'-ends (TcNPR1-5'-SmaI, CCCGGGATGGATAACAGAAATGGCTT; TcNPR1_3'-KpnI, GGTACCGACCGCCCCTACCACTACCAGTTAG) was first cloned into pGEM T-Easy (pGEM-TcNPR1-EGFP). The sequence was verified, the DNA fragment was excised with SmaI and KpnI and blunt ends ligated into the blunt-ended NcoI site of pXCGH positioned between the E12-Ω promoter and at the 5'end of the EGFP coding sequence to generate the binary vector p35S:TcNPR1:EGFP.
Binary vector p35S:AtNPR1:EGFP - The pGEM-AtNPR1 containing AtNPR1 coding sequence was digested with NcoI and the fragment was ligated into the NcoI site of pE2113 as described above. The 3.6 kb fragment containing the AtNPR1- EGFP fusion gene cassette was digested with SalI and EcoRI and cloned into the SalI and EcoRI sites of pCAMBIA-1300.
Arabidopsis Transformation
The binary vectors described above were introduced into Agrobacterium tumefaciens strain AGL1 by electroporation, as previously described in [87]. Arabidopsis Col-0 plants were grown in a Conviron growth chamber (Model No. MTPS144) maintained at 22°C, under a 16:8::L:D cycle. Light intensity was maintained at 200 μM/m2·s with Octron 4100K Ecologic bulbs (Sylvania, Danvers MA). To increase the number of inflorescences, plants were cut back after bolting, and allowed to re-grow. The floral dip method was used to transform Arabidopsis as described previously [88]. Briefly, Agrobacterium cultures were grown at 25° on a platform shaker (200 rpm) to an OD600 = 1.2. Cells were centrifuged at 1,500 × g for 6 minutes and re-suspended in 300 mls of a solution containing 2.15 g L-1 MS salts, 5% sucrose, 0.02% Silwet-77. The flowers were dipped in the solution for three seconds, domed to remain humidity and covered with black cloth. The cloth was removed the next day and plants were regularly watered until seed maturation.
Following seed set, seeds were collected from nine plants for each independent transgenic event. Seeds from 5 individual lines were soaked in 0.1% Tween-20 for 2 minutes and sterilized with 50% bleach for 10 minutes at room temperature. Seeds were then washed five times with 1 ml of sterile water. To select for transformants, seeds were planted on 1/2 MS media, agar plates (pH 5.7) supplemented with 25 μg ml-1 hygromycin B. Plates were place in a Conviron growth chamber under the same light and temperature conditions as above. After 10 days, germinated seedlings were examined for leaf development and root elongation. Those seedlings that showed root elongation were transferred to soil and allowed to grow. Transformations were performed with the following vectors: p35S:TcNPR1, p35S:AtNPR1, p35S:TcNPR1:EGFP, and p35S:AtNPR1:EGFP constructed as described above, and control vectors p35S: EGFP (pGH00.0126, [89])and pCambia 1300.
Salicylic acid (SA) Arabidopsis induction assay
Four week-old wild type Arabidopsis Col-0 and npr1-2 mutants and five independent transgenic lines growing in soil were sprayed with 1 mM SA, along with water-treated control plants. Three biological replicates, each containing leaves from 5 individual plants were collected 24 hrs after treatment. Total RNA was isolated from treated and control samples using RNeasy plant mini kit (QIAGEN, Valencia CA). cDNA was generated as described above. Semi-quantitative RT-PCR was performed as described above to measure the expression of TcNPR1 and AtPR1. Arabidopsis Ubiquitin served as cDNA loading and normalization control. The following primer sets and conditions were employed:
TcNPR1-5': ATGGATTCCCGTCTGGAACTTGGT; TcNPR1-3': TCTGGAGTGTCATTTCCTCCGCAT (27 cycles of 94°C for 30 sec., 56°C for 30 sec., 72°C for 1 min). AtPR1-5': CTCGAAAGCTCAAGATAGCCCACA; AtPR1-3': CTTCTCGTTCACATAATTCCCACG (25 cycles of 94°C for 30 sec., 54°C for 30 sec., 72°C for 1 min). Ubiquitin-5': ACCGGCAAGACCATCACTCT; Ubiquitin-3': AGGCCTCAACTGGTTGCTGT (22 cycles of 94°C for 30 sec., 54°C for 30 sec., 72°C for 1 min). The conditions of PCR were determined by cycle titration to avoid saturating conditions. The relative expression levels were determined as described above.
