Abstract
The design of polyvalent molecules, consisting of multiple copies of a biospecific ligand attached to a suitable scaffold, represents a promising approach to inhibit pathogens and oligomeric microbial toxins. Despite the increasing interest in structure-based drug design, few polyvalent inhibitors based on this approach have shown efficacy in vivo. Here we demonstrate the structure-based design of potent biospecific heptavalent inhibitors of anthrax lethal toxin. Specifically, we illustrate the ability to design potent polyvalent ligands by matching the pattern of binding sites on the biological target. We used a combination of experimental studies based on mutagenesis and computational docking studies to identify the binding site for an inhibitory peptide on the heptameric subunit of anthrax toxin. We developed an approach based on copper-catalyzed azide-alkyne cycloaddition (click-chemistry) to facilitate the attachment of seven copies of the inhibitory peptide to a β-cyclodextrin core via a polyethylene glycol linker of an appropriate length. The resulting heptavalent inhibitors neutralized anthrax lethal toxin both in vitro and in vivo and showed appreciable stability in serum. Given the inherent biocompatibility of cyclodextrin and polyethylene glycol, these potent well-defined heptavalent inhibitors show considerable promise as anthrax anti-toxins.
Keywords: oligovalent, inhibitor, toxin, anthrax, docking
INTRODUCTION
Anthrax toxin, responsible for the major symptoms and death associated with anthrax, is composed of three proteins: lethal factor (LF) and edema factor (EF) are enzymes that act in the mammalian cell cytosol; protective antigen (PA) binds to cell-surface receptors and mediates toxin internalization 1. PA is proteolytically processed into a 63 kDa fragment, PA63, which oligomerizes into heptamers ([PA63]7) that bind EF and LF. The assembled toxin complex is internalized by receptor-mediated endocytosis and the enzymes are translocated into the cytosol when the toxin reaches an acidic compartment. The enzymatic activities of these proteins cause a variety of cellular dysfunctions that contribute to disease progression 2. Anthrax toxin is, therefore, an important therapeutic target and inhibitors have been described that block different steps in this intoxication pathway 3-16.
A promising approach to designing potent ligands for oligomeric targets involves the design of inhibitors that are oligovalent, as oligovalency or polyvalency can enhance the affinity of interactions by several orders of magnitude 17-19. Oligovalent inhibitors have been designed without prior knowledge of the spatial relationships among the binding sites on the target 9, 19. The rational structure-based design of oligovalent inhibitors 20-24 that provide structurally and compositionally well-defined molecules may, however, be better suited for pre-clinical and clinical development. There are but a few examples of the structure-based design of oligovalent inhibitors that have shown efficacy in vivo 7, 23.
Here, we describe the rational structure-based design of an anthrax anti-toxin based on the heptavalent display of a biospecific ligand. Our strategy to optimize the design of the inhibitor was to attach peptides to a defined scaffold that would present the peptides in a spatial orientation matching the peptide-binding sites of the toxin. For the core of the scaffold, we chose β-cyclodextrin because it has 7-fold symmetry, as does [PA63]7 (Fig. 1), and because cyclodextrins are widely used as pharmaceutical agents to enhance the solubility, bioavailability, and stability of drug molecules. Furthermore, cyclodextrin has been found to partially block the pore of oligomeric staphylococcal alpha-hemolysin 25 and small cationic β-cyclodextrin derivatives can bind to and block the [PA63]7 pore via electrostatic interactions 7. To graft the inhibitory peptides to the β-cyclodextrin core, we selected polyethylene glycol as a linker. We chose an appropriate linker length using information on the location of the peptide-binding sites on [PA63]7, obtained as described below. The well-defined heptavalent inhibitor not only neutralizes anthrax lethal toxin in vitro, but also protect animals from a toxin challenge.
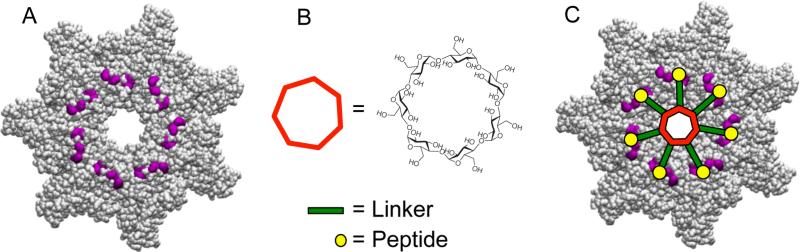
Figure 1.
