SUMMARY
Accurate genome segregation depends on cohesion mechanisms that link duplicated sister chromatids, thereby allowing their tension-dependent biorientation in metaphase. In Saccharomyces cerevisiae, cohesion is established during DNA replication when Eco1 acetylates the cohesin subunit Smc3. Cohesion establishment is restricted to S phase of the cell cycle, but the molecular basis of this regulation is unknown. Here we show that Eco1 is negatively regulated by the protein kinase Cdk1. Phosphorylation of Eco1 after S phase targets it to SCFCdc4 for ubiquitination and subsequent degradation. A nonphosphorylatable mutant of Eco1 establishes cohesion after DNA replication, suggesting that Cdk1-dependent phosphorylation of Eco1 is a key factor limiting establishment to S phase. We also show that deregulation of Eco1 results in chromosome separation defects in anaphase. We conclude that this regulatory mechanism helps optimize the level of sister chromatid cohesion, ensuring a robust and efficient anaphase.
INTRODUCTION
The accurate transfer of genetic information to the next generation is necessary for the propagation of life. Multiple mechanisms therefore exist in the cell division cycle to ensure proper duplication and segregation of chromosomes (Morgan, 2007). Following DNA replication in S phase, sister chromatid pairs are bioriented on the mitotic spindle, with one sister attached to each pole. Biorientation requires a means of discerning which chromatids are sisters; a mechanism has therefore evolved to hold sister chromatids together from S phase to mitosis (Onn et al., 2008; Nasmyth and Haering, 2009; Uhlmann, 2009). When sister chromatid pairs are attached to microtubules from opposite poles, sister-chromatid cohesion resists the spindle forces pulling the chromatids apart. The resulting tension is used both to strengthen microtubule attachments and as a readout for the spindle assembly checkpoint, which delays anaphase until correct biorientation is achieved. When all chromosomes are correctly bioriented, sister-chromatid cohesion is removed, allowing sisters to separate and be pulled apart by the spindle.
Sister-chromatid cohesion depends primarily on a protein complex called cohesin. In Saccharomyces cerevisiae, the core components of this complex are Smc1, Smc3, Scc1/Mcd1, and Scc3 (Onn et al., 2008; Nasmyth and Haering, 2009; Uhlmann, 2009). Smc1 and Smc3 are large coiled-coil proteins of the highly conserved ‘Structural Maintenance of Chromosomes’ (SMC) family. These two subunits form a large ring structure that is linked tightly at one end by globular hinge domains, and at the other end by the Scc1 subunit, together with Scc3. At the onset of anaphase, cleavage of Scc1 by the protease separase opens the ring, resulting in cohesin dissociation and thus sister separation (Uhlmann et al., 2000).
Sister-chromatid cohesion is achieved in two steps: cohesin loading and establishment. Beginning in G1 and continuing throughout the cell cycle, cohesin is constitutively loaded onto chromosomes by a poorly understood mechanism that requires the proteins Scc2 and Scc4 (Ciosk et al., 2000). Loaded cohesin appears to have a low affinity for chromosomes (Gerlich et al., 2006) and is thus unable to hold sister chromatids together. As the chromosomes are duplicated in S phase, sister chromatid cohesion is ‘established’ when a fraction of the loaded cohesin molecules is converted into a form that tightly links the newly formed sister chromatids, providing cohesion that can resist the forces of the spindle (Uhlmann and Nasmyth, 1998; Skibbens et al., 1999; Toth et al., 1999).
Among the many proteins known to be required for cohesion establishment, the best understood is the highly conserved acetyltransferase Eco1/Ctf7 (Skibbens et al., 1999; Toth et al., 1999; Ivanov et al., 2002). Eco1 promotes cohesion by acetylating the cohesin subunit Smc3 during S phase. Smc3 acetylation appears to counteract an ‘anti-establishment’ activity of the cohesin-associated proteins Wpl1/Rad61 and Pds5, as mutation of either of these proteins renders Eco1 non-essential for viability (Rolef Ben-Shahar et al., 2008; Unal et al., 2008; Zhang et al., 2008; Rowland et al., 2009; Sutani et al., 2009).
The establishment of cohesion is tightly regulated both spatially and temporally. In a normal cell cycle, establishment is strictly confined to S phase; cohesin loaded onto chromatin in a metaphase-arrested cell is not able to establish cohesion (Uhlmann and Nasmyth, 1998; Haering et al., 2004). The restriction of establishment to S phase is believed to depend in part on the recruitment of Eco1 to the replication fork by the processivity clamp PCNA (Lengronne et al., 2006; Moldovan et al., 2006; Rolef Ben-Shahar et al., 2008). However, association with the replication fork cannot be the only factor limiting cohesion establishment, since certain mutants (such as wpl1Δ) are able to establish cohesion post-replication (Heidinger-Pauli et al., 2009), suggesting that the necessary components are present after S phase but are somehow inhibited.
Indeed, cohesion can also be established after S phase in cells experiencing DNA damage (Strom et al., 2004; Strom et al., 2007; Unal et al., 2007). Damage-induced cohesion is necessary for efficient DNA repair (Sjogren and Nasmyth, 2001; Strom et al., 2004; Unal et al., 2004) and genetic evidence suggests that it might be established by acetylation of the Scc1 subunit of cohesin by Eco1, which antagonizes the inhibitory activity of Wpl1 (Heidinger-Pauli et al., 2009). The S phase and damage acetylation marks appear to be distinct, as neither is able to compensate for loss of the other at the appropriate stage (Heidinger-Pauli et al., 2009).
Despite its critical role in the control of sister chromatid cohesion, the regulation of Eco1 is largely unexplored. In HeLa cells, Esco1 and Esco2 are known to undergo post-translational changes during the cell cycle: chromatin-bound Esco1 is phosphorylated during mitosis, and Esco2 levels are reduced from G2 through the end of M phase (Hou and Zou, 2005). In Xenopus laevis, Eco1 and Eco2 are phosphorylated upon entry into mitosis, and Eco2 is ubiquitinated in late mitosis by the ubiquitin ligase APCCdh1 (Lafont et al., 2010). The functions of these regulatory events are not known. Intriguingly, over-expression of Eco1 allows yeast cells to establish cohesion in metaphase (Unal et al., 2007), suggesting that Eco1 activity is a limiting factor for post-replicative cohesion establishment, and that Eco1 is somehow inhibited after S phase.
The tight coupling of cohesion establishment to the cell cycle suggests that a cell cycle component regulates Eco1 activity. Indeed, yeast Eco1 is known to be a target of the cyclin-dependent kinase Cdk1, a key regulator of cell cycle progression (Ubersax et al., 2003), and mutations that reduce Cdk1 activity are synthetically lethal with an eco1 mutation (Brands and Skibbens, 2008). However, mutation of the four Cdk1 consensus phosphorylation sites in Eco1 does not affect cell viability or sensitivity to DNA-damaging agents (Brands and Skibbens, 2008), and thus the role of Cdk1 in Eco1 regulation is not clear.
Here, we demonstrate that Cdk1-dependent phosphorylation of Eco1 from late S phase through mitosis leads to its degradation. This helps limit the establishment of cohesion to S phase, as stabilization of Eco1, either through mutation of its Cdk1 sites or activation of the DNA damage pathway, permits ectopic establishment of cohesion in metaphase. Finally, we show that anaphase progression is defective in cells that fail to decrease Eco1 activity after S phase, illustrating the importance of this regulation for robust chromosome segregation.
RESULTS
Phosphorylation of Eco1 by Cdk1
Cdk1 has previously been implicated in Eco1 regulation in vitro (Ubersax et al., 2003) and in genetic studies (Brands and Skibbens, 2008). However, mutation of the four Cdk1 consensus phosphorylation motifs in Eco1 (the ECO1-4A mutant, see Figure S1A) has no apparent effect on cell viability or DNA damage sensitivity (Brands and Skibbens, 2008). Given that Eco1 activity is limiting in metaphase (Unal et al., 2007), when Clb-Cdk1 activity is highest, we speculated that Cdk1 phosphorylation is inhibitory and that Eco1-4A is therefore overactive. This could explain why ECO1-4A cells display no obvious phenotype, as overexpression of Eco1 has no detectable effect on viability (Borges et al., 2010). Also, overactive Eco1 might be genetically equivalent to wpl1Δ, which also has no viability defect (Rolef Ben-Shahar et al., 2008; Sutani et al., 2009).
