Abstract
Background
Proline (Pro) accumulation is a widespread response of prokaryotic and eukaryotic cells subjected to osmotic stress or dehydration. When the cells are released from stress, Pro is degraded to glutamate by Pro-dehydrogenase (ProDH) and Pyrroline-5-carboxylate dehydrogenase (P5CDH), which are both mitochondrial enzymes in eukaryotes. While P5CDH is a single copy gene in Arabidopsis, two ProDH genes have been identified in the genome. Until now, only ProDH1 (At3g30775) had been functionally characterised.
Results
We demonstrate vasculature specific expression of the Arabidopsis ProDH2 gene (At5g38710) as well as enzymatic activity and mitochondrial localisation of the encoded protein. Expression levels of ProDH2 are generally low, but increased in senescent leaves and in the abscission zone of floral organs. While sucrose represses ProDH2 expression, Pro and NaCl were identified as inducers. Endogenous ProDH2 expression was not able to overcome Pro sensitivity of ProDH1 mutants, but overexpression of a GFP-tagged form of ProDH2 enabled the utilisation of Pro as single nitrogen source for growth. Amongst two intronic insertion mutants, one was identified as a null allele, whereas the other still produced native ProDH2 transcripts.
Conclusions
Arabidopsis possesses two functional ProDHs, which have non-redundant, although partially overlapping physiological functions. The two ProDH isoforms differ with respect to spatial, developmental and environmental regulation of expression. While ProDH1 appears to be the dominant isoform under most conditions and in most tissues, ProDH2 was specifically upregulated during salt stress, when ProDH1 was repressed. The characterisation of ProDH2 as a functional gene requires a careful re-analysis of mutants with a deletion of ProDH1, which were so far considered to be devoid of ProDH activity. We hypothesise that ProDH2 plays an important role in Pro homeostasis in the vasculature, especially under stress conditions that promote Pro accumulation.
Background
Arabidopsis, like most plant species, accumulates free proline (Pro) to high concentration in response to salt and drought stress. The physiological function of Pro is subject to controversial debate, and Arabidopsis plants with a reduced capacity to accumulate Pro showed only a moderate decrease or no change in stress tolerance [1-3]. Pro is suggested to act as a compatible osmolyte, a stabilising agent for macromolecules, a radical scavenger or as a nitrogen and energy store for the recovery phase [4,5]. Other hypotheses attribute the beneficial function to the process of Pro metabolism rather than the mere accumulation of Pro. Along this line, energy transfer between cellular compartments, regeneration of electron acceptors or signalling functions have been proposed [6,7].
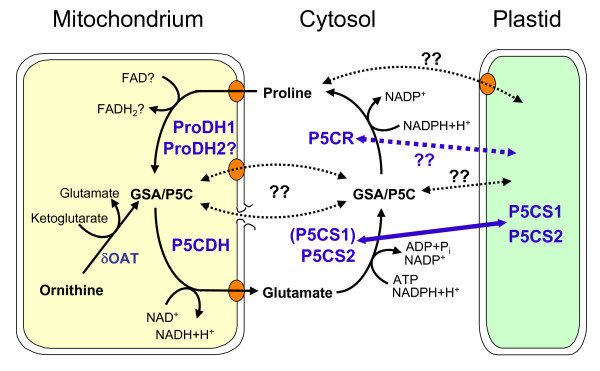
The biochemistry of Pro biosynthesis and degradation has been intensively studied and most of the participating proteins and the genes encoding for these enzymes were characterised with respect to temporal and spatial expression patterns and enzymatic properties (Fig. 1; [5]). The first step of Pro synthesis is catalysed by Pyrroline-5-carboxylate synthetase (P5CS), which uses glutamate, NADPH and ATP to generate glutamate-γ-semialdehyde (GSA). GSA is in spontaneous equilibrium with the cyclic pyrroline-5-carboxylate (P5C), which is converted to Pro by P5C reductase (P5CR), again consuming NADPH. Arabidopsis contains two isoforms of P5CS that are differentially regulated. P5CS2 (At3g55610) expression was consistent with a housekeeping function, and the P5CS2 protein was found to be localised in the cytosol under normal conditions while being partially re-located to plastids during stress [3,8,9]. Pro accumulation under stress conditions is primarily contingent upon induction of P5CS1 (At2g39800) expression. Also P5CS1 was observed in the cytosol, partly as presumably inactive aggregates, and re-localised to plastids during stress [3]. P5CR seems to be predominantly localised in the cytosol, while some activity was also associated with plastid preparations [10,11]. However, direct investigations of the localisation and enzymatic properties of Arabidopsis P5CR (At5g14800) are still pending. A second pathway for Pro synthesis from ornithine had also been postulated, but characterisation of Ornithine-δ-aminotransferase indicated that ornithine is degraded to glutamate inside the mitochondria prior to its conversion to Pro by the standard pathway [12,13]. Loss or gain of function mutants provided valuable evidence for the physiological role of Pro accumulation, although the interpretation of the mutant phenotypes was always hampered by the primary function of Pro in protein biosynthesis [4].
Figure 1.
Proline metabolism in Arabidopsis. Schematic illustration of the current knowledge on Pro metabolism and its intracellular distribution. Black arrows indicate metabolic fluxes, enzymes are given in bold blue letters, for abbreviations see the main text. The molecular identity of mitochondrial and plastidic Pro and glutamate transporters is currently not known. Alternating localisation between cytosol and plastids was demonstrated for P5CS1 and P5CS2, the localisation of P5CR requires further detailed analysis. Export or leakage of GSA/P5C from mitochondria was postulated but experimental evidence is lacking.
For degradation, Pro is imported into the mitochondria, where it is converted back to glutamate by the tightly coupled activities of Pro dehydrogenase (ProDH) and P5C dehydrogenase (P5CDH [14,15]). In Arabidopsis, one ProDH (ProDH1; At3g30775) and one P5CDH (At5g62530) have been characterised at the molecular level. ProDH1 and P5CDH expression is repressed by osmotic stress and upregulated by Pro, with the changes being more pronounced for ProDH1 [4]. Also in flowers, strong expression of ProDH1 was observed in the stigma and in pollen, while P5CDH is only upregulated in pollen [16,17]. Mutants with defects in Pro degradation grew without obvious phenotypic differences under greenhouse conditions, only a slight decrease in seed quality was noted for p5cdh mutants [16,18-20]. Common to the p5cdh mutants and the ProDH1 mutant pdh1-1 were an enhanced and prolonged accumulation of Pro during stress and an unexpected hypersensitivity to external Pro supply in the absence of stress. In wildtype plants, external application of harmful amounts of Pro caused the loss of mitochondrial and plastid integrity [21]. Hare et al. (2002) hypothesised that excess Pro degradation would cause misdirection of electrons to O2 in mitochondria and plastids due to electron overflow or acceptor limitation, respectively. Recent results confirmed the Pro-dependent production of reactive oxygen species (ROS) in mitochondria, an effect that was more pronounced in ProDH overexpressing or p5cdh mutant plants [22].
The only obvious gap in the molecular characterisation of Pro metabolism in Arabidopsis remains a second isoform of ProDH (ProDH2, At5g38710), which has been identified as an expressed gene with high homology to ProDH1 [1,23]. The predicted pre-proteins of ProDH1 and ProDH2 share 75% identical amino acids (aa). Hanson et al. (2008) demonstrated a direct induction of ProDH2 by the sucrose-repressed transcription factor bZIP11 [24]. However, until now the enzymatic functionality of ProDH2 had not been addressed. Here, we demonstrate that ProDH2 expressed in yeast can mediate Pro degradation equal to ProDH1. Overexpression of a ProDH2-GFP construct in Arabidopsis was able to rescue the Pro-hypersensitive phenotype of the pdh1-1 knockout mutant. We detected ProDH2-GFP in the mitochondria and analysis of GUS expression under control of the ProDH2 promoter demonstrated specific expression in the vascular tissue and in the abscission zone of petals, sepals and stamina. In striking contrast to ProDH1, ProDH2 expression was induced by salt stress and was absent in reproductive tissues.