Structure-based design of heptavalent anthrax toxin inhibitors. A) Structure of the LF-binding face of [PA63]7. Residues 184, 187, 197, and 200, which form part of the peptide-binding site are shown in purple. B) Structure of the core, β-cyclodextrin. C) Scheme illustrating the binding of a heptavalent inhibitor, synthesized by the attachment of seven inhibitory peptides to the β-cyclodextrin core via an appropriate polyethylene glycol linker, to [PA63]7
MATERIALS AND METHODS
Preparation of toxin components
Mutations in PA were generated using Quickchange mutagenesis according to the manufacturer's instructions (Stratagene). PA and LF were purified as described previously 26.
Cytotoxicity Assay
RAW264.7 cells were seeded in 96-well plates and incubated overnight. The cells were treated with 10-8 M PA and 10-9 M LF in the absence or presence of inhibitors. After an incubation period of 4 h, cell viability was assessed using the MTS assay according to the manufacturer's instructions (Promega).
Serum Stability
The heptavalent inhibitor was dissolved in PBS and incubated with mouse serum (Sigma, St. Louis, MO, USA) (80% v/v) at 37 °C. Samples were withdrawn at different time intervals and tested in a cytotoxicity assay as described above.
Rat Intoxication
Animal experiments were performed under University of Toronto ethical guidelines. A mixture of purified PA (40 μg) and LF (8 μg) mixed with either PBS, heptavalent inhibitor (300 nmol on a per-peptide basis), or control thioglycerol-functionalized heptavalent molecules, was used for the rat intoxication experiments. Fisher 344 rats (Charles River Laboratories) were injected intravenously in the tail vein. Seven rats were used per group, and the appearance of symptoms of intoxication was monitored over 4 h. The rats were euthanized to avoid unnecessary distress when the symptoms became pronounced.
RESULTS AND DISCUSSION
Identification of the Binding site for the Inhibitory Peptide on [PA63]7 by Mutagenesis
Mourez et al. 9 used phage display to identify an inhibitory 12-mer peptide, HTSTYWWLDGAP, that competes with LF for binding [PA63]7. A subsequent screen identified a related peptide, HYTYWWLD, that also contains the TYWWLD sequence, which we’ve shown is both necessary and sufficient for competing with LF 27. These experiments suggested that the peptides interact with [PA63]7 at or near the LF-binding site, so we decided to mutate this surface of [PA63]7 to map the binding site of the inhibitory peptides.
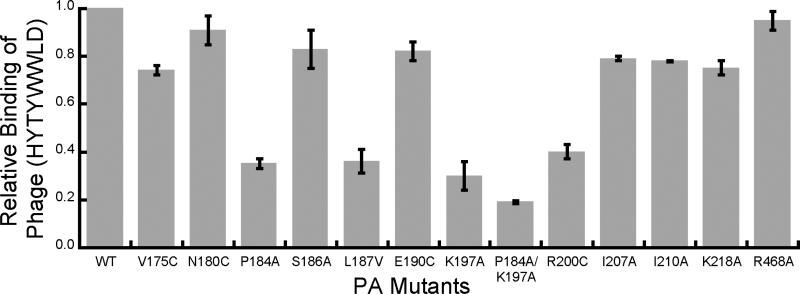
We first prepared a panel of PA mutants in which a single amino acid was mutated to either cysteine, alanine, or valine. We then used ELISA to assess the interaction between phage presenting the sequence HYTYWWLD to wild-type and mutant [PA63]7 (Fig. 2). The mutants P184A, L187V, K197A, and R200C showed a significant reduction in the binding of the phage, with an even greater reduction in binding observed for the double mutant P184A/K197A. Other mutations (V175C, N180C, S186A, E190C, F202C, I207A, I210A, K218A, and R468A) had a minimal effect in the ELISA. Notably, mutation of amino acids 197 and 200 was shown previously to interfere with binding to LF28, indicating that these two amino acids are important for both toxin assembly and binding of the inhibitory peptide.
Figure 2.
Mutations in PA63 diminish binding of phage displaying HYTYWWLD. Phage displaying HYTYWWLD peptide were added to wells containing adsorbed mutant or wild-type (WT) [PA63]7 as indicated. Unbound phage were removed by washing and the bound phage were detected with an anti-M13 phage antibody conjugated to horseradish peroxidase.
Computational Studies of the Binding of the Peptide to [PA63]7
Next, we used docking studies to characterize the interactions between the inhibitory peptides and [PA63]7 at a molecular level. This computational approach consisted of two steps: 1) determining the dominant clusters of structures adopted by the peptide in the absence of toxin; and 2) docking representative structures to [PA63]7.
