Abstract
Background
The genus Arachis comprises 80 species and it is subdivided into nine taxonomic sections (Arachis, Caulorrhizae, Erectoides, Extranervosae, Heteranthae, Procumbentes, Rhizomatosae, Trierectoides, and Triseminatae). This genus is naturally confined to South America and most of its species are native to Brazil. In order to provide a better understanding of the evolution of the genus, we reconstructed the phylogeny of 45 species using the variation observed on nucleotide sequences in internal transcribed spacer regions (ITS1 and ITS2) and 5.8 S of nuclear ribosomal DNA.
Results
Intraspecific variation was detected, but in general it was not enough to place accessions of the same species in different clades. Our data support the view that Arachis is a monophyletic group and suggested Heteranthae as the most primitive section of genus Arachis. The results confirmed the circumscriptions of some sections (Caulorrhizae, Extranervosae), but raised questions about others. Sections Erectoides, Trierectoides and Procumbentes were not well defined, while sections Arachis and Rhizomatosae seem to include species that could be moved to different sections. The division of section Arachis into A and B genome species was also observed in the phylogenetic tree and these two groups of species may not have a monophyletic origin. The 2n = 2x = 18 species of section Arachis (A. praecox, A. palustris and A. decora) were all placed in the same clade, indicating they are closely related to each other, and their genomes are more related to B genome than to the A genome. Data also allowed insights on the origin of tetraploid A. glabrata, suggesting rhizome appeared twice within the genus and raising questions about the placement of that species in section Rhizomatosae.
Conclusion
The main clades established in this study in general agreed with many other studies that have used other types of evidences and sets of species, being some of them included in our study and some not. Thus, the relationships established can be a useful framework for future systematic reviews of genus Arachis and for the selection of species to pre-breeding programs.
Background
The genus Arachis originated in South America, where all the cultivated and wild species are found, and includes 80 described species [1,2]. Groundnut, the allotetraploid species A. hypogaea (genome formula AABB), is the most important species of the genus because it is cultivated as an oilseed crop and as a direct source of human food. The genus also includes species such as A. glabrata (section Rhizomatosae) and A. pintoi (section Caulorrhizae), which are frequently used in cultivated pastures.
Many studies have focused on the taxonomy of genus Arachis (Table 1). The first published classification divided the genus into six sections, some of which were sub-divided in series [3]. A taxonomic revision of this genus resulted in the inclusion of section Triseminalae [4]. Krapovickas [5] divided the genus into eight sections and classified one of the series of section Erectoides, established by Gregory et al. [4], as a new section called Procumbensae. The genus is currently divided into nine sections (Arachis, Caulorrhizae, Erectoides, Extranervosae, Heteranthae, Procumbentes, Rhizomatosae, Trierectoides, and Triseminatae) based on morphology, geographical distribution and crossability [1]. In this last revision, two species with trifoliolate leaves from section Erectoides were transferred to a new section called Trierectoides.
Table 1.
Classifications of genus Arachis*.
| Krapovickas (1969) |
Krapovickas (1975) |
Gregory et al. (1973, 1980) |
Krapovickas (1990) |
Krapovickas and Gregory (1994) |
Number of described species | Number of species analyzed | Genomes |
|---|---|---|---|---|---|---|---|
| AXONOMORPHAE | ARACHIS | AXONOMORPHAE | ARACHIS | ARACHIS | 31 | 25 | A,B,D,F,K, AB |
| Annuae | Annuae | ||||||
| Perennes | Perennes | ||||||
| Amphiploides | Amphiploides | ||||||
| ERECTOIDES | TRIERECTOIDES | ERECTOIDES | ERECTOIDES | ERECTOIDES | 14 | 4 | E |
| Trifoliolatae | Tetrafoliolate | ||||||
| TETRAERECTOIDES | Tetrafoliolate | Trifoliolatae | TRIERECTOIDES | 2 | 2 | E | |
| Procumbensae | PROCUMBENSAE | PROCUMBENTES | 10 | 5 | E | ||
| CAULORRHIZAE | CAULORRHIZAE | CAULORRHIZAE | CAULORRHIZAE | CAULORRHIZAE | 2 | 2 | C |
| RHIZOMATOSAE | RHIZOMATOSAE | RHIZOMATOSAE | RHIZOMATOSAE | RHIZOMATOSAE | 2 | 2 | R |
| Prorhizomatosae | Prorhizomatosae | Prorhizomatosae | |||||
| Eurhizomatosae | Eurhizomatosae | Eurhizomatosae | |||||
| EXTRANERVOSAE | EXTRANERVOSAE | EXTRANERVOSAE | EXTRANERVOSAE | EXTRANERVOSAE | 10 | 3 | Ex |
| AMBINERVOSAE | AMBINERVOSAE | PSEUDAXONOMORPHAE | AMBINERVOSAE | HETERANTHAE | 6 | 1 | Am |
| TRISEMINALAE | TRISEMINALAE | TRISEMINATAE | 1 | 1 | T |
*Modified from Valls and Simpson [69].
The systematic relationships among Arachis species have been inferred using different molecular markers, such as RAPDs [6], storage proteins [7,8], isozymes [9,10], variation on sequence of desaturase genes [11], RFLP [12], microsatellites [13-16], AFLPs [17], cytogenetic and molecular data from AFLP and the trnT-F plastid region [18], FISH and GISH [19-21]. However, most of these studies included only species belonging to section Arachis. Just recently one study that included species from all sections was published [22].
Understanding the phylogenetic relationships among Arachis species would contribute to the systematics of the genus, comprehension of the origins and evolution of species and sections and the use of species of genus Arachis. For instance, the circumscriptions of some sections are based in criterions that may not reflect phylogenetic relationships. The maintenance of species associated respectively to the A and the B genomes of the cultivated peanut in a single section does not seem to be natural and it may be an artificial construction derived from the existence of the peanut, a fixed amphidiploid gathering genetic material from species that when crossed to each other produce unfertile hybrids at the diploid level. Also, section Rhizomatosae, a very important group from the standpoint of forage production, comprises polyploid species (A. glabrata, A. pseudovillosa, A. nitida) and one diploid (A. burkartii) that RAPD [23] and microsatellite data [24] showed to be very distinct from the other species in this section. As mentioned before, phylogenetic information will also have great impact in the utilization of the species, mainly those from sections that comprise cultivated species. For instance, for many years A. batizocoi was considered the donor of the B genome of A. hypogaea and that species was used with moderated success in pre-breeding programs. However, molecular and cytogenetic evidences showed that A. ipaënsis was the most probable donor of B genome of A. hypogaea and A. duranensis the donor of the A genome [25]. That information was corroborated based on molecular cytogenetics [19] and crossability [26]. The number of accessions of A. duranensis is very large [1] comprising large variability that could be used to improve A. hypogaea through introgression into its A genome. Besides, the relationships between A genome species are well defined and they show very good crossability to each other [1]. On the other hand, there is an unique accession of A. ipaënsis available and the relationships among species of section Arachis that do not have the A genome is not well defined. The non A genome species group is very diverse comprising species with different degrees of affinity to the B genome of A. hypogaea [18,27]. Phylogeny and genomic data also allow a better understanding of the evolution in section Arachis. For instance, recently the first comparative genomic study between the genomes of A. hypogaea using microsatellite markers and two map populations resulting from crosses between two A genome species and two B genome species was published [28]. The comparison between the B genome and A genome maps revealed a high degree of synteny. The development of genetic maps for Arachis diploid wild species with A and B genomes associated to phylogenetic studies effectively is a significant advance towards the construction of a transferable reference map for Arachis.