Results
At5g38710 encodes a protein with ProDH activity
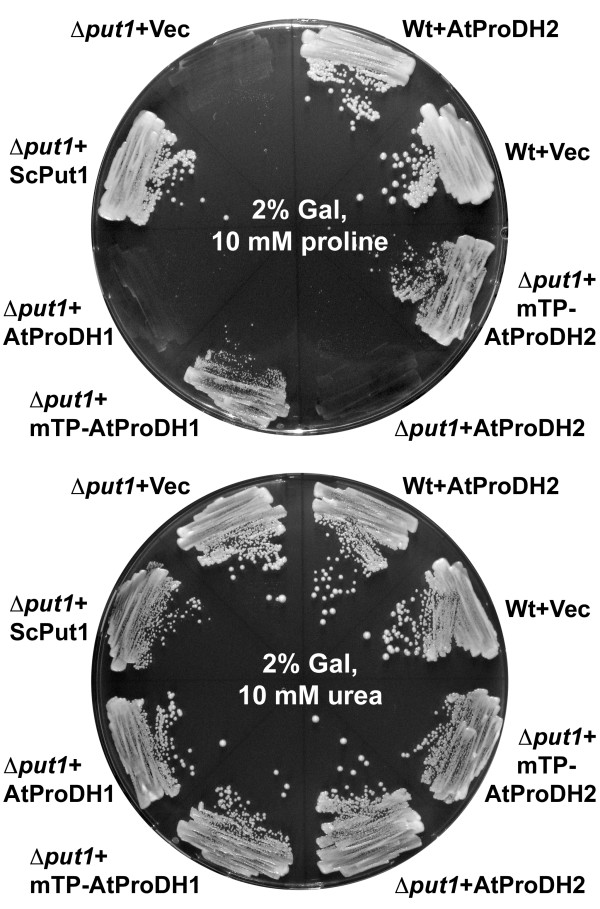
Reports on successful determination of ProDH activity from plant mitochondrial membranes are very rare. A variety of papers ascribe a soluble activity of Pro dependent NAD+ reduction at pH 10.3 to ProDH, but this is more likely to be the reverse reaction of P5CR (Giuseppe Forlani, unpublished results). Therefore we choose to test the enzymatic function of the putative ProDH2 (At5g38710) by heterologous expression in yeast. To obtain a suitable test system, we generated a mutant yeast strain in which the Put1 gene, encoding the single ProDH, was replaced by a kanamycin and geneticin resistance cassette by homologous recombination. The resulting geneticin resistant strain Δput1 was unable to catabolise Pro as a source of nitrogen for growth (Fig. 2). Transformation of the Δput1 strain with a plasmid harbouring the full-length cDNA of At5g38710 did not enable the cells to metabolise Pro. A similar observation for ProDH1 has been described by Kiyosue et al. (1996) and was confirmed by our results [25]. Exchange of the predicted mitochondrial transit peptide (mTP) of the putative ProDH2 (aa 1-29) against the mTP of the flavoprotein subunit of yeast succinate dehydrogenase (ScSdh1) lead to the expression of a protein that rescued the Pro metabolism defect of the Δput1 strain. Also an exchange of the first 39 amino acid residues of ProDH1 against the ScSdh1-mTP enabled the expression of a functional protein. The ScSdh1-mTP was chosen because it is unlikely to interfere with the functionality test for the Arabidopsis ProDHs. From these results we concluded that the name ProDH2 was justified for At5g38710. Under non-inducing conditions, with glucose instead of galactose as carbon source, none of the ProDH1 or ProDH2 expression constructs enabled the Δput1 strain to catabolise Pro (data not shown). Expression of the full-length ProDH2 cDNA in wildtype yeast did not alter the growth properties and western blot analysis of cells expressing a ProDH2 protein with 6×His-tag confirmed the presence of the recombinant protein (Fig. 2 and data not shown). These findings indicate that the native ProDH2 protein is not harmful to yeast, but does not have sufficient ProDH activity to enable utilisation of Pro as nitrogen source.
Figure 2.
ProDH2 expression complements a yeast Δput1 mutant. Yeast wildtype or Δput1 mutant with various ProDH expression constructs were grown for 4 d on minimal medium with 2% (w/v) galactose (Gal) as inducing carbon source and 10 mM Pro (upper plate) or 10 mM urea (lower plate) as the sole nitrogen source. Expression of ScPut1 confers wildtype growth to the Δput1 mutant. Native AtProDH pre-proteins do not complement the Pro utilisation deficiency of the Δput1 mutant. Replacement of the predicted Arabidopsis mTPs by the mTP of the yeast Sdh1 gene confers functional expression and the capability to metabolise external Pro as N-source (mTP-AtProDH1 and mTP-AtProDH2).
ProDH2 is expressed specifically in the vascular tissue
The presence of two ProDH genes in the Arabidopsis genome, which both encode functional ProDHs, raised the question of the specific function of ProDH2. Expression analysis of ProDH1 indicated ubiquitous expression in all organs, with the highest expression levels detected in pollen grains and in the stigma of the carpel [17]. To compare the expression of ProDH1 and ProDH2, we fused 1500 bp of upstream sequence of the ProDH2 gene to a β-glucuronidase (GUS) gene. Bioinformatic analysis of 3000 bp of genomic sequence upstream of the start codon of ProDH2 with the Athena analysis tool did not reveal any potential transcription factor binding sites that were more abundant than the expectations for random distribution would predict [26,27]. However, consensus motifs for an abscisic acid responsive element and a drought responsive element were identified at -880 and -1395 bp, respectively. Most of the putative binding motifs that were found between -1500 and -3000 bp were also present in the region between -1 and -1500 bp. Therefore we chose to include only the region up to -1500 bp in the GUS-construct. Transgenic plants harbouring this construct expressed GUS activity exclusively in the vascular tissue (Fig. 3A-C). Of the 30 analysed primary transformants, all showed a very similar expression pattern, albeit the amount of GUS activity detected by X-Gluc staining varied considerably (Data not shown). Staining was generally weak in the youngest leaves and strongest in the older leaves. For further analysis, plants with intermediate activity in mature leaves were chosen. In inflorescences, the ProDH2-promoter induced GUS expression only in the abscission zone immediately below the pistils or siliques. This high expression persisted through silique maturation, demonstrating a partial spatial separation of ProDH1 and ProDH2 expression in flowers. Northern blot analysis of RNA isolated from various tissues of soil grown Arabidopsis wildtype plants confirmed that the expression of ProDH2 is stronger in floral organs (Fig. 3D). In young (less than 7 mm long) and mature leaves (more that 3 cm long), no ProDH2 transcripts were detected, presumably due to the low relative content of vascular tissue, whereas a strong signal in senescent leaves (showing anthocyanin accumulation and visible reduction in chlorophyll content) confirmed age-dependent regulation of ProDH2 expression. In petioles from mature leaves, no ProDH2 expression was detected by northern blot although this part of the leaf should be enriched in vascular tissue. Also in roots and leaf-derived cell cultures, a strong expression of ProDH2 was detected. In contrast, ProDH1 was expressed also in a root-derived cell culture and was only slightly induced in senescent leaves. In the inflorescence, ProDH1 expression reached its peak before ProDH2. P5CDH was detected in all organs analysed. However, a surprisingly low transcript level of P5CDH was observed in roots, where both ProDH isoforms were strongly expressed.
Figure 3.