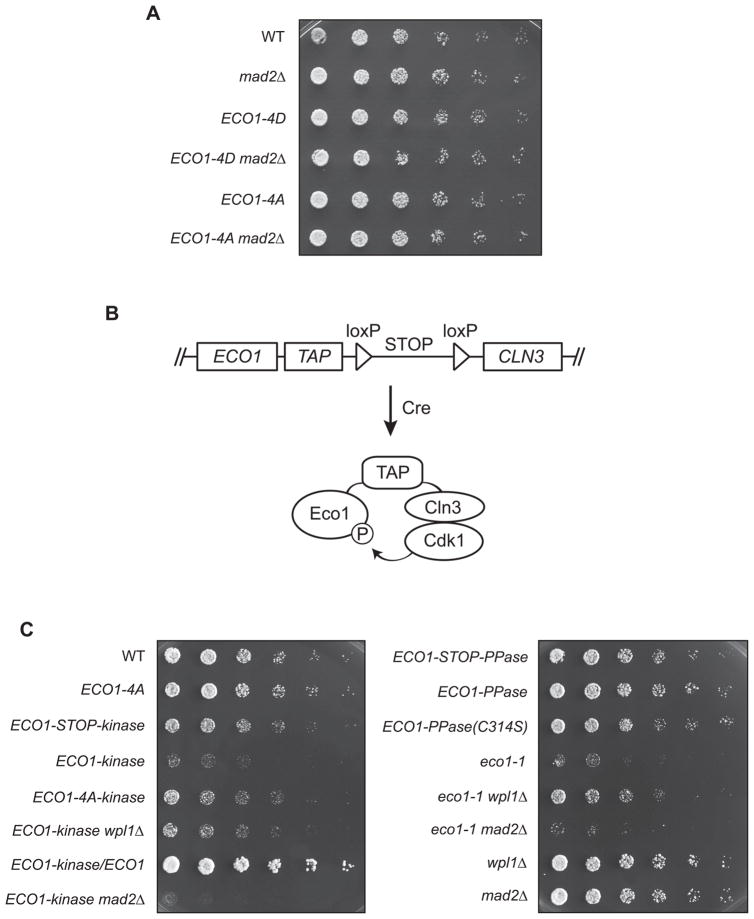
According to these arguments, a constitutively phosphorylated form of Eco1 should be less active and therefore more likely to display a phenotype, since Eco1 is essential. In an attempt to test this, we constructed a ‘phosphomimic’ version of Eco1 in which the Cdk1 consensus sites were mutated to aspartate or glutamate (ECO1-4D) to imitate the negative charge of the phosphorylated state. This mutant, like ECO1-4A, displayed neither a loss of viability nor a genetic interaction with mutation of the spindle checkpoint (mad2Δ, Figure 1A), which is known to enhance the phenotype of Eco1 loss-of-function mutants (see Figure 1C below).
Figure 1. Eco1 phosphomutant fusions reveal a phenotype of constitutive phosphorylation.
(A) Three-fold serial dilutions of the indicated yeast strains were grown at 30°C on a YPD plate.
(B) Diagram of fusion protein method (see Figure S1B for details). After looping out the stop codon with the Cre recombinase, ECO1-TAP is attached to the kinase (CLN3-1), as shown here, or phosphatase (MmCDC14B).
(C) Three-fold serial dilutions of the indicated strains were grown at room temperature.
Development of a Phosphomutant Fusion Approach
Our negative results with the ECO1-4D mutant illustrate a common problem in the study of phosphoregulation: no amino acid substitution can perfectly mimic a phosphorylated residue. To address this problem, we developed a method for rapidly creating proteins that are constitutively phosphorylated (or dephosphorylated) in vivo. We modified the target substrate to include a kinase or phosphatase fused to the C-terminus (Figure 1B) to promote constant phosphorylation or dephosphorylation, respectively. To direct the phosphorylation of Cdk1 sites, we fused target proteins to the G1 cyclin Cln3, which recruits Cdk1. For the phosphatase fusion we used the mouse protein Cdc14B, a homolog of a yeast phosphatase, Cdc14, that is known to specifically dephosphorylate Cdk1 consensus motifs (Gray et al., 2003). To reduce the impact of ectopic regulation on the substrate, both fused proteins were truncated versions lacking localization and degradation motifs (Nash et al., 1988; Gray et al., 2003).
We designed the fusions to be attached to the end of a tandem affinity purification (TAP) tag, to provide a flexible linker between the substrate and enzyme. The availability of a library of yeast strains with every ORF tagged with a TAP tag (Ghaemmaghami et al., 2003) allows this fusion technique to be applied to any protein using the same DNA construct, facilitating the rapid creation of large numbers of different phosphomutants. In addition, to allow the study of lethal mutants, a loxP-STOP-loxP system with a galactose-inducible Cre recombinase was used to allow conditional generation of the protein fusions (Figure 1B; see more detailed diagram in Figure S1B).
In pilot experiments with numerous Cdk substrates, we found that cellular levels of substrate proteins were greatly reduced, to widely varying degrees, by the integration of the fusion construct, even prior to Cre induction of the fusion (data not shown). Although we were unable to solve this problem, we could control for the effects of protein levels by analysis of additional mutants: in the case of cyclin fusions, mutation of Cdk1 consensus phosphorylation sites in the substrate should reduce the phenotype, and for phosphatase fusions, the phenotype should be reduced by mutation of the catalytic cysteine, C314, in Cdc14B.
Constitutive Phosphorylation Inhibits Eco1 Function
We used the fusion technique to address the phosphoregulation of Eco1 by Cdk1. Fusion of the phosphatase Cdc14B to Eco1 (ECO1-PPase) had no effect on cell viability, while fusion to Cln3 (ECO1-kinase) caused a severe growth defect (Figure 1C). The kinase fusion, but not the phosphatase fusion, was expressed at very low levels (data not shown). Importantly, the viability defect in ECO1-kinase cells was reduced by mutation of the four Cdk1 consensus phosphorylation sites in Eco1 (ECO1-4A-kinase), implying that the defect was due in part to the phosphorylation state of the fused substrate and not simply to nonspecific disruption of Eco1 function or abundance.
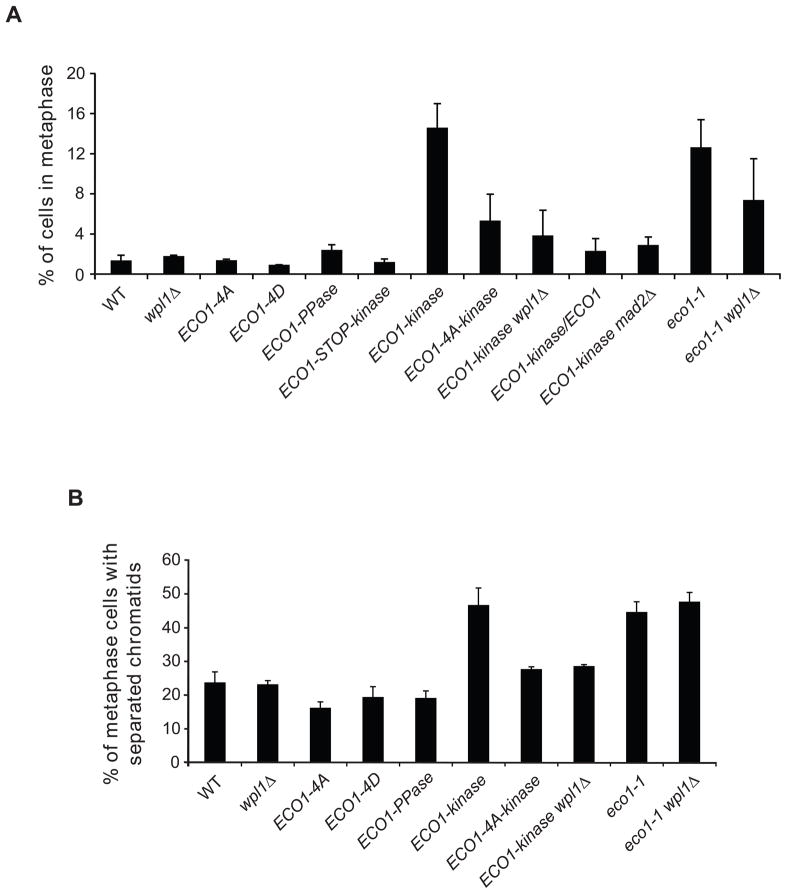
The growth defect of the ECO1-kinase strain was also reduced by deletion of WPL1, mutation of which is known to rescue loss-of-function mutants such as the temperature-sensitive mutant eco1-G211D (eco1-1, Figure 1C) (Rolef Ben-Shahar et al., 2008). This result, along with the observation that the kinase fusion is heterozygous recessive (Figure 1C), suggested that the constitutively phosphorylated form of Eco1 is inactive. Indeed, ECO1-kinase cells phenocopied the metaphase arrest seen in eco1-1 cells (Figure 2A). This metaphase accumulation was reduced by mutation of the four Cdk1 sites in Eco1, wpl1Δ, or heterozygosity (Figure 2A). The arrest was due to activation of the spindle checkpoint, as ECO1-kinase mad2Δ double mutants lacking a functional checkpoint did not arrest and were synthetically sick (Figures 1C, 2A), indicative of a cohesion loss mutant (Skibbens et al., 1999).
Figure 2. Phosphorylation inhibits the cohesion establishment function of Eco1.
(A) Cells of the indicated genotype were grown asynchronously at room temperature and examined by immunofluorescence microscopy, staining for tubulin and DAPI. Cells were scored for cell cycle stage, defining a metaphase cell as large-budded with a single DNA mass and short bipolar spindles. Over 200 cells were counted in two independent experiments; error bars represent the standard error of the mean (SEM).
(B) Strains containing a GFP-tagged URA3 locus were arrested in metaphase at 30°C using nocodazole, and scored for one GFP focus (maintenance of cohesion) or two foci (loss of cohesion). Over 200 metaphase-arrested cells were counted, and an average was calculated from two or three independent experiments; error bars represent SEM.