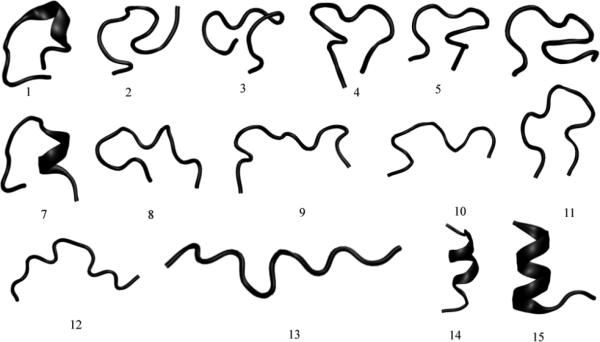
The ensemble of structures sampled by the HTSTYWWLDGAP peptide were first studied using replica exchange molecular dynamics simulations (REMD) 29, 30 using the Amber force field (amber94). The REMD trajectories were analyzed to extract 15 clusters representative of the ensemble of configurations sampled by the peptide. Each cluster represents the main structural trends of the configurations around it, making each cluster a unique conformation of the peptide. The backbone conformations of the 15 clusters are shown in Figure 3. As seen in the figure, the clusters show structures with diverse conformations containing coils, turns and alpha helical regions.
Figure 3.
Clusters representing conformations sampled by the HTSTYWWLDGAP peptide, obtained using replica exchange molecular dynamics simulations. The figure was generated using the program VMD31.
For docking calculations, we used the 1TZO.pdb crystal structure to model [PA63]732. The crystal structure file contains two heptameric molecules, with a total of 14 protein chains. To make the docking calculations tractable, we used only one of the heptameric molecules; we also removed residues irrelevant for peptide-PA63 binding from each of the seven chains. The structures used for the docking calculations consisted of seven chains, each containing amino acids 174 to 259, 372 to 381, 402 to 406, 454 to 483, 514 to 523, and 564 to 583. These amino acids include the solvent-exposed residues of the heptamer on the face distal to the cell membrane (i.e., the LF-binding face).
Docking was carried out using the Rosetta++ software package 33. During the docking all side chains were optimized via a Monte Carlo procedure, while maintaining backbone configurations fixed for both protein and the peptide. The 15 conformational clusters (Fig. 3) were each docked with the heptamer 1000 times. Each docking iteration produced a docked peptide-protein complex, which was then ranked using the Rossetta++ scoring function. The score function is a number with arbitrary units that indicates the strength of non-covalent interactions between the protein and the peptide. Table 1 shows the Rossetta++ scores for the top five bound conformations – the ones having the lowest scores – for each of the 15 clusters.
Table 1.
Characterization of peptide-[PA63]7 interactions by docking. The five best (i.e., lowest) Rossetta++ scores for the docking runs with each of the 15 clusters shown in Figure 3. The 5 best scores from all of the 15,000 docking runs are marked bold.
| Cluster | Rosetta++ Score | ||||
|---|---|---|---|---|---|
| First | Second | Third | Fourth | Fifth | |
| 1 | -753.42 | -753.35 | -753.04 | -752.83 | -752.61 |
| 2 | -758.88 | -757.32 | -757.19 | -757.19 | -757.15 |
| 3 | -755.77 | -755.29 | -755.01 | -754.82 | -754.57 |
| 4 | -761.80 | -760.53 | -760.17 | -760.02 | -759.92 |
| 5 | -754.97 | -753.60 | -752.88 | -752.14 | -751.88 |
| 6 | -758.39 | -758.38 | -758.30 | -758.00 | -757.59 |
| 7 | -759.13 | -758.53 | -758.29 | -757.83 | -757.63 |
| 8 | -754.09 | -753.50 | -752.37 | -752.13 | -752.11 |
| 9 | -751.86 | -751.36 | -750.46 | -750.01 | -749.94 |
| 10 | -755.24 | -754.45 | -753.99 | -753.97 | -753.95 |
| 11 | -755.88 | -755.79 | -755.53 | -755.50 | -755.40 |
| 12 | -752.98 | -751.92 | -751.54 | -751.29 | -750.83 |
| 13 | -752.93 | -752.61 | -752.42 | -752.14 | -751.97 |
| 14 | -760.53 | -759.95 | -759.94 | -759.80 | -759.69 |
| 15 | -758.29 | -758.21 | -758.11 | -758.00 | -757.84 |
We stress that the peptide was not constrained to bind to a specific location on the LF-binding face of the heptamer during the docking runs. Nevertheless, two of the top 5 scores obtained from the unbiased docking runs – the 3rd and 4th ranked structures for cluster 4 (referred to as 4-3 and 4-4 respectively) – represent complexes in which the peptide makes contact with heptamer residues identified as being important for binding by mutagenesis (Fig. 4). Moreover, most of the contacts with the heptamer for complexes 4-3 and 4-4 involved the TYWWLD region of the peptide (Table S1 and S2) – one that we have previously identified as being important for binding to the heptamer 27. Thus, 4-3 and 4-4 represent potential conformations for the peptide-protein complex. Interestingly, the contact residues on the heptamer for these two bound conformations are at similar distances from the center of the heptamer, suggesting that an appropriately designed heptavalent inhibitor would bind equally well at either site (Fig. 1).
