SUMMARY
The mechanism by which protein folding is coupled to biosynthesis is a critical, but poorly understood aspect of protein conformational diseases. Here we use FRET to characterize tertiary structural transitions of nascent polypeptides and show that the first nucleotide-binding domain (NBD1) of human CFTR, whose folding is defective in cystic fibrosis, folds via a cotranslational multi-step pathway as it is synthesized on the ribosome. Folding begins abruptly as NBD1 residues 389–500 emerge from the ribosome exit tunnel, thereby initiating compaction of a small, N-terminal α/β-subdomain. Real-time kinetics of synchronized nascent chains revealed that subdomain folding is rapid, occurs coincident with synthesis, and is facilitated by direct ATP binding to the nascent polypeptide. These findings localize the major CF defect late in the NBD1 folding pathway and establish a paradigm wherein a cellular ligand promotes vectorial domain folding by facilitating an energetically favored local peptide conformation.
INTRODUCTION
In cells, many proteins begin to acquire tertiary structure during their synthesis as the nascent polypeptide emerges vectorially, N- to C-terminus, from the ribosome exit tunnel (Cabrita et al., 2010; Clark, 2004; Fedorov and Baldwin, 1997). Cotranslational protein folding, in contrast to in vitro refolding, is therefore constrained by the rate and vectorial nature of translation (Fedorov and Baldwin, 1997; Siller et al., 2010), geometry of the ribosome exit tunnel (Lu and Deutsch, 2005; Voss et al., 2006; Ziv et al., 2005), molecular crowding (Ellis, 2001), and interaction with cellular chaperones (Ellis, 2007; Frydman, 2001; Hartl and Hayer-Hartl, 2009). The sequential appearance of a nascent polypeptide with limited conformational freedom can therefore influence the de novo folding funnel by restricting conformational diversity (Clark, 2004; Fedorov and Baldwin, 1999; Netzer and Hartl, 1997) and enabling the nascent chain to achieve local on-pathway energy minima (Clark and King, 2001; Evans et al., 2008). These factors may contribute to the higher yields of native protein generated in vivo (Fedorov and Baldwin, 1999). While cotranslational folding is conceptually intuitive for low contact order (C.O.) proteins, which exhibit predominantly local sequence contacts and less compact folding transition states, it represents a formidable challenge for high C.O. substrates that must adopt metastable conformational intermediates prior to the synthesis of C-terminal regions required for the native fold (Plaxco et al., 1998). Despite recent advances in evaluating biochemical properties of nascent chains (Komar, 2009) and structural analyses using Cryo EM (Becker et al., 2009) and NMR spectroscopy (Cabrita et al., 2009; Eichmann et al., 2010; Hsu et al., 2007), the mechanism by which folding information is vectorially integrated during protein synthesis remains poorly understood and limited by the lack of robust methods to identify and discriminate conformational changes of ribosome-bound folding intermediates.
De novo folding is particularly relevant in human diseases where inherited mutations produce abnormal conformers that either accumulate in the cell or are rapidly degraded (Balch et al., 2008; Barral et al., 2004; Luheshi et al., 2008). For example, the cystic fibrosis transmembrane conductance regulator (CFTR) is a multidomain ATP Binding Cassette (ABC) transporter that forms an ATP-dependent chloride channel and regulates water and ion transport in epithelial tissues (Riordan, 2008). The most common inherited mutation in cystic fibrosis, F508del, introduces a temperature sensitive defect that disrupts CFTR folding at its site of synthesis in the endoplasmic reticulum (Cheng et al., 1990; Riordan, 2008). Phe508 resides in the first nucleotide-binding domain (NBD1) of CFTR where it interacts with cytosolic extensions of CFTR transmembrane domains (TMDs) and facilitates the coupling of ATP binding and hydrolysis to transmembrane helix movement during channel gating (Dawson and Locher, 2006; Mendoza and Thomas, 2007; Serohijos et al., 2008). In addition to disrupting critical inter-domain contacts, F508del increases local dynamics and decreases stability of the NBD1 domain (Lewis et al., 2010), but has minimal effect on its native structure (Du et al., 2005; Lewis et al., 2005; Thibodeau et al., 2005). Rather, the mutation is predicted to alter, through poorly understood mechanisms, kinetics and/or structural intermediates of the native NBD1 folding pathway (He et al., 2010; Pissara et al., 2008; Qu et al., 1997; Thibodeau et al., 2010; Wang et al., 2010).