Pseudomonas syringae infection assay of Arabidopsis transgenic plants overexpressing NPR1 genes
Pseudomonas syringae pv. tomato DC3000 (P.s.t.) was grown on Difco Pseudomonas agar (PA) (Becton, Dickinson and Company, http://www.bdbioscience.com/) supplemented with rifampicin (100 μg ml-1) and kanamycin (25 μg ml-1) at 25°C for 48 hrs. Cells were scraped from plates using a bacterial inoculating loop and re-suspended in water. Plant infection assays and bacterial growth assays were carried out as described previously in [90]. Five individual transgenic npr1-2 mutant overexpressing TcNPR1 coding sequence were infected with P.s.t. at OD600 = 0.002. Briefly, three days after inoculation leaf disks from treated leaves of 2 independent replicate plants were pooled for a single sample. Data represents means ± SE (cfu/mg FW) of three biological replicates per treatment and statistical differences were determined by Single factor ANOVA analysis.
Nuclear translocation of TcNPR1 in transgenic Arabidopsis plants
For observations of green leaves, four week-old soil-grown transgenic plants containing one of transgenes 35S:AtNPR1:EGFP, 35S:TcNPR1:EGFP, 35S:EGFP and plants transformed with empty binary vector pCambia 1300 were sprayed with either a 1 mM solution of SA in water or water. For root observations, control and transgenic seed were germinated on MS agar or MS agar supplemented with 0.5 mM SA [59] and seedlings were grown for 10 days. Leaves and roots were placed in a drop of water on a standard microscope glass slide and overlaid with a cover slip. The samples were imaged with an inverted Olympus FV1000 Laser Scanning Confocal Microscope (Olympus America Inc., Melville, NY). For imaging EGFP, tissues were excited with a blue argon laser (488 nm) and emission wavelengths of 500-600 nm were detected through a variable bandpass filter positioned in front of the photomultiplier tube. Tissues were observed using 40× and 10× objectives for leaf cells and root cells, respectively, each with numerical apertures and 1.15. FV10-ASW version 1.6 software (OLYMPUS, Pittsburgh, PA) was used to collect images, select slices, and create intensity projections over the Z axis.
SA and JA combination treatment of Arabidopsis transgenic plants overexpressing TcNPR1
Four weeks old soil-grown wild type Arabidopsis Col-0, npr1-2 mutants and five independent transgenic lines containing p35S:TcNPR1 were sprayed with a combination of 1 mM SA and 0.1 mM MeJA in 0.015% Silwet L-77. Plants treated with 1 mM SA alone in water, 0.1 mM MeJA alone in 0.015% Silwet L-77 and water with 0.015% Silwet L-77 served as control treatment. Three biological replicates each consisting of leaves from 5 individual plants were collected at 48 hrs after treatment, total RNA was isolated, cDNA was synthesized and semi-quantitative RT-PCR was performed as described above to determine the transcripts level of TcNPR1 and AtPR1. For expression analysis of VSP2 and PDF1.2, following primer sets and conditions were used to maintain the reaction in its linear amplification range. VSP2 Forward: TACGGTCTCGGCATCCGTTC; VSP2 Reverse: CCTCAAGTTCGAACCATTAGGCT (21 cycles of 94°C for 30 sec., 58°C for 30 sec., 72°C for 1 min). PDF1.2 Forward: TCATCATGGCTAAGTTTGCTTCCATC; PDF1.2 Reverse: TGTCATAAAGTTACTCATAGAGTGAC (27 cycles of 94°C for 30 sec., 60°C for 30 sec., 72°C for 1 min). The PCR products were analyzed on 1% agarose gel, stained with ethidium bromide. The expression values of AtVSP2 and AtPDF1.2 were quantified using ImageQuant software (Molecular Dynamics, Amersham Bioscience) as described in [84] and relative expression of two genes was calculated by comparing with the expression of AtUbiquitin.
Accession numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative, GenBank/EMBL databases or Esttik database http://esttik.cirad.fr/ under the following accession numbers: At1g64280 (NPR1), At2g14610 (PR1), At5g24770 (VSP2), At5g44420 (PDF1.2), At3g52590 (ubiquitin), HM117159 (TcNPR1) and CL33contig2 (TcActin).