The DNA sequence variation observed in the internal transcribed spacers (ITS1 and ITS2) of nuclear rDNA, located between the 18 S and 26 S rDNA coding regions, has been largely used for phylogenetic analysis at plant genus and species discrimination levels [29-34]. The sequences are relatively easy to align because few length variations have been observed at the genus level in flowering plants, they are long enough to offer a sufficient number of potential characters for phylogenetic reconstruction, and are flanked by regions that are highly conserved within genera, thus simplifying the isolation and sequencing of the region through the use of universal primers [35].
The objective of this work was to establish the phylogenetic relationships among species of the genus Arachis. The polymorphism in sequences of internal transcribed spacers ITS1 and ITS2 and 5.8 S rDNA coding region was used to determine relationships among 45 species of genus Arachis.
Results and Discussion
We have analyzed 55 accessions encompassing 45 Arachis species and the nine taxonomical sections. Consensus sequences were obtained for each accession using four to ten reads. The number of reads per accession varied because their quality also varied. For some species only four reads were necessary to get a good quality consensus sequence and for some ten reads were necessary.
In spite of the large number of studies using ITS to infer phylogeny in a very large number of genus and families, some authors have criticized the use of these genomic regions. ITS data may cause incongruence due to the various mechanisms that can influence ITS variation [33]. Among the most prevalent complications for phylogenetic inference is the existence in many plant genomes of extensive sequence variation, arising from ancient or recent array duplication events, genomic harboring of pseudogenes in various states of decay, and/or incomplete intra- or interarray homogenization [32]. Despite that, we have used ITS region to infer phylogeny in genus Arachis and, to have insights in how much the variation in this region may have interfered in the species relationship establishment, we included more than one individual from six species (A. pintoi: two accessions; A. major: two accessions; A. paraguariensis: two accessions; A. magna: two accessions, A. kuhlmannii: four accessions, A. hoehnei: three accessions). We also included species that are very related to each other based on different types of evidences, such as A. hypogaea and A. monticola [12,16,36] and A. repens and A. pintoi [37,38] and included groups of species whose high affinity was demonstrated by many different methods, such as the A genome species [21].
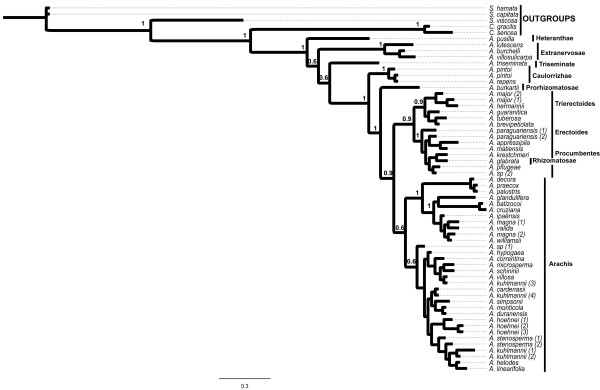
As it can be seen in Figure 1, there was variation between accessions of the same species which may have been caused by the ITS variation cited above. The variation found for three species (A. pintoi, A. major, A. hoehnei) was not enough to place their accessions away from each other. Arachis magna accessions were not placed together but they were in the same clade. Arachis paraguariensis and A. kuhlmannii accessions were, in general, placed away from each other.
Figure 1.
Phylogenetic tree obtained from Bayesian Inference Analysis. Numbers ahead nodes are the posterior probabilities support values. Three branchs are proportional to mutational events sampled in sequences alignment.
The two accessions of A. paraguariensis analyzed belong to different subspecies (A. paraguariensis subsp. paraguariensis and A. paraguariensis subsp. capibarensis). The differentiation between these two subspecies based on morphology is very difficult but they are considered as different subspecific taxa based on their distinct geographic distribution [1] and SAT chromosome morphology [39].
Four accessions of A. kuhlmannii were analyzed (VPoBi 9375 - A. kuhlmannii 1; VSGr 6404 - A. kuhlmannii - 2; VPzRcSgSv 13530 - A. kuhlmannii 3; VSPmSv 13721 - A. kuhlmannii 4). All of them were placed in the A genome clade, but into three different subclades. Arachis kuhlmannii 1 and 2 were placed close to each other in a clade that also included A. helodes and A. linearifolia. Arachis kuhlmannii 3 was placed in a subclade with A. hypogaea, A. correntina, A. microsperma, A. schininii and A. villosa, while A. kuhlmannii 4 in a clade with A. cardenasii, A. simpsonii, A. monticola and A. duranensis. Accessions of A. kulhmannii were also placed in different subclusters based on microsatellite and AFLP data [16,17]. In a dendrogram obtained using RAPD data, A. kuhlmannii 1, 2 and 4 grouped together and A. kuhlmannii 3 was placed in a different group that comprised accessions from Mato Grosso do Sul State in Brazil [40]. Arachis kuhlmannii 4 was collected in Brazil near the frontier with Bolivia, and probably is more distinct from the other A. kuhlmannii accessions and more related to A. cardenasii. In the RAPD dendrogram, A. kuhlmannii 4 grouped to A. simpsonii accession VSPmSv 13728, which was collected in Bolivia, in the same population where the species typus was collected.
Thus, the placement of A. paraguariensis and A. kuhlmannii accessions may have been due to factors not related exclusively to the variation found in ITS regions since our data corroborates previous studies demonstrating high variability within these species.
The closely related allopolyploid species A. hypogaea and A. monticola were placed in different clades. It is known that A. monticola is the closest related species to A. hypogaea [1]. Despite that, A. monticola was considered as a distinct species from A. hypogaea [1]. However, some doubts about its classification still remain, because this species has a high crossability with A. hypogaea [1,41]. It is also an allotetraploid and both species have identical genomes [19,36,39]. Furthermore part of their geographic distribution overlaps [1]. Molecular marker data also confirmed the very close genetic relationship between these two species [6,11,13,15,17,42,43]. In our case, despite the causes of variation in ITS regions, the observed placement of A. hypogaea and A. monticola was certainly influenced by the fact that these species have ITS regions from A and B genomes which our results showed to be different and seem to be specific for each genome type found in genus Arachis. The sequences of A. hypogaea and A. monticola included in this study represent one of the genomes and may even be a mixture of the sequences from both. As mentioned above the close relationships between these two species have been demonstrated using different evidences. Thus, our data is limited to infer phylogenetic relationships between the allopolyploids as well as their relationships to the wild diploid species.
Section Heteranthae showed a basal position, followed by Extranervosae, Triseminatae, Caulorrhizae, and Rhizomatosae Ser. Prorhizomatosae, Procumbentes/Trierectoides/Erectoides/Rhizomatosae and Arachis. Despite the lack of dating analysis, the position of clades in relation to the outgroups agreed with DNA content analysis showing that species with greater DNA content were included in sections believed to have a more recent origin (Procumbentes, Caulorrhizae, Rhizomatosae and Arachis), whereas those with lower DNA content in the most primitive or ancient sections (Extranervosae, Heteranthae and Triseminatae) [44]. Sections Extranervosae, Heteranthae and Triseminatae are believed to be among the oldest sections in genus Arachis based on their affinity with genus Stylosanthes [1].
Section circumscriptions
The results confirmed the circumscriptions of sections Caulorrhizae and Extranervosae and suggested some sections may not be natural groups. The circumscriptions of sections Erectoides, Trierectoides and Procumbentes were not well defined, suggesting that these three sections could be grouped in one or two sections. On the other hand, our data suggested sections Arachis and Rhizomatosae could be divided into two new sections each.