Crystal structures of ABC transporter NBDs have revealed a highly conserved topography consisting of three canonical subdomains: an antiparallel three-strand ABC β-subdomain, an α-helical subdomain, and a central F1-like ATP-binding core (Lewis et al., 2004). CFTR NBD1 also contains a unique unstructured regulatory insertion (RI, residues 405–436) and a C-terminal regulatory extension (RE, residues 656–673) that control channel gating through NBD1–NBD2 interactions in response to phosphorylation (Aleksandrov et al., 2010; Lewis et al., 2004). Interestingly, the NBD fold has a very high C.O. (24.2) and is characterized by a preponderance of long-distance intra-chain interactions that are predicted to slow kinetics and impart cooperativity in domain folding (Lewis et al., 2010; Plaxco et al., 1998). While folding likely begins cotranslationally (Kleizen et al., 2005) after topology of the first transmembrane domain is established (Carveth et al., 2002), the pathway by which NBD1 acquires its native structure in the cellular environment remains entirely unknown. We therefore investigated de novo NBD1 folding by characterizing stepwise conformational transitions as the nascent polypeptide emerges from the ribosome. Our results identify a multistep pathway that is initiated during synthesis by rapid cotranslational compaction of a small (~112 residue) N-terminal subdomain. This polypeptide region contains a minimal ATP binding site, and its cotranslational folding is stimulated by ATP. These findings suggest a revised classification for NBD subdomain organization, localize the F508del defect to a late stage of the NBD1 folding pathway, and provide an example in which a ubiquitious cellular ligand promotes vectorial domain folding by facilitating an energetically favored local peptide conformation.
RESULTS
Strategy
Förster resonance energy transfer (FRET) (Stryer, 1978) provides a useful tool to monitor conformational changes on a macromolecular scale that are typically associated with protein folding (Miyake-Stoner et al., 2009; Woolhead et al., 2004), structural dynamics (Kajihara et al., 2006; Taraska and Zagotta, 2007), and chaperone interaction (Kaiser et al., 2006). FRET involves the transfer of excited state energy from a donor fluorophore to an acceptor fluorophore (or chromophore) without emission of a photon. Because FRET efficiency (EFRET) is highly dependent on the distance (R) between the donor and acceptor probes (EFRET = R06/(R06 + R6) where R0 is the Förster distance at which EFRET is 50%), it can readily distinguish structural changes that take place as a protein folds from an extended conformation to a compact tertiary structure (Woolhead et al., 2004).
To measure conformational changes that occur cotranslationally during protein synthesis, we incorporated a donor fluorophore, cyan fluorescent protein (CFP), via an in-frame N-terminal fusion, and a small acceptor dye, 7-nitrobenz-2-oxa-1, 3-diazole (NBD) at an engineered ‘UAG’ stop codon using a synthetic amber suppressor tRNA, εNBD-[14C]Lys-tRNAamb (Figure S1). Nascent polypeptides were then synthesized in vitro from truncated mRNA templates to generate uniform cohorts of folding intermediates that are kinetically trapped at a specific stage of synthesis (Cabrita et al., 2010; Johnson, 2005). Because the donor (CFP) is located at the N-terminus, CFP-containing nascent chains that terminate prematurely at the UAG codon are removed during ribosome isolation. However, if the UAG codon is translated by the suppressor tRNA, polypeptides that remain tethered to the ribosome by a covalent peptidyl-tRNA ester bond contain both a donor and acceptor fluorophore as well as a single [14C]Lys in 1:1:1 stoichiometry. Since the donor dye, acceptor dye, and [14C]Lys concentrations are equal (in the absence of ribosomal stalling), the precise concentration of nascent chains present in the sample can be determined, thereby allowing EFRET to be calculated directly from the acceptor-induced decrease in donor fluorescence intensity (see Experimental Procedures and (Woolhead et al., 2004)).