Abbreviations
NPR1: non expressor of PR genes 1; SA: salicylic acid; INA: 2,6-dichloroisonicotic acid; BTH: benzothiadiazole; BTB/POZ: broad complex, tramtrack and bric a brac/pox virus and zinc finger; JA: jasmonic acid; PR: pathogenesis related; SAR: systemic acquired resistance; NLS: nuclear localization signal; MEJA: methyl jasmonate; VSP2: vegetative storage protein 2; PDF1.2: plant defensin 1.2; QTL: quantitative trait locus.
Authors' contributions
ZS performed most of the experiments, ie, sequence analysis, gene expression studies, phenotypic analysis of transgenic Arabidopsis plants, confocal microscopy observations and drafted the manuscript. SNM participated in the design of the study, directed the transformation vector construction and transgenic lines generation, and participated in drafting of the manuscript. YL participated in transgenic Arabidopsis plants analysis and helped to analyze the sequence. JV cloned the TcNPR1 gene. MJG conceived the study, drafted the manuscript and gave advice on experimental design, data analysis and execution. All authors read and approved the final manuscript.
Contributor Information
Zi Shi, Email: zxs128@psu.edu.
Siela N Maximova, Email: snm104@psu.edu.
Yi Liu, Email: yzl122@psu.edu.
Joseph Verica, Email: joeverica@gmail.com.
Mark J Guiltinan, Email: mjg9@psu.edu.
Acknowledgements
We would like to thank Nicole Zembower at cytometry facility for providing help for confocal microscopy imaging. Thanks to the people in the Guiltinan lab, especially Ann Young and Sharon Pishak for their technical assistance in construction the transformation vectors and generation of transgenic plants. This work is supported in part by The Pennsylvania State University, The Huck Institutes of Life Sciences and American Research Institute Penn State Endowed Program in the Molecular Biology of Cacao.
References
- Heath MC. Hypersensitive response-related death. Plant Mol Biol. 2000;44(3):321–334. doi: 10.1023/A:1026592509060. [DOI] [PubMed] [Google Scholar]
- Durrant WE, Dong X. Systemic acquired resistance. Annu Rev Phytopathol. 2004;42:185–209. doi: 10.1146/annurev.phyto.42.040803.140421. [DOI] [PubMed] [Google Scholar]
- Attaran E, Zeier TE, Griebel T, Zeier J. Methyl salicylate production and jasmonate signaling are not essential for systemic acquired resistance in Arabidopsis. Plant Cell. 2009;21(3):954–971. doi: 10.1105/tpc.108.063164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mur LA, Kenton P, Lloyd AJ, Ougham H, Prats E. The hypersensitive response; the centenary is upon us but how much do we know? J Exp Bot. 2008;59(3):501–520. doi: 10.1093/jxb/erm239. [DOI] [PubMed] [Google Scholar]
- Dong X. NPR1, all things considered. Curr Opin Plant Biol. 2004;7(5):547–552. doi: 10.1016/j.pbi.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Mukhtar MS, Nishimura MT, Dangl J. NPR1 in plant defense: It's not over 'til It's turned over. Cell. 2009;137(5):804–806. doi: 10.1016/j.cell.2009.05.010. [DOI] [PubMed] [Google Scholar]
- Cao H, Bowling SA, Gordon AS, Dong X. Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell. 1994;6(11):1583–1592. doi: 10.1105/tpc.6.11.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Marathe R, Dinesh-Kumar SP. Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 2002;30(4):415–429. doi: 10.1046/j.1365-313X.2002.01297.x. [DOI] [PubMed] [Google Scholar]
- Fitzgerald HA, Chern MS, Navarre R, Ronald PC. Overexpression of (At)NPR1 in rice leads to a BTH- and environment-induced lesion-mimic/cell death phenotype. Mol Plant Microbe Interact. 2004;17(2):140–151. doi: 10.1094/MPMI.2004.17.2.140. [DOI] [PubMed] [Google Scholar]
- Vlot AC, Klessig DF, Park SW. Systemic acquired resistance: the elusive signal(s) Curr Opin Plant Biol. 2008;11(4):436–442. doi: 10.1016/j.pbi.2008.05.003. [DOI] [PubMed] [Google Scholar]