Sections Trierectoides, Erectoides and Procumbentes seem to be a monophyletic group. Species from these sections were distributed in two sub-clades. One of them comprised three species of section Erectoides (A. major, A. hermannii and A. brevipetiolata) and two species of section Trierectoides (A. guaranitica and A. tuberosa). Arachis brevipetiolata was more related to Trierectoides than to the other two species of Erectoides. The other sub-clade comprised one species of section Erectoides (A. paraguariensis), four species of section Procumbentes (A. appressipila, A. kretschmeri, A. matiensis, A. pflugeae, and A. sp. 2) and one species of section Rhizomatosae (A. glabrata). As can be seen in Table 1, species from sections Trierectoides, Erectoides and Procumbentes were all put together in the same section for many years and after three taxonomic revisions of genus Arachis. Only in 1990, some species were put in a new section called Procumbensae that is the actual Procumbentes section. Our data supported the first classification [3].
Section Arachis comprises species of three types of genomes (A, B and D). The crossability and inter-specific hybrid fertility between A genome species and between some B genome species are very high [12,18]. However, crosses between A and B genome species result in unfertile hybrids [1]. The cultivated peanut is fertile just because it had its chromosomes duplicated, having a diploid-like meiosis, with no pairing of chromosomes from different genomes. Thus, based on the use of crossability for the establishment of the taxonomic sections it may be considered that, if the peanut have not evolved, most Arachis specialists would certainly assign the A genome and B genome species to distinct sections.
The presence of rhizomes is the main reason for a species to be in section Rhizomatosae, which comprises three polyploidy species (A. glabrata, A. pseudovillosa, A. nitida) and one diploid (A. burkartii). RAPD [23] and microsatellite data [24] showed that the diploid species is very distinct from the other species of this section. The different ploidy levels and variation in the ITS could result in the misplacement of A. glabrata or A. burkartii. However, we observed in section Arachis, which also includes diploid and polyploidy species that ITS variation may result in some unexpected placement of species, but that is not enough for their placement in clades of different sections.
Sections Triseminatae and Heteranthae had just one species analyzed but they were placed in the tree in individual branches suggesting these sections are also natural groups. Arachis burkartii had a placement very similar to those species from sections Triseminatae and Heteranthae. Thus, based on ours and previous data [23,24] the establishment of a new section for A. burkartii should be considered.
Section Arachis
The two genomes presented in domesticated peanut differ by one striking feature: one of the genomes (A) has a pair of chromosomes, called A, which is conspicuously smaller than the other chromosomes, while the other genome lacks this small chromosome [25]. The species of section Arachis are therefore classified as having A or B genomes based on the presence or absence of the A pair. A third genome was identified in the genus, the D genome, which is only found in A. glandulifera [45].
The species of section Arachis were placed in two clades. The first one was divided into sub-clades. The first sub-clade comprised A. hoehnei (1,2,3), A. stenosperma (1,2), A. kuhlmannii (1,2), A. helodes and A. linearifolia; the second A. cardenasii, A. kuhlmannii (4), A. simpsonii, A. monticola and A. duranensis and the third comprised A. hypogaea, A. correntina, A. microsperma, A. schininii, A. villosa and A. kuhlmannii (3). Arachis sp (1) was very related to the A genome species but it was not included in any of the above clades. The second clade of section Arachis species was divided into two sub-clades. The first included 2n = 2x = 20 species and the other, species with 2n = 2x = 18 (A. decora, A. praecox and A. palustris). The species with 2n = 20 were separated into two sister subclades, one of them being formed only by B genome species (A. magna, A. valida, A. ipaënsis and A. williamsii) and the other included A. batizocoi (B genome), A. cruziana (B genome) and A. glandulifera (D genome). Arachis batizocoi and A. cruziana were very recently described as having a K genome based on FISH mapping of rDNA loci and heterochromatin detection [21].
The first clade of section Arachis included all the A genome species and A. hoehnei. It was believed that A. hoehnei did not present the small "A" chromosome pair [39]. Arachis hoehnei also grouped to A genome species based on the polymorphism of trnT-F region [18] and RAPD markers [27], but grouped to other B genome species based on microsatellite markers [13], and to the aneuploid (2n = 18) species with AFLP markers [18]. In our study, three different accessions of this species were included in the analysis and all of them were placed close to the A genome species, confirming that this species was correctly placed on the phylogenetic tree. The cytogenetical analysis of A. hoehnei was recently re-done and it was verified that this species has the A chromosome pair [21]. Thus, our data suggested A genome species are monophyletic.
The second clade comprised all species of section Arachis that do not have the A chromosome pair. This clade included the 2n = 20 species that are classified as B genome species and the ones that possess 2n = 18 chromosomes (A. decora, A. palustris, and A. praecox). The 2n = 2x = 18 species lack the small pair of chromosomes characteristic of the A genome species [46]. Lavia [47] suggested that A. palustris was derived from a species with × = 10 chromosomes and these species are phylogenetically related to the B genomes. Analyses based on AFLP [18] and microsatellite [48] data placed those 2n = 2x = 18 species closely to A genome species. However, microsatellite markers [13] and sequencing of the trnT-F region [18] and also our data showed those species are more closely related to B genome species. Thus, our study corroborates previous ones [13,18] and suggested these species originated from B genome species.
As mentioned above, the non A genome species from section Arachis with 2n = 20 chromosomes were placed in two clades. Molecular evidences based on markers such as RFLPs [42,49], RAPD [6,50], AFLP [17], and microsatellites [48] suggested that section Arachis diploids lacking the small A chromosome pair comprise a very diverse group, of which A. ipaënsis, A. magna, A. williamsii, and A. valida, closely linked in the present analysis, plus A. gregoryi [12] are those more closely associated to the B genome of A. hypogaea/A. monticola. Hybrids between A. ipaënsis and A. magna have shown 84% viable pollen [1]. Other kinds of evidences, such as molecular [6,28,43] and morphological [51] data, showed that these two species are closely related. Crossings between A. williamsii and A. ipaënsis resulted in hybrids with 66.9% of pollen stainability [52]. If the latter is the donor of the B genome to A. hypogaea [25], our data suggest that A. ipaënsis, A. williamsii, A. magna and A. valida could be used for the improvement of the B genome of cultivated peanut. This would increase the variability available for this purpose since a single accession of A. ipaënsis is available.
Arachis batizocoi was very close related to A. cruziana. Arachis batizocoi is considered a good genetic bridge to transfer genes to cultivated peanut [53,54]. If the phylogenetic relationships were correlated with the crossability, as observed among the A genome species [55,56], A. cruziana would have some crossability with A. batizocoi and could also be used as bridge for gene introgression in A. hypogaea. In fact, F1 hybrids between these two species had 36.4% of pollen viability (male fertility) and 0.3 I (univalents) and 9.9 II (bivalents) [18]. Thus, our data suggested that A. cruziana also can be a source of genes to A. hypogaea since it is very related to A. batizocoi.
Arachis glandulifera is classified as D genome since it does not cross with A. hypogaea and it has the most asymmetrical karyotype in genus Arachis [45]. Arachis glandulifera, like the B genome species, does not show the small pair of chromosomes found in the A genome species. Isoenzyme [10], RFLP [42], RAPD [6], AFLP [18] and cytogenetical [57] data also showed that A. batizocoi and A. glandulifera were closely related. Our data suggest that A. glandulifera may be derived from a B genome ancestor species.
Through the analysis of the heterochromatic bands and 45 S rDNA loci patterns, the species previously classified as B genome were arranged into three groups called B (A. ipaënsis, A. magna, A. gregoryi, A. valida and A. williamsii), K (A. batizocoi, A. cruziana, and A. krapovickasii) and F (A. benensis and A. trinitensis) [21]. Our data supported the classification in B and K genomes. We have not analyzed species of the new F genome.