FRET-based analysis of ribosome-bound nascent chains
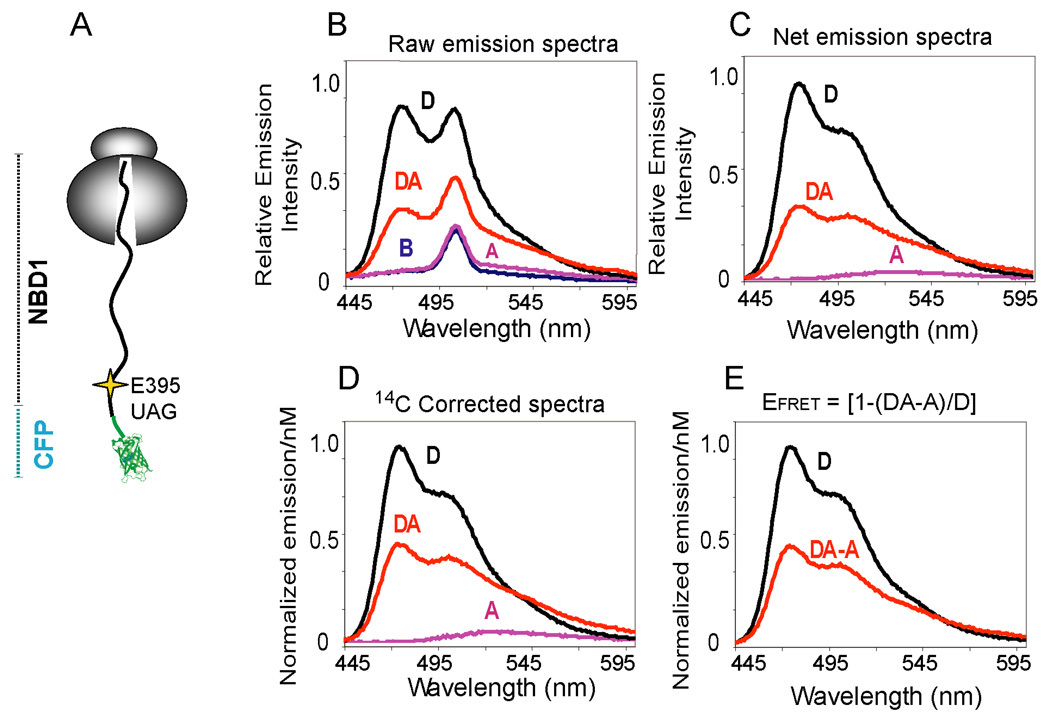
To develop this approach, a truncated form of CFP (Figure S2) was fused in-frame to CFTR residue 389 at the N-terminus of NBD1, and the acceptor dye was incorporated six residues C-terminal to the fusion site (Figure 1A). Four parallel programmed translation reactions, Blank (B), Acceptor only (A), Donor only (D), and Donor + Acceptor (DA) were performed, and raw CFP emission spectra were obtained from intact ribosome nascent chain complexes (RNCs) (Figure 1B and Figure S1). Spectra were corrected for background signal (Figure 1B,C), and normalized to the nascent chain concentration based on [14C]Lys incorporation (Figure 1D). The fluorescent signal due to direct acceptor excitation, which is very small relative to CFP, was then subtracted to generate two net corrected CFP emission spectra that differ only by the presence or absence of the acceptor dye (Figure 1E). Using the equation EFRET = [1-(FDA-A)/FD], these results yielded an EFRET of 51% where FD and FD-DA are the maximal fluorescence emission intensity (λmax = 475nm) for each sample.
Figure 1.
FRET detection on a ribosome bound nascent chain. (A) Cartoon representation of a ribosome attached CFP-NBD1 fusion protein truncated at CFTR residue 460 shows location of the acceptor dye incorporation site (Thr395). (B) Raw CFP emission spectra (λex = 430 nm, λem = 445–600 nm) obtained from ribosome nascent chain complexes isolated from B, A, D, and DA samples. The background emission peak at ~510 nm is due to water Raman Scatter. (C) Net CFP emission spectra after background subtraction and (D) correction for donor dye concentration based on [14C]Lys incorporation. Normalized emission intensities are shown per nM of donor dye concentration. (E) Corrected and normalized emission spectra after subtracting fluorescence of A from DA to correct for direct excitation of acceptor.