- Yuan Y, Zhong S, Li Q, Zhu Z, Lou Y, Wang L, Wang J, Wang M, Yang D, He Z. Functional analysis of rice NPR1-like genes reveals that OsNPR1/NH1 is the rice orthologue conferring disease resistance with enhanced herbivore susceptibility. Plant Biotechnol J. 2007;5(2):313–324. doi: 10.1111/j.1467-7652.2007.00243.x. [DOI] [PubMed] [Google Scholar]
- Chen YY, Lin YM, Chao TC, Wang JF, Liu AC, Ho FI, Cheng CP. Virus-induced gene silencing reveals the involvement of ethylene-, salicylic acid- and mitogen-activated protein kinase-related defense pathways in the resistance of tomato to bacterial wilt. Physiol Plant. 2009;136(3):324–335. doi: 10.1111/j.1399-3054.2009.01226.x. [DOI] [PubMed] [Google Scholar]
- Makandar R, Essig JS, Schapaugh MA, Trick HN, Shah J. Genetically engineered resistance to Fusarium head blight in wheat by expression of Arabidopsis NPR1. Mol Plant Microbe Interact. 2006;19(2):123–129. doi: 10.1094/MPMI-19-0123. [DOI] [PubMed] [Google Scholar]
- Le Henanff G, Heitz T, Mestre P, Mutterer J, Walter B, Chong J. Characterization of Vitis vinifera NPR1 homologs involved in the regulation of pathogenesis-related gene expression. BMC Plant Biol. 2009;9:54. doi: 10.1186/1471-2229-9-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney T, Friedrich L, Vernooij B, Negrotto D, Nye G, Uknes S, Ward E, Kessmann H, Ryals J. Requirement of salicylic acid for the induction of systemic acquired resistance. Science. 1993;261(5122):754–756. doi: 10.1126/science.261.5122.754. [DOI] [PubMed] [Google Scholar]
- Rochon A, Boyle P, Wignes T, Fobert PR, Despres C. The coactivator function of Arabidopsis NPR1 requires the core of its BTB/POZ domain and the oxidation of C-terminal cysteines. Plant Cell. 2006;18(12):3670–3685. doi: 10.1105/tpc.106.046953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loake G, Grant M. Salicylic acid in plant defence--the players and protagonists. Curr Opin Plant Biol. 2007;10(5):466–472. doi: 10.1016/j.pbi.2007.08.008. [DOI] [PubMed] [Google Scholar]
- Shah J, Tsui F, Klessig DF. Characterization of a salicylic acid-insensitive mutant (sai1) of Arabidopsis thaliana, identified in a selective screen utilizing the SA-inducible expression of the tms2 gene. Mol Plant Microbe Interact. 1997;10(1):69–78. doi: 10.1094/MPMI.1997.10.1.69. [DOI] [PubMed] [Google Scholar]
- Delaney TP, Friedrich L, Ryals JA. Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc Natl Acad Sci USA. 1995;92(14):6602–6606. doi: 10.1073/pnas.92.14.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J, Rogers EE, Ausubel FM. Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics. 1996;143(2):973–982. doi: 10.1093/genetics/143.2.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals J, Weymann K, Lawton K, Friedrich L, Ellis D, Steiner HY, Johnson J, Delaney TP, Jesse T, Vos P. et al. The Arabidopsis NIM1 protein shows homology to the mammalian transcription factor inhibitor I kappa B. Plant Cell. 1997;9(3):425–439. doi: 10.1105/tpc.9.3.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Li X, Dong X. Generation of broad-spectrum disease resistance by overexpression of an essential regulatory gene in systemic acquired resistance. Proc Natl Acad Sci USA. 1998;95(11):6531–6536. doi: 10.1073/pnas.95.11.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stogios PJ, Downs GS, Jauhal JJ, Nandra SK, Prive GG. Sequence and structural analysis of BTB domain proteins. Genome Biol. 2005;6(10):R82. doi: 10.1186/gb-2005-6-10-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin AS. The NF-kappa B and I kappa B proteins: new discoveries and insights. Annu Rev Immunol. 1996;14:649–683. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- Mou Z, Fan W, Dong X. Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell. 2003;113(7):935–944. doi: 10.1016/S0092-8674(03)00429-X. [DOI] [PubMed] [Google Scholar]