Sections Trierectoides, Erectoides and Procumbentes
As mentioned before the circumscriptions of these three sections were not clear and because of that their results were presented and discussed together. Our results agreed with the classification proposed by Krapovickas [3] which had all species from those three sections in only one section, called Erectoides. The data partially agreed also to a more recent classification proposed by the same author [5], that divided section Erectoides in two sections (Erectoides and Procumbensae), since as it can be seen in Figure 1, species of Erectoides and Trierectoides were all in a same sub-clade and species of Procumbentes were placed all together with A. paraguariensis and A. glabrata.
Arachis paraguariensis shows low genetic affinity to the other species of section Erectoides and because it shows some morphological peculiarities in the root, flowers and fruits it was suggested that this species should be segregated in an independent section [1]. Our results also raised doubt about the classification of this species, and suggested that A. paraguariensis might be classified as belonging to section Procumbentes. The segregation of A. paraguariensis from other Erectoides species was also observed in phylogeny based on ITS data using parsimony analysis [22].
The placement of A. glabrata in a clade with Procumbentes and Erectoides species will be discussed in the section Rhizomatosae item.
The clade formed by sections Erectoides, Trierectoides and Procumbentes was the most related to section Arachis. Crossability data between members of sections Arachis and Erectoides suggest that these two sections are more phylogenetically related to each other than to the other sections of the genus since pollination has lead to fertilization. Although there has been no development of the resulting proembryos [58], members of the other sections do not even show such a degree of success on crossings with section Arachis [55]. Isozyme and protein pattern data [8,10] suggested that section Erectoides and Procumbentes are closely related to the section Arachis.
Section Caulorrhizae
The morphological traits that have been traditionally used to distinguish the two type specimens of A. pintoi (GK 12787) and A. repens (GKP10538) are not sufficient to differentiate all the accessions collected, as they show intermediate phenotypes between the extreme types [59].
The clade was formed by the two species of section Caulorrhizae, confirming that A. pintoi and A. repens are very closely related. There were no differences between the sequences of A. pintoi (CIAT 22237 = W132) and A. repens (V 5868), and few differences of these two in relation to accession V 6791, also considered as A. pintoi. RAPD data also suggested they are very closely related [37]. F1 hybrids between accession GK 12787 of A. pintoi and accession GKP 10538 of A. repens, which represent the extreme types, had 86.8% pollen fertility [55], which is higher than the level of pollen fertility found in intraespecific hybrids of crosses between accessions of some other Arachis species [45,60].
Section Triseminatae
The phylogenetic data showed that A. triseminata was not closely related to species of any other sections, in agreement with its placement in a separate section (Triseminatae) [1]. No successful crossings among A. triseminata and members of other sections were obtained [1,55], showing its genetic isolation from other sections of genus Arachis.
Section Heteranthae
Arachis pusilla, the only species of section Heteranthae included in this study, had a basal position, on the first radiation of genus Arachis (Figure 1) and closely related to Extranervosae, which formed the second radiation in the genus. The affinity of Heteranthae and Extranervosae sections agreed with floral morphology, geographical distribution and crossability data, since the only known intersectional hybrid of Extranervosae was obtained from a cross with Heteranthae [1].
Section Rhizomatosae
Cytogenetic data suggested that the origins of Arachis tetraploid species (A. glabrata and A. hypogaea) were independent [61]. An analysis using RFLP markers also showed that A. glabrata was very distinct from A. hypogaea [49]. In the present study, A. glabrata and A. hypogaea were placed in different clades, confirming that the polyploid species evolved independently in the genus.
Arachis glabrata was more related to species of section Procumbentes than to A. burkartii which is also traditionally allocated in section Rhizomatosae, although in a series of its own. These species differed on the ploidy level, since A. glabrata is a tetraploid (2n = 4x = 40) and A. burkartii a diploid species. Crossings between A. glabrata and diploid species from other sections resulted in hybrids, in contrast to A. burkartii, for which no hybrids were obtained from numerous attempted crosses [55]. The other tetraploid species analyzed (A. hypogaea) was placed close to diploid species that have similar genomes and are certainly involved in its origin [1,25]. Thus, the data indicated that the referred species of section Rhizomatosae did not have a monophyletic origin.
Arachis glabrata was placed in a clade with species from section Erectoides and Procumbentes. Tetraploid species of section Rhizomatosae were classified as EERR [55], and at that time the EE crossing group was attributed to section Erectoides, which comprised all species that, according to the last classification [1], are distributed in sections Procumbentes, Erectoides and Trierectoides. Thus, based in our and previous data we suggested the following hypothesis to the origin of A. glabrata: 1) A. glabrata originated from species of the Erectoides group, with rhizomes appearing twice, independently, in the evolution of genus Arachis; 2) A. glabrata is an allopolyploid EERR, as previously suggested [55], that resulted from a cross between one species from section Erectoides and one species from section Rhizomatosae, that had a genome not similar to the one found in A. burkartii.
Conclusion
The main clades established in this study in general agreed with many other studies that have used other types of evidences (morphological, crossability, biochemical, cytogenetical and molecular) and different species, being some of them included in our study and some not. Thus, the relationships established do reflects the affinity of the species, and that can be a useful framework for future systematic reviews of genus Arachis and for the selection of species to pre-breeding programs.
Methods
Plant material
A total of 55 accessions, which represent 45 species and the nine sections of the genus Arachis were analyzed (Table 2). These accessions were obtained from the Brazilian Arachis Germplasm Collection, maintained at Embrapa Recursos Genéticos e Biotecnologia - CENARGEN (Brasília-DF, Brazil). All plants were grown from seed or cuttings (sections Rhizomatosae and Caulorrhizae) under greenhouse conditions prior to DNA extraction. Stylosanthes capitata, S. hamata, S. viscosa, Chapmannia gracilis and C. sericea were used as outgroups, because these genera are considered to be closely related to Arachis [1,30,31,62].
Table 2.
Accessions of genus Arachis analyzed in this study.