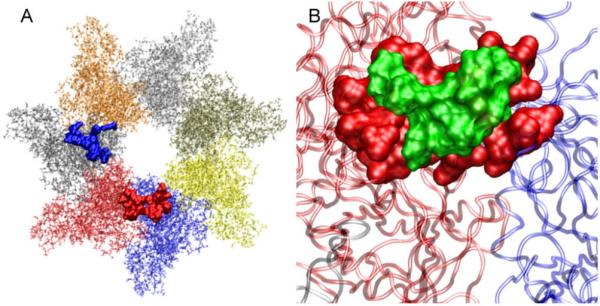
Figure 4.
(A) Residues of [PA63]7 that are contacted by the peptide in docked complexes 4-3 (blue) and 4-4 (red) are shown as space-filled atoms. Individual protein chains of the heptamer (each colored differently) are also shown as transparent background. Both 4-3 and 4-4 dock at the interface of two monomeric units and their contact sites include amino acid residues identified as important by mutagenesis. (B) Close-up view of the docked conformation 4-4 showing HTSTYWWLDGAP peptide (green) and the [PA63]7 contact residues (red) as space-filled atoms. Two of the chains in the heptameric unit that 4-4 is in contact with are also shown as transparent background.
Structure-based Design of Heptavalent Inhibitor
We developed a synthetic strategy for attaching the peptide to the β-cyclodextrin core via a polyethylene glycol linker (Fig. 5). Given the difficulties inherent in attaching seven polymeric linkers to a single core with high yield, we developed an approach based on “click chemistry” in which the copper-catalyzed 1,3-dipolar cycloaddition of an azide to a terminal alkyne results in the production of a triazole with high yield 34-37. Briefly, the secondary hydroxyl groups of the β-cyclodextrin core were first methylated and seven terminal alkyne groups were introduced by the reaction of the primary hydroxyl groups with propargyl bromide. Seven polyethylene glycol linkers were attached to the core by the reaction of the alkynes with O-(2-Aminoethyl)-O′-(2-azidoethyl)decaethylene glycol (H2N-CH2CH2-(OCH2CH2)11-N3, MW = 570.7 Da) (PEG11) – a commercially available, monodisperse, azidefunctionalized polyethylene glycol derivative. Chloroacetylation of the free terminal amine, followed by reaction with the peptide HTSTYWWLDGAPC, a cysteine-derivatized version of the inhibitory 12-mer peptide, yielded a heptavalent inhibitor (6, Fig. 5).
Figure 5.
Synthesis scheme of heptavalent anthrax toxin inhibitor. (a) TBDMSCl, pyridine, 0 °C - rt. (b) NaH, MeI, THF (c) NH4F, MeOH, reflux (d) NaH, propargyl bromide, DMF, 0 °C - rt. (e) CuSO4, sodium ascorbate, THF:H2O:BuOH (0.5:1:1), 80 °C (f) Chloroacetic anhydride, triethylamine (g) Peptide, DMF, DBU, triethylamine.
The selection of the polyethylene glycol linker, PEG11, was based on the location of the peptide-binding site (Fig. 2). The distance between the peptide-binding residues (P184, L187, K197, and R200) and the center of the lumen of [PA63]7 is ca. 30-40 Å. Recognition and inhibition of the toxin would be facilitated by choosing a linker such that the root-mean-square distance from the center of the cyclodextrin core to the end of the linker matched the distance from the center of the lumen of [PA63]7 to the peptide-binding residues 21, 38-41, i.e., by statistically matching the heptavalent inhibitor with the heptavalent target 12. For our inhibitor, the linker consisted of a rigid cyclodextrin core, a flexible region (consisting primarily of ethylene glycol repeat units 42), and the amino acids CPAG (since the TYWWLD residues of the HTSTYWWLDGAPC are necessary and sufficient for binding 27). Using the method of Krishnamurthy et al. 41, we estimated that the root-mean-square distance from the center of the cyclodextrin core to the end of the linker was ca. 30 Å for inhibitors based on the PEG11 linker, which was consistent with the distance from the center of the lumen to the peptide-binding residues on [PA63]7.