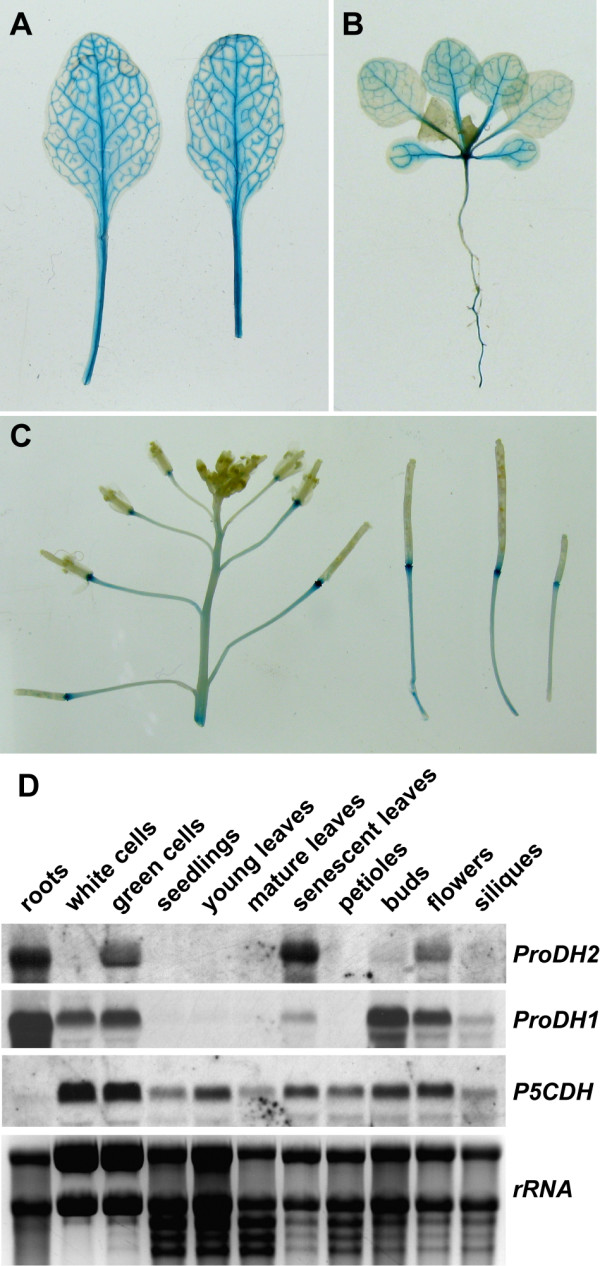
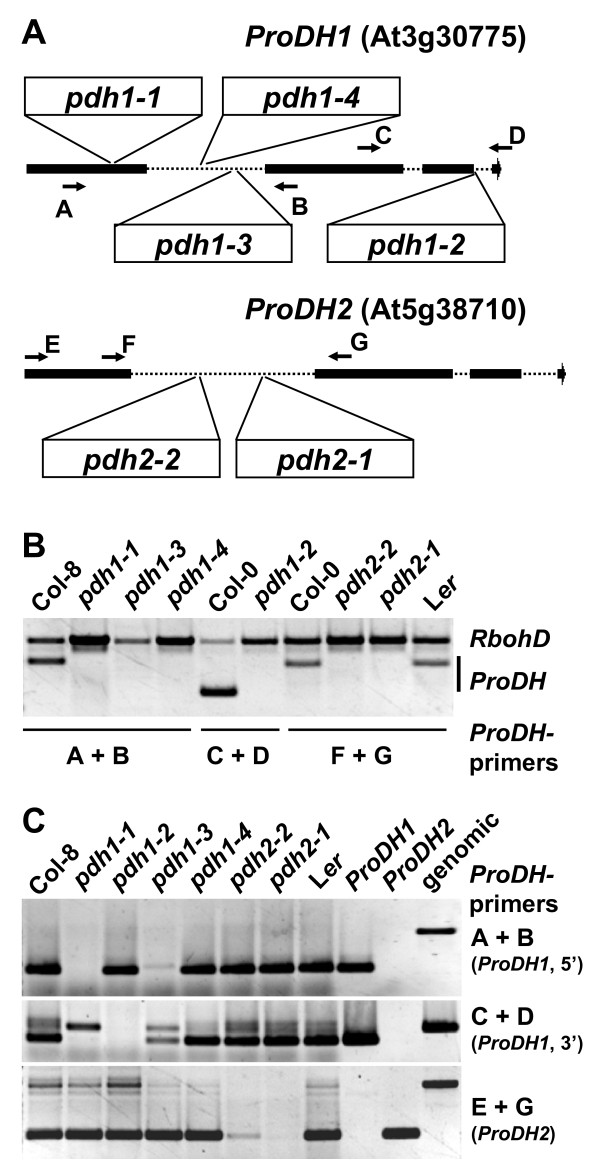
Tissue distribution of ProDH2 expression. A-C: ProDH2-promoter activity visualised in transgenic plants carrying a ProDH2-promoter-Gus fusion construct. Tissues were stained for 6 h in 1 mM X-Gluc and destained in 80% (v/v) EtOH. A: Two mature leaves of a 6-week-old plant. B: Three-week-old seedling showing enhanced staining with progressing leaf age. C: Inflorescence and young siliques showing GUS activity in the abscission zone between the pistils and the floral stalk. D: Northern blot analysis of transcript levels of Pro degradation genes in various tissues and two different cell cultures. The picture of the EtBr stained gel is shown to demonstrate RNA integrity and equal loading.
ProDH2 is induced by proline and salt but repressed by sugar
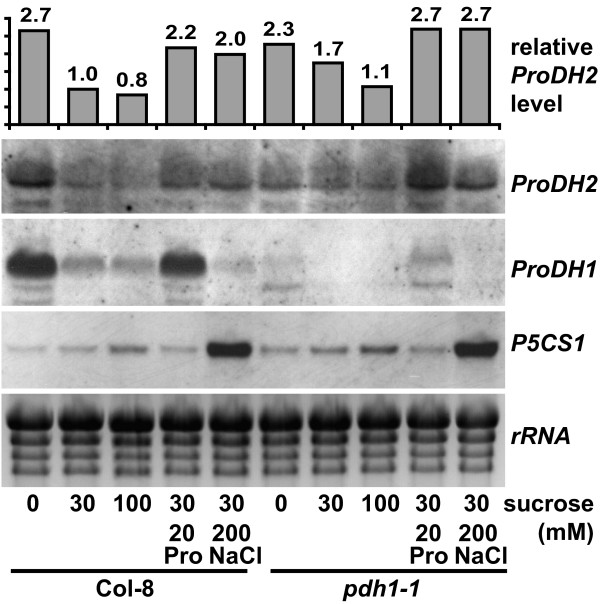
Since the variation of GUS activity between different transgenic lines was high, we used northern blot analysis to investigate the regulation of ProDH2 expression. Hanson et al. (2008) identified ProDH2 as a direct target of the transcription factor bZIP11 [24]. In agreement with these findings, ProDH2 transcript levels were higher when plants were incubated in medium without sucrose compared to treatment with 30 or 100 mM sucrose (Fig. 4). Also incubation for 6 h in medium containing 20 mM Pro or 200 mM NaCl in addition to 30 mM sucrose enhanced ProDH2 expression slightly. These concentrations were found to be sub-lethal during the period of the treatment and effective in triggering changes in Pro metabolism. Induction of ProDH2 by salt stress was surprising, since ProDH1 is repressed under these conditions. Induction of P5CS1 expression by 200 mM NaCl demonstrated that the treatment was efficient in triggering Pro accumulation. In general, the signals obtained for ProDH2 were near the detection limit, while ProDH1 detection yielded much stronger signals. This observation was confirmed by analysis of publicly available microarray data, in which ProDH2 expression is frequently not clearly detected (data not shown [28,29]). For example, in the AtGenExpress stress series, the maximal signal intensity for ProDH1 is 7.7-fold higher than for ProDH2. In the same dataset, the ProDH1 expression pattern was more similar to P5CDH than to ProDH2. Pathogen challenge and drought stress were among the rare cases, in which microarray analyses detected a stronger expression of ProDH2 than of ProDH1.
Figure 4.
ProDH2 is induced by Pro and repressed by sucrose. Three-week-old wildtype (Col-8) or ProDH1-knockout mutant (pdh1-1) seedlings from sterile culture were transferred for 6 h to liquid half-strength MS medium with the indicated supplements prior to RNA extraction and northern blot analysis. The same membrane was hybridised consecutively with specific probes for ProDH2, P5CS1 and ProDH1. During the detection of P5CS1, no remainders of the ProDH2 specific signal were observed. The histogram above the northern blot panel indicates the intensity of the ProDH2-specific signal normalised to the level of Col-8 in medium with 30 mM sucrose.
We also analysed potential compensatory changes of ProDH2 expression in the ProDH1 knockout mutant pdh1-1 [20]. Basal levels and sugar-dependent regulation of ProDH2 were only mildly affected in the pdh1-1 mutant. Induction of ProDH2 by Pro was slightly enhanced, presumably by the higher intracellular Pro content caused by lack of the ProDH1 isoform, which seems to be predominant in most tissues of wildtype plants. P5CS1 transcript levels were the same in wildtype and pdh1-1 mutant plants, while ProDH1 transcripts were mostly undetectable in the mutant. Under conditions of ProDH1 induction, some aberrant transcripts were also detected in pdh1-1.