To directly test the effect of phosphorylation on Eco1 cohesion establishment activity, we performed sister chromatid cohesion assays using a ura3::lacO256, GFP-LacI strain, which contains a GFP dot at a specific chromosomal locus. In a metaphase arrest, properly joined sister chromatids display a single GFP focus, while loss of cohesion results in two separate foci. ECO1-kinase cells showed a marked loss of cohesion, to the same extent as eco1-1 cells at the restrictive temperature (Figure 2B). This effect was rescued by mutation of the four phosphorylation sites or deletion of WPL1. Interestingly, wpl1Δ did not rescue the loss of cohesion in eco1-1 cells at the restrictive temperature (Figure 2B), as seen in previous studies with eco1Δ cells (Rolef Ben-Shahar et al., 2008; Rowland et al., 2009; Sutani et al., 2009), perhaps indicating that WPL1 deletion does not fully compensate for a severe loss of ECO1 function, and that the kinase fusion retains a low level of activity. Consistent with this possibility, deletion of WPL1 rescued the metaphase arrest in eco1-1 cells at a permissive temperature (Figure 2A), where Eco1-1 possesses a small amount of acetyltransferase activity (Rowland et al., 2009).
All ECO1-kinase phenotypes were absent in the ECO1-4D mutant, demonstrating the usefulness of the fusion technique for uncovering effects that cannot be detected using traditional phosphomimics.
Cdk1 Phosphorylates Eco1 in vivo
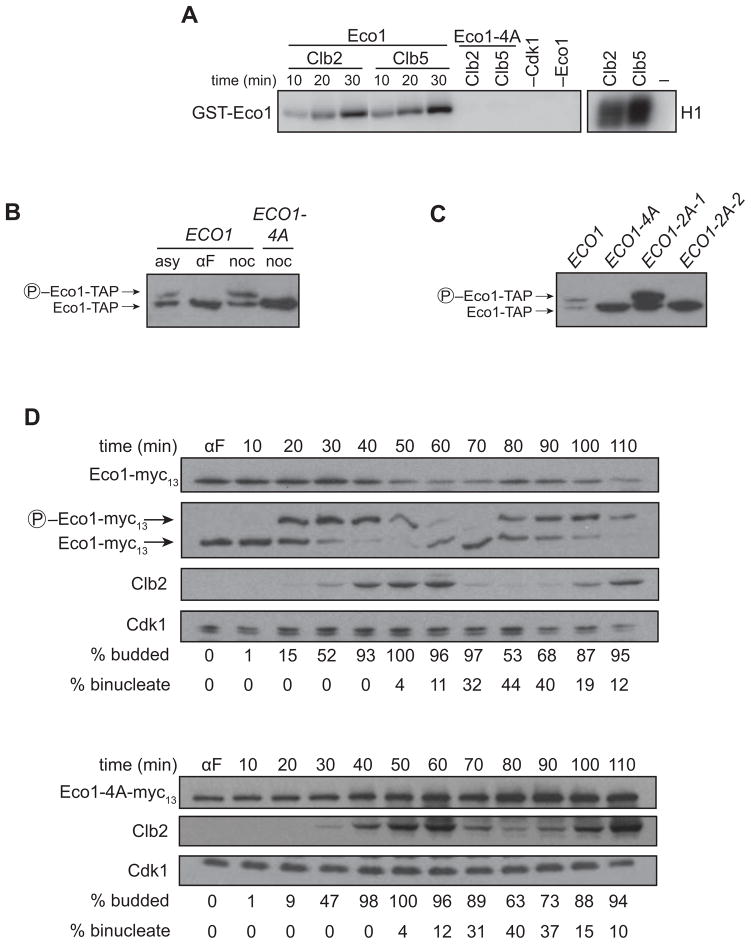
Our results suggest that Eco1 function in the cell is inhibited by phosphorylation at one or more of its four Cdk1 consensus sites. We next confirmed that these sites are targets of Cdk1 in vitro and in vivo. As in our previous work (Ubersax et al., 2003; Loog and Morgan, 2005), we found that purified Eco1 is phosphorylated in vitro by purified complexes of Cdk1 with the S-phase cyclin Clb5 or the M-phase cyclin Clb2 (Figure 3A). This phosphorylation was abolished by the 4A mutations, indicating that Cdk1 phosphorylates one or more of these sites in vitro.
Figure 3. Phosphorylation in late S phase reduces Eco1 levels.
(A) GST-Eco1 or GST-Eco1-4A, purified from bacteria, was incubated for the indicated times with purified Clb2-Cdk1 or Clb5-Cdk1 in the presence of γ-32P-ATP, and analyzed by SDS-PAGE and autoradiography. Negative controls lacking phosphorylation sites (Eco1-4A), enzyme (−Cdk1), or substrate (−Eco1) were reacted for 30 min. Right panel shows activity toward the general substrate histone H1.
(B) Lysates from ECO1-TAP and ECO1-4A-TAP cells growing asynchronously (asy) or arrested in G1 with alpha factor (αF) or in mitosis with nocodazole (noc) were analyzed on an SDS-PAGE gel containing Phos-tag reagent to retard the mobility of phosphorylated protein, followed by western blotting for the TAP tag.
(C) Lysates from the indicated cells arrested in mitosis with nocodazole were analyzed on an SDS-PAGE gel containing Phos-tag reagent, followed by western blotting for the TAP tag.
(D) ECO1-myc13 and ECO1-4A-myc13 cells were arrested with alpha factor (αF), washed and released. Samples were removed at the indicated times for western blot analysis. Wild-type Eco1 was also analyzed on a separate gel containing Phos-tag reagent. Progression through the cell cycle was monitored by western blotting for Clb2, as well as quantification of budding (entry into S phase) and anaphase binucleate formation (staining DNA with DAPI). Cdk1 was used as a gel loading control.
Eco1 was phosphorylated at equivalent rates by Clb2-Cdk1 and Clb5-Cdk1, after normalization for activity toward a general substrate, histone H1 (Figure 3A). Because the intrinsic activity of Clb5-Cdk1 is ten-fold lower than that of Clb2-Cdk1 (Loog and Morgan, 2005), normalization for general activity requires the use of at least ten-fold more Clb5-Cdk1 protein. Thus, Eco1 behaves like a general Cdk1 substrate that is expected to be poorly phosphorylated in S phase by Clb5-Cdk1 and more effectively phosphorylated by Clb2-Cdk1 in mitosis.
We next confirmed that the Cdk1 sites on Eco1 are phosphorylated in vivo. We found that Eco1 from metaphase cells, but not from G1 cells, displayed a mobility shift on an SDS-PAGE gel supplemented with a phosphate-binding reagent (Figure 3B) (Kinoshita et al., 2006). This shift was abolished in the ECO1-4A mutant, demonstrating that Eco1 in mitotic cells is phosphorylated at one or more of the Cdk1 consensus sites.
Studies of additional point mutants revealed that the phosphate-dependent gel mobility shift was not affected by mutation to alanine of the first two Cdk1 sites in the Eco1 sequence (ECO1-2A-1; see Figure S1A) but was abolished by mutation of the nearby third and fourth sites (ECO1-2A-2) (Figure 3C). Thus, the mobility shift is caused by phosphorylation at the third and/or fourth sites. Given that phosphorylation does not always result in mobility shifts, these results do not rule out phosphorylation at the first two sites; indeed, results discussed below clearly support phosphorylation at these sites.
Phosphorylation Promotes Eco1 Degradation
We noticed that the steady-state levels of Eco1-4A and 2A proteins in metaphase were higher than those of the wild-type protein (Figures 3B, C), suggesting that phosphorylation causes a decrease in Eco1 abundance. If so, Eco1 levels should drop at cell cycle stages where Cdk1 activity is high. Indeed, there is previous evidence that Eco1 levels oscillate in the cell cycle (Toth et al., 1999; Borges et al., 2010). To address whether these oscillations are due to phosphorylation by Cdk1, we measured Eco1 and Eco1-4A levels in cells progressing synchronously through the cell cycle (Figure 3D). Upon release from a G1 arrest, levels of wild-type Eco1 rose slightly at the G1/S transition and dropped in late S phase through mitosis. Entry into S phase was accompanied by partial phosphorylation, which became more complete as Clb2 levels rose just before mitosis. This timing of phosphorylation is consistent with the biochemical data above (Figure 3A). Levels of Eco1 declined as peak mitotic Clb2 levels were reached. Eco1-4A levels did not oscillate but simply accumulated throughout the time course, suggesting that phosphorylation by Clb2-Cdk1 promotes the decrease in Eco1 levels after S phase.
ECO1 mRNA levels oscillate slightly during the cell cycle, peaking in early S phase (Spellman et al., 1998). It was therefore possible that oscillations in Eco1 protein levels are due in part to variations in transcription rates. However, cells in which the ECO1 promoter was replaced with that of the constitutively-transcribed CYC1 gene still showed a difference in protein levels between asynchronous and metaphase-arrested cultures, and this difference depended on the presence of Cdk1 sites (Figure S2). Thus, cell cycle variation in Eco1 levels depends primarily on post-transcriptional mechanisms.