- Pieterse CM, Van Loon LC. NPR1: the spider in the web of induced resistance signaling pathways. Curr Opin Plant Biol. 2004;7(4):456–464. doi: 10.1016/j.pbi.2004.05.006. [DOI] [PubMed] [Google Scholar]
- Feys BJ, Parker JE. Interplay of signaling pathways in plant disease resistance. Trends Genet. 2000;16(10):449–455. doi: 10.1016/S0168-9525(00)02107-7. [DOI] [PubMed] [Google Scholar]
- Despres C, DeLong C, Glaze S, Liu E, Fobert PR. The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell. 2000;12(2):279–290. doi: 10.1105/tpc.12.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JM, Trifa Y, Silva H, Pontier D, Lam E, Shah J, Klessig DF. NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR-1 gene required for induction by salicylic acid. Mol Plant Microbe Interact. 2000;13(2):191–202. doi: 10.1094/MPMI.2000.13.2.191. [DOI] [PubMed] [Google Scholar]
- Ndamukong I, Abdallat AA, Thurow C, Fode B, Zander M, Weigel R, Gatz C. SA-inducible Arabidopsis glutaredoxin interacts with TGA factors and suppresses JA-responsive PDF1.2 transcription. Plant J. 2007;50(1):128–139. doi: 10.1111/j.1365-313X.2007.03039.x. [DOI] [PubMed] [Google Scholar]
- Johnson C, Mhatre A, Arias J. NPR1 preferentially binds to the DNA-inactive form of Arabidopsis TGA2. Biochim Biophys Acta. 2008;1779(10):583–589. doi: 10.1016/j.bbagrm.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Fan W, Dong X. In vivo interaction between NPR1 and transcription factor TGA2 leads to salicylic acid-mediated gene activation in Arabidopsis. Plant Cell. 2002;14(6):1377–1389. doi: 10.1105/tpc.001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle P, Le Su E, Rochon A, Shearer HL, Murmu J, Chu JY, Fobert PR, Despres C. The BTB/POZ domain of the Arabidopsis disease resistance protein NPR1 interacts with the repression domain of TGA2 to negate its function. Plant Cell. 2009;21(11):3700–3713. doi: 10.1105/tpc.109.069971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel SH, Koornneef A, Claessens SM, Korzelius JP, Van Pelt JA, Mueller MJ, Buchala AJ, Metraux JP, Brown R, Kazan K. et al. NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell. 2003;15(3):760–770. doi: 10.1105/tpc.009159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon-Reyes A, Spoel SH, De Lange ES, Abe H, Kobayashi M, Tsuda S, Millenaar FF, Welschen RA, Ritsema T, Pieterse CM. Ethylene modulates the role of NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1 in cross talk between salicylate and jasmonate signaling. Plant Physiol. 2009;149(4):1797–1809. doi: 10.1104/pp.108.133926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef A, Pieterse CM. Cross talk in defense signaling. Plant Physiol. 2008;146(3):839–844. doi: 10.1104/pp.107.112029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malnoy M, Jin Q, Borejsza-Wysocka EE, He SY, Aldwinckle HS. Overexpression of the apple MpNPR1 gene confers increased disease resistance in Malus x domestica. Mol Plant Microbe Interact. 2007;20(12):1568–1580. doi: 10.1094/MPMI-20-12-1568. [DOI] [PubMed] [Google Scholar]
- Sakamoto K, Tada Y, Yokozeki Y, Akagi H, Hayashi N, Fujimura T, Ichikawa N. Chemical induction of disease resistance in rice is correlated with the expression of a gene encoding a nucleotide binding site and leucine-rich repeats. Plant Mol Biol. 1999;40(5):847–855. doi: 10.1023/A:1006244323934. [DOI] [PubMed] [Google Scholar]
- Yang Y, Qi M, Mei C. Endogenous salicylic acid protects rice plants from oxidative damage caused by aging as well as biotic and abiotic stress. Plant J. 2004;40(6):909–919. doi: 10.1111/j.1365-313X.2004.02267.x. [DOI] [PubMed] [Google Scholar]
- Chern M, Fitzgerald HA, Canlas PE, Navarre DA, Ronald PC. Overexpression of a rice NPR1 homolog leads to constitutive activation of defense response and hypersensitivity to light. Mol Plant Microbe Interact. 2005;18(6):511–520. doi: 10.1094/MPMI-18-0511. [DOI] [PubMed] [Google Scholar]