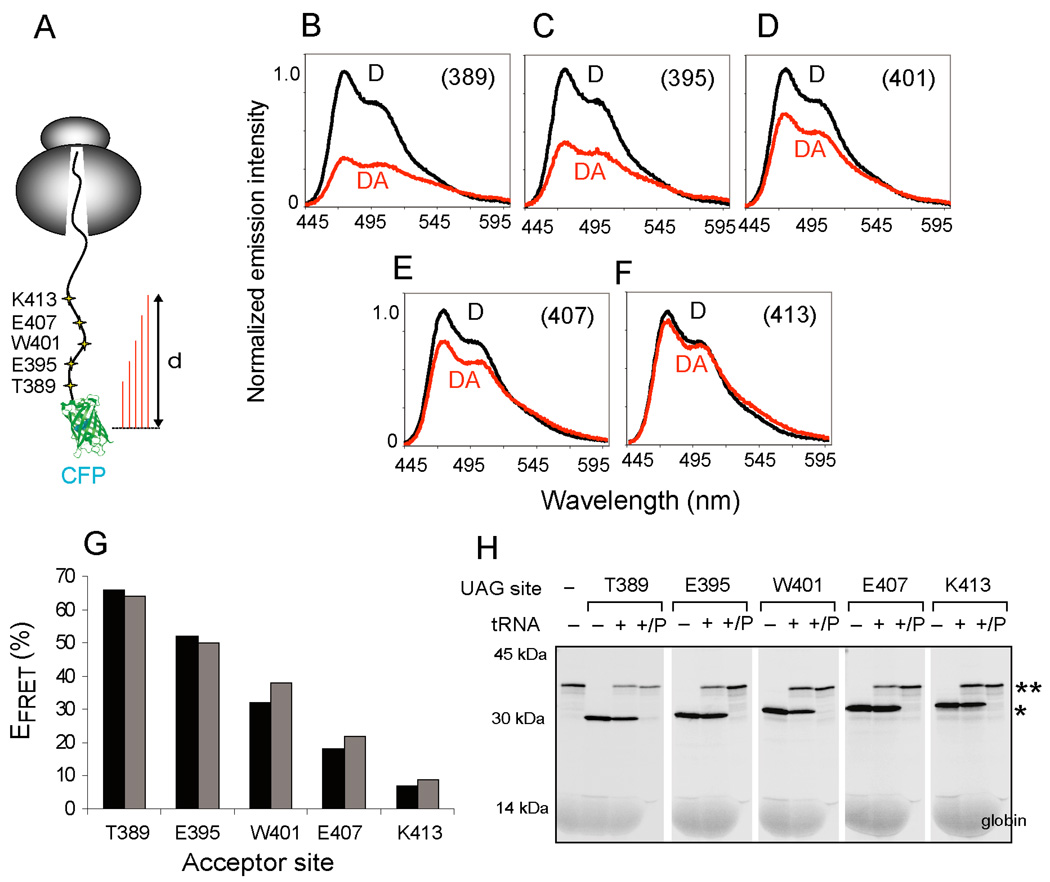
We reasoned that if the acceptor-induced decrease in CFP fluorescence were indeed caused by FRET, then it should be highly sensitive to the position of acceptor and donor probes in the nascent chain. The acceptor probe was therefore incorporated at six-residue intervals (from NBD1 Thr389 to Lys413, Figure 2A) located 9 to 33 residues beyond the last strand of the CFP β-barrel, in a polypeptide region that is predicted to be predominantly unstructured (Lewis et al., 2004). mRNA was truncated at codon 460, and FRET was determined based on the corrected emission spectra obtained from D and DA samples (Figure 2B–F). The highest EFRET (65%) was observed when the acceptor dye was located 9 residues from the CFP barrel, and EFRET decreased 9-fold to ~8% as the acceptor site was moved 24 residues more distally (Figure 2G). Parallel analysis of 35S-labeled polypeptides confirmed that translation termination at the UAG codon released the nascent chain from the ribosome, whereas addition of suppressor tRNA enabled stop codon readthrough and formation of stable RNCs (Figure 2H).
Figure 2.
FRET dependence on donor-acceptor distance. (A) Schematic of ribosome-bound CFP-NBD1 nascent chains truncated at CFTR residue 460. UAG codons were positioned at 6 residue intervals starting at Thr389. (B–F) Background corrected and 14C normalized CFP emission spectra for D and DA samples obtained for CFP-NBD1 proteins with εNBD-Lys incorporated at the indicated residues. (G) Calculated EFRET obtained from constructs shown in B–F. Results from two independent experiments are shown. (H) Autoradiogram showing 35S-labeled total translation products or pelleted RNCs (P) expressed in presence or absence of [14C]Lys-tRNAamb as indicated. Polypeptides that terminate or readthrough the UAG codon are indicated by single or double asterisk, respectively.