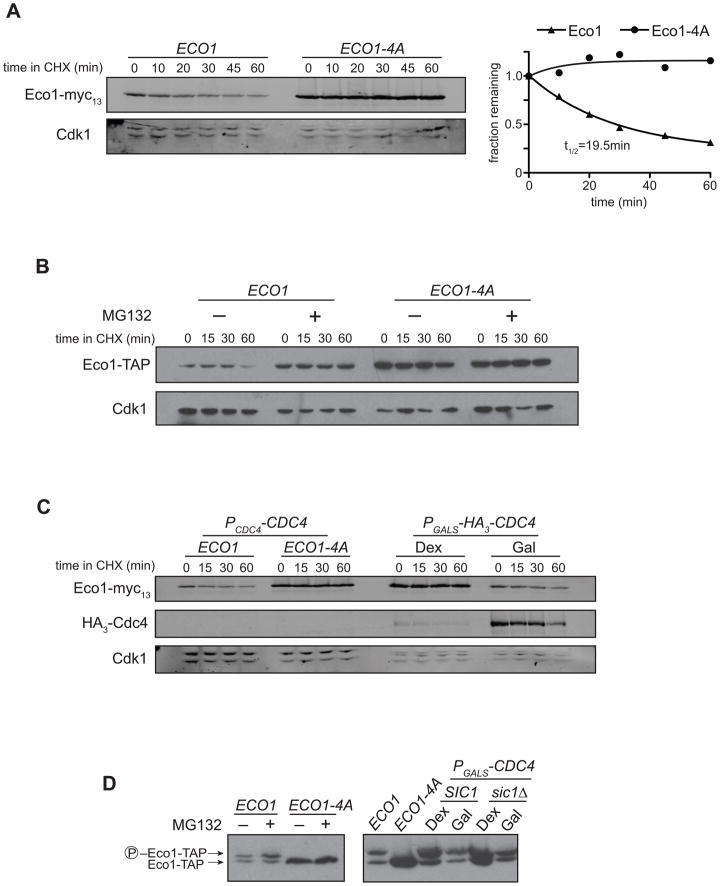
We next tested whether phosphorylation affects Eco1 stability. We measured the half-life of Eco1 in metaphase-arrested cells, using cycloheximide to inhibit new protein synthesis (Figure 4A). Wild-type Eco1 was rapidly degraded in metaphase-arrested cells, with a half-life of about 20 minutes. Eco1-4A was greatly stabilized, demonstrating that degradation depends on phosphorylation by Cdk1. Inhibition of the proteasome also stabilized Eco1 (Figure 4B), implying that phosphorylated Eco1 is targeted for destruction by the proteasome.
Figure 4. Eco1 is targeted for proteasomal destruction by SCFCdc4-mediated ubiquitination.
(A) ECO1-myc13 and ECO1-4A-myc13 strains were arrested in nocodazole for 3 h, followed by addition of 100 μg/ml cycloheximide (CHX). Lysates were analyzed by western blotting using fluorophore-conjugated secondary antibodies, and the relative amount of protein at each timepoint was quantified and plotted at right.
(B) ECO1-TAP pdr5Δ and ECO1-4A-TAP pdr5Δ strains were arrested in nocodazole and treated with 100 μM MG132 (+) or DMSO (−) for 1 h before addition of 100 μg/ml cycloheximide. Samples were analyzed by standard western blotting.
(C) ECO1-myc13 strains in which the endogenous CDC4 was replaced with PGALS-HA3-CDC4 were arrested in nocodazole and then supplemented with 2% dextrose or galactose to repress or activate CDC4 expression, respectively. Cycloheximide was then added for the indicated time prior to sample removal for western blot analysis using fluorescent secondary antibodies.
(D) Lysates of nocodazole-arrested cells treated as in panels B and C were analyzed by Phos-tag-supplemented SDS-PAGE and western blotting.
Based on our kinase assays in vitro (Figure 3A), together with the correlation between Clb2 levels and the decline in Eco1 levels during the cell cycle (Figure 3D), we propose that the primary regulator of Eco1 abundance is the Clb2-Cdk1 complex (and perhaps complexes of Cdk1 with the other late cyclins Clb1, 3, and 4). Our results with Eco1-2A mutants further suggest that Eco1 must be phosphorylated at multiple sites to be degraded. Interestingly, gel mobility shifts in our cell cycle experiments indicate that Eco1 is partially phosphorylated starting at the beginning of S phase, long before it is degraded. It seems likely that this partial phosphorylation is catalyzed by the relatively low activity of Clb5-Cdk1 and is not sufficient for degradation. Consistent with this possibility, the Eco1-2A-1 mutant is stabilized despite undergoing a mobility shift (Figure 3C), indicating that phosphorylation of the third and/or fourth Cdk1 sites causes a mobility shift without causing degradation. The degradation of Eco1 therefore resembles that of the Cdk inhibitor Sic1, which is degraded only when phosphorylated at multiple sites (Nash et al., 2001).
Eco1 Destruction Depends on SCFCdc4
The interplay of phosphorylation and ubiquitin-mediated degradation is a common theme in cell cycle regulation. For instance, many Cdk1 substrates are also substrates of the ubiquitin ligase SCF, a modular complex that employs F-box adaptor subunits to target different substrates for ubiquitination (Willems et al., 2004). The only F-box protein essential for cell cycle regulation in budding yeast is Cdc4, which is required for the ubiquitination of the Cdk1 inhibitors Sic1 and Far1, the DNA replication factor Cdc6, and the transcription factor Swi5 (Jonkers and Rep, 2009). Although SCFCdc4 activity is constant throughout the cell cycle, it is only known to recognize phosphorylated substrates, allowing protein destruction to be coordinated with cell cycle events.
Because Eco1 degradation requires both phosphorylation and proteasome activity (Figure 4A, 4B), we hypothesized that Eco1 is an SCF substrate. We initially attempted to measure Eco1 half-life in temperature-sensitive SCF mutants, but found that Eco1 is intrinsically thermolabile independent of its phosphorylation state (Figure S3). We therefore constructed a strain in which CDC4 was placed under the control of the dextrose-repressible GALS promoter, allowing us to conditionally turn off SCFCdc4 activity. When we arrested cells in metaphase and shut off CDC4 expression, Eco1 was stabilized to a similar degree as Eco1-4A (Figure 4C). Phosphorylated Eco1 accumulated in the presence of MG132 and in a CDC4 shutoff (Figure 4D), consistent with the model of phosphorylation-mediated degradation. The increased abundance of Eco1 in the absence of Cdc4 was not due to an indirect effect caused by stabilization of the Cdk1 inhibitor Sic1, as Eco1 was equally stabilized and phosphorylated when CDC4 was shut off in a sic1Δ background (Figure 4D). Our earlier finding that degradation requires multiple phosphorylation sites (Figure 3C) is also consistent with Eco1 being a substrate of SCFCdc4, as recognition of many Cdc4 substrates requires multiple phosphates (Nash et al., 2001; Hao et al., 2007).
Nuclear Localization of Eco1 Enhances Its Degradation
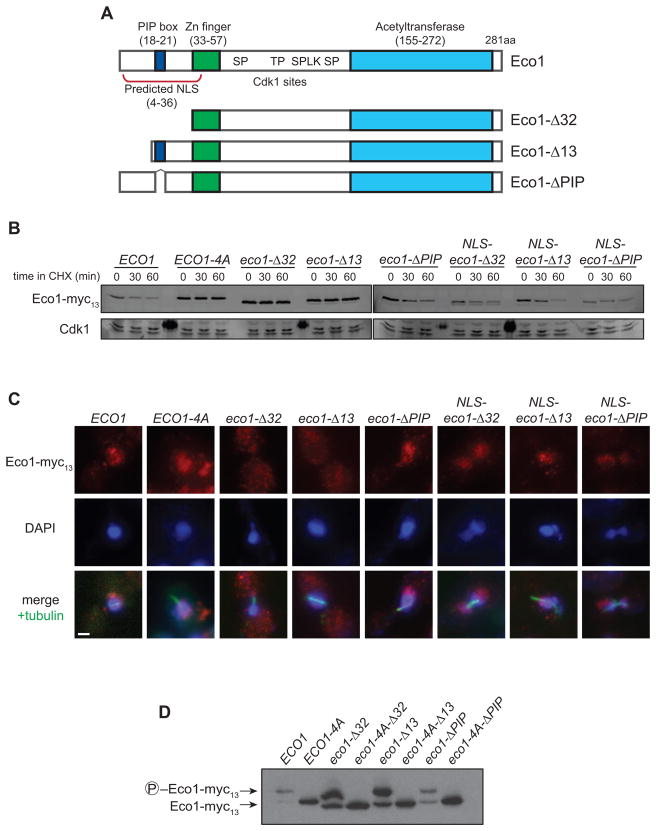
Eco1 is known to interact during S phase with the DNA replication clamp PCNA, and this interaction depends on a PCNA-interacting motif (‘PIP box’, QxKL; Figure S1A) near the N-terminus of Eco1 (Moldovan et al., 2006). Deletion of the N-terminal 32 residues (eco1-Δ32, Figure 5A), including the PIP box, was previously shown to abolish the PCNA interaction and cause an increase in Eco1 levels (Moldovan et al., 2006). To investigate further the role of the N-terminal region in protein stability, we tested the effects of a series of mutations in this region. Mutations of the N-terminus are lethal (Figure S4) (Moldovan et al., 2006), so experiments were performed in a suppressing wpl1Δ background, which does not affect Eco1 stability or localization (data not shown). Deletion of the N-terminal 32 residues resulted in complete protein stabilization, consistent with the previous study, while deletion of just the PIP box (eco1-ΔPIP) conferred partial stabilization (Figure 5B).
Figure 5. Eco1 contains a functional NLS that is necessary for its degradation.
(A) Diagram of known Eco1 protein domains; numbers indicate amino acid position. See Figure S1A for sequence of N-terminus.