- Hebbar PK. Cacao diseases: a global perspective from an industry point of view. Phytopathology. 2007;97(12):1658–1663. doi: 10.1094/PHYTO-97-12-1658. [DOI] [PubMed] [Google Scholar]
- Purdy LH, Schmidt RA. STATUS OF CACAO WITCHES' BROOM: biology, epidemiology, and management. Annu Rev Phytopathol. 1996;34:573–594. doi: 10.1146/annurev.phyto.34.1.573. [DOI] [PubMed] [Google Scholar]
- Wood G, Lass R. Cocoa. New York: Longman Scientific & Technical Press; 1985. [Google Scholar]
- Evans H, Priori C. Cocoa pod diseases: causal agents and control. Outlook on Agriculture. 1987;16:35–41. [Google Scholar]
- Phillips-Mora W, Wilkinson MJ. Frosty pod of cacao: a disease with a limited geographic range but unlimited potential for damage. Phytopathology. 2007;97(12):1644–1647. doi: 10.1094/PHYTO-97-12-1644. [DOI] [PubMed] [Google Scholar]
- Bailey BA, Bae H, Strem MD, Antunez de Mayolo G, Guiltinan MJ, Verica JA, Maximova SN, Bowers JH. Developmental expression of stress response genes in Theobroma cacao leaves and their response to Nep1 treatment and a compatible infection by Phytophthora megakarya. Plant Physiol Biochem. 2005;43(6):611–622. doi: 10.1016/j.plaphy.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Bailey BA, Strem MD, Bae H, de Mayolo GA, Guiltinan MJ. Gene expression in leaves of Theobroma cacao in response to mechanical wounding, ethylene, and/or methyl jasmonate. Plant Science. 2005;168(5):1247–1258. doi: 10.1016/j.plantsci.2005.01.002. [DOI] [Google Scholar]
- Buddenhagen IW. Resistance and Vulnerability of Tropical Crops in Relation to Their Evolution and Breeding. Annals of the New York Academy of Sciences. 1977;287(Feb25):309–326. doi: 10.1111/j.1749-6632.1977.tb34249.x. [DOI] [Google Scholar]
- Guiltinan M, Verica J, Zhang D, Figueira A. In: Genomics of Tropical Crop Plants. Moore PM, Ming R, editor. New York: Springer; 2008. Genomics of Theobroma cacao, "The foods of the Gods". [Google Scholar]
- Schnell RJ, Kuhn DN, Brown JS, Olano CT, Phillips-Mora W, Amores FM, Motamayor JC. Development of a marker assisted selection program for cacao. Phytopathology. 2007;97(12):1664–1669. doi: 10.1094/PHYTO-97-12-1664. [DOI] [PubMed] [Google Scholar]
- Bae H, Kim SH, Kim MS, Sicher RC, Lary D, Strem MD, Natarajan S, Bailey BA. The drought response of Theobroma cacao (cacao) and the regulation of genes involved in polyamine biosynthesis by drought and other stresses. Plant Physiol Biochem. 2008;46(2):174–188. doi: 10.1016/j.plaphy.2007.10.014. [DOI] [PubMed] [Google Scholar]
- Maximova SN, Marelli JP, Young A, Pishak S, Verica JA, Guiltinan MJ. Over-expression of a cacao class I chitinase gene in Theobroma cacao L. enhances resistance against the pathogen, Colletotrichum gloeosporioides. Planta. 2006;224(4):740–749. doi: 10.1007/s00425-005-0188-6. [DOI] [PubMed] [Google Scholar]
- Verica JA, Maximova SN, Strem MD, Carlson JE, Bailey BA, Guiltinan MJ. Isolation of ESTs from cacao (Theobroma cacao L.) leaves treated with inducers of the defense response. Plant Cell Rep. 2004;23(6):404–413. doi: 10.1007/s00299-004-0852-5. [DOI] [PubMed] [Google Scholar]
- Jones PG, Allaway D, Gilmour DM, Harris C, Rankin D, Retzel ER, Jones CA. Gene discovery and microarray analysis of cacao (Theobroma cacao L.) varieties. Planta. 2002;216(2):255–264. doi: 10.1007/s00425-002-0882-6. [DOI] [PubMed] [Google Scholar]
- Argout X, Fouet O, Wincker P, Gramacho K, Legavre T, Sabau X, Risterucci AM, Da Silva C, Cascardo J, Allegre M. et al. Towards the understanding of the cocoa transcriptome: Production and analysis of an exhaustive dataset of ESTs of Theobroma cacao L. generated from various tissues and under various conditions. BMC Genomics. 2008;9:512. doi: 10.1186/1471-2164-9-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albagli O, Dhordain P, Deweindt C, Lecocq G, Leprince D. The BTB/POZ domain: a new protein-protein interaction motif common to DNA- and actin-binding proteins. Cell Growth Differ. 1995;6(9):1193–1198. [PubMed] [Google Scholar]