| Species | Accession | Genome 1 | Chromosome number/ Ploidy level 2 |
GenBank Accession Numbers |
|---|---|---|---|---|
| Sect. Arachis | ||||
| A. cardenasii Krapov. & W.C.Greg. | GKP 10017 | A | 2n = 2x = 20 | AY615236 |
| A. correntina (Burkart) Krapov. & W.C.Greg. | CIAT 22249 | A | 2n = 2x = 20 | AF203554 |
| A. duranensis Krapov. & W.C.Greg. | VNvEc 14167 | A | 2n = 2x = 20 | AY615240 |
| A. helodes Mart. ex Krapov. & Rigoni | VSGr 6325 | A | 2n = 2x = 20 | AY615241 |
| A. kuhlmannii (1) Krapov. & W.C.Greg. | VPoBi 9375 | A | 2n = 2x = 20 | AY615232 |
| A. kuhlmannii (2) | VSGr 6404 | A | 2n = 2x = 20 | AY615219 |
| A. kuhlmannii (3) | VPzRcSgSv 13530 | A | 2n = 2x = 20 | AY615238 |
| A. kuhlmannii (4) | VSPmSv 13721 | A | 2n = 2x = 20 | AY615243 |
| A. sp (1) | VSPmSv 13736 | A | 2n = 2x = 20 | AY615226 |
| A. linearifolia Valls, Krapov. & C.E.Simpson | VPoBi 9401 | A | 2n = 2x = 20 | AY615242 |
| A. microsperma Krapov., W.C.Greg. & Valls | VRGeSv 7681 | A | 2n = 2x = 20 | AY615221 |
| A. schininii Krapov., Valls & C.E.Simpson | VSW 9923 | A | 2n = 2x = 20 | AY615248 |
| A. simpsonii Krapov. & W.C.Greg. | VSPmSv 13728 | A | 2n = 2x = 20 | AY615247 |
| A. stenosperma (1) Krapov. & W.C.Greg. | Lm 1 | A | 2n = 2x = 20 | AY615252 |
| A. stenosperma (2) | VSPmW 13844 | A | 2n = 2x = 20 | AY615227 |
| A. villosa Benth. | VGoMrOv 12812 | A | 2n = 2x = 20 | AY615215 |
| A. hypogaea L. | Mf 1560 | AB | 2n = 4x = 40 | AY615267 |
| A. monticola Krapov. & Rigoni | VOa 14165 | AB | 2n = 4x = 40 | AY615239 |
| A. batizocoi Krapov. & W.C.Greg. | K 9484 | B | 2n = 2x = 20 | AY615256 |
| A. cruziana Krapov., W.C.Greg. & C.E.Simpson | WiSVg 1302 | B | 2n = 2x = 20 | AY615259 |
| A. hoehnei (1) Krapov. & W.C.Greg. | KG 30006 | A | 2n = 2x = 20 | AY615223 |
| A. hoehnei (2) Krapov. & W.C.Greg. | VPoBi 9146 | A | 2n = 2x = 20 | AY615224 |
| A. hoehnei (3) Krapov. & W.C.Greg. | VPoBi 9140 | A | 2n = 2x = 20 | AY615222 |
| A. ipaënsis Krapov. & W.C.Greg. | KGBPSSc 30076 | B | 2n = 2x = 20 | AY615257 |
| A. magna (1) Krapov., W.C.Greg. & C.E.Simpson | KGSSc 30097 | B | 2n = 2x = 20 | AY615230 |
| A. magna (2) | VSPmSv 13760 | B | 2n = 2x = 20 | AY615231 |
| A. williamsii Krapov. & W.C.Greg. | WiDc 1118 | B | 2n = 2x = 20 | AY615255 |
| A. valida Krapov., & W.C.Greg. | VPoBi 9153 | B | 2n = 2x = 20 | AY615244 |
| A. glandulifera Stalker | VSPmSv 13738 | D | 2n = 2x = 20 | AY615258 |
| A. decora Krapov., W.C.Greg. & Valls | VSPmPzRs 13290 | Unknown | 2n = 2x = 18 | AY615237 |
| A. palustris Krapov., W.C.Greg. & Valls | VPmSv 13023 | Unknown | 2n = 2x = 18 | AY615238 |
| A. praecox Krapov., W.C.Greg. & Valls | VSGr 6416 | Unknown | 2n = 2x = 18 | AY615234 |
| Sect. Erectoides Krapov. & W.C.Greg. | ||||
| A. brevipetiolata Krapov. & W.C.Greg. | VMPzW 13959 | E | 2n = 2x = 20 | AY615251 |
| A. hermannii Krapov. & W.C.Greg. | VPoJSv 10390 | E | 2n = 2x = 20 | AY615260 |
| A. major (1) Krapov. & W.C.Greg. | VRGeSv 7644 | E | 2n = 2x = 20 | AY615229 |
| A. major (2) | VRGeSv 7632 | E | 2n = 2x = 20 | AY615228 |
| A. paraguariensis Chodat & Hassl. (1) subsp. capibarensis Krapov. & W.C.Greg. | VMPzW 14024 | E | 2n = 2x = 20 | AY615217 |
| A. paraguariensis (2) subsp. paraguariensis Chodat & Hassl. | VRGeSv 7677 | E | 2n = 2x = 20 | AY615218 |
| Sect. Trierectoides Krapov. & W.C.Greg. | ||||
| A. guaranitica Chodat & Hassl. | VRcSgSv 13600 | E | 2n = 2x = 20 | AY615261 |
| A. tuberosa Bong. ex Benth | VRGeSv 7607 | E | 2n = 2x = 20 | AY615235 |
| Sect. Procumbentes Krapov. & W.C.Greg. | ||||
| A. appressipila Krapov. & W.C.Greg. | GKP 10002 | E | 2n = 2x = 20 | AY615254 |
| A. kretschmeri Krapov. & W.C.Greg. | KrRy s/n (IRFL 2273) | E | 2n = 2x = 20 | AY615220 |
| A. matiensis Krapov., W.C.Greg. & C.E.Simpson | VSPmSv 13718 | E | 2n = 2x = 20 | AY615249 |
| A. pflugeae C.E.Simpson, Krapov. & Valls | VRcSgSv 13589 | E | 2n = 2x = 20 | AY615233 |
| A. sp (2) | VMPzW 14044 | E | 2n = 2x = 20 | AY615225 |
| Sect. Rhizomatosae Krapov. & W.C.Greg. | ||||
| Ser. Prorhizomatosae Krapov. & W.C.Greg. | ||||
| A. burkartii Handro | VZnMrOvW 12322 | R | 2n = 2x = 20 | AY615245 |
| Ser. Rhizomatosae | ||||
| A. glabrata var. glabrata Benth. | Cv. Florigraze | R | 2n = 4x = 40 | AY615250 |
| Sect. Caulorrhizae Krapov. & W.C.Greg. | ||||
| A. pintoi Krapov. & W.C.Greg. (1) | VSWSa 6791 | C | 2n = 2x = 20 | AY615263 |
| A. pintoi (2) | CIAT 22237 = W132 | C | 2n = 2x = 20 | AF203551 |
| A. repens Handro | V 5868 | C | 2n = 2x = 20 | AY615264 |
| Sect. Triseminatae Krapov. & W.C.Greg. | ||||
| A. triseminata Krapov. & W.C.Greg. | W 195 | T | 2n = 2x = 20 | AY615253 |
| Sect. Heteranthae Krapov. & W.C.Greg. | ||||
| A. pusilla Benth. | VRSv 10833 | AM | 2n = 2x = 20 | AY615216 |
| Sect. Extranervosae Krapov. & W.C.Greg. | ||||
| A. burchellii Krapov. & W.C.Greg. | VGaRoSv 12618 | EX | 2n = 2x = 20 | AY615262 |
| A. lutescens Krapov. & Rigoni | VSStGdW 7741 | EX | 2n = 2x = 20 | AY615246 |
| A. villosulicarpa Hoehne | VKSSv 8816 | EX | 2n = 2x = 20 | AY615265 |
| Outgroups | ||||
| Chapmannia gracilis Balf.f Thullin | Miller & Alexander14039(E) | Not described in the source | Not described in the source | AF203545 |
| Chapmannia sericea: Thulin & Mc Kean | Miller & Alexander14241(E) | Not described in the source | Not described in the source | AF203548 |
| Stylosanthes capitata Vogel | CIAT1693 | Not described in the source | 2n = 4x (40) | AF203549 |
| Stylosanthes hamata | Beyra M 595 (MONT) | Not described in the source | 2n = 2x(20) | AF203550 |
| Stylosanthes viscosa Sw. | Clemente J C | Not described | 2n = 2x(20) | AY6152141 |
Collectors: B = D.J.Banks; Bi = L.B.Bianchetti; Dc = D. Claure; Ec = E.D.Cruz; G = W.C.Gregory; Ga = M.L.Galgaro; Gd = I.J.Godoy; Ge = M.A.N.Gerin; Go = K.E.Gomes; Gr = A.Gripp; J = L.Jank; K = A.Krapovickas; Kr = A.Kretschmer Jr.; Lm = L.Monçato; M = J.P.Moss; Mf = Est.Exp.Agr. Manfredi, Córdoba, Argentina; Mr = C.O.C.Moraes; Nv = L.Novara; Oa = O.Ahumada; Ov = J.C.Oliveira; P = J.Pietrarelli; Pm = R.N.Pittman; Po = A.Pott; Pz = E.A.Pizarro; R = V.R.Rao; Rc = R.C.Oliveira; Ro = D.M.S.Rocha; Rs = R.C.Santos; Ry = P.R.Rayman; S = C.E.Simpson; Sa = J.M.Santos; Sc = A. Schinini; Sg = A.K.Singh; St = H.T.Stalker; Sv = G.P.Silva; V = J.F.M.Valls; Vg = I.Vargas; W = W.L.Werneck; Wi = D.E.Williams; Zn = A.Zanin
1 - Genome designations follow abreviations proposed by Smartt & Stalker [61] for taxonomic sections and group of species in section Arachis [53].
