Abstract
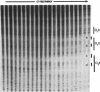
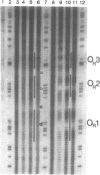
The peroxonitrite anion (ONOO-) is a stable species in alkaline solution that quickly generates a strong oxidant at neutral pH. A convenient procedure for the preparation of ONOOK has been developed based on the procedure of Keith & Powell [(1969) J. Chem. Soc. A, 90], which when added to a sample of duplex DNA buffered at neutral pH rapidly generates a strong oxidant capable of nonspecifically cleaving the DNA present. We show that this solution containing ONOOK can be used to hydroxyl radical footprint the binding the cl-repressor (cl) of phage lambda with the right operator, OR. In addition, we show that the individual-site binding isotherms determined by quantitative DNase I, Fe-EDTA and ONOOK footprinting are identical within experimental error. The identical isotherms obtained with the three different reagents with greatly differing sampling times indicates that the sampling time of the footprinting probe need not be short relative to the kinetic dissociation constants that govern protein-DNA interactions.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ackers G. K., Johnson A. D., Shea M. A. Quantitative model for gene regulation by lambda phage repressor. Proc Natl Acad Sci U S A. 1982 Feb;79(4):1129–1133. doi: 10.1073/pnas.79.4.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackers G. K., Shea M. A., Smith F. R. Free energy coupling within macromolecules. The chemical work of ligand binding at the individual sites in co-operative systems. J Mol Biol. 1983 Oct 15;170(1):223–242. doi: 10.1016/s0022-2836(83)80234-4. [DOI] [PubMed] [Google Scholar]
- Backman K., Ptashne M., Gilbert W. Construction of plasmids carrying the cI gene of bacteriophage lambda. Proc Natl Acad Sci U S A. 1976 Nov;73(11):4174–4178. doi: 10.1073/pnas.73.11.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckman J. S., Beckman T. W., Chen J., Marshall P. A., Freeman B. A. Apparent hydroxyl radical production by peroxynitrite: implications for endothelial injury from nitric oxide and superoxide. Proc Natl Acad Sci U S A. 1990 Feb;87(4):1620–1624. doi: 10.1073/pnas.87.4.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenowitz M., Senear D. F., Shea M. A., Ackers G. K. Quantitative DNase footprint titration: a method for studying protein-DNA interactions. Methods Enzymol. 1986;130:132–181. doi: 10.1016/0076-6879(86)30011-9. [DOI] [PubMed] [Google Scholar]
- Buchanan C. E., Gustafson A. Mutagenesis and mapping of the gene for a sporulation-specific penicillin-binding protein in Bacillus subtilis. J Bacteriol. 1992 Aug;174(16):5430–5435. doi: 10.1128/jb.174.16.5430-5435.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhoff A. M., Tullius T. D. The unusual conformation adopted by the adenine tracts in kinetoplast DNA. Cell. 1987 Mar 27;48(6):935–943. doi: 10.1016/0092-8674(87)90702-1. [DOI] [PubMed] [Google Scholar]
- Dreyer G. B., Dervan P. B. Sequence-specific cleavage of single-stranded DNA: oligodeoxynucleotide-EDTA X Fe(II). Proc Natl Acad Sci U S A. 1985 Feb;82(4):968–972. doi: 10.1073/pnas.82.4.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson L. H., Borg L. A. A spectrophotometric method for determination of catalase activity in small tissue samples. Anal Biochem. 1988 Oct;174(1):331–336. doi: 10.1016/0003-2697(88)90554-4. [DOI] [PubMed] [Google Scholar]
- Johnson A. D., Pabo C. O., Sauer R. T. Bacteriophage lambda repressor and cro protein: interactions with operator DNA. Methods Enzymol. 1980;65(1):839–856. doi: 10.1016/s0076-6879(80)65078-2. [DOI] [PubMed] [Google Scholar]
- Kim K., Rhee S. G., Stadtman E. R. Nonenzymatic cleavage of proteins by reactive oxygen species generated by dithiothreitol and iron. J Biol Chem. 1985 Dec 15;260(29):15394–15397. [PubMed] [Google Scholar]
- Klein S. M., Cohen G., Cederbaum A. I. Production of formaldehyde during metabolism of dimethyl sulfoxide by hydroxyl radical generating systems. Biochemistry. 1981 Oct 13;20(21):6006–6012. doi: 10.1021/bi00524a013. [DOI] [PubMed] [Google Scholar]
- Koblan K. S., Ackers G. K. Energetics of subunit dimerization in bacteriophage lambda cI repressor: linkage to protons, temperature, and KCl. Biochemistry. 1991 Aug 6;30(31):7817–7821. doi: 10.1021/bi00245a022. [DOI] [PubMed] [Google Scholar]
- Lomonossoff G. P., Butler P. J., Klug A. Sequence-dependent variation in the conformation of DNA. J Mol Biol. 1981 Jul 15;149(4):745–760. doi: 10.1016/0022-2836(81)90356-9. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oefner C., Suck D. Crystallographic refinement and structure of DNase I at 2 A resolution. J Mol Biol. 1986 Dec 5;192(3):605–632. doi: 10.1016/0022-2836(86)90280-9. [DOI] [PubMed] [Google Scholar]
- Price P. A. The essential role of Ca2+ in the activity of bovine pancreatic deoxyribonuclease. J Biol Chem. 1975 Mar 25;250(6):1981–1986. [PubMed] [Google Scholar]
- Prütz W. A., Mönig H., Butler J., Land E. J. Reactions of nitrogen dioxide in aqueous model systems: oxidation of tyrosine units in peptides and proteins. Arch Biochem Biophys. 1985 Nov 15;243(1):125–134. doi: 10.1016/0003-9861(85)90780-5. [DOI] [PubMed] [Google Scholar]
- Radi R., Beckman J. S., Bush K. M., Freeman B. A. Peroxynitrite oxidation of sulfhydryls. The cytotoxic potential of superoxide and nitric oxide. J Biol Chem. 1991 Mar 5;266(7):4244–4250. [PubMed] [Google Scholar]
- Reiner P., Brenowitz M. Quantitative densitometry of autoradiograms: digital images representative of optical density. Comput Appl Biosci. 1991 Jul;7(3):337–340. doi: 10.1093/bioinformatics/7.3.337. [DOI] [PubMed] [Google Scholar]
- Sluka J. P., Horvath S. J., Bruist M. F., Simon M. I., Dervan P. B. Synthesis of a sequence-specific DNA-cleaving peptide. Science. 1987 Nov 20;238(4830):1129–1132. doi: 10.1126/science.3120311. [DOI] [PubMed] [Google Scholar]
- Tullius T. D., Dombroski B. A., Churchill M. E., Kam L. Hydroxyl radical footprinting: a high-resolution method for mapping protein-DNA contacts. Methods Enzymol. 1987;155:537–558. doi: 10.1016/0076-6879(87)55035-2. [DOI] [PubMed] [Google Scholar]
- Tullius T. D., Dombroski B. A. Hydroxyl radical "footprinting": high-resolution information about DNA-protein contacts and application to lambda repressor and Cro protein. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5469–5473. doi: 10.1073/pnas.83.15.5469. [DOI] [PMC free article] [PubMed] [Google Scholar]