(B) ECO1-myc13 strains containing the indicated N-terminal mutations were arrested in metaphase with nocodazole. After addition of cycloheximide (CHX) for the indicated times, lysates were analyzed by western blotting using fluorophore-conjugated secondary antibodies. All N-terminal truncations are wpl1Δ. ‘NLS’ indicates that the SV40 Large T antigen nuclear localization signal (PKKKRKVG) was inserted immediately after the start codon. Dark spots on the Cdk1 loading control are due to fluorescence of a molecular weight marker.
(C) Asynchronous ECO1-myc13 cultures were analyzed by immunofluorescence microscopy using antibodies to Myc and alpha-tubulin, and stained for DNA using DAPI. Images from mitotic cells are shown (based on cell and spindle morphology, and DNA mass separation), and are representative of all cell cycle stages. Bar = 1 μm.
(D) Lysates of nocodazole-arrested cells were analyzed by Phos-tag-containing SDS-PAGE and western blotting.
We wondered what other features of the N-terminus might be contributing to degradation. Using a recently developed method (Kosugi et al., 2009), we found that residues 4-36 of Eco1 contain a potential bipartite nuclear localization signal (NLS; Figure 5A, S1A). To verify this, we deleted the first 13 residues of Eco1, comprising half of the predicted NLS (eco1-Δ13), and examined its subcellular localization. While Eco1, Eco1-4A and Eco1-ΔPIP all appeared constitutively nuclear, Eco1-Δ32 and Eco1-Δ13 were present in both the cytoplasm and nucleus (Figure 5C). This localization defect was rescued by addition of an ectopic NLS (from the SV40 Large T antigen) to the N-terminus of these mutants (NLS-eco1-Δ32 and NLS-eco1-Δ13; Figure 5C). The ectopic NLS also rescued the lethality of the eco1-Δ13 mutation (Figure S4), demonstrating that Eco1 nuclear localization is essential for viability.
Deletion of just the NLS (the Eco1-Δ13 mutant) stabilized the protein to the same extent as Eco1-Δ32 (Figure 5B). Attachment of the SV40 NLS to Eco1-Δ13 and Eco1-Δ32 restored the normal rapid rate of degradation (Figure 5B), suggesting that the stability of these mutants is due entirely to their localization defect. We also tested whether deletion of the PIP box, which contains basic residues within the NLS, has a subtle effect on localization despite appearing mostly nuclear. Addition of an ectopic NLS to Eco1-ΔPIP decreased its stability (Figure 5B), implying that this mutant does have a slight localization defect and that binding to PCNA is not contributing significantly to Eco1 destruction. Both eco1-Δ32 and eco1-ΔPIP mutants are inviable (Figure S4) and are not rescued by forcing nuclear localization with the SV40 NLS. This shows that PCNA binding is essential beyond the contribution of the PIP box residues to subcellular localization, as suggested previously (Moldovan et al., 2006).
The dependence of Eco1 degradation on nuclear localization is consistent with Eco1 being a substrate of SCFCdc4, as Cdc4 is entirely nuclear (Blondel et al., 2000; Pries et al., 2002). Consistent with this, all N-terminal mutants displayed a gel mobility shift in a metaphase arrest, and this shift depended on their Cdk1 sites (Figure 5D). Thus, the stabilization conferred by these mutants is downstream of Cdk1 phosphorylation, likely at the level of SCF recognition.
Eco1 Degradation Prevents Post-Replication Cohesion Establishment
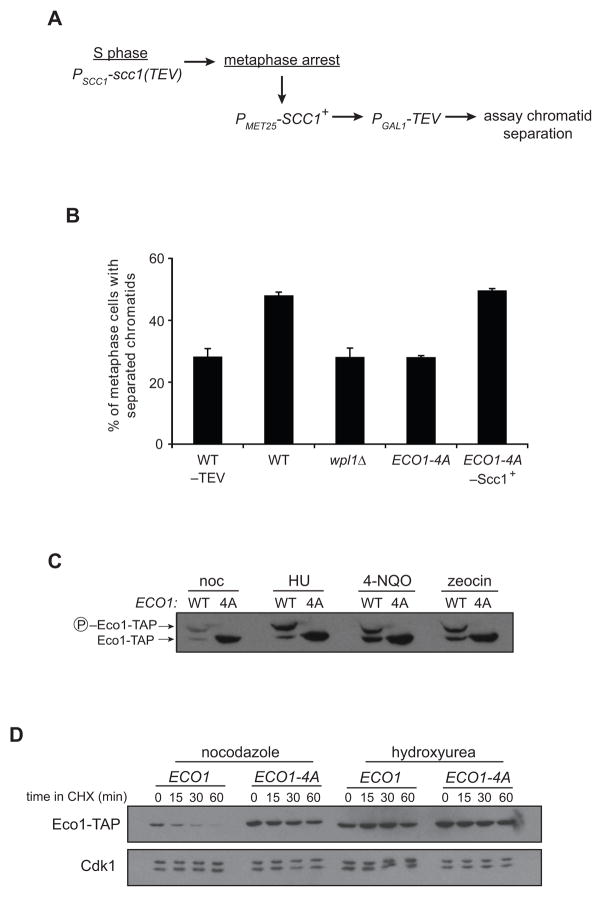
We hypothesized that the degradation of Eco1 upon Cdk1 phosphorylation provides an explanation for previous evidence that limited Eco1 activity prevents cohesion establishment after S phase (Unal et al., 2007). Cells expressing the nonphosphorylatable Eco1 mutant should therefore be competent to establish cohesion even after DNA replication. To test this, we assayed cohesion establishment in metaphase-arrested cells by modifying a previously published approach (Figure 6A) (Unal et al., 2007) to avoid the use of high temperatures, due to Eco1 thermolability (Figure S3). In this assay, cells establish cohesion in S phase with a version of Scc1 containing a TEV protease recognition site (Scc1-TEV) and are then arrested in metaphase with nocodazole. Expression of wild-type Scc1 (without a TEV site) is then induced, followed by induction of TEV protease to dissolve pre-established S phase cohesion, leaving only the metaphase-expressed cohesin to hold sister chromatids together. The readout for establishment of cohesion is the same as that used in Figure 2B: GFP-tagged chromosomes appearing as one or two foci.
Figure 6. Unphosphorylated Eco1 can establish cohesion after S phase, and Eco1 is stabilized in the presence of DNA damage.
(A) Experimental setup for measurement of cohesion establishment in metaphase. Cohesion in these strains is established normally in S phase with Scc1(TEV). Cultures are arrested in metaphase, wild-type Scc1 is induced for 1 h, TEV is induced for 2 h, and cohesion of GFP-tagged chromatids is analyzed by microscopy.
(B) Cells were treated as indicated in panel A, and separation of sister chromatids was measured in over 200 metaphase cells in each of two experiments; bars indicate mean +/− SEM. Controls were included in which S-phase cohesion was not cleaved (−TEV) or wild-type Scc1 was not induced in metaphase (−Scc1+). The difference between WT and ECO1-4A is statistically significant (P=0.004); for all other differences, P≤0.02.
(C) Cultures of ECO1-TAP (WT) or ECO1-4A-TAP (4A) were treated with nocodazole (noc), hydroxyurea (HU), 4-nitroquinoline 1-oxide (4-NQO), or zeocin for 3 h. Lysates were analyzed by western blotting, using Phos-tag reagent to measure phosphorylation state.
(D) ECO1-TAP and ECO1-4A-TAP strains were arrested by treatment with nocodazole or hydroxyurea, and Eco1 half-life was analyzed by addition of CHX.
In otherwise wild-type cells, which cannot establish new cohesion in metaphase, induction of TEV resulted in an increased frequency of chromatid separation (Figure 6B). Strikingly, we found that cells expressing Eco1-4A were able to establish cohesion in a metaphase-arrested cell, to the same extent as a wpl1Δ positive control (Heidinger-Pauli et al., 2009) and dependent on the expression of metaphase cohesin. These results demonstrate that Cdk1 activity towards Eco1 helps limit the establishment of cohesion after replication.
DNA Damage Stabilizes Eco1
Double-strand DNA breaks are known to trigger cohesion establishment outside of S phase (Strom et al., 2004; Strom et al., 2007; Unal et al., 2007). Given that the reactivation of cohesion establishment after damage depends on Eco1 (Unal et al., 2007), we wondered whether DNA damage affects Eco1 stability. Indeed, Eco1 levels were markedly increased in cells treated with the DNA damaging agents 4-nitroquinoline 1-oxide (4-NQO, a UV mimetic), zeocin (which generates DNA breaks), or the ribonucleotide reductase inhibitor hydroxyurea (HU, which induces replication stress) (Figure 6C). Analysis of Eco1 half-life confirmed that Eco1 was dramatically stabilized in HU-arrested cells (Figure 6D).
This stabilization occurred despite the fact that DNA damage triggers a mitotic arrest with high Clb2-Cdk1 activity. Indeed, we found that Eco1 was extensively phosphorylated at Cdk1 sites in damage arrests (Figure 6C). Thus, Eco1 is stabilized after DNA damage despite being phosphorylated at the destabilizing Cdk1 sites. We conclude that upon DNA damage, ubiquitination of phospho-Eco1 by SCF is somehow prevented, thereby allowing the establishment of new, replication-independent cohesion.