While it is not possible to determine absolute distances in this non-rigid system, the EFRET measured from these programmed RNCs is in good agreement with theoretical values. The calculated R0 for CFP and εNBD-Lys fluorophores in our system is 38.5Å (see Supplementary Experimental Procedures). Thus the effective distance from the CFP chromophore to the acceptor dye attached at residue 389 of NBD1 or residue 233 in CFP (Figures S2, S3) is 34–35 Å (EFRET = 65–70% and R = R0[(1-E)/E]1/6). Considering that the center of the CFP chromophore is ~15 Å from the last strand of the β-barrel (ala227, pdb 1CV7), then the estimated length of 6–8 residues from the barrel to the acceptor probe site is approximately 2.5–3.2 Å/aa. Although this calculation does not account for the nonlinearity of FRET over the distance distribution of the acceptor dye caused by tethering, the result is remarkably consistent with a relatively unstructured polypeptide for these short tether lengths (See also Supplementary Experimental Procedures). Changes in EFRET could also potentially reflect parameters that affect the Förster distance (R0) (e.g. quantum yield of the donor, spectral overlap of donor and acceptor, or transition dipole orientation). However, this is unlikely in the system used here (see Supplementary Experimental Procedures, Tables S1, S2). In addition, a very similar FRET pattern was observed for corresponding acceptor sites when CFP was fused to an unrelated peptide sequence derived from the CFTR N-terminus (Figure S3). Thus, our results indicate that the major factor influencing EFRET is the linear distance between the acceptor and donor probes in the nascent polypeptide.
NBD1 folds cotranslationally on the ribosome
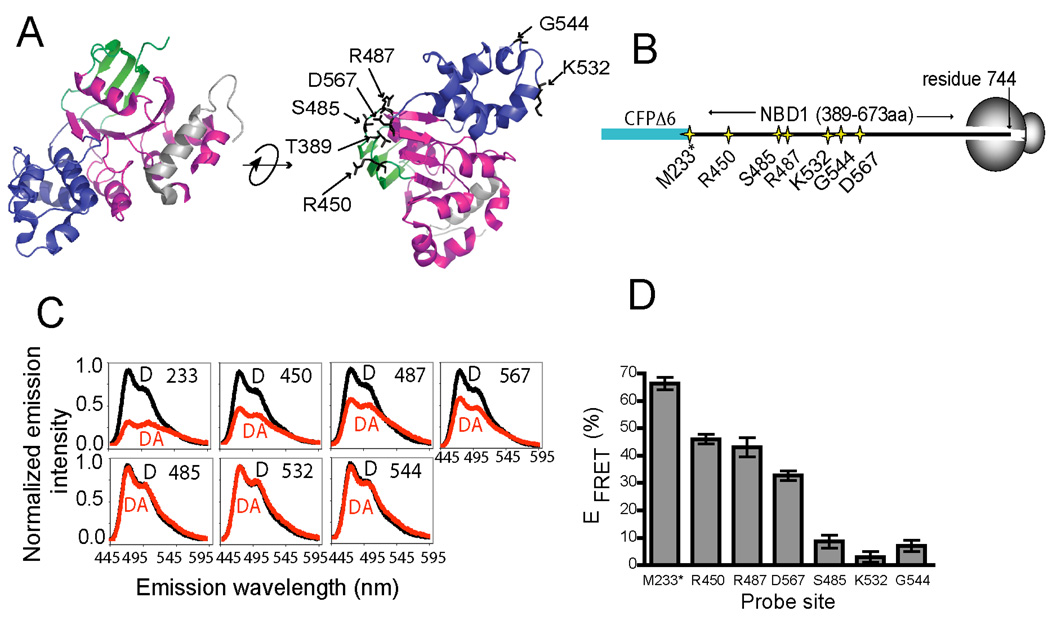
We next used FRET to determine whether in vitro-translated NBD1 might fold cotranslationally and achieve a native conformation as it is synthesized on the ribosome. For these experiments, the acceptor dye was incorporated individually at multiple surface-exposed residues that are variably distant in primary sequence but predicted to be adjacent to the fusion site in the folded domain (Figure 3A,B). Two residues (Lys532 and Gly544) on the opposite side of the domain were used as controls. mRNA was then truncated at CFTR residue 744 to ensure that the entire NBD1 domain (residues 389–673) would reside outside the ribosome tunnel.
Figure 3.
FRET-detected cotranslational NBD1-folding. (A) Conventional subdomain organization of hCFTR NBD1 (residues 389–674, PDB ID: 2BBO) showing: ABC β- (green), ABC α- (blue), and F1-type ATP-binding core (magenta) subdomains, and regulatory extension (grey). Acceptor dye incorporation sites are indicated. (B) Schematic of ribosome-bound CFP-NBD1 fusion proteins. Asterisk denotes residue Met233 at the CFP C-terminus. (C) Corrected CFP fluorescence emission spectra from D (black), or DA (red) samples. (D) Calculated EFRET for nascent polypeptides (from C), n = 3 ± s.e.m.