2 - Chromosome numbers are the observed for each species and compiled by Fernández & Krapovickas [39], Lavia & Fernández [44], Lavia et al [46,70]. Eventually, counts were obtained by the accessions listed.
DNA extraction and PCR amplification
DNA was extracted from young leaflets of single plants, using a procedure previously described [63]. Primers ITS5 (5'GGAAGTAAAAGTCGTAACAAGG3') and ITS4 (5'TCCTCCGCTTATTGATATGC3') were used to amplify the two internal transcribed regions, ITS1 and ITS2, and the 5.8 S gene [64]. Each amplification reaction contained 12 μl of water, 1.5 μl of magnesium chloride (50 mM), 2.6 μl of 10× Taq polymerase reaction buffer, 1.5 μl of each primer (10 mM), 5.0 μl of a 5 ng/μl DNA dilution, 2.2 μl of dNTPs (2.5 μM each) and 0.2 μl of Taq DNA polymerase (5 U/μl). The reactions were performed on a PTC 100 (MJ Research) using the following program: an initial denaturing step (2 min at 94°C) followed by 35 cycles of the following steps: denaturing (1 min at 95°C), annealing (1 min at 55°C) and extension (1.30 min at 72°C); and a final extension step of 10 min at 72°C. The PCR products were purified using the kit Concert™Rapid PCR Purification System (Life Technologies) before sequencing.
DNA sequencing
PCR products were sequenced using the procedure proposed by Sanger et al. [65]. Each sequence reaction contained: 2 μl of Big Dye™Terminator (Applied Biosystems), 1.5 μl of PCR product (5 ng/μl), 0.5 μl of primer solution (0.25 mM) and water up to 10 μl. The primers used on the sequencing reactions were the same used on the amplification of the target fragments. The sequencing reactions were performed on a PTC 100 (M.J. Research) using the following program: 1 min at 96°C, 40 cycles of 10 sec at 96°C; 10 sec at 55°C; 4 min at 60°C. The sequencing was performed in an ABI PRISM 377 Automated DNA Sequencer (Perkin-Elmer/Applied Biosystems). Each DNA strand was sequenced at least twice to ensure the accuracy of the results.
Sequence and phylogenetic analysis
The phylogenetic analysis was the Bayesian Inference with the MCMC calculations implemented by Mr. Bayes 3.1 [66]. The best evolution model (GTR+G) was selected using mrmodeltest [67] and PAUP 4b10 [68]. The model parameters were set in the alignment nexus file and then ran in Mr. Bayes 3.1, which performed 20,000,000 of generations, sampling trees in each 100 generations. The first 1250 trees were eliminated as the burn-in. The 50% majority consesus trees was inspected and prepared in figtree software.
Authors' contributions
All authors read and approved the final manuscript. MDB carried out the data collection and analysis and drafted the manuscript. MCM participated in the drafting of the manuscript. DAP and JPM participated in the sequencing and sequence analysis. MBJ and JMJ participated in the phylogenetic analysis. JFMV participated in the conception of the project and provided the germplasm. CRL and MAG participated in conceiving the study and analysis, and participated in drafting the manuscript.
Contributor Information
Marcelo D Bechara, Email: marcelodibi@itelefonica.com.br.
Márcio C Moretzsohn, Email: marciocm@cenargen.embrapa.br.
Darío A Palmieri, Email: darioap@assis.unesp.br.
Jomar P Monteiro, Email: jomarmont@hotmail.com.
Maurício Bacci, Jr, Email: mbacci@rc.unesp.br.
Joaquim Martins, Jr, Email: martins.jr.bio@gmail.com.
José FM Valls, Email: valls@cenargen.embrapa.br.
Catalina R Lopes, Email: dtcatalina@terra.com.br.
Marcos A Gimenes, Email: gimenes@cenargen.embrapa.br.
Acknowledgements
This work was funded by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP). JFMV, CRL, MAG thank CNPq for their fellowships.
References
- Krapovickas A, Gregory WC. Taxonomía del género Arachis (Leguminosae) Bonplandia. 1994;8:1–186. [Google Scholar]
- Valls JFM, Simpson CE. New species of Arachis L. (Leguminosae) from Brazil, Paraguay and Bolivia. Bonplandia. 2005;14:35–64. [Google Scholar]
- Krapovickas A. In: The domestication and exploitation of plants and animals. Ucko J, Dimbleby C, editor. Duckworth, London; 1969. The origin, variability and spread of the peanut (Arachis hypogaea) pp. 427–440. [Google Scholar]
- Gregory WC, Krapovickas A, Gregory MP. In: Advances in Legume Science. Summerfiled RJ, Bunting AH, editor. Kew: Royal Botanic Gardens; 1980. Structure, variation, evolution and classification in Arachis; pp. 469–481. [Google Scholar]
- Krapovickas A. A taxonomic summary of the genus Arachis. International Board for Plant Genetic Resources (IBPRG) - International Crop Network Series, 2. Report of a workshop on the genetic resources of wild species: Including preliminary descriptors for Arachis (IBPGR/ICRISAT) Rome, Italy. Appendix III. 1990. p. 9.
- Halward TM, Stalker HT, LaRue EA, Kochert G. Use of single primer DNA amplification in genetic studies of peanut. Plant Mol Biol. 1992;18:315–325. doi: 10.1007/BF00034958. [DOI] [PubMed] [Google Scholar]
- Lanham PG, Forster BP, McNicol P, Moss JP, Powell W. Seed storage protein variation in Arachis species. Genome. 1994;37:487–496. doi: 10.1139/g94-068. [DOI] [PubMed] [Google Scholar]
- Singh AK, Gurtu S, Jambunathan R. Phylogenetic relationships in the genus Arachis based on seed protein profiles. Euphytica. 1994;74:219–225. doi: 10.1007/BF00040404. [DOI] [Google Scholar]
- Lu J, Pickersgill B. Isozyme variation and species relationships in peanut and its wild relatives (Arachis L. - Leguminosae) Theor Appl Genet. 1993;85:550–560. doi: 10.1007/BF00220913. [DOI] [PubMed] [Google Scholar]
- Stalker HT, Phillips TD, Murphy JP, Jones TM. Variation of isozyme patterns among Arachis species. Theor Appl Genet. 1994;87:746–755. doi: 10.1007/BF00222901. [DOI] [PubMed] [Google Scholar]
- Jung S, Tate PL, Horn R, Kochert G, Moore K, Abbott AG. The Phylogenetic relationship of possible progenitors of the cultivated peanut. J Hered. 2003;94:334–340. doi: 10.1093/jhered/esg061. [DOI] [PubMed] [Google Scholar]
- Burow MD, Simpson CE, Faries W, Starr JL, Paterson AH. Molecular biogeography study of recently described B- and A-genome Arachis species, also providing new insights into the origins of cultivated peanut. Genome. 2009;52:107–119. doi: 10.1139/G08-094. [DOI] [PubMed] [Google Scholar]
- Moretzsohn MC, Hopkins MS, Mitchell SE, Kresovich S, Valls JFM, Ferreira ME. Genetic diversity of peanut (Arachis hypogaea L.) and its wild relatives based on the analysis of hypervariable regions of the genome. BMC Plant Biol. 2004;4:11. doi: 10.1186/1471-2229-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkley NA, Dean RE, Pittman RN, Wang ML, Holbrook CC, Pederson GA. Genetic diversity of cultivated and wild-type peanuts evaluated with M13-tailed SSR markers and sequencing. Genet Res. 2007;89:93–106. doi: 10.1017/S0016672307008695. [DOI] [PubMed] [Google Scholar]