- Li J, Mahajan A, Tsai MD. Ankyrin repeat: a unique motif mediating protein-protein interactions. Biochemistry. 2006;45(51):15168–15178. doi: 10.1021/bi062188q. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Fan W, Kinkema M, Li X, Dong X. Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc Natl Acad Sci USA. 1999;96(11):6523–6528. doi: 10.1073/pnas.96.11.6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinkema M, Fan W, Dong X. Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell. 2000;12(12):2339–2350. doi: 10.1105/tpc.12.12.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999;27(1):297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouze P, Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30(1):325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rombauts S, Dehais P, Van Montagu M, Rouze P. PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 1999;27(1):295–296. doi: 10.1093/nar/27.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina C, Grotewold E. Genome wide analysis of Arabidopsis core promoters. BMC Genomics. 2005;6(1):25. doi: 10.1186/1471-2164-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Bolotin E, Jiang T, Sladek FM, Martinez E. Prevalence of the initiator over the TATA box in human and yeast genes and identification of DNA motifs enriched in human TATA-less core promoters. Gene. 2007;389(1):52–65. doi: 10.1016/j.gene.2006.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebel E, Heifetz P, Thorne L, Uknes S, Ryals J, Ward E. Functional analysis of regulatory sequences controlling PR-1 gene expression in Arabidopsis. Plant J. 1998;16(2):223–233. doi: 10.1046/j.1365-313x.1998.00288.x. [DOI] [PubMed] [Google Scholar]
- Yu D, Chen C, Chen Z. Evidence for an important role of WRKY DNA binding proteins in the regulation of NPR1 gene expression. Plant Cell. 2001;13(7):1527–1540. doi: 10.1105/tpc.13.7.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Chen C, Wang Z, Fan B, Chen Z. A pathogen- and salicylic acid-induced WRKY DNA-binding activity recognizes the elicitor response element of the tobacco class I chitinase gene promoter. Plant J. 1999;18(2):141–149. doi: 10.1046/j.1365-313X.1999.00437.x. [DOI] [Google Scholar]
- Faleiro F, Queiroz V, Lopes U, Guimarães C, Pires J, Yamada M, Araújo I, Pereira M, Schnell R, Filho G. et al. Mapping QTLs for witches' broom (Crinipellis Perniciosa) resistance in cacao (Theobroma Cacao L) Euphytica. 2006;149(1):227–235. doi: 10.1007/s10681-005-9070-7. [DOI] [Google Scholar]
- Cao H, Glazebrook J, Clarke JD, Volko S, Dong X. The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell. 1997;88(1):57–63. doi: 10.1016/S0092-8674(00)81858-9. [DOI] [PubMed] [Google Scholar]
- Spoel SH, Mou Z, Tada Y, Spivey NW, Genschik P, Dong X. Proteasome-mediated turnover of the transcription coactivator NPR1 plays dual roles in regulating plant immunity. Cell. 2009;137(5):860–872. doi: 10.1016/j.cell.2009.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Chen S, Mou Z. Nuclear localization of NPR1 is required for regulation of salicylate tolerance, isochorismate synthase 1 expression and salicylate accumulation in Arabidopsis. J Plant Physiol. 2010;167(2):144–148. doi: 10.1016/j.jplph.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Penninckx IA, Thomma BP, Buchala A, Metraux JP, Broekaert WF. Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell. 1998;10(12):2103–2113. doi: 10.1105/tpc.10.12.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endah R, Beyene G, Kiggundu A, van den Berg N, Schluter U, Kunert K, Chikwamba R. Elicitor and Fusarium-induced expression of NPR1-like genes in banana. Plant Physiol Biochem. 2008;46(11):1007–1014. doi: 10.1016/j.plaphy.2008.06.007. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang X, Cheng C, Gao Q, Liu J, Guo X. Molecular cloning and characterization of GhNPR1, a gene implicated in pathogen responses from cotton (Gossypium hirsutum L) Biosci Rep. 2008;28(1):7–14. doi: 10.1042/BSR20070028. [DOI] [PubMed] [Google Scholar]