Characterization of Heptavalent Inhibitors
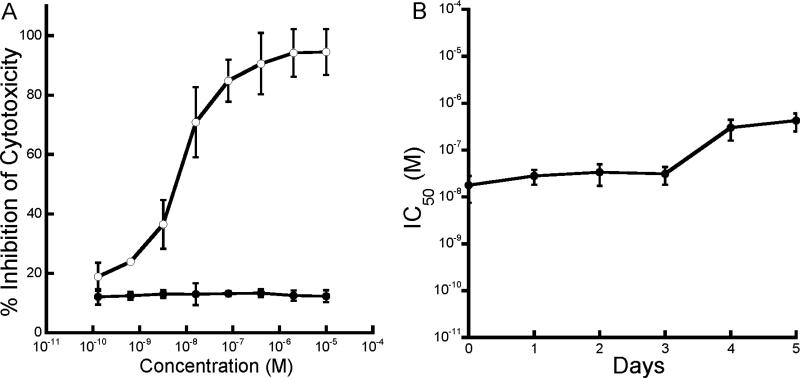
We tested the ability of this well-defined heptavalent inhibitor (6, Fig. 5) to neutralize anthrax lethal toxin in vitro by incubating RAW264.7 cells with a mixture of PA and LF in the presence of several concentrations of the inhibitor. The heptavalent molecule could inhibit cytotoxicity with a half-maximal inhibitory concentration (IC50) of ca. 10 nM on a per-peptide basis (Fig. 6A). Heptavalent molecules presenting only thioglycerol showed no inhibitory activity (Fig. 6A), and the monovalent peptide did not inhibit cytotoxicity at concentrations as high as 2 mM. The heptavalent inhibitor therefore provided a more than 100,000-fold enhancement in the activity of this peptide. To test whether the well-defined heptavalent inhibitor based on the PEG11 linker was resistant to proteolytic degradation, we also incubated the inhibitor with 80% serum at 37 °C. Samples were withdrawn at various time intervals and their inhibitory activity was determined using the cytotoxicity assay. As seen in Figure 6B, the heptavalent inhibitor did not show any significant loss in activity over a three day period.
Figure 6.
Characterization of a well-defined heptavalent anthrax toxin inhibitor. A) Inhibition of anthrax toxin-induced cytotoxicity of RAW264.7 cells by heptavalent inhibitors presenting HTSTYWWLDGAP (open circles) or control thioglycerol-functionalized molecules (closed circles). B) Stability of heptavalent inhibitor in serum. Heptavalent inhibitor was incubated in 80% serum for indicated times and the IC50 was determined in a RAW264.7 cell cytotoxicity assay.
Next, we decided to probe the effect of the structure of the heptavalent inhibitor on its potency. To that end, we synthesized a series of inhibitors with polyethylene glycol linkers of different molecular weights (and therefore, different lengths), and tested their activity in the cytotoxicity assay (Fig. 7). As seen in Figure 7, heptavalent inhibitors based on short PEG linkers (n ranging from 2 - 6) did not show significant activity in the cytotoxicity assay. In contrast, the inhibitory activity did not significantly change with further increase in the number of polyethylene glycol units beyond 11 (i.e., for n > 11), in the range of molecular weights tested (Fig. 7). When the root-mean-square distance from the center of the cyclodextrin to the end of the linker was greater than or equal to the distance from the lumen of the heptamer to the peptide-binding site, effective inhibition was observed, consistent with both experimental and theoretical studies 38, 39, 41, 43.
Figure 7.
Influence of linker length on the activity of heptavalent inhibitors. The indicated PEG linkers were used to join the HTSTYWWLDGAP peptide to β-cyclodextrin and the IC50 values of the resulting inhibitors were measured in a RAW264.7 cell cytotoxicity assay. The error bars represent the standard deviation from four separate experiments. Asterisks indicate that inhibitory activity was not detected.
Finally, we tested the ability of the well-defined heptavalent inhibitors to neutralize anthrax toxin in vivo, in Fisher 344 rats. Six of seven rats that were injected intravenously with anthrax lethal toxin (a mixture of 40 μg of PA and 8 μg of LF) and six of seven rats that were co-injected with toxin and thioglycerol-functionalized heptavalent molecules became moribund (Table 2). Co-injection of the heptavalent inhibitor based on the PEG11 linker (6, Fig. 5) with the toxin, however, prevented six of seven animals from becoming moribund.