Four of the five acceptor sites predicted to reside near the CFP chromophore, residue 233 (in CFP), and residues 450, 487, and 567 (in NBD1) showed high EFRET (66 ± 2%, 46 ± 2%, 43 ± 4% and 33 ± 2%) even though they are separated from the fusion site by 0, 63, 100, and 180 residues, respectively (Figure 3C,D). In contrast, residues 532 and 544 yielded low EFRET (3 ± 2% and 7 ± 2%, respectively). Residue 485, also yielded low EFRET (9 ± 2%) probably due to the disruptive effect of replacing the inward facing Ser485 hydroxyl side chain with the εNBD-Lys moiety. Interestingly, this residue is also associated with a CF-causing mutation (S485C, www.genet.sickkids.on.ca), The slightly reduced EFRET values obtained for residues 450, 487 and 567 (compared to residues 233 in CFP and 389) was likely caused by subtle differences in the acceptor site location or a minor population of RNCs that contain ribosomes stalled between the CFP C-terminus and the UAG codon (as determined by fluorescence per nM of [14C]Lys, data not shown). While we cannot conclude with certainty that ribosome-bound NBD1 achieves a completely native fold, or that all nascent chains fold to the same extent, the FRET pattern obtained here is highly unlikely to result from a random distribution or non-specific association of acceptor dye with the ribosome or CFP. Rather, the strong concordance of measured and predicted EFRET values indicates that multiple residues at multiple locations in nascent NBD1 exhibit a spatial organization that is consistent with a native-like fold.
NBD1 folds in a stepwise and length-dependent manner
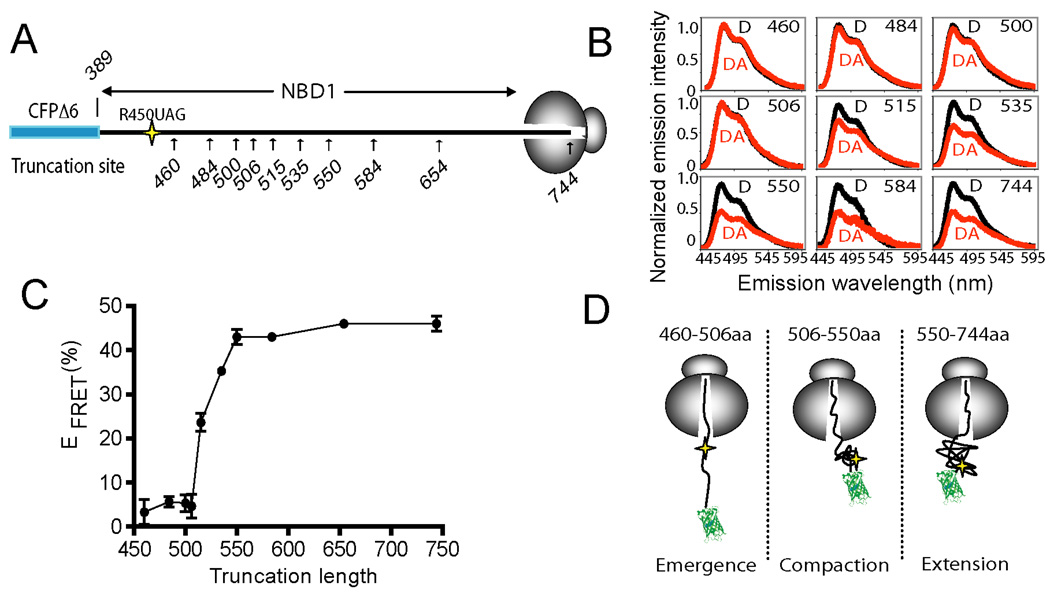
Proteins retained on the ribosome by an intact peptidyl-tRNA ester bond provide snapshots of the elongation process and thereby mimic structural and dynamic properties of nascent chains at a specific stage of synthesis (Cabrita et al., 2010; Eichmann et al., 2010; Johnson, 2005). We therefore envisage (from Figure 3) that NBD1 folding must begin at some point as the polypeptide exits from the ribosome tunnel. To determine the earliest stage at which this occurs, the acceptor dye was incorporated at residue 450, 63 aa C-terminal to the CFP fusion, and EFRET was measured in a series of constructs truncated between residues 460 and 744, each of which emulate a different stage of the cotranslational folding process (Figure 4A). Because donor and acceptor fluorophores are located at identical sites in these nascent chains of different length, the linear separation between the fluorophores must be dictated primarily by the conformation of the intervening polypeptide (Figure 4A).
