Abstract
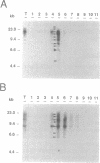
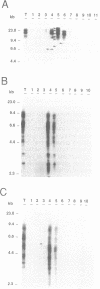
We have studied the genomic distribution of five different families of plant transposable elements by analyzing their location in DNA fractions from maize and tobacco genomes fractionated according to base composition. The results show that each family of elements is preferentially integrated in one specific fraction of its respective host genome. This demonstrates that the distribution of transposable elements in the nuclear genome of plants is not random but compartmentalized, i.e., the elements are located in specific genomic compartments characterized by having a specific G+C content and representing a small proportion of the genomes. Furthermore, these compartments seem to correspond to the genomic regions where most of the plant genes are also located, suggesting a preferential integration of transposable elements in the transcriptionally active regions of the plant genome. The implications of these results on the current applications of transposon tagging techniques are discussed.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bennetzen J. L. Transposable element Mu1 is found in multiple copies only in Robertson's Mutator maize lines. J Mol Appl Genet. 1984;2(6):519–524. [PubMed] [Google Scholar]
- Bernardi G., Olofsson B., Filipski J., Zerial M., Salinas J., Cuny G., Meunier-Rotival M., Rodier F. The mosaic genome of warm-blooded vertebrates. Science. 1985 May 24;228(4702):953–958. doi: 10.1126/science.4001930. [DOI] [PubMed] [Google Scholar]
- Bernardi G. The isochore organization of the human genome. Annu Rev Genet. 1989;23:637–661. doi: 10.1146/annurev.ge.23.120189.003225. [DOI] [PubMed] [Google Scholar]
- Bureau T. E., Wessler S. R. Tourist: a large family of small inverted repeat elements frequently associated with maize genes. Plant Cell. 1992 Oct;4(10):1283–1294. doi: 10.1105/tpc.4.10.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis E. S., Gerlach W. L., Pryor A. J., Bennetzen J. L., Inglis A., Llewellyn D., Sachs M. M., Ferl R. J., Peacock W. J. Molecular analysis of the alcohol dehydrogenase (Adh1) gene of maize. Nucleic Acids Res. 1984 May 11;12(9):3983–4000. doi: 10.1093/nar/12.9.3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner H. K., Belachew A. Transposition Pattern of the Maize Element Ac from the Bz-M2(ac) Allele. Genetics. 1989 Jun;122(2):447–457. doi: 10.1093/genetics/122.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döring H. P., Starlinger P. Molecular genetics of transposable elements in plants. Annu Rev Genet. 1986;20:175–200. doi: 10.1146/annurev.ge.20.120186.001135. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Finnegan D. J. Eukaryotic transposable elements and genome evolution. Trends Genet. 1989 Apr;5(4):103–107. doi: 10.1016/0168-9525(89)90039-5. [DOI] [PubMed] [Google Scholar]
- Flavell A. J., Smith D. B., Kumar A. Extreme heterogeneity of Ty1-copia group retrotransposons in plants. Mol Gen Genet. 1992 Jan;231(2):233–242. doi: 10.1007/BF00279796. [DOI] [PubMed] [Google Scholar]
- Gierl A., Saedler H. Plant-transposable elements and gene tagging. Plant Mol Biol. 1992 May;19(1):39–49. doi: 10.1007/BF00015605. [DOI] [PubMed] [Google Scholar]
- Grandbastien M. A. Retroelements in higher plants. Trends Genet. 1992 Mar;8(3):103–108. doi: 10.1016/0168-9525(92)90198-d. [DOI] [PubMed] [Google Scholar]
- Grandbastien M. A., Spielmann A., Caboche M. Tnt1, a mobile retroviral-like transposable element of tobacco isolated by plant cell genetics. Nature. 1989 Jan 26;337(6205):376–380. doi: 10.1038/337376a0. [DOI] [PubMed] [Google Scholar]