A role of ProDH2 in Pro tolerance
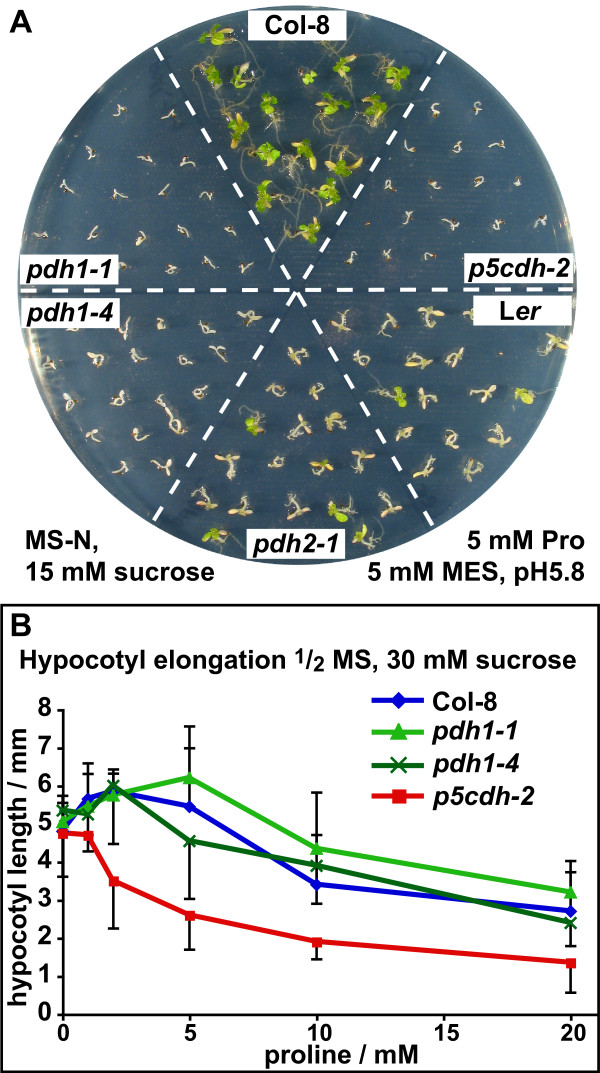
The low and spatially limited expression of ProDH2 posed the question of whether the ProDH2 protein can make a significant contribution to Pro metabolism. Therefore we directly compared the Pro sensitivity of pdh1-1, pdh1-4 (see below) and p5cdh-2 mutants, reasoning that Pro degradation is completely blocked in p5cdh-2 mutants, while pdh1 mutants still contain the active ProDH2 isoform. As described earlier, neither of these mutants was able to use 5 mM Pro as the sole source of nitrogen (Fig. 5A). Seedlings of both mutant genotypes did not expand properly and stayed etiolated, while wildtype seedlings turned green and developed true leaves. Compared to Col-8, the Ler ecotype showed a decreased ability to de-etiolate and grow with Pro as the only nitrogen source. A transposon insertion mutant devoid of ProDH2 (pdh2-1; see below) did not show increased sensitivity compared to the parental Ler ecotype.
Figure 5.
p5cdh-2 mutants are more sensitive to external Pro supply than pdh1 mutants. A: Col-8 and Ler wildtypes, pdh1-1, pdh1-4, pdh2-1 and p5cdh-2 seedlings were cultivated for 2 weeks on MS medium with 5 mM Pro as the sole source of nitrogen and 15 mM sucrose as carbon source, buffered to pH 5.8 with 5 mM MES. B: Hypocotyl elongation of Col-8 wildtype, pdh1-1, pdh1-4 and p5cdh-2 seedlings cultivated on MS medium supplemented with 30 mM sucrose and varying concentrations of Pro. Error bars indicate SD of ≥ 10 seedlings per genotype. The experiments were repeated with similar results.
When pdh1 and p5cdh mutants were assayed for Pro sensitivity in the presence of mineral nitrogen, only p5cdh-2 mutants showed a dose dependent inhibitory effect of Pro on hypocotyl elongation (cell elongation) and fresh weight accumulation (cell growth) that was different from the wildtype (Fig 5B and data not shown). These findings indicate that expression of ProDH2 is not sufficient to compensate fully for the lack of ProDH1, but may still be able to confer limited Pro tolerance in the presence of mineral nitrogen. Contrary to the loss of ProDH1 or P5CDH expression, loss of ProDH2 did not result in Pro hypersensitivity.
Robust splicing of T-DNAs in the first intron of ProDH1 and ProDH2
To enable a direct comparison of mutants completely devoid of ProDH activity to the p5cdh-2 mutant, we screened the public T-DNA insertion lines for pdh2 mutants. Several lines (SALK_108179, GABI_918D08, SAIL_90_G05, GABI_187C05) with T-DNA insertions immediately upstream or downstream of the ProDH2 coding sequence had little or no effect on transcript abundance (data not shown). Two lines were identified with a T-DNA or a transposon in the first intron of ProDH2 (GABI_328G05 and GT1788, respectively). By northern blot and RT-PCR analysis we were unable to detect native transcripts in homozygous GT1788 plants, while both aberrant and a small amount of native transcripts were present in GABI_328G05 (Fig. 6 and data not shown). Therefore we termed these two insertion-lines pdh2-1 and pdh2-2, respectively. Histochemical analysis revealed no detectable GUS activity in pdh2-1 plants, indicating that the GUS gene of gene-trap construct contained in the transposon is not in frame with the first ProDH2 exon after potential splicing events (data not shown). A similar analysis of ProDH1 insertion lines confirmed the absence of native transcripts in pdh1-1, SALK_081276 (pdh1-2) and GABI_308F08 (pdh1-3). In line SALK_119334 (pdh1-4), which carries an inverted tandem repeat of the T-DNA in the first intron of ProDH1, a mixture of aberrant and native transcripts was detected, similar to the pdh2-2 insertion line. When grown under greenhouse conditions, all ProDH1 or ProDH2 insertion lines showed no phenotypic differences to the corresponding wildtypes (data not shown). The production of useful pdh1/pdh2 double mutants is impeded by the fact that the insertion line pdh2-1 is in Ler background, in which stress-responsive Pro accumulation and Pro sensitivity differs from Col-8 (Fig. 5A and data not shown). Therefore, double mutants will have to be compared to multiple families of Col-8/Ler hybrids or crossed several times to one of the parental ecotypes.
Figure 6.
Knockout lines for ProDH1 and ProDH2 and T-DNA excision during splicing. A: Graphic representation of the exon-intron structure of the ProDH1 and ProDH2 genes with the position of T-DNA insertions in the analysed mutant lines (see material and methods section). Arrows with letters indicate the binding sites of primers used for PCR analyses in B and C. B: PCR on genomic DNA with two primer pairs, one for RbohD (At5g47910 as internal control, fragment size 1272 bp) and one ProDH1 or ProDH2 specific primer pair. Absence of ProDH specific PCR products demonstrates that all mutant plants were homozygous for the respective T-DNA insertion. C: RT-PCR analysis of ProDH1 and ProDH2 expression in the mutant lines demonstrates the presence of native transcripts in pdh1-4 and pdh2-2. PCR reactions with cloned cDNAs and genomic DNA (leftmost three lanes) demonstrate specificity of the PCR products. cDNAs for C and genomic DNA for B were obtained from the same samples, a slight contamination of the RNA samples with genomic DNA caused the additional amplification of intron-containing PCR products in the RT-PCRs.
ProDH2 is localised to mitochondria
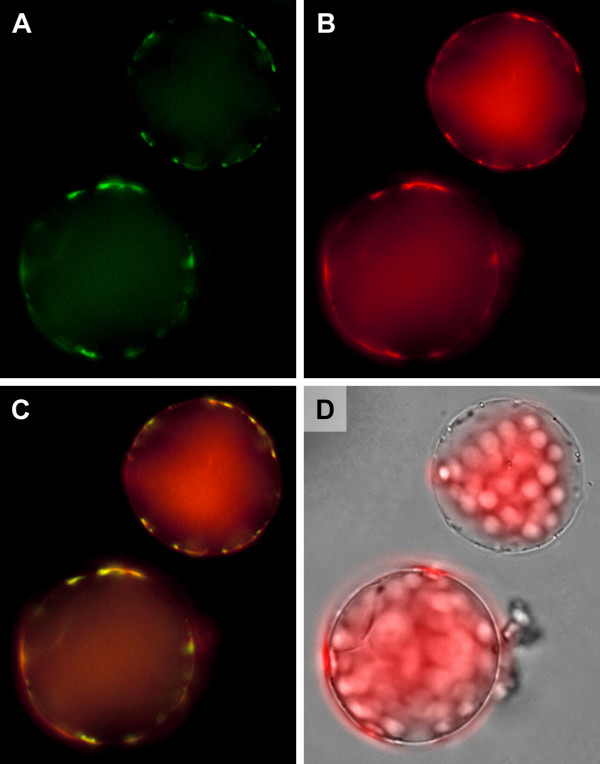
The reason for the inability of ProDH1 mutants to utilise Pro as nitrogen source, despite a functional ProDH2 copy, could be that ProDH2 is not in the correct subcellular localisation to support Pro degradation. Prediction programs clearly indicated a mitochondrial localisation, which we analysed with the help of transgenic plants expressing a ProDH2-GFP fusion protein. The GFP signals obtained from such plants were much weaker compared to ProDH1-GFP expression driven by the same promoter, but both ProDH2-GFP and ProDH1-GFP were clearly localised in mitochondria (Fig. 7 and Additional File 1). Subcellular localisation and enzymatic functionality of ProDH2 indicated that the low expression level and/or the tissue specificity were responsible for the inability of pdh1-1 mutants to utilise Pro as nitrogen source.