Table 2.
Inhibition of anthrax toxin action in a rat intoxication model.
| Inhibitor | Amount of peptide (nmol) | Moribund/ Total |
|---|---|---|
| None | 0 | 6/7 |
| Negative Control | 0 | 6/7 |
| Heptavalent inhibitor | 300 | 1/7 |
CONCLUSION
In conclusion, we have demonstrated the structure-based design of potent biospecific oligovalent inhibitors of anthrax lethal toxin. We used experimental and computational studies to identify the binding site for an inhibitory peptide on the heptameric subunit of anthrax toxin. We developed a click-chemistry-based approach for the efficient attachment of a suitable polymeric linker to a cyclodextrin core. Subsequent functionalization with the peptide yielded a well-defined heptavalent inhibitor that neutralized anthrax lethal toxin both in vitro and in vivo and showed appreciable stability in serum. Given the inherent biocompatibility of cyclodextrin and polyethylene glycol, these potent well-defined heptavalent anti-toxins might serve as valuable adjuncts to antibiotics for the treatment of anthrax. The approach outlined in this work might also be broadly applicable to designing well-defined oligovalent molecules that inhibit pathogens or other microbial toxins in vivo.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by NIH grant U01 AI056546 and NSF grant DMR-0642573. J.M. holds the Canada Research Chair in Bacterial Pathogenesis.
Footnotes
SUPPORTING INFORMATION AVAILABLE
Information on the synthesis and characterization of inhibitors is included. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Young JA, Collier RJ. Anthrax toxin: receptor binding, internalization, pore formation, and translocation. Annu. Rev. Biochem. 2007;76:243–265. doi: 10.1146/annurev.biochem.75.103004.142728. [DOI] [PubMed] [Google Scholar]
- 2.Moayeri M, Leppla SH. Cellular and systemic effects of anthrax lethal toxin and edema toxin. Mol. Aspects Med. 2009;30:439–455. doi: 10.1016/j.mam.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basha S, Rai P, Poon V, Saraph A, Gujraty K, Go MY, Sadacharan S, Frost M, Mogridge J, Kane RS. Polyvalent inhibitors of anthrax toxin that target host receptors. Proc Natl Acad Sci U S A. 2006;103:13509–13. doi: 10.1073/pnas.0509870103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradley KA, Mogridge J, Mourez M, Collier RJ, Young JAT. Identification of the cellular receptor for anthrax toxin. Nature. 2001;414:225–229. doi: 10.1038/n35101999. [DOI] [PubMed] [Google Scholar]
- 5.Collier RJ, Young JAT. Anthrax Toxin. Annu. Rev. Cell Dev. Biol. 2003;19:45–70. doi: 10.1146/annurev.cellbio.19.111301.140655. [DOI] [PubMed] [Google Scholar]
- 6.Joshi A, Saraph A, Poon V, Mogridge J, Kane RS. Synthesis of potent inhibitors of anthrax toxin based on poly-L-glutamic acid. Bioconjug. Chem. 2006;17:1265–1269. doi: 10.1021/bc060042y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karginov VA, Nestorovich EM, Moayeri M, Leppla SH, Bezrukov SM. Blocking anthrax lethal toxin at the protective antigen channel by using structure-inspired drug design. Proc. Natl. Acad. Sci. USA. 2005;102:15075–15080. doi: 10.1073/pnas.0507488102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maynard JA, Maassen CBM, Leppla SH, Brasky K, Patterson JL, Iverson BL, Georgiou G. Protection against anthrax toxin by recombinant antibody fragments correlates with antigen affinity. Nat. Biotechnol. 2002;20:597–601. doi: 10.1038/nbt0602-597. [DOI] [PubMed] [Google Scholar]
- 9.Mourez M, Kane RS, Mogridge J, Metallo S, Deschatelets P, Sellman BR, Whitesides GM, Collier RJ. Designing a polyvalent inhibitor of anthrax toxin. Nat. Biotech. 2001;19:958–961. doi: 10.1038/nbt1001-958. [DOI] [PubMed] [Google Scholar]
- 10.Numa MMD, Lee LV, Hsu C-C, Bower KE, Wong C-H. Identification of novel anthrax lethal factor inhibitors generatead by combinatorial pictet-spengler reaction followed by screening in situ. ChemBioChem. 2005;6:1002–1006. doi: 10.1002/cbic.200500009. [DOI] [PubMed] [Google Scholar]