Excess Cohesion in ECO1-4A Cells Leads to Anaphase Defects
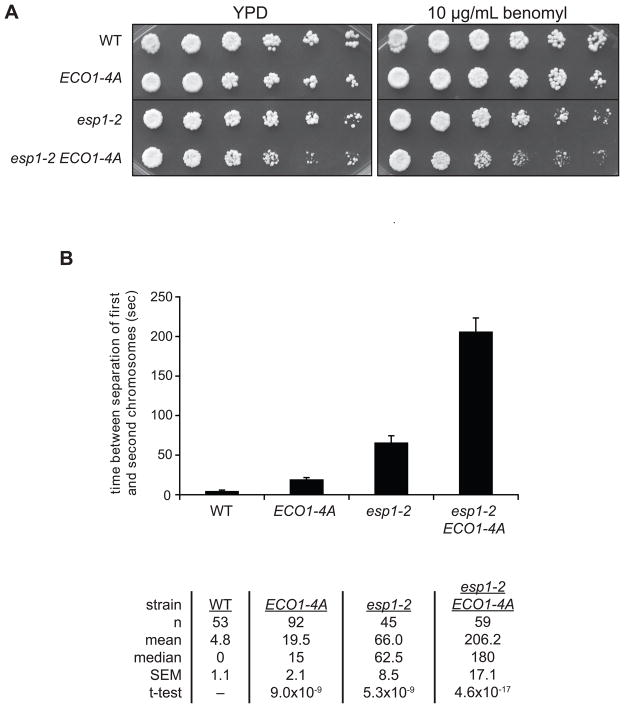
Our studies reveal a mechanism that appears to limit the establishment of excess sister-chromatid cohesion by reducing Eco1 activity after S phase, a mechanism that can also be circumvented when extra cohesion is desirable (i.e. after DNA damage). We next sought to understand the importance of this mechanism by determining whether Eco1-4A, by generating excess cohesion, causes defects in sister chromatid behavior during anaphase.
If Eco1-4A generates excess cohesion, then anaphase defects might occur in cells with a reduced ability to remove cohesin. However, we found that there is only a minor genetic interaction between the ECO1-4A mutation and a temperature-sensitive separase mutant (esp1-2) at the permissive temperature (Figure 7A). Thus, the low separase activity in these cells seems sufficient to remove any increased cohesion in ECO1-4A cells. We next hypothesized that a spindle checkpoint delay would allow ECO1-4A cells to accumulate higher levels of cohesion that are less readily removed by separase. Indeed, we observed a subtle but reproducible synthetic defect in esp1-2 ECO1-4A cells growing in the presence of the spindle poison benomyl (Figure 7A).
Figure 7. Unregulated Eco1 activity perturbs anaphase dynamics.
(A) The indicated strains were serially diluted in 3-fold increments onto YPD or YPD plus 10 μg/ml benomyl at room temperature.
(B) Strains containing GFP-tagged TRP1 and URA3 loci on chromosomes IV and V, respectively, were grown asynchronously at room temperature, affixed to slides, and live single-cell microscopy movies were taken to monitor sister chromatid separation in cells passing through mitosis. Times between the separation of the two chromosomes were measured. Bars indicate mean +/− SEM. The table lists the values for each parameter. See Figure S5 for details.
Although ECO1-4A cells appear normal in crude assessments of colony growth, we wanted to use a more refined approach to test the possibility that these cells exhibit anaphase defects that do not result in gross viability problems. Our approach was to use a single-cell microscopy assay to measure the synchrony of sister chromatid separation (Holt et al., 2008). This method employs a yeast strain with lacO arrays at two loci, TRP1 and URA3, 12 and 35 kilobases from the centromeres of chromosomes IV and V, respectively (note that this is a different strain from that used in our previous studies; see supplemental experimental procedures for details). Expression of LacI-GFP in these cells generates GFP foci on two different chromosomes, and the time between the separation of the two chromosomes provides an indication of the synchrony of sister chromatid separation. Our previous work indicated that this method provides a useful measure of the rates of separase activation and cohesin cleavage (Holt et al., 2008).
In a wild-type background, the two GFP foci separated almost simultaneously during anaphase, with a mean time of about 5 seconds between chromatid separation events (Figures 7B, S5). A hypomorphic mutant of separase (esp1-2) increased the mean time between separations by over an order of magnitude (Figure 7B, S5), presumably because a decreased rate of cohesin cleavage prolongs anaphase, resulting in greater differences in the time at which each sister pair is separated.
Most importantly, ECO1-4A cells showed a significant four-fold increase in the average time between chromatid separation events (Figure 7B). This increased average resulted from a skewing of the distribution of separation times towards longer intervals (Figure S5). We observed many ECO1-4A cells in which the second chromosome separated well over 40 seconds after the first, a phenomenon that was also seen in esp1-2 but never in wild-type cells. These data are consistent with the idea that unregulated Eco1 activity leads to excess cohesion, which delays but does not completely disrupt anaphase progression.
A combination of ECO1-4A and esp1-2 mutations resulted in a severe loss of anaphase synchrony, with chromosome separation times on the scale of minutes (Figures 7B, S5). We observed many ECO1-4A esp1-2 cells in which neither chromosome had separated but both were repeatedly pulled back and forth between poles, possibly reflecting the initiation of late anaphase spindle dynamics prior to full cleavage of cohesin. Thus, despite the near-normal growth of ECO1-4A esp1-2 cells on plates (Figure 7A), the timing and robustness of sister chromatid separation is clearly compromised, illustrating that proper cohesion regulation is important for the precise coordination of anaphase events.
DISCUSSION
The cycle of sister chromatid cohesion is tightly woven into the fabric of the cell division cycle. In this study we demonstrate the direct regulation of cohesion establishment by the master cell cycle regulator Cdk1. Our results suggest that Clb2-Cdk1-dependent phosphorylation of Eco1 from late S phase to mitosis targets nuclear Eco1 for ubiquitination by SCFCdc4 and subsequent degradation by the proteasome. This reduces the amount of Eco1 acetyltransferase activity below some threshold required to establish cohesion, which, together with reduced ECO1 transcription in G1 (Spellman et al., 1998), prevents the establishment of new cohesion until DNA replication occurs in S phase of the next cell cycle. In cells containing a nonphosphorylatable Eco1, or upon activation of the DNA damage response, Eco1 is stabilized and cohesion can be established in metaphase.
The regulation of cohesion establishment, like that of all critical processes, is a finely tuned balance of opposing positive and negative influences. In this case, the acetyltransferase activity of Eco1 during DNA replication facilitates the establishment of sister chromatid cohesion. In metaphase, however, Cdk1 kinase activity inhibits Eco1, allowing the anti-establishment activities of Wpl1 and Pds5 to predominate, thus preventing establishment after replication. By modulating the positive input (establishment), Cdk1 helps generate a level of chromosomal cohesion that is optimal for efficient sister chromatid separation in anaphase.
We uncovered the regulation of Eco1 by Cdk1 using a new method of creating constitutively phosphorylated mutants. Linking Cdk1 to Eco1 (via Cln3) inhibited Eco1 function, greatly reducing cell viability. Significantly, ECO1-4D did not mimic the phosphorylated state, revealing the usefulness of the fusion technique for detecting phenotypes that are not seen with traditional phosphomimics. The effect of Cdk1 phosphorylation on the majority of its substrates is unknown; the technique presented here, combined with previously created yeast library strains (Ghaemmaghami et al., 2003) and the recent identification of hundreds of Cdk1 substrates (Ubersax et al., 2003; Holt et al., 2009), could facilitate a better understanding of the role of Cdk1 in regulating cell cycle events.
Eco1 undergoes an interesting pattern of phosphorylation by Cdk1 over the cell cycle (Figure 3D). Given that Eco1 is not a Clb5-specific substrate (Figure 3A) (Loog and Morgan, 2005), the phosphorylation seen in early S phase is likely to be incomplete. This partial phosphorylation might not be sufficient for degradation, given that Cdc4 only has appreciable affinity for multiply phosphorylated substrates. Instead, early phosphorylation by Clb5-Cdk1 may prime Eco1 to be rapidly phosphorylated when Clb2 activity rises in late S phase. It is also possible that phosphorylated Eco1 exists in S phase cells because some other mechanism prevents its degradation at that stage of the cell cycle.
In a cell experiencing double-strand DNA breaks (DSBs), new cohesion aids DNA repair and is established by the reactivation of Eco1 (Sjogren and Nasmyth, 2001; Strom et al., 2004; Unal et al., 2004; Unal et al., 2007). This appears to occur by two separate mechanisms: the stabilization of Eco1 (Figure 6D) and the phosphorylation of Scc1 by Chk1 (Heidinger-Pauli et al., 2008). Either mechanism is sufficient to establish cohesion in metaphase independent of DNA damage signaling (Figure 6B) (Heidinger-Pauli et al., 2009). Once the mechanism of Eco1 stabilization in damage is elucidated, it will be possible to test whether stabilization, like Scc1 phosphorylation (Heidinger-Pauli et al., 2008), is necessary for establishment of damage-induced cohesion. Interestingly, we found that Eco1 is stabilized not only after induction of DSBs, but also following inhibition of DNA replication with hydroxyurea (Figure 6D). Perhaps Eco1 stabilization facilitates cohesion establishment upon removal of replication stress.