Figure 7.
Subcellular localisation of ProDH2. False colour images of protoplasts isolated from Arabidopsis plants stably transformed with a 35S:ProDH2:GFP fusion construct and stained with MitoTracker Orange. A: GFP fluorescence depicted in green; B: MitoTracker fluorescence depicted in red; C: Overlay of A and B demonstrating co-localisation of ProDH2-GFP with mitochondria. D: Overlay of a transmitted light picture with chlorophyll autofluorescence.
ProDH2-GFP overexpression rescues the pdh1-1 mutant
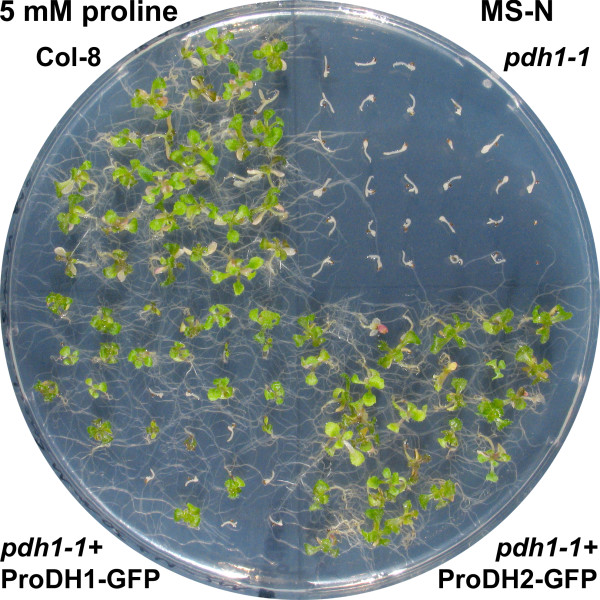
To test the functional equivalence of ProDH2 and ProDH1, also the pdh1-1 mutant was transformed with the constructs for the 35S-driven expression of ProDH1-GFP and ProDH2-GFP. Overexpression of ProDH2-GFP or ProDH1-GFP restored the capability of pdh1-1 mutants to use Pro as the sole source of nitrogen (Fig. 8). These findings indicate that tissue specificity and expression levels are the main determinants of the different functionality of the two ProDH isoforms in Arabidopsis.
Figure 8.
ProDH-GFP expression rescues the pdh1-1 mutant. Wildtype Col-8, pdh1-1 and pdh1-1 transformed with 35S:ProDH1:GFP or 35S:ProDH2:GFP were cultivated for 3 weeks on MS-N medium with 30 mM sucrose and 5 mM Pro as the only source of nitrogen. The transformants are segregating T2 populations, therefore not all seedlings are able to utilise Pro as nitrogen source.
Discussion
Functional expression of Arabidopsis ProDHs in yeast
In the initial characterisation of ProDH1, two groups reported conflicting results concerning the expression of a functional protein from the native Arabidopsis gene in yeast. The contra group circumvented the problem by using the mTP of the yeast ProDH gene Put1, which might in itself interfere with the functionality test [25]. The data provided by the pro group lack proper positive and negative controls, for which it is impossible to tell if there is some strain specificity in the use of non-native mTPs [30]. Both studies suffer from the use of tryptophan auxotrophic strains, which necessitated the addition of low amounts of tryptophan, a potential nitrogen source. We circumvented these limitations by generating a Δput1 deletion strain in the 23344c background, which is wildtype except for the ura3 mutation that served as a selection marker for the expression plasmids. In this background, the native cDNAs of either ProDH gene from Arabidopsis did not confer sufficient ProDH activity to enable Pro utilisation. Exchange of the predicted mTPs for the N-terminus of a metabolically unrelated mitochondrial protein from yeast allowed the expression of functional ProDHs as evidenced by complementation of the Pro utilisation defect of the Δput1 strain. From this we concluded that both ProDH genes from Arabidopsis encode proteins that mediate Pro catabolism to P5C/GSA. Functional expression of the hybrid genes in yeast indicated that protein folding and co-factor insertion of the Arabidopsis proteins worked correctly in the heterologous system. Mitochondrial localisation of ProDH1-GFP and ProDH2-GFP fusion proteins in Arabidopsis demonstrated functionality of the ProDH-mTPs in the native organism. Complementation of the Pro hypersensitive phenotype of Arabidopsis pdh1-1 mutant by overexpression of ProDH1-GFP or ProDH2-GFP provided further evidence that both proteins have the same enzymatic activity. It remains to be investigated, why the native Arabidopsis ProDH genes do not enable the expression of sufficient amounts of active and correctly targeted protein, whereas the Arabidopsis P5CDH gene produced a functional protein with its native mTP [19].
Spatial, temporal and metabolic regulation of ProDH2 expression
Analysis of the tissue specificity of ProDH2 expression by promoter-Gus fusion demonstrated that ProDH2 expression is restricted to the vascular tissue. This finding is in agreement with regulation of ProDH2 expression by the transcription factor bZIP11, which is also spatially confined to the vasculature [24,31]. Translation of bZIP11 is repressed by sucrose, which resulted in reduced transcript levels of ProDH2 in the presence of high sucrose concentrations. High sucrose concentrations (3%) might also be the reason for the absence of ProDH2 transcripts in the root-derived heterotrophic cell culture. The medium of the mixotrophic leaf-derived cell culture contained 1% sucrose (roughly corresponding to 30 mM), which is not sufficient to switch off ProDH2 expression. ProDH2-promoter driven Gus expression increased with leaf age and was strongest in the abscission zone of the floral organs, indicating that developmental signals also play a role in ProDH2 regulation. ProDH1, which had also enhanced promoter activity in the abscission zone, was additionally strongly expressed in pollen, in the stigma and in developing embryos, where no ProDH2 expression was detected [17]. In roots, both ProDH1 and ProDH2 were strongly detected, while P5CDH was only weakly expressed. Potentially, arginine catabolism, which is the second task of P5CDH, is less prominent in roots. Alternatively, the strong ProDH expression could result from the preparation of the roots by extensive washing in (hypotonic) tap water. Expression changes of ProDH1 are reportedly very fast, while regulation of P5CDH was slower [25,32]. Increased ProDH2 expression in old leaves was confirmed by northern blot analysis and evaluation of publicly available microarray data [28,29]. The spatial expression pattern was the most striking difference between ProDH1 and ProDH2 expression but also NaCl treatment caused down-regulation only of ProDH1. Similarly, Ribarits et al. (2007) found a differential regulation of two ProDH isoforms during dehydration in tobacco, while under these conditions the Arabidopsis ProDHs were co-regulated [23]. Over the broad range of different treatments documented in the microarray data collection, co-regulation of ProDH1 and ProDH2 is only sparsely observed, supporting our findings that the two ProDH isoforms fulfil at least partially different physiological functions. Up-regulation of ProDH2 expression during salt or drought stress might indicate a special need for proline degradation in the vasculature, while down-regulation of ProDH1 occurs in the remaining tissues and enables proline accumulation. These findings imply that proline accumulation might not be favourable in the vascular tissue and the continued degradation of proline may provide energy and glutamate for other metabolic pathways or for long distance transport.
ProDH2 cannot fully compensate the lack of ProDH1
Loss-of-function mutants of ProDH1 were identified and characterised as sensitive to external Pro supply and heat treatment [20,33]. The existence and potential activity of ProDH2 was neglected in these studies. Mutants in P5CDH, which is a single copy gene in Arabidopsis, were completely unable to degrade Pro [16]. In a direct comparison of pdh1, pdh2 and p5cdh mutants, we observed that pdh1 and p5cdh mutants were unable to utilise Pro as the sole nitrogen source, whereas pdh2 mutants grew equally well as the corresponding wildtype. When pdh1-1, pdh1-4 and p5cdh-2 mutants were grown on medium containing mineral nitrogen, only p5cdh-2 was more sensitive to growth-inhibition by external Pro than the wildtype. Preliminary characterisation of the pdh2-1 mutant showed a slightly enhanced Pro accumulation in response to salt stress but not after external Pro application (data not shown). Pro sensitivity and stress induced Pro accumulation differed between the Col-8 and Ler wildtypes, confirming that a useful double mutant can only be produced after extensive backcrossing. From these data we conclude that the slightly enhanced ProDH2 expression in Pro treated pdh1 mutants could not fully compensate the loss of ProDH1, while it may be able to diminish the toxic effects of Pro. Alternatively, the toxic effects of Pro supply might result from P5C, which is only expected to accumulate in the p5cdh-2 mutant. Recent results presented by Miller et al. (2009) suggest that ROS production by ProDH rather than P5C accumulation is the cause of the toxic effects of Pro application [22]. Unfortunately, Miller et al. (2009) did not analyse ProDH mutants and from the data presented it is not possible to determine, whether ROS production is the cause or a consequence of the loss of mitochondrial integrity that was observed in earlier studies [21]. Our identification of pdh2 mutants and the possibility to generate double mutants completely devoid of ProDH activity opens new possibilities to dissect the contribution of Pro degradation to Pro toxicity and stress tolerance. The detailed characterisation of the pdh2 mutants will also help to identify the specific physiological function of ProDH2 in the vascular tissue.