- Greenblatt I. M. A chromosome replication pattern deduced from pericarp phenotypes resulting from movements of the transposable element, modulator, in maize. Genetics. 1984 Oct;108(2):471–485. doi: 10.1093/genetics/108.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J. D., Carland F., Lim E., Ralston E., Dooner H. K. Preferential transposition of the maize element Activator to linked chromosomal locations in tobacco. Plant Cell. 1990 Aug;2(8):701–707. doi: 10.1105/tpc.2.8.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matassi G., Melis R., Macaya G., Bernardi G. Compositional bimodality of the nuclear genome of tobacco. Nucleic Acids Res. 1991 Oct 25;19(20):5561–5567. doi: 10.1093/nar/19.20.5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero L. M., Filipski J., Gil P., Capel J., Martínez-Zapater J. M., Salinas J. The distribution of 5-methylcytosine in the nuclear genome of plants. Nucleic Acids Res. 1992 Jun 25;20(12):3207–3210. doi: 10.1093/nar/20.12.3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero L. M., Salinas J., Matassi G., Bernardi G. Gene distribution and isochore organization in the nuclear genome of plants. Nucleic Acids Res. 1990 Apr 11;18(7):1859–1867. doi: 10.1093/nar/18.7.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacken W. K., Piotrowiak R., Saedler H., Sommer H. The transposable element Tam1 from Antirrhinum majus shows structural homology to the maize transposon En/Spm and has no sequence specificity of insertion. Mol Gen Genet. 1991 Aug;228(1-2):201–208. doi: 10.1007/BF00282466. [DOI] [PubMed] [Google Scholar]
- Natsoulis G., Thomas W., Roghmann M. C., Winston F., Boeke J. D. Ty1 transposition in Saccharomyces cerevisiae is nonrandom. Genetics. 1989 Oct;123(2):269–279. doi: 10.1093/genetics/123.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planckaert F., Walbot V. Molecular and genetic characterization of Mu transposable elements in Zea mays: behavior in callus culture and regenerated plants. Genetics. 1989 Nov;123(3):567–578. doi: 10.1093/genetics/123.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlman R. F., Fedoroff N. V., Messing J. The nucleotide sequence of the maize controlling element Activator. Cell. 1984 Jun;37(2):635–643. doi: 10.1016/0092-8674(84)90395-7. [DOI] [PubMed] [Google Scholar]
- Rynditch A., Kadi F., Geryk J., Zoubak S., Svoboda J., Bernardi G. The isopycnic, compartmentalized integration of Rous sarcoma virus sequences. Gene. 1991 Oct 15;106(2):165–172. doi: 10.1016/0378-1119(91)90196-i. [DOI] [PubMed] [Google Scholar]
- Salinas J., Matassi G., Montero L. M., Bernardi G. Compositional compartmentalization and compositional patterns in the nuclear genomes of plants. Nucleic Acids Res. 1988 May 25;16(10):4269–4285. doi: 10.1093/nar/16.10.4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas J., Zerial M., Filipski J., Bernardi G. Gene distribution and nucleotide sequence organization in the mouse genome. Eur J Biochem. 1986 Nov 3;160(3):469–478. doi: 10.1111/j.1432-1033.1986.tb10063.x. [DOI] [PubMed] [Google Scholar]
- Scherdin U., Rhodes K., Breindl M. Transcriptionally active genome regions are preferred targets for retrovirus integration. J Virol. 1990 Feb;64(2):907–912. doi: 10.1128/jvi.64.2.907-912.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz-Sommer Z., Leclercq L., Göbel E., Saedler H. Cin4, an insert altering the structure of the A1 gene in Zea mays, exhibits properties of nonviral retrotransposons. EMBO J. 1987 Dec 20;6(13):3873–3880. doi: 10.1002/j.1460-2075.1987.tb02727.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih C. C., Stoye J. P., Coffin J. M. Highly preferred targets for retrovirus integration. Cell. 1988 May 20;53(4):531–537. doi: 10.1016/0092-8674(88)90569-7. [DOI] [PubMed] [Google Scholar]
- Voytas D. F., Ausubel F. M. A copia-like transposable element family in Arabidopsis thaliana. Nature. 1988 Nov 17;336(6196):242–244. doi: 10.1038/336242a0. [DOI] [PubMed] [Google Scholar]