- Gimenes MA, Hoshino AA, Barbosa AVG, Palmieri DA, Lopes CR. Characterization and transferability of microsatellite markers of the cultivated peanut (Arachis hypogaea) BMC Plant Biol. 2007;7:9. doi: 10.1186/1471-2229-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppolu R, Upadhyaya HD, Dwivedi SL, Hoisington DA, Varshney RK. Genetic relationships among seven sections of genus Arachis studied by using SSR markers. BMC Plant Biol. 2010;10:15. doi: 10.1186/1471-2229-10-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milla SR, Isleib TG, Stalker HT. Taxonomic relationships among Arachis sect. Arachis species as revealed by AFLP markers. Genome. 2005;48:1–11. doi: 10.1139/g04-089. [DOI] [PubMed] [Google Scholar]
- Tallury SP, Hilu KW, Milla SR, Friend SA, Alsaghir M, Stalker HT, Quandt D. Genomic affinities in Arachis section Arachis (Fabaceae): molecular and cytogenetic evidence. Theor Appl Genet. 2005;111:1229–1237. doi: 10.1007/s00122-005-0017-0. [DOI] [PubMed] [Google Scholar]
- Seijo GJ, Lavia GI, Fernandez A, Krapovickas A, Ducasse D, Moscone EA. Physical mapping of the 5 S and 18S-25 S rRNA genes by FISH as evidence that Arachis duranensis and A. ipaënsis are the wild diploid progenitors of A. hypogaea (Leguminosae) Am J Bot. 2004;91:1294–1303. doi: 10.3732/ajb.91.9.1294. [DOI] [PubMed] [Google Scholar]
- Robledo G, Lavia GI, Seijo G. Species relations among wild Arachis species with the A genome as revealed by FISH mapping of rDNA loci and heterochromatin detection. Theor Appl Genet. 2009;118:1295–1307. doi: 10.1007/s00122-009-0981-x. [DOI] [PubMed] [Google Scholar]
- Robledo G, Seijo G. Species relationships among the wild B genome of Arachis species (section Arachis) based on FISH mapping of rDNA loci and heterochromatin detection: a new proposal for genome arrangement. Theor Appl Genet. 2010. [DOI] [PubMed]
- Wang CT, Wang XZ, Tang YY, Chen DX, Cui FG, Zhang JC, Yu SL. Phylogeny of Arachis based on internal transcribed spacer sequences. Genet Resour Crop Evol. 2010.
- Nóbile PM, Gimenes MA, Valls JFM, Lopes CR. Genetic variation within and among species of genus Arachis, section Rhizomatosae. Genet Resour Crop Evol. 2004;51:299–307. doi: 10.1023/B:GRES.0000024015.98151.66. [DOI] [Google Scholar]
- Angelici CMLCD, Hoshino AA, Nóbile PM, Palmieri DA, Valls JFM, Gimenes MA, Lopes CR. Genetic diversity in section Rhizomatosae of the genus Arachis (Fabaceae) based on microsatellite markers. Genet Mol Biol. 2008;31:79–88. doi: 10.1590/S1415-47572008000100016. [DOI] [Google Scholar]
- Kochert G, Stalker HT, Gimenes MA, Galgaro L, Lopes CR, Moore K. RFLP and cytogenetic evidence on the origin and evolution of allotetraploid domesticated peanut Arachis hypogaea (Leguminosae) Am J Bot. 1996;83:1282–1291. doi: 10.2307/2446112. [DOI] [Google Scholar]
- Fávero AP, Simpson CE, Valls FMJ, Velho NA. Study of evolution of cultivated peanut trough crossability studies among Arachis ipaënsis, A duranensis and A hypogaea. Crop Sci. 2006;46:1546–1552. doi: 10.2135/cropsci2005.09-0331. [DOI] [Google Scholar]
- Cunha FB, Nóbile PM, Hoshino AA, Moretzsohn MC, Lopes CR, Gimenes MA. Genetic relationships among Arachis hypogaea L. (AABB) and diploid species with AA and BB genomes. Genet Resour Crop Evol. 2008;55:15–20. doi: 10.1007/s10722-007-9209-6. [DOI] [Google Scholar]
- Moretzsohn MC, Barbosa AVG, Alves-Freitas DMT, Teixeira C, Leal-Bertioli SCM, Guimarães PM, Pereira RW, Lopes CR, Cavallari MM, Valls JFM, Bertioli DJ, Gimenes MA. A linkage map for the B-genome of Arachis (Fabaceae) and its synteny to the A-genome. BMC Plant Biol. 2009;9:40. doi: 10.1186/1471-2229-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lashermes P, Combes MC, Trouslot P, Charrier A. Phylogenetic relationships of coffee-tree species (Coffea L.) as inferred from ITS sequences of nuclear ribossomal DNA. Theor Appl Genet. 1997;94:947–955. doi: 10.1007/s001220050500. [DOI] [Google Scholar]
- Lavin M, Thulin M, Labat JN, Pennington RT. Africa, the old man out: molecular biogeography of dalbergioid legumes (Fabaceae) suggests otherwise. Syst Bot. 2000;25:449–467. doi: 10.2307/2666689. [DOI] [Google Scholar]
- Lavin M, Pennington RT, Klitgaard BB, Sprent JI, Lima HC, Gasson PE. The dalbergioid legumes (Fabaceae): delimitation of a Pantropical monophyletic clade. Am J Bot. 2001;50:550–560. [PubMed] [Google Scholar]
- Álvarez I, Wendel JF. Ribosomal ITS sequences and plant phylogenetic inference. Mol Phylogenet Evol. 2003;29:417–434. doi: 10.1016/S1055-7903(03)00208-2. [DOI] [PubMed] [Google Scholar]
- Nieto Feliner G, Rosselló JA. Better the devil you know? Guidelines for insightful utilization of nrDNA ITS in species-level evolutionary studies in plants. Mol Phylogenet Evol. 2007;44:911–919. doi: 10.1016/j.ympev.2007.01.013. [DOI] [PubMed] [Google Scholar]
- Zhu X-Y, Cai D-T, Ding Y. Molecular and cytological characterization of 5 S rDNA in Oryza species: genomic organization and phylogenetic implications. Genome. 2008;51:332–340. doi: 10.1139/G08-016. [DOI] [PubMed] [Google Scholar]
- Bayer RJ, Soltis DE, Soltis PS. Phylogenetic inferences in Antennaria (Asteraceae: Gnaphlilae: Cassiinae) based on sequences from nuclear ribossomal DNA internal transcribed spacers (ITS) Am J Bot. 1996;83:516–527. doi: 10.2307/2446220. [DOI] [Google Scholar]
- Seijo G, Lavia GI, Fernandez A, Krapovickas A, Ducasse DA, Bertioli DJ, Moscone EA. Genomic relationships between the cultivated peanut (Arachis hypogaea, Leguminosae) and its close relatives revealed by double GISH. Amer J Bot. 2007;94:1963–1971. doi: 10.3732/ajb.94.12.1963. [DOI] [PubMed] [Google Scholar]
- Gimenes MA, Lopes CR, Galgaro ML, Valls JFM, Kochert G. Genetic variation and phylogenetic relationships based on RAPD analysis in section Caulorrhizae, genus Arachis (Leguminosae) Euphytica. 2000;116:187–195. doi: 10.1023/A:1004025619704. [DOI] [Google Scholar]
- Palmieri DA, Bechara MD, Curi RA, Monteiro JP, Valente SES, Gimenes MA, Lopes CR. Genetic diversity analysis in the section Caulorrhizae (genus Arachis) using microsatellite markers. Gen Mol Biol. 2010;33:109–118. doi: 10.1590/S1415-47572010005000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández A, Krapovickas A. Cromosomas y evolución en Arachis (Leguminosae) Bonplandia. 1994;8:187–200. [Google Scholar]