- 11.Panchal R, Hermone AR, Nguyen TL, Wong TY, Schwarzenbacher R, Schmidt J, Lane D, McGrath C, Turk B, Burnett J, Aman MJ, Little S, Sausville E, Zaharevitz D, Cantley L, Liddington R, Gussio R, Bavari S. Identification of small molecule inhibitors of anthrax lethal factor. Nat. Struc. Mol. Bio. 2004;11:67–72. doi: 10.1038/nsmb711. [DOI] [PubMed] [Google Scholar]
- 12.Rai P, Padala C, Poon V, Saraph A, Basha S, Kate S, Tao K, Mogridge J, Kane RS. Statistical pattern matching facilitates the design of polyvalent inhibitors of anthrax and cholera toxins. Nat. Biotechnol. 2006;24:582–6. doi: 10.1038/nbt1204. [DOI] [PubMed] [Google Scholar]
- 13.Rai PR, Saraph A, Ashton R, Poon V, Mogridge J, Kane RS. Raftlike polyvalent inhibitors of the anthrax toxin: modulating inhibitory potency by formation of lipid microdomains. Angew. Chem. Int. Ed. Engl. 2007;46:2207–9. doi: 10.1002/anie.200604317. [DOI] [PubMed] [Google Scholar]
- 14.Rainey GJA, Wigelsworth DJ, Ryan PL, Scobie HM, Collier RJ, Young JAT. Receptor-specific requirements for anthrax toxin delivery into cells. Proc. Natl. Acad. Sci. USA. 2005;102:13278–13283. doi: 10.1073/pnas.0505865102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scobie HM, Rainey GJA, Bradley KA, Young JAT. Human capillary morphogenesis protein 2 functions as an anthrax toxin receptor. Proc. Natl. Acad. Sci. USA. 2003;100:5170–5174. doi: 10.1073/pnas.0431098100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sellman BR, Mourez M, Collier RJ. Dominant-negative mutants of a toxin subunit: An approach to therapy of anthrax. Science. 2001;292:695–697. doi: 10.1126/science.109563. [DOI] [PubMed] [Google Scholar]
- 17.Badjic JD, Nelson A, Cantrill SJ, Turnbull WB, Stoddart JF. Multivalency and cooperativity in supramolecular chemistry. Acc. Chem. Res. 2005;38(9):723–732. doi: 10.1021/ar040223k. [DOI] [PubMed] [Google Scholar]
- 18.Kiessling LL, Strong LE, Gestwicki JE. Principles for multivalent ligand design. Annu. Rep. Med. Chem. 2000;35:321–330. [Google Scholar]
- 19.Mammen M, Choi S-K, Whitesides GM. Polyvalent Interactions in Biological Systems: Implications for Design and Use of Multivalent Ligands and Inhibitors. Angew. Chem. Int. Ed. Engl. 1998;37:2754–2794. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 20.Fan E, Merritt EA, Verlinde CLMJ, Hol WGJ. AB5 toxins: structures and inhibitor design. Curr. Opin. Struct. Biol. 2000;10:680–686. doi: 10.1016/s0959-440x(00)00152-4. [DOI] [PubMed] [Google Scholar]
- 21.Fan E, Zhang Z, Minke WE, Hou Z, Verlinde CLMJ, Hol WGJ. High-affinity pentavalent ligands of Escherichia coli heat-labile enterotoxin by modular structure-based design. J. Am. Chem. Soc. 2000;122:2663–2664. [Google Scholar]
- 22.Kitov PI, Sadowska JM, Mulvey G, Armstrong GD, Ling H, Pannu NS, Read RJ, Bundle DR. Shiga-like toxins are neutralized by tailored multivalent carbohydrate ligands. Nature. 2000;403:669–672. doi: 10.1038/35001095. [DOI] [PubMed] [Google Scholar]
- 23.Mulvey GL, Marcato P, Kitov PI, Sadowska J, Bundle DR, Armstrong GD. Assessment in mice of the therapeutic potential of tailored, multivalent Shiga toxin carbohydrate ligands. J. Infect. Dis. 2003;187:640–649. doi: 10.1086/373996. [DOI] [PubMed] [Google Scholar]
- 24.Polizzotti BD, Maheshwari R, Vinkenborg J, Kiick KL. Effects of Saccharide Spacing and Chain Extension on Toxin Inhibition by Glycopolypeptides of Well-Defined Architecture. Macromolecules. 2007;40:7103–7110. doi: 10.1021/ma070725o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gu LQ, Braha O, Conlan S, Cheley S, Bayley H. Stochastic sensing of organic analytes by a pore-forming protein containing a molecular adapter. Nature. 1999;398:686–690. doi: 10.1038/19491. [DOI] [PubMed] [Google Scholar]