Conclusions
Despite numerous biochemical and molecular approaches, the protective mechanism of Pro accumulation remains unclear. Each new piece of experimental evidence added to our picture of Pro metabolism in plants requires careful re-interpretation of previous results. Only by a thorough analysis of all components and also non-obvious side effects it will become possible to understand the seemingly dual nature of Pro as a protective and toxic compound. The functional characterisation of ProDH2 opens up a whole new set of experimental approaches to understand the function of Pro in the stress tolerance of plants.
Methods
Plant material and growth conditions
Arabidopsis (Arabidopsis thaliana (L.) Heynh. ecotype Col-8 or Ler) and T-DNA or transposon insertion lines were obtained from the NASC (SALK_108179, GABI_918D08, SAIL_90_G05, GABI_328G05 (pdh2-2), GT1788 (pdh2-1), SALK_081276 (pdh1-2), GABI_308F08 (pdh1-3), SALK_119334 (pdh1-4) and Salk_018453 (p5cdh-2)), from Bernd Weisshaar (GABI_187C05; GABI-Kat program, Cologne, Germany [34], or from the KAZUSA research institute, Kisarazu, Japan (pdh1-1). Presence of the T-DNA and allelic status were verified by PCR and sequencing of the T-DNA flanking sequences. Gene and T-DNA-specific primers are listed in Table 1. All physiological experiments with pdh1 and p5cdh mutants were performed with homozygous progeny of plants backcrossed three times to Col-8. For the pdh2-1 mutant (in Ler background), segregation analysis demonstrated the presence of a single transposon. Plants were cultivated axenically in 9 cm Petri dishes on commercial MS medium (Duchefa, Haarlem, Netherlands) or self-made MS medium, in which KNO3 and NH4NO3 were replaced by 20 mM KCl [35]. Media were supplemented with sucrose and nitrogen containing compounds as indicated for each experiment and solidified with 8 g/l purified agar (BD Biosciences, San Jose, CA, USA) when appropriate. Seeds were surface sterilised by sequential treatment with 70% (v/v) EtOH and 1% (w/v) NaOCl/0,01% (v/v) Triton-X-100 and vernalised for 24 h at 4°C in 0.1% (w/v) agarose. Plants were cultivated in a climatised room with short day (9 h) light period and a light intensity of 110 μmol photons*s-1 *m-2 from mixed fluorescence tubes (Biolux and Fluora, Osram, Munich, Germany) at a constant temperature of 23°C. For induction experiments, axenically grown seedlings were transferred to liquid medium and incubated in the light with agitation at 100 rpm for 6 h. Induction in liquid medium was chosen because wildtype plants metabolised proline very quickly when it was provided via the solid culture medium and only marginal increases in proline content were observed in the leaves. For seed production, plants were kept in a greenhouse with a light period of at least 16 h. A heterotrophic (white) culture was obtained from Giuseppe Forlani, Ferrara, Italy and maintained in liquid MS medium supplemented with 3% (w/v) sucrose, 1× Gamborg's vitamin mix, 0.5 mg/l 2,4-dichlorophenoxyacetic acid, 0.5 mg/l benzylaminopurine and 0,2% (v/v) Plant Preservative Medium (Plant Cell Technology, Washington, DC, USA) under constant agitation in dim light. A mixotrophic, green cell culture was obtained from Silke Robatzek (Cologne, Germany and maintained in liquid MS medium supplemented with 1% (w/v) sucrose, 4× Gamborg's vitamin mix, 0,5 mg/l 2-(1-naphthyl)acetic acid and 0,1 mg/l kinetin at 110 μmol photons*s-1 *m-2.
Table 1.
PCR primers used in this study
| Name1 | Sequence (5'->3') | Used for |
|---|---|---|
| Pdh1-LP1 (A) | tctcctctatcccaacctctg | pdh1-1, GABI_308F08 (pdh1-3), SALK_119334 (pdh1-4) |
| Pdh1-RP1 (B) | gatcgctcactcgtttcagaag | |
| Pdh1-LP2 (C) | aagttggtgagaggggcttac | SALK_081276 (pdh1-2) |
| Pdh1-NS-r (D) | cgcaatcccggcgattaatctc | ProDH1 cloning |
| Pdh1-f | caccataATGgcaacccgtcttctc2 | |
| Pdh2-LP | gtaaccagcccctaaacctc | SALK_108179, GABI_918D08, SAIL_90_G05 |
| Pdh2-RP | ggagtactagatcgcgtgtaac | |
| Pdh2-LP2 (F) | aaaccctaccttcgtctcac | GT1788 (pdh2-1), GABI_328G05 (pdh2-2) |
| Pdh2-RP2 (G) | cactaacccgttttaggacattc | |
| Pdh2-LP3 | gagaagagttatggcttggtg | GABI_187C05 |
| Pdh2-RP3 | atgtccctttgtaatctgaattgg | |
| Pdh2-f (E) | caccataATGgcaaaccgtttcctc2 | ProDH2 cloning |
| Pdh2-NSr | ccaagccataactcttctcttaag | |
| Pdh2-Prom-f | catttggatccttaccatccac | ProDH2 promoter cloning |
| Pdh2-Prom-r | cgggatccgtttgcCATttaaactc2 | |
| Salk_LBb2 | ttcggaaccaccatcaaacag | SALK lines |
| Gabi-LB | cccatttggacgtgaatgtagacac | GABI lines |
| Sail-LB2 | gcttcctattatatcttcccaaattaccaataca | SAIL_90_G05 |
| Gus5'rev | atttcacgggttggggtttc | GT1788 |
| Put1KO-f | aaacatcgctacatagtaataacactaacgcacgcta gaaCGGATCCCCGGGTTAATTAA3 |
Put1 knockout |
| Put1KO-r | ttggtttgtctttgaaattggagtatatattatagtcctcG AATTCGAGCTCGTTTAAAC3 |
|
| SDH-f | caccacgagaataaagATGctatcgct2 | SDH-MTP |
| SDH-Pdh1-r | cttgttgtccaaaggagagCTCGTGATCTATTATGTG4 | SDH-MTP ProDH1 fusion |
| SDH-Pdh2-r | ggttggtcaaaggaaaggatCTCGTGATCTATTATGTG4 | SDH-MTP ProDH2-fusion |
| Put1-f | caccctagaaATGatagcttcc2 | Put1 cloning |
| Put1-r | taggcctactctttttggaatc | |
| Pdh1-pr-r | atggtcataaaacgtacttttcac | northern probes (with Pdh1-f or Pdh2-f) |
| Pdh2-pr-r | gactcatacacgctactcac | |
| P5CS1-f | aatgagaggaaaaggacaag | P5CS1 probe [8] |
| P5CS1-r | gataggatatgagtactaagcagag | |
| P5CDH-f | tggacagaagtgttctgcac | P5CDH probe |
| P5CDH-r | gcttccaacactagaggaag | |
1 Letters in brackets refer to the abbreviations in Figure 6; 2 capital letters indicate the translation start codon; 3 capital letters indicate the nucleotides specific for the resistance cassette, lower case letters correspond to Put1 5' and 3' sequences; 4 capital letters correspond to the 3' sequence of the SDH-MTP, lower case letters correspond to the nucleotides encoding the N-terminus of the predicted mature Arabidopsis ProDH proteins.
Promoter-GUS construct and histochemical analysis
A 1500 bp promoter fragment including the native start codon of ProDH2 was amplified from Arabidopsis genomic DNA with primers introducing BamHI restriction sites (Tab. 1). The PCR-product was subcloned into pCR-blunt and sequenced. The promoter fragment was excised with BamHI and inserted into pCB308 [36]. Agrobacterium tumefaciens strain GV3101 was used for floral dip transformation of Arabidopsis [37,38]. Transformants were selected by spraying soil grown seedlings with 50 mg/l BASTA or by addition of 10 mg/l BASTA to the culture medium. Histochemical GUS staining was performed according to [39].