The substrate of Eco1 during S phase is the Smc3 subunit of cohesin, whereas damage-induced Eco1 activity is thought to target the Scc1 subunit (Heidinger-Pauli et al., 2009). It is likely that Eco1-4A also targets Scc1, since the metaphase cohesion that results from ECO1 overexpression depends on lysines in Scc1, not Smc3 (Heidinger-Pauli et al., 2009), and does not promote excess Smc3 acetylation (Borges et al., 2010). Mechanisms therefore exist to focus Eco1 activity on Smc3 in S phase and on Scc1 thereafter. The molecular basis of this selectivity is not known, although it seems likely that other factors, such as the replication fork or mitotic chromatin structure, influence the choice of targets. Structural changes in cohesin or cohesin-associated factors throughout the cell cycle are also possible. Interestingly, acetylation of Smc3 in S phase can occur even when Eco1 levels are severely reduced (Breslow et al., 2008) or when replication is delayed until late S phase (Beckouet et al., 2010), possibly due to a high local concentration of Eco1 at the replication fork. Scc1 acetylation, on the other hand, is more sensitive to enzyme concentration, as the degradation promoted by Cdk1 in mitosis is sufficient to prevent cohesion establishment at this stage.
Given that excess cohesion establishment activity is not grossly detrimental to yeast cell viability, why has an intricate system evolved to inhibit it? Since cells establish new genome-wide cohesion upon DNA damage, the cohesion removal system has likely developed a tolerance to some excess cohesion. This could be achieved by an excess of separase activity, for example. Indeed, we found that a reduction in separase activity (the esp1-2 mutation) had little effect on the viability of ECO1-4A cells (Figure 7A). However, we did observe defects in ECO1-4A esp1-2 cells delayed in mitosis by the spindle checkpoint, suggesting that separase activity becomes limiting when ECO1-4A cells accumulate too much cohesion during a metaphase delay. Moreover, we observed a loss of sister separation synchrony in ECO1-4A cells that was greatly enhanced when ECO1-4A was combined with a separase mutation. Yeast cells have therefore evolved both an excess of separase activity and a mechanism to temporally inhibit cohesion establishment to ensure robust and efficient cohesion removal at the onset of anaphase.
The regulatory mechanism we uncovered might be conserved in other eukaryotes. Vertebrate homologs of Eco1 are phosphorylated during mitosis (Hou and Zou, 2005; Lafont et al., 2010; Olsen et al., 2010). The positions of Cdk1 consensus sites are not precisely conserved beyond the most closely related yeasts, but multiple Cdk1 motifs are present at some location in the N-terminus of most homologs. Given that the phosphorylation of Eco1 seems to serve only as an interaction motif for Cdc4, the exact position of the Cdk1 sites would be under less selective pressure to remain fixed through evolution (Holt et al., 2009). If the regulation of cohesion establishment in the cell cycle is conserved, it would be interesting to study the effect of an Eco1 phosphomutant in organisms that use a prophase pathway to remove cohesion from chromosome arms prior to anaphase (Sumara et al., 2000), as these cells may be more sensitive to persistent establishment activity. Additionally, because mutations in human Eco1 homologs have been implicated in certain cancers (Ryu et al., 2007; Luedeke et al., 2009), it will also be important to pursue the intriguing possibility that defects in Eco1 regulation generate chromosomal instability and thereby facilitate tumor evolution.
EXPERIMENTAL PROCEDURES
General methods
Yeast strains were derivatives of S288C and are listed in Table S1, along with a list of plasmid constructs. Genetic manipulations were performed using standard techniques (Longtine et al., 1998). Construction of kinase and phosphatase fusion proteins is described in Figure S1B. Cell cycle arrests were done with 20 μg/ml nocodazole, 10 μg/ml alpha factor, 200 mM hydroxyurea, 2 μM 4-nitroquinoline 1-oxide, or 33 μg/ml zeocin for 3 h at 30 C unless otherwise stated. All arrests were confirmed by flow cytometric analysis of DNA content. Protein half-lives were analyzed by addition of 100 μg/ml cycloheximide. To inhibit proteasome activity, 100 μM MG132 was added to cells with a PDR5 deletion to increase retention of the drug. For the CDC4 shutoff experiment, cells were grown in YEP media containing 2% raffinose and 0.04% galactose, then treated with nocodazole for 1.5 h before the addition of 2% galactose or dextrose for 4 h to allow for complete turnover of Cdc4. To analyze Eco1 phosphorylation, resolving portions of SDS-PAGE gels were supplemented with 50 μM MnCl2 and 10 μM Phos-tag reagent (Kinoshita et al., 2006) made using standard chemical techniques, with two-fold excess ammonium persulfate and TEMED.
Post-Replication Cohesion Assay
PSCC1-scc1(TEV268)-HA3:hphNT1:PSCC1:natNT2:PMET25-GFP-SCC1, trp1::TRP1:PGAL1-NLS-myc6-TEV-NLS2, ura3::lacO256:LEU2, his3::PCUP1-1-GFP-LacI:HIS3 strains were grown in YEP media plus 2% raffinose and arrested for 3 h in nocodazole. Cells were then washed and switched to synthetic media lacking methionine (plus nocodazole) for 1 h to induce wild-type Scc1, or kept in complete media to maintain Scc1 repression. Scc1(TEV) was then cleaved by the addition of 2% galactose to induce expression of TEV protease (or 2% dextrose to keep TEV repressed) for 2 h. 100 μM CuSO4 was also added to fully induce expression of GFP-LacI.
Anaphase Synchrony Assay
ura3::lacO256:LEU2, trp1::lacO256:TRP1, his3::PCUP1-1-GFP-LacI:HIS3 cultures were grown in synthetic media at room temperature in the presence of 100 μM CuSO4 to mid-log phase. Cells were then affixed to slides using concanavalin A and imaged using a spinning disk confocal microscope. Images spanning about 4 microns in the z plane were taken every 2.5 s for 10–15 min, except ECO1-4A esp1-2 images, which were taken every 10 s to allow longer movies (up to 28 min) without photobleaching.
Supplementary Material
Acknowledgments
We thank L. Holt for ideas and assistance with the phosphomutant fusions, S. Foster, F. Uhlmann, and G. Yaakov for comments on the manuscript, and F. Uhlmann, K. Shokat, D. Toczyski, and S. Gasser for strains and reagents. Spinning disk confocal microscopy was performed with the valuable assistance of K. Thorn at the UCSF Nikon Imaging Center. This work was supported by funding from the National Institute of General Medical Sciences (GM069901).
Footnotes
Supplemental data include five figures, one table, supplemental experimental procedures, and supplemental references.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Beckouet F, Hu B, Roig MB, Sutani T, Komata M, Uluocak P, Katis VL, Shirahige K, Nasmyth K. An smc3 acetylation cycle is essential for establishment of sister chromatid cohesion. Mol Cell. 2010;39:689–699. doi: 10.1016/j.molcel.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel M, Galan JM, Chi Y, Lafourcade C, Longaretti C, Deshaies RJ, Peter M. Nuclear-specific degradation of Far1 is controlled by the localization of the F-box protein Cdc4. EMBO J. 2000;19:6085–6097. doi: 10.1093/emboj/19.22.6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges V, Lehane C, Lopez-Serra L, Flynn H, Skehel M, Rolef Ben-Shahar T, Uhlmann F. Hos1 deacetylates smc3 to close the cohesin acetylation cycle. Mol Cell. 2010;39:677–688. doi: 10.1016/j.molcel.2010.08.009. [DOI] [PubMed] [Google Scholar]
- Brands A, Skibbens RV. Sister chromatid cohesion role for CDC28-CDK in Saccharomyces cerevisiae. Genetics. 2008;180:7–16. doi: 10.1534/genetics.108.092288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow DK, Cameron DM, Collins SR, Schuldiner M, Stewart-Ornstein J, Newman HW, Braun S, Madhani HD, Krogan NJ, Weissman JS. A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Nat Methods. 2008;5:711–718. doi: 10.1038/nmeth.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciosk R, Shirayama M, Shevchenko A, Tanaka T, Toth A, Nasmyth K. Cohesin’s binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol Cell. 2000;5:243–254. doi: 10.1016/s1097-2765(00)80420-7. [DOI] [PubMed] [Google Scholar]
- Gerlich D, Koch B, Dupeux F, Peters JM, Ellenberg J. Live-cell imaging reveals a stable cohesin-chromatin interaction after but not before DNA replication. Curr Biol. 2006;16:1571–1578. doi: 10.1016/j.cub.2006.06.068. [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Gray CH, Good VM, Tonks NK, Barford D. The structure of the cell cycle protein Cdc14 reveals a proline-directed protein phosphatase. EMBO J. 2003;22:3524–3535. doi: 10.1093/emboj/cdg348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haering CH, Schoffnegger D, Nishino T, Helmhart W, Nasmyth K, Lowe J. Structure and stability of cohesin’s Smc1-kleisin interaction. Mol Cell. 2004;15:951–964. doi: 10.1016/j.molcel.2004.08.030. [DOI] [PubMed] [Google Scholar]
