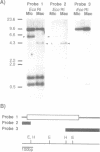
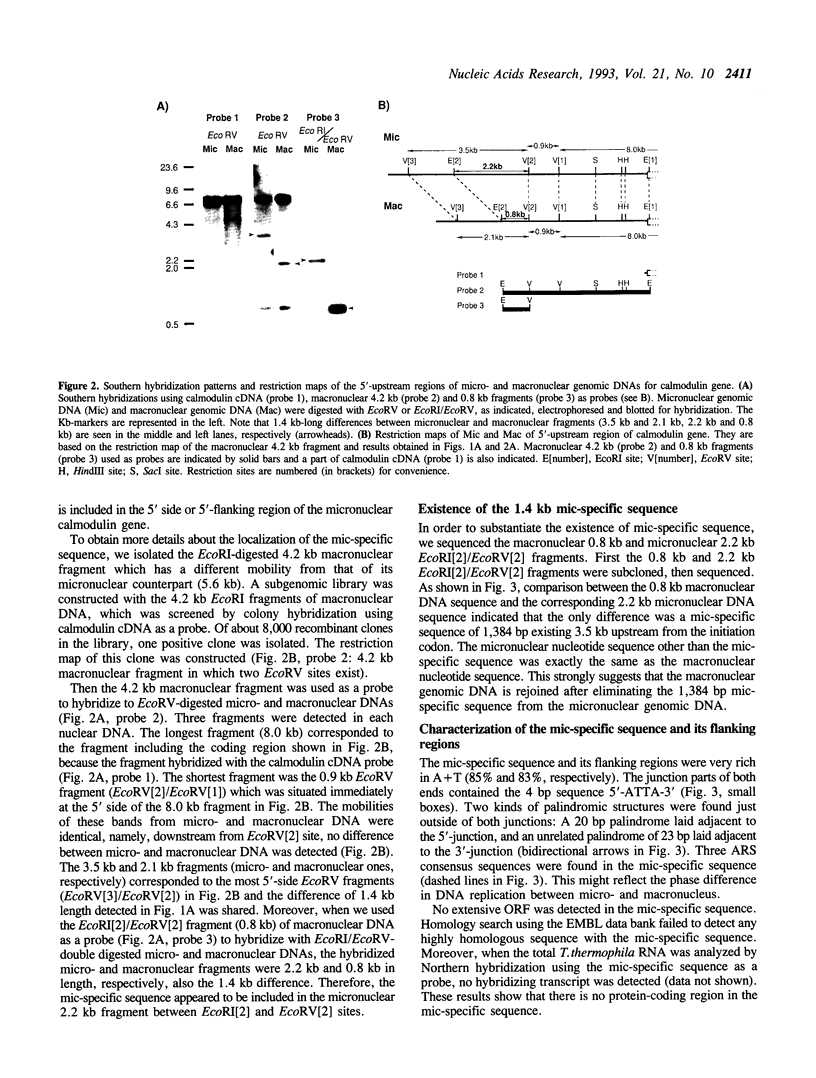
Abstract
Tetrahymena thermophila possesses a transcriptionally inactive micronucleus and an active macronucleus. Both nuclei are developed from micronucleus-derived germ nuclei during conjugation. Extensive DNA rearrangement and transcriptional activation are known to be involved in macronuclear development, but little has been known about these processes in a particular functional gene. Therefore the micro- and macronuclear genomic DNAs for calmodulin gene were analyzed. A 1,384 bp micronucleus-specific sequence located about 3.5 kb upstream of calmodulin gene has been found, suggesting DNA rearrangement during macronuclear development. The micronucleus-specific sequence had 85% A + T, no extensive ORF, ATTAs at both ends, and two palindromic structures just outside of both ends. Interestingly, the micronucleus-specific sequence included a T-rich tract, T16CT5, in the middle, and a nearly complementary A-rich tract, A5TA10GA5, existed 7 bp upstream from the initiation codon. In addition, there was a 20 bp repetitive sequence TAAT(TAAC)4 about 100 bp upstream of the micronucleus-specific sequence and also in the promoter region of calmodulin gene. Although the functional significance of the micronucleus-specific sequence remains unclear, T16CT5 and TAAT(TAAC)4 elements might exert an influence on transcription of the calmodulin gene. Stringent Southern hybridization revealed that this micronucleus-specific sequence or very similar sequence(s) were abundant in the Tetrahymena micronuclear genome.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bantock C. R. Experiments on chromosome elimination in the gall midge, Mayetiola destructor. J Embryol Exp Morphol. 1970 Sep;24(2):257–286. [PubMed] [Google Scholar]
- Beermann S. The diminution of Heterochromatic chromosomal segments in Cyclops (Crustacea, Copepoda). Chromosoma. 1977 Apr 20;60(4):297–344. doi: 10.1007/BF00292858. [DOI] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Callahan R. C., Shalke G., Gorovsky M. A. Developmental rearrangements associated with a single type of expressed alpha-tubulin gene in Tetrahymena. Cell. 1984 Feb;36(2):441–445. doi: 10.1016/0092-8674(84)90237-x. [DOI] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Engberg J., Andersson P., Leick V., Collins J. Free ribosomal DNA molecules from Tetrahymena pyriformis GL are giant palindromes. J Mol Biol. 1976 Jun 25;104(2):455–470. doi: 10.1016/0022-2836(76)90281-3. [DOI] [PubMed] [Google Scholar]
- Godiska R., Yao M. C. A programmed site-specific DNA rearrangement in Tetrahymena thermophila requires flanking polypurine tracts. Cell. 1990 Jun 29;61(7):1237–1246. doi: 10.1016/0092-8674(90)90688-b. [DOI] [PubMed] [Google Scholar]
- Hattori M., Hidaka S., Sakaki Y. Sequence analysis of a KpnI family member near the 3' end of human beta-globin gene. Nucleic Acids Res. 1985 Nov 11;13(21):7813–7827. doi: 10.1093/nar/13.21.7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karrer K. M. Germ line-specific DNA sequences are present on all five micronuclear chromosomes in Tetrahymena thermophila. Mol Cell Biol. 1983 Nov;3(11):1909–1919. doi: 10.1128/mcb.3.11.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraut H., Lipps H. J., Prescott D. M. The genome of hypotrichous ciliates. Int Rev Cytol. 1986;99:1–28. doi: 10.1016/s0074-7696(08)61422-9. [DOI] [PubMed] [Google Scholar]
- McKnight S. L., Kingsbury R. Transcriptional control signals of a eukaryotic protein-coding gene. Science. 1982 Jul 23;217(4557):316–324. doi: 10.1126/science.6283634. [DOI] [PubMed] [Google Scholar]
- Mita T., Shiomi H., Iwai K. Isolation of nuclei from exponentially growing Tetrahymena pyriformis. Exp Cell Res. 1966 Oct;43(3):696–699. doi: 10.1016/0014-4827(66)90049-8. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemasa T., Takagi T., Edamatsu M., Watanabe Y. Calmodulin cDNAs from two species of Tetrahymena. Biochim Biophys Acta. 1992 Sep 24;1132(2):219–221. doi: 10.1016/0167-4781(92)90017-t. [DOI] [PubMed] [Google Scholar]
- Takemasa T., Takagi T., Kobayashi T., Konishi K., Watanabe Y. The third calmodulin family protein in Tetrahymena. Cloning of the cDNA for Tetrahymena calcium-binding protein of 23 kDa (TCBP-23). J Biol Chem. 1990 Feb 15;265(5):2514–2517. [PubMed] [Google Scholar]