Expression analysis
Total RNA was extracted from snap-frozen material with phenol/guanidine thiocyanate reagent according to each manufacturer's recommendations. Per lane, 15 μg of total RNA was separated by denaturing agarose gel electrophoresis and transferred to a positively charged nylon membrane by capillary transfer. ProDH1, ProDH2, P5CS1 and P5CDH transcripts were detected by hybridisation with digoxigenin-labelled PCR products obtained with primers listed in Tab. 1, followed by detection with alkaline phosphatase coupled anti-DIG antibodies and the chemiluminescent substrate CDP-star (Roche, Basel, Switzerland). Densitometric quantification was performed with the ImageJ software. For RT-PCR, 4 μg of total RNA was converted to cDNA using random hexamer primers and the Transcriptor cDNA synthesis Kit (Roche). One μl of the cDNA preparation was used as a template for endpoint RT-PCR. Public microarray data collection were evaluated with the Expression Browser of the Bio-Array Resource and Genvestigator v3 [28,29,40,41].
ProDH1-GFP and ProDH2-GFP constructs and imaging
The open reading frames of ProDH1 and ProDH2 without the stop codon were amplified by PCR from EST clones 38H5 and full-length ORF clone U66465, respectively (ABRC, Columbus, OH, USA). Sequences of PCR primers are given in Tab. 1. The resulting PCR fragments were purified and integrated into pENTR by directional TOPO cloning (Invitrogen, Carlsbad, CA, USA). Subsequently, the ProDH1 and ProDH2 cDNAs were transferred from pENTR to pEarleyGate103 (CD3-685, ABRC) or pGWB5 [42] by LR-recombination (Invitrogen). Both plant transformation vectors yielded the same results. Sequencing of the resulting constructs demonstrated in-frame fusion of the ProDH cDNAs to the GFP gene and revealed that EST 38H5 is not derived from Col-0 but most likely from C24 ecotype, which carries 12 silent or conservative single nucleotide polymorphisms in the ProDH1 coding sequence. These nucleotide exchanges are present in a number of Arabidopsis wildtype accessions. Transgenic plants were produced as described above except for selection of plants that carried pGWB5 derived constructs, which was performed in axenic culture.
Protoplasts from transformed leaves were obtained by overnight incubation with cellulase and macerase (Serva, Heidelberg, Germany), stained with MitoTracker Orange (Invitrogen) and viewed under an Olympus BX51 epifluorescence microscope equipped with a Nikon DXM1200 digital camera system (Olympus Europe, Hamburg, Germany). Chlorophyll autofluorescence, MitoTracker and GFP fluorescence of the cells were dissected using the filter sets U-MWSG2 (Olympus), 41007 and 41020 (Chroma Technology Corp, Rockingham, VT, USA), respectively. False colouring and overlay of images was performed using AxioVision software (Zeiss, Oberkochen, Germany).
Yeast strains, growth conditions and expression plasmids
Saccharomyces cerevisiae strain 23344c (Matα ura3; [43]) was received from Giuseppe Forlani, Ferrara, Italy. For the deletion of the Put1 gene, the kanamycin resistance cassette from pFA6a-KanMX6 was amplified with primers adding 40 bp of Put1 5' and 3' sequences (Tab. 1, [44]). After transformation of 23344c with the PCR product, geneticin resistant colonies were selected and tested for their ability to utilise Pro or urea as the sole source of nitrogen. Replacement of the Put1 coding sequence by the resistance cassette was verified by PCR. The Put1 coding sequence and the Sdh1-mTP (bases 1-156 of the coding sequence) were amplified with the primers listed in Tab. 1 from genomic DNA isolated from 23344c. The reverse primers for the Sdh1-mTP introduce overlaps to the coding sequences of ProDH1 and ProDH2 that were used to fuse the coding sequences by PCR. All final PCR products were introduced into pENTR as described above and the coding sequences were transferred to pYES-Dest52 by LR-recombination (Invitrogen). Plasmids were introduced into yeast by the LiAc/PEG method [45] and transformants were selected on synthetic medium with 20 g/l glucose and 5 g/l NH4SO4 supplemented with a mixture of amino acids but no uracil (BD biosciences). For growth tests on organic nitrogen sources, yeast strains were washed once in sterile water, resuspended in 0.1% agarose and streaked on plates containing synthetic minimal medium, 20 g/l galactose and no other additives except the indicated nitrogen sources.
Authors' contributions
GM analysed the expression pattern of ProDH2. SE generated the yeast expression constructs. DF designed the study, performed the other experiments and compiled the manuscript. All authors contributed to and approved the final manuscript.
Supplementary Material
ProDH1-GFP is localised in mitochondria. False colour images of a protoplast expressing a ProDH1-GFP fusion protein under control of the CaMV-35S promoter and stained with MitoTracker Orange.
Contributor Information
Dietmar Funck, Email: dietmar.funck@uni-konstanz.de.
Sonja Eckard, Email: sonja.wenndorf@uni-konstanz.de.
Gudrun Müller, Email: gudrun.2.mueller@uni-konstanz.de.
Acknowledgements
We would like to thank Giuseppe Forlani, University of Ferrara, Italy, for communicating unpublished results, for the 23344c yeast strain, for the white Arabidopsis cell culture and for helpful comments on the manuscript. Marc Erhardt, University of Konstanz, is acknowledged for the pFA6a-KanMX6 plasmid and for helpful advice on the construction of the Δput1 knockout strain. We are also thankful to Tsuyoshi Nakagawa, Shimane University, Japan, for the pGWB5 vector. Regina Grimm, Silvia Kuhn, Roswitha Miller-Sulger and the gardeners of the University of Konstanz are acknowledged for technical assistance in the lab and for excellent plant care, respectively. We are indebted to Iwona Adamska for providing a functional working environment and for critical reading of the manuscript. Additional thanks to Helen Gunter, who made helpful suggestions concerning the use of the English language. The University of Konstanz provided financial support for this study and GM is grateful for a fellowship by the Stiftung der deutschen Wirtschaft.
References
- Mani S, Cotte B Van De, Van Montagu M, Verbruggen N. Altered levels of proline dehydrogenase cause hypersensitivity to proline and its analogs in Arabidopsis. Plant Physiol. 2002;128(1):73–83. doi: 10.1104/pp.010572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanjo T, Kobayashi M, Yoshiba Y, Sanada Y, Wada K, Tsukaya H, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K. Biological functions of proline in morphogenesis and osmotolerance revealed in antisense transgenic Arabidopsis thaliana. Plant J. 1999;18(2):185–193. doi: 10.1046/j.1365-313X.1999.00438.x. [DOI] [PubMed] [Google Scholar]
- Szekely G, Abraham E, Cseplo A, Rigo G, Zsigmond L, Csiszar J, Ayaydin F, Strizhov N, Jasik J, Schmelzer E. et al. Duplicated P5CS genes of Arabidopsis play distinct roles in stress regulation and developmental control of proline biosynthesis. Plant Journal. 2008;53(1):11–28. doi: 10.1111/j.1365-313X.2007.03318.x. [DOI] [PubMed] [Google Scholar]
- Verbruggen N, Hermans C. Proline accumulation in plants: a review. Amino Acids. 2008;35(4):753–759. doi: 10.1007/s00726-008-0061-6. [DOI] [PubMed] [Google Scholar]
- Szabados L, Savoure A. Proline: a multifunctional amino acid. Trends Plant Sci. 2010;15(2):89–97. doi: 10.1016/j.tplants.2009.11.009. [DOI] [PubMed] [Google Scholar]
- Hare PD, Cress WA. Metabolic implications of stress-induced proline accumulation in plants. Plant Growth Regulation. 1997;21(2):79–102. doi: 10.1023/A:1005703923347. [DOI] [Google Scholar]
- Phang JM, Donald SP, Pandhare J, Liu Y. The metabolism of proline, a stress substrate, modulates carcinogenic pathways. Amino Acids. 2008;35(4):681–690. doi: 10.1007/s00726-008-0063-4. [DOI] [PubMed] [Google Scholar]
- Strizhov N, Abraham E, Okresz L, Blickling S, Zilberstein A, Schell J, Koncz C, Szabados L. Differential expression of two P5CS genes controlling proline accumulation during salt-stress requires ABA and is regulated by ABA1, ABI1 and AXR2 in Arabidopsis. Plant J. 1997;12(3):557–569. doi: 10.1046/j.1365-313X.1997.00557.x. [DOI] [PubMed] [Google Scholar]