Figure 4.
Vectorial folding of NBD1. (A) Schematic of CFP-NBD1 fusion protein showing approximate location of acceptor dye (residue 450) and truncation sites used to generate RNCs. (B) Selected corrected CFP emission spectra for D (black) or DA (red) samples as in (A), truncated at the indicated residues. (C) EFRET between CFP and residue 450 plotted as a function of nascent chain length (n = 3, +/− s.e.m. where shown, or average of two experiments for truncations 535, 584 and 654). (D) Schematic showing experimentally derived stages of cotranslational NBD1 folding.
Low EFRET was observed for all truncations between residues 460 and 506 (Average EFRET = 4.7 ± 1%, Figure 4B,C). Thus, the nascent chain remains in an extended conformation until after the acceptor probe has exited the ribosome. As residues 515 to 550 were synthesized, however, EFRET abruptly increased to approximately 43 ± 2% and remained at this level while rest of the domain was synthesized. These results identify three distinct stages of NBD1 folding as determined by the spatial positioning of residue 450 (Figure 4D). In the first stage, N-terminal residues emerge from the ribosome in an extended conformation (truncations 460–506). They then undergo an abrupt compaction for truncations 515–550 as residue 450 is positioned proximal to the CFP fusion site (Stage 2), and maintain this spatial relationship during synthesis of residues 550 to 744 (Stage 3). This suggests that the N-terminal region of NBD1 folds early in the process of NBD1 synthesis and independently of the C-terminal region. Such behavior was initially surprising, as we expected NBD1 folding to occur cooperatively based on its high C.O. and complex topological organization. However, the N-terminus of NBD1 (residues 389–500) has a lower C.O. than the full-length domain (18.9 vs. 24.2) suggesting that local contacts might contribute to local folding of this peptide region.
The NBD1 N-terminal subdomain binds ATP
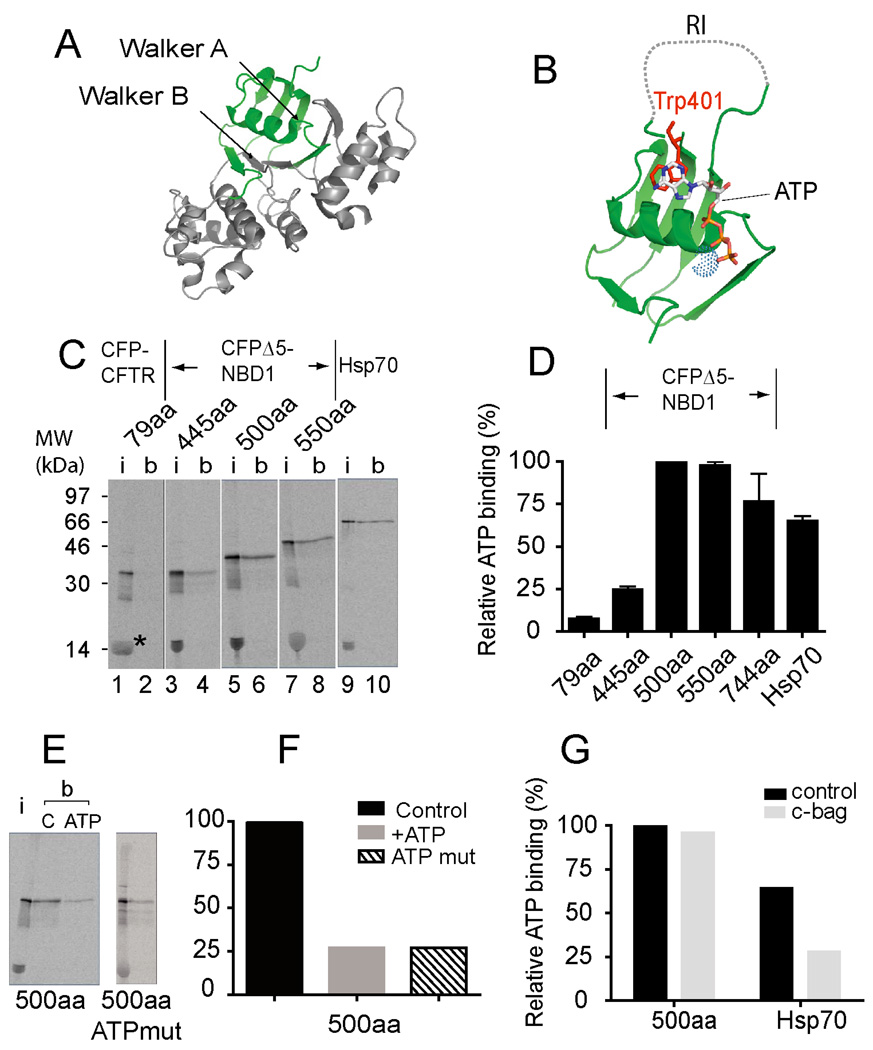
Given that the ribosome can accommodate 30–50 residues of nascent polypeptide (Ban et al., 2000; Hardesty and Kramer, 2001), the stage of NBD1 synthesis at which EFRET increases (truncations 530–550) corresponds approximately to egress of residues 480–510 from the ribosome exit tunnel. This peptide region contains the three-strand ABC β-domain, the RI, 2 β-strands from the F1-type core, and several key elements of the ATP-binding site including: the Walker A motif (Lys464, Thr465), which interacts with the γ-phosphate of ATP, the Q loop (Gln493), Tryp401, which forms pi-electron stacking interactions with the adenine ring, and additional residues predicted to contact the adenine base and ribose (Phe409, Asp425, Leu428, and Phe430)(Lewis et al., 2004; Lewis et al., 2010) (Figure 5A,B). We therefore investigated whether the sharp increase in EFRET might reflect physiological folding of a minimal N-terminal ATP-binding subdomain.