- Fávero AP. Caracterização morfológica, citogenética e molecular de acessos de germoplasma da espécie Arachis Kuhlmannii Krapov. & W.C. Gregory (Secção Arachis) (Mestrado). UNESP: Botucatu; 1999. p. 159. [Google Scholar]
- Raman VS. Studies in the genus Arachis. IV. Hybrid between A. hypogaea and A. monticola. Indian Oilseeds J. 1958;1:20–23. [Google Scholar]
- Kochert G, Halward T, Branch WD, Simpson CE. RFLP variability in peanut (Arachis hypogaea L.) cultivars and wild species. Theor Appl Genet. 1991;81:565–570. doi: 10.1007/BF00226719. [DOI] [PubMed] [Google Scholar]
- Raina SN, Mukai Y. Genomic in situ hybridization in Arachis (Fabaceae) identifies the diploid wild progenitors of cultivated (A. hypogaea) and related wild (A. monticola) peanut species. Plant Syst Evol. 1999;214:251–262. doi: 10.1007/BF00985743. [DOI] [Google Scholar]
- Lavia GI, Fernández A. Genome Size in wild and cultivated peanut germplasm. Pl Syst Evol. 2008;272:1–10. doi: 10.1007/s00606-007-0632-0. [DOI] [Google Scholar]
- Stalker HT. A new species in section Arachis of peanuts with a D genome. Am J Bot. 1991;78:630–637. doi: 10.2307/2445084. [DOI] [Google Scholar]
- Lavia GI, Ortíz AM, Fernández A. Karyotypic studies in wild germplasm of Arachis (Leguminosae) Genet Resour Crop Evol. 2009;56:755–764. doi: 10.1007/s10722-008-9399-6. [DOI] [Google Scholar]
- Lavia GI. Estúdios cromosómicos en Arachis (Leguminosae) Bonplandia. 1996;9:111–120. [Google Scholar]
- Bravo JP, Hoshino AA, Angelici CMLCD, Lopes CR, Gimenes MA. Transferability and use of microsatellite markers for the genetic analysis of the germplasm of some Arachis section species of the genus Arachis. Genet Mol Biol. 2006;29:516–524. doi: 10.1590/S1415-47572006000300021. [DOI] [Google Scholar]
- Halward TM, Stalker HT, LaRue EA, Kochert G. Genetic variation detectable with molecular markers among unadapted germplasm resources of cultivated peanut and wild species. Genome. 1991;34:1013–1020. [Google Scholar]
- Hilu KW, Stalker HT. Genetic relationships between peanut and wild species of Arachis sect. Arachis (Fabaceae): evidence from RAPDs. Plant Syst Evol. 1995;198:167–178. doi: 10.1007/BF00984735. [DOI] [Google Scholar]
- Stalker HT. A morphological apprasial of wild species in section Arachis of peanuts. Peanut Sci. 1990;17:117–122. doi: 10.3146/i0095-3679-17-2-17. [DOI] [Google Scholar]
- Simpson CE, Faries MJ. Simpósio de Recursos Genéticos para a América Latina e Caribe: 19-22 November 2001. Londrina, Brazil. Instituto Agronômico do Paraná; 2001. Advances in the characterization of diversity in section Arachis: archeological evidence, crossing results and their relationship in the understanding the origins of Arachis hypogaea L; pp. 103–104. [Google Scholar]
- Singh AK, Moss JP. Utilization of wild relatives in the genetic improvement of Arachis hypogaea L 8. Synthetic amphidiploids and their importance in interspecific breeding. Theor Appl Genet. 1986;72:433–439. doi: 10.1007/BF00289523. [DOI] [PubMed] [Google Scholar]
- Burow MD, Simpson CE, Starr JL, Paterson AH. Transmission genetics of chromatin from a synthetic amphidiploid to cultivated peanut (Arachis hypogaea L.): Broadening the gene pool of a monophyletic polyploid species. Genetics. 2001;159:823–837. doi: 10.1093/genetics/159.2.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory MP, Gregory WC. Exotic germ plasm of Arachis L. interspecific hybrids. J Hered. 1979;70:185–193. [Google Scholar]
- Singh AK, Stalker HT, Moss JP. In: Chromosome Engineering in Plants: Genetics, Breeding, Evolution. Part B. Tsuchiya T, Gupta PK, editor. Amsterdam: Elsevier Science Publishers; 1990. Cytogenetics and uses of alien genetic variation in groundnut improvement; pp. 65–76. [Google Scholar]
- Robledo G, Seijo G. Characterization of the Arachis (Leguminosae) D genome using fluorescence in situ hybridization (FISH) chromosome markers and total genome DNA hybridization. Genet Mol Biol. 2008;31:717–724. doi: 10.1590/S1415-47572008000400019. [DOI] [Google Scholar]
- Singh AK. Hybridization barriers among the species of Arachis L., namely of the sections Arachis (including the groundnut) and Erectoides. Genet Resour Crop Evol. 1998;45:41–45. doi: 10.1023/A:1008681020851. [DOI] [Google Scholar]
- Valls JFM. In: Identifying Germplasm for Successful Forage Legume-Grass Interactions. Proceedings of a Symposium of the Crop Science Society of America. Springer TL, Pittman RN, editor. USDA, Washington; 1996. Variability in the genus Arachis and potential forage uses; pp. 15–27. [Google Scholar]
- Stalker HT, Dhesi JS, Kochert G. Variation within the species Arachis duranensis Krap & W.C. Gregory, a possible progenitor of cultivated peanut. Genome. 1995;38:1201–1212. doi: 10.1139/g95-158. [DOI] [PubMed] [Google Scholar]
- Smartt J, Stalker HT. In: Peanut science and technology. Pattee HE, Young CT, editor. Yoakun: American Peanut Research Education Society; 1982. Speciation and cytogenetics in Arachis; pp. 21–49. [Google Scholar]
- Vander Stappen J, De Laet J, Gama-López S, Van Campenhout S, Volckaert G. Phylogenetic analysis of Stylosanthes (Fabaceae) based on the internal transcribed spacer region (ITS) of nuclear ribosomal DNA. Plant Syst Evol. 2002;234:27–51. doi: 10.1007/s00606-002-0193-1. [DOI] [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh life tissue. Phytochem Bull. 1987;19:11–15. [Google Scholar]
- White TJ, Bruns TD, Lee SB, Taylor JW. In: PCR Protocols: A Guide to Methods and Applications. Innis M, Gelfand D, Sninsky J, White T, editor. San Diego: Academic Press; 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics; pp. 315–322. [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5468. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogeny. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Nylander JAA. Program distributed by the author. Evolutionary Biology Centre, Uppsala University; 2004. MrModeltest v2. [Google Scholar]
- Swofford DL. PAUP* Phylogenetic analysis using parsimony (*and other methods), v 4b10. Sinauer Sunderland; 2002. [Google Scholar]
- Valls JFM, Simpson CE. In: Biology and Agronomy of Forage Arachis. Kerridge PC, Hardy B, editor. Cali: CIAT; 1994. Taxonomy, natural distribution, and attributes of Arachis; pp. 1–18. [Google Scholar]
- Lavia G, Fernández A, Seijo J. In: Plant Genome: Biodiversity and Evolution, Phanerogams-Angiosperm. Sharma A, editor. 1E. Enfield, NH: Science Publishers; 2008. Cytogenetic and molecular evidences on the evolutionary relationships among Arachis species; pp. 101–134. [Google Scholar]