- 26.Liao KC, Mogridge J. Expression of Nlrp1b inflammasome components in human fibroblasts confers susceptibility to anthrax lethal toxin. Infect. Immun. 2009:4455–4462. doi: 10.1128/IAI.00276-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gujraty K, Sadacharan S, Frost M, Poon V, Kane RS, Mogridge J. Functional characterization of peptide-based anthrax toxin inhibitors. Mol. Pharm. 2005;2:367–372. doi: 10.1021/mp050040f. [DOI] [PubMed] [Google Scholar]
- 28.Cunningham K, Lacy DB, Mogridge J, Collier RJ. Mapping the lethal factor and edema factor binding sites on oligomeric anthrax protective antigen. Proc. Natl. Acad. Sci. USA. 2002;99:7049–7053. doi: 10.1073/pnas.062160399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garcia AE, Sanbonmatsu KY. Exploring the energy landscape of a beta hairpin in explicit solvent. Proteins. 2001;42:345–354. doi: 10.1002/1097-0134(20010215)42:3<345::aid-prot50>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 30.Sugita Y, Okamoto Y. Replica-exchange molecular dynamics methods for protein folding. Chem. Phys. Lett. 1999;314:141–151. [Google Scholar]
- 31.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–8. 27–8. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 32.Lacy DB, Wigelsworth DJ, Melnyk RA, Harrison SC, Collier RJ. Structure of heptameric protective antigen bound to an anthrax toxin receptor: a role for receptor in pH-dependent pore formation. Proc. Natl. Acad. Sci. USA. 2004;101:13147–13151. doi: 10.1073/pnas.0405405101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gray JJ, Moughon S, Wang C, Schueler-Furman O, Kuhlman B, Rohl CA, Baker D. Protein-protein docking with simultaneous optimization of rigid-body displacement and side-chain conformations. J Mol Biol. 2003;331:281–299. doi: 10.1016/s0022-2836(03)00670-3. [DOI] [PubMed] [Google Scholar]
- 34.Kolb HC, Finn MG, Sharpless KB. Click Chemistry: Diverse Chemical Function from a Few Good Reactions. Angew. Chem. Int. Ed. Engl. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 35.Lutz J-F. 1,3-Dipolar Cycloadditions of Azides and Alkynes: a Universal Ligation Tool in Polymer and Materials Science. Angew. Chem. Int. Ed. Engl. 2007;46:1018–1025. doi: 10.1002/anie.200604050. [DOI] [PubMed] [Google Scholar]
- 36.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. A stepwise huisgen cycloaddition process: copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew. Chem. Int. Ed. Engl. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 37.Tornoe CW, Christensen C, Meldal M. Peptidotriazoles on solid phase: [1,2,3]-triazoles by regiospecific copper(i)-catalyzed 1,3-dipolar cycloadditions of terminal alkynes to azides. J. Org. Chem. 2002;67:3057–3064. doi: 10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]
- 38.Gargano JM, Ngo T, Kim JY, Acheson DWK, Lees WJ. Multivalent inhibition of AB5 toxins. J. Am. Chem. Soc. 2001;123:12909–12910. doi: 10.1021/ja016305a. [DOI] [PubMed] [Google Scholar]
- 39.Kane RS. Thermodynamics of multivalent interactions: influence of the linker. Langmuir. 2010;26:8636–8640. doi: 10.1021/la9047193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kramer RH, Karpen JW. Spanning binding sites on allosteric proteins with polymer-linked ligand dimers. Nature. 1998;395:710–713. doi: 10.1038/27227. [DOI] [PubMed] [Google Scholar]
- 41.Krishnamurthy VM, Semetey V, Bracher PJ, Shen N, Whitesides GM. Dependence of effective molarity on linker length for an intramolecular protein-ligand system. J. Am. Chem. Soc. 2007;129:1312–1320. doi: 10.1021/ja066780e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Knoll D, Hermans J. Polymer-protein interactions. Comparison of experiment and excluded volume theory. J. Biol. Chem. 1983;258:5710–5715. [PubMed] [Google Scholar]
- 43.Diestler DJ, Knapp EW. Statistical thermodynamics of the stability of multivalent ligand-receptor complexes. Phys. Rev. Lett. 2008;100:178101–178104. doi: 10.1103/PhysRevLett.100.178101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.