- Mattioli R, Falasca G, Sabatini S, Altamura MM, Costantino P, Trovato M. The proline biosynthetic genes P5CS1 and P5CS2 play overlapping roles in Arabidopsis flower transition but not in embryo development. Physiologia Plantarum. 2009;137(1):72–85. doi: 10.1111/j.1399-3054.2009.01261.x. [DOI] [PubMed] [Google Scholar]
- Rayapati PJ, Stewart CR, Hack E. Pyrroline-5-Carboxylate Reductase Is in Pea (Pisum sativum L.) Leaf Chloroplasts. Plant Physiol. 1989;91(2):581–586. doi: 10.1104/pp.91.2.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szoke A, Miao GH, Hong Z, Verma DP. Subcellular Location of Delta-Pyrroline-5-Carboxylate Reductase in Root/Nodule and Leaf of Soybean. Plant Physiol. 1992;99(4):1642–1649. doi: 10.1104/pp.99.4.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams E, Frank L. Metabolism of proline and the hydroxyprolines. Annu Rev Biochem. 1980;49:1005–1061. doi: 10.1146/annurev.bi.49.070180.005041. [DOI] [PubMed] [Google Scholar]
- Funck D, Stadelhofer B, Koch W. Ornithine-delta-aminotransferase is essential for arginine catabolism but not for proline biosynthesis. BMC Plant Biol. 2008;8:40. doi: 10.1186/1471-2229-8-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elthon TE, Stewart CR. Proline Oxidation in Corn Mitochondria: Involvement of NAD, Relationship to Ornithine Metabolism, and Sidedness on the Inner Membrane. Plant Physiol. 1982;70(2):567–572. doi: 10.1104/pp.70.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elthon TE, Stewart CR. Submitochondrial Location and Electron Transport Characteristics of Enzymes Involved in Proline Oxidation. Plant Physiol. 1981;67(4):780–784. doi: 10.1104/pp.67.4.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuschle K, Funck D, Forlani G, Stransky H, Biehl A, Leister D, Graaff E van der, Kunze R, Frommer WB. The role of [Delta]1-pyrroline-5-carboxylate dehydrogenase in proline degradation. Plant Cell. 2004;16(12):3413–3425. doi: 10.1105/tpc.104.023622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Satoh R, Kiyosue T, Yamaguchi-Shinozaki K, Shinozaki K. A gene encoding proline dehydrogenase is not only induced by proline and hypoosmolarity, but is also developmentally regulated in the reproductive organs of Arabidopsis. Plant Physiol. 1998;118(4):1233–1241. doi: 10.1104/pp.118.4.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell. 2005;123(7):1279–1291. doi: 10.1016/j.cell.2005.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuschle K, Funck D, Hellmann H, Daschner K, Binder S, Frommer WB. A nuclear gene encoding mitochondrial Delta-pyrroline-5-carboxylate dehydrogenase and its potential role in protection from proline toxicity. Plant J. 2001;27(4):345–356. doi: 10.1046/j.1365-313X.2001.01101.x. [DOI] [PubMed] [Google Scholar]
- Nanjo T, Fujita M, Seki M, Kato T, Tabata S, Shinozaki K. Toxicity of free proline revealed in an arabidopsis T-DNA-tagged mutant deficient in proline dehydrogenase. Plant Cell Physiol. 2003;44(5):541–548. doi: 10.1093/pcp/pcg066. [DOI] [PubMed] [Google Scholar]
- Hare PD, Cress WA, van Staden J. Disruptive effects of exogenous proline on chloroplast and mitochondrial ultrastructure in Arabidopsis leaves. South African Journal of Botany. 2002;68(3):393–396. [Google Scholar]
- Miller G, Honig A, Stein H, Suzuki N, Mittler R, Zilberstein A. Unraveling delta1-pyrroline-5-carboxylate-proline cycle in plants by uncoupled expression of proline oxidation enzymes. J Biol Chem. 2009;284(39):26482–26492. doi: 10.1074/jbc.M109.009340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribarits A, Abdullaev A, Tashpulatov A, Richter A, Heberle-Bors E, Touraev A. Two tobacco proline dehydrogenases are differentially regulated and play a role in early plant development. Planta. 2007;225(5):1313–1324. doi: 10.1007/s00425-006-0429-3. [DOI] [PubMed] [Google Scholar]
- Hanson J, Hanssen M, Wiese A, Hendriks MM, Smeekens S. The sucrose regulated transcription factor bZIP11 affects amino acid metabolism by regulating the expression of ASPARAGINE SYNTHETASE1 and PROLINE DEHYDROGENASE2. Plant J. 2008;53(6):935–949. doi: 10.1111/j.1365-313X.2007.03385.x. [DOI] [PubMed] [Google Scholar]
- Kiyosue T, Yoshiba Y, Yamaguchi-Shinozaki K, Shinozaki K. A nuclear gene encoding mitochondrial proline dehydrogenase, an enzyme involved in proline metabolism, is upregulated by proline but downregulated by dehydration in Arabidopsis. Plant Cell. 1996;8(8):1323–1335. doi: 10.1105/tpc.8.8.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athena promoter visualization tool. http://www.bioinformatics2.wsu.edu/Athena
- O'Connor TR, Dyreson C, Wyrick JJ. Athena: a resource for rapid visualization and systematic analysis of Arabidopsis promoter sequences. Bioinformatics. 2005;21(24):4411–4413. doi: 10.1093/bioinformatics/bti714. [DOI] [PubMed] [Google Scholar]
- BAR The Bio-Array Resource for Arabidopsis Functional Genomics. http://bbc.botany.utoronto.ca/
- Genevestigator v3. http://www.genevestigator.com
- Peng Z, Lu Q, Verma DP. Reciprocal regulation of delta 1-pyrroline-5-carboxylate synthetase and proline dehydrogenase genes controls proline levels during and after osmotic stress in plants. Mol Gen Genet. 1996;253(3):334–341. doi: 10.1007/pl00008600. [DOI] [PubMed] [Google Scholar]
- Rook F, Gerrits N, Kortstee A, van Kampen M, Borrias M, Weisbeek P, Smeekens S. Sucrose-specific signalling represses translation of the Arabidopsis ATB2 bZIP transcription factor gene. Plant J. 1998;15(2):253–263. doi: 10.1046/j.1365-313X.1998.00205.x. [DOI] [PubMed] [Google Scholar]
- Hellmann H, Funck D, Rentsch D, Frommer WB. Hypersensitivity of an Arabidopsis sugar signaling mutant toward exogenous proline application. Plant Physiol. 2000;123(2):779–789. doi: 10.1104/pp.123.2.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkindale J, Vierling E. Core genome responses involved in acclimation to high temperature. Plant Physiol. 2008;146(2):748–761. doi: 10.1104/pp.107.112060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B. An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol. 2003;53(1-2):247–259. doi: 10.1023/B:PLAN.0000009297.37235.4a. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15(15):473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- Xiang C, Han P, Lutziger I, Wang K, Oliver DJ. A mini binary vector series for plant transformation. Plant Mol Biol. 1999;40(4):711–717. doi: 10.1023/A:1006201910593. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Wydro M, Kozubek E, Lehmann P. Optimization of transient Agrobacterium-mediated gene expression system in leaves of Nicotiana benthamiana. Acta Biochim Pol. 2006;53(2):289–298. [PubMed] [Google Scholar]
- Martin T, Hellmann H, Schmidt R, Willmitzer L, Frommer WB. Identification of mutants in metabolically regulated gene expression. The Plant Journal. 1997;11(1):53–62. doi: 10.1046/j.1365-313X.1997.11010053.x. [DOI] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics. 2008;2008:420747. doi: 10.1155/2008/420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toufighi K, Brady SM, Austin R, Ly E, Provart NJ. The Bio-Array Resource for Plant Functional Genomics. Plant J. 2005;43(1):153–163. doi: 10.1111/j.1365-313X.2005.02437.x. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. Journal of Bioscience and Bioengineering. 2007;104(1):34. doi: 10.1263/jbb.104.34. [DOI] [PubMed] [Google Scholar]
- Grenson M. The utilization of exogenous pyrimidines and the recycling of uridine-5'-phosphate derivatives in Saccharomyces cerevisiae, as studied by means of mutants affected in pyrimidine uptake and metabolism. Eur J Biochem. 1969;11(2):249–260. doi: 10.1111/j.1432-1033.1969.tb00767.x. [DOI] [PubMed] [Google Scholar]
- Achim W, Arndt B, Rainer P, Peter P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10(13):1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/S0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ProDH1-GFP is localised in mitochondria. False colour images of a protoplast expressing a ProDH1-GFP fusion protein under control of the CaMV-35S promoter and stained with MitoTracker Orange.