- Irish VF, Yamamoto YT. Conservation of floral homeotic gene function between Arabidopsis and antirrhinum. Plant Cell. 1995;7(10):1635–1644. doi: 10.1105/tpc.7.10.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maizel A, Busch MA, Tanahashi T, Perkovic J, Kato M, Hasebe M, Weigel D. The floral regulator LEAFY evolves by substitutions in the DNA binding domain. Science. 2005;308(5719):260–263. doi: 10.1126/science.1108229. [DOI] [PubMed] [Google Scholar]
- Dong X, Braun EL, Grotewold E. Functional conservation of plant secondary metabolic enzymes revealed by complementation of Arabidopsis flavonoid mutants with maize genes. Plant Physiol. 2001;127(1):46–57. doi: 10.1104/pp.127.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Amornsiripanitch N, Dong X. A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog. 2006;2(11):e123.. doi: 10.1371/journal.ppat.0020123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C, Boden E, Arias J. Salicylic acid and NPR1 induce the recruitment of trans-activating TGA factors to a defense gene promoter in Arabidopsis. Plant Cell. 2003;15(8):1846–1858. doi: 10.1105/tpc.012211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argout X, Salse J, Aury J, Guiltinan M, Droc G, Gouzy J, Allegre M, Chaparro C, Legavre T, Maximova S, The Genome of Theobroma cacao. Nature Genetics. 2010. in press . [DOI] [PubMed]
- Ploetz RC. Cacao diseases: important threats to chocolate production worldwide. Phytopathology. 2007;97(12):1634–1639. doi: 10.1094/PHYTO-97-12-1634. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnick RL, Zidack NK, Bailey BA, Maximova SN, Guiltinan M, Backman PA. Bacterial endophytes: Bacillus spp. from annual crops as potential biological control agents of black pod rot of cacao. Biological Control. 2008;46(1):46–56. doi: 10.1016/j.biocontrol.2008.01.022. [DOI] [Google Scholar]
- Brechenmacher L, Weidmann S, van Tuinen D, Chatagnier O, Gianinazzi S, Franken P, Gianinazzi-Pearson V. Expression profiling of up-regulated plant and fungal genes in early and late stages of Medicago truncatula-Glomus mosseae interactions. Mycorrhiza. 2004;14(4):253–262. doi: 10.1007/s00572-003-0263-4. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol. 1994;25(6):989–994. doi: 10.1007/BF00014672. [DOI] [PubMed] [Google Scholar]
- Mitsuhara I, Ugaki M, Hirochika H, Ohshima M, Murakami T, Gotoh Y, Katayose Y, Nakamura S, Honkura R, Nishimiya S. et al. Efficient promoter cassettes for enhanced expression of foreign genes in dicotyledonous and monocotyledonous plants. Plant Cell Physiol. 1996;37(1):49–59. doi: 10.1093/oxfordjournals.pcp.a028913. [DOI] [PubMed] [Google Scholar]
- Lin JJ. Electrotransformation of Agrobacterium. Methods Mol Biol. 1995;47:171–178. doi: 10.1385/0-89603-310-4:171. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Maximova S, Miller C, Antunez de Mayolo G, Pishak S, Young A, Guiltinan MJ. Stable transformation of Theobroma cacao L. and influence of matrix attachment regions on GFP expression. Plant Cell Rep. 2003;21(9):872–883. doi: 10.1007/s00299-003-0596-7. [DOI] [PubMed] [Google Scholar]
- Katagiri F, Thilmony R, He S. The Arabidopsis Book. The American Society of Plant Biologists; 2002. The Arabidopsis thaliana-Pseudomonas syringae interaction. [DOI] [PMC free article] [PubMed] [Google Scholar]