- Hao B, Oehlmann S, Sowa ME, Harper JW, Pavletich NP. Structure of a Fbw7-Skp1-Cyclin E Complex: Multisite-Phosphorylated Substrate Recognition by SCF Ubiquitin Ligases. Mol Cell. 2007;26:131–143. doi: 10.1016/j.molcel.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Heidinger-Pauli JM, Unal E, Guacci V, Koshland D. The kleisin subunit of cohesin dictates damage-induced cohesion. Mol Cell. 2008;31:47–56. doi: 10.1016/j.molcel.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Heidinger-Pauli JM, Unal E, Koshland D. Distinct targets of the Eco1 acetyltransferase modulate cohesion in S phase and in response to DNA damage. Mol Cell. 2009;34:311–321. doi: 10.1016/j.molcel.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt LJ, Krutchinsky AN, Morgan DO. Positive feedback sharpens the anaphase switch. Nature. 2008;454:353–357. doi: 10.1038/nature07050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt LJ, Tuch BB, Villen J, Johnson AD, Gygi SP, Morgan DO. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science. 2009;325:1682–1686. doi: 10.1126/science.1172867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou F, Zou H. Two human orthologues of Eco1/Ctf7 acetyltransferases are both required for proper sister-chromatid cohesion. Mol Biol Cell. 2005;16:3908–3918. doi: 10.1091/mbc.E04-12-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov D, Schleiffer A, Eisenhaber F, Mechtler K, Haering CH, Nasmyth K. Eco1 is a novel acetyltransferase that can acetylate proteins involved in cohesion. Curr Biol. 2002;12:323–328. doi: 10.1016/s0960-9822(02)00681-4. [DOI] [PubMed] [Google Scholar]
- Jonkers W, Rep M. Lessons from fungal F-box proteins. Eukaryot Cell. 2009;8:677–695. doi: 10.1128/EC.00386-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita E, Kinoshita-Kikuta E, Takiyama K, Koike T. Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol Cell Proteomics. 2006;5:749–757. doi: 10.1074/mcp.T500024-MCP200. [DOI] [PubMed] [Google Scholar]
- Kosugi S, Hasebe M, Tomita M, Yanagawa H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc Natl Acad Sci U S A. 2009;106:10171–10176. doi: 10.1073/pnas.0900604106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafont AL, Song J, Rankin S. Sororin cooperates with the acetyltransferase Eco2 to ensure DNA replication-dependent sister chromatid cohesion. Proc Natl Acad Sci U S A. 2010;107:20364–20369. doi: 10.1073/pnas.1011069107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengronne A, McIntyre J, Katou Y, Kanoh Y, Hopfner KP, Shirahige K, Uhlmann F. Establishment of sister chromatid cohesion at the S. cerevisiae replication fork. Mol Cell. 2006;23:787–799. doi: 10.1016/j.molcel.2006.08.018. [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Loog M, Morgan DO. Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature. 2005;434:104–108. doi: 10.1038/nature03329. [DOI] [PubMed] [Google Scholar]
- Luedeke M, Linnert CM, Hofer MD, Surowy HM, Rinckleb AE, Hoegel J, Kuefer R, Rubin MA, Vogel W, Maier C. Predisposition for TMPRSS2-ERG fusion in prostate cancer by variants in DNA repair genes. Cancer Epidemiol Biomarkers Prev. 2009;18:3030–3035. doi: 10.1158/1055-9965.EPI-09-0772. [DOI] [PubMed] [Google Scholar]
- Moldovan GL, Pfander B, Jentsch S. PCNA controls establishment of sister chromatid cohesion during S phase. Mol Cell. 2006;23:723–732. doi: 10.1016/j.molcel.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Morgan DO. The Cell Cycle: Principles of Control. London: New Science Press; 2007. [Google Scholar]
- Nash P, Tang X, Orlicky S, Chen Q, Gertler FB, Mendenhall MD, Sicheri F, Pawson T, Tyers M. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature. 2001;414:514–521. doi: 10.1038/35107009. [DOI] [PubMed] [Google Scholar]
- Nash R, Tokiwa G, Anand S, Erickson K, Futcher AB. The WHI1+ gene of Saccharomyces cerevisiae tethers cell division to cell size and is a cyclin homolog. EMBO J. 1988;7:4335–4346. doi: 10.1002/j.1460-2075.1988.tb03332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K, Haering CH. Cohesin: its roles and mechanisms. Annu Rev Genet. 2009;43:525–558. doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, Gnad F, Cox J, Jensen TS, Nigg EA, et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal. 2010;3:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- Onn I, Heidinger-Pauli JM, Guacci V, Unal E, Koshland DE. Sister chromatid cohesion: a simple concept with a complex reality. Annu Rev Cell Dev Biol. 2008;24:105–129. doi: 10.1146/annurev.cellbio.24.110707.175350. [DOI] [PubMed] [Google Scholar]
- Pries R, Bomeke K, Irniger S, Grundmann O, Braus GH. Amino acid-dependent Gcn4p stability regulation occurs exclusively in the yeast nucleus. Eukaryot Cell. 2002;1:663–672. doi: 10.1128/EC.1.5.663-672.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolef Ben-Shahar T, Heeger S, Lehane C, East P, Flynn H, Skehel M, Uhlmann F. Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science. 2008;321:563–566. doi: 10.1126/science.1157774. [DOI] [PubMed] [Google Scholar]
- Rowland BD, Roig MB, Nishino T, Kurze A, Uluocak P, Mishra A, Beckouet F, Underwood P, Metson J, Imre R, et al. Building sister chromatid cohesion: smc3 acetylation counteracts an antiestablishment activity. Mol Cell. 2009;33:763–774. doi: 10.1016/j.molcel.2009.02.028. [DOI] [PubMed] [Google Scholar]
- Ryu B, Kim DS, Deluca AM, Alani RM. Comprehensive expression profiling of tumor cell lines identifies molecular signatures of melanoma progression. PLoS One. 2007;2:e594. doi: 10.1371/journal.pone.0000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjogren C, Nasmyth K. Sister chromatid cohesion is required for postreplicative double-strand break repair in Saccharomyces cerevisiae. Curr Biol. 2001;11:991–995. doi: 10.1016/s0960-9822(01)00271-8. [DOI] [PubMed] [Google Scholar]
- Skibbens RV, Corson LB, Koshland D, Hieter P. Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev. 1999;13:307–319. doi: 10.1101/gad.13.3.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom L, Karlsson C, Lindroos HB, Wedahl S, Katou Y, Shirahige K, Sjogren C. Postreplicative formation of cohesion is required for repair and induced by a single DNA break. Science. 2007;317:242–245. doi: 10.1126/science.1140649. [DOI] [PubMed] [Google Scholar]
- Strom L, Lindroos HB, Shirahige K, Sjogren C. Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol Cell. 2004;16:1003–1015. doi: 10.1016/j.molcel.2004.11.026. [DOI] [PubMed] [Google Scholar]
- Sumara I, Vorlaufer E, Gieffers C, Peters BH, Peters JM. Characterization of vertebrate cohesin complexes and their regulation in prophase. J Cell Biol. 2000;151:749–762. doi: 10.1083/jcb.151.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutani T, Kawaguchi T, Kanno R, Itoh T, Shirahige K. Budding yeast Wpl1(Rad61)-Pds5 complex counteracts sister chromatid cohesion-establishing reaction. Curr Biol. 2009;19:492–497. doi: 10.1016/j.cub.2009.01.062. [DOI] [PubMed] [Google Scholar]
- Toth A, Ciosk R, Uhlmann F, Galova M, Schleiffer A, Nasmyth K. Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 1999;13:320–333. doi: 10.1101/gad.13.3.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubersax JA, Woodbury EL, Quang PN, Paraz M, Blethrow JD, Shah K, Shokat KM, Morgan DO. Targets of the cyclin-dependent kinase Cdk1. Nature. 2003;425:859–864. doi: 10.1038/nature02062. [DOI] [PubMed] [Google Scholar]
- Uhlmann F. A matter of choice: the establishment of sister chromatid cohesion. EMBO Rep. 2009;10:1095–1102. doi: 10.1038/embor.2009.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F, Nasmyth K. Cohesion between sister chromatids must be established during DNA replication. Curr Biol. 1998;8:1095–1101. doi: 10.1016/s0960-9822(98)70463-4. [DOI] [PubMed] [Google Scholar]
- Uhlmann F, Wernic D, Poupart MA, Koonin EV, Nasmyth K. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell. 2000;103:375–386. doi: 10.1016/s0092-8674(00)00130-6. [DOI] [PubMed] [Google Scholar]
- Unal E, Arbel-Eden A, Sattler U, Shroff R, Lichten M, Haber JE, Koshland D. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol Cell. 2004;16:991–1002. doi: 10.1016/j.molcel.2004.11.027. [DOI] [PubMed] [Google Scholar]
- Unal E, Heidinger-Pauli JM, Kim W, Guacci V, Onn I, Gygi SP, Koshland DE. A molecular determinant for the establishment of sister chromatid cohesion. Science. 2008;321:566–569. doi: 10.1126/science.1157880. [DOI] [PubMed] [Google Scholar]
- Unal E, Heidinger-Pauli JM, Koshland D. DNA double-strand breaks trigger genome-wide sister-chromatid cohesion through Eco1 (Ctf7) Science. 2007;317:245–248. doi: 10.1126/science.1140637. [DOI] [PubMed] [Google Scholar]
- Willems AR, Schwab M, Tyers M. A hitchhiker’s guide to the cullin ubiquitin ligases: SCF and its kin. Biochim Biophys Acta. 2004;1695:133–170. doi: 10.1016/j.bbamcr.2004.09.027. [DOI] [PubMed] [Google Scholar]
- Zhang J, Shi X, Li Y, Kim BJ, Jia J, Huang Z, Yang T, Fu X, Jung SY, Wang Y, et al. Acetylation of Smc3 by Eco1 is required for S phase sister chromatid cohesion in both human and yeast. Mol Cell. 2008;31:143–151. doi: 10.1016/j.molcel.2008.06.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.