Figure 5.
The NBD1 N-terminal sub-domain binds ATP. (A) hNBD1 crystal structure (2BBO) highlighting residues 389–495 (green) and (B) N-terminal subdomain showing ATP, Trp401 (red) and Mg+2 (dotted sphere). (C) Autoradiogram of [35S] labeled translation products showing input (i = 17% of total) and protein bound (b) to γ-ATP coupled beads. Lanes show CFP fused to the first 79aa of CFTR (1–2), CFP-NBD1 truncated at residue 445 (3–4), residue 500 (5–6), residue 550 (7–8). Lanes 9–10 show human Hsp70. Asterisk indicates RRL globin band. (D) Quantification of samples from (C), also with NBD1 truncation 744 (n=3 +/− s.e.m.) Truncation 500 is set at 100%. (E,F), Competitive inhibition of wild type CFP-NBD1 ATP binding (truncation 500, C = control, ATP = addition of exogenous ATP) and CFP-NBD1 subdomain containing mutations in the ATP-binding site. (G) Binding of CFP-NBD1 (truncation 500) and Hsp70 to γ–ATP beads as in panel D but translated in the presence and absence of recombinant c-Bag protein.
Consistent with this hypothesis, when in vitro translated [35S]Met-labeled CFP-NBD1 polypeptides were truncated at CFTR residue 500, 550 or 744 and released from the ribosome, they bound γ–ATP-conjugated polyacrylamide beads with similar efficiency as a known ATP-binding protein, Hsp70 (Figure 5C,D). Binding was specific, and inhibited (>70%) by excess Mg2+ ATP (Figure 5E,F). In contrast, a shorter truncation containing only NBD1 residues 389–445, a CFP fusion containing CFTR N-terminal residues 1–79, and endogenous RRL globin all exhibited weak or undetectable ATP binding. Thus, the length-dependent acquisition of ATP-binding competence mirrors the stage of synthesis at which initial polypeptide compaction is observed by FRET. To distinguish whether the newly synthesized protein bound directly to ATP or indirectly via an associated chaperone, NBD1 was truncated at residue 500 and translated in the presence of a C-terminal fragment of Bag1 (c-Bag), which stimulates nucleotide exchange and Hsp70 release from client proteins ((Young et al., 2003) and our unpublished observations). Although c-Bag inhibited Hsp70 binding to ATP-coupled beads by more than 50%, it had no effect on NBD1 (Figure 5G). In addition, mutations that inhibit ATP binding to NBD1 in full length CFTR (W401A, A462F & K464A, (Berger et al., 2005)) also inhibited binding to truncated NBD1 (Figure 5E,F). We therefore propose that NBD1 residues 389–500, which reflect the transition boundary with the ABC α-subdomain, comprise a previously unappreciated N-terminal subdomain that folds autonomously to form a minimal ATP binding site.
Kinetics of subdomain folding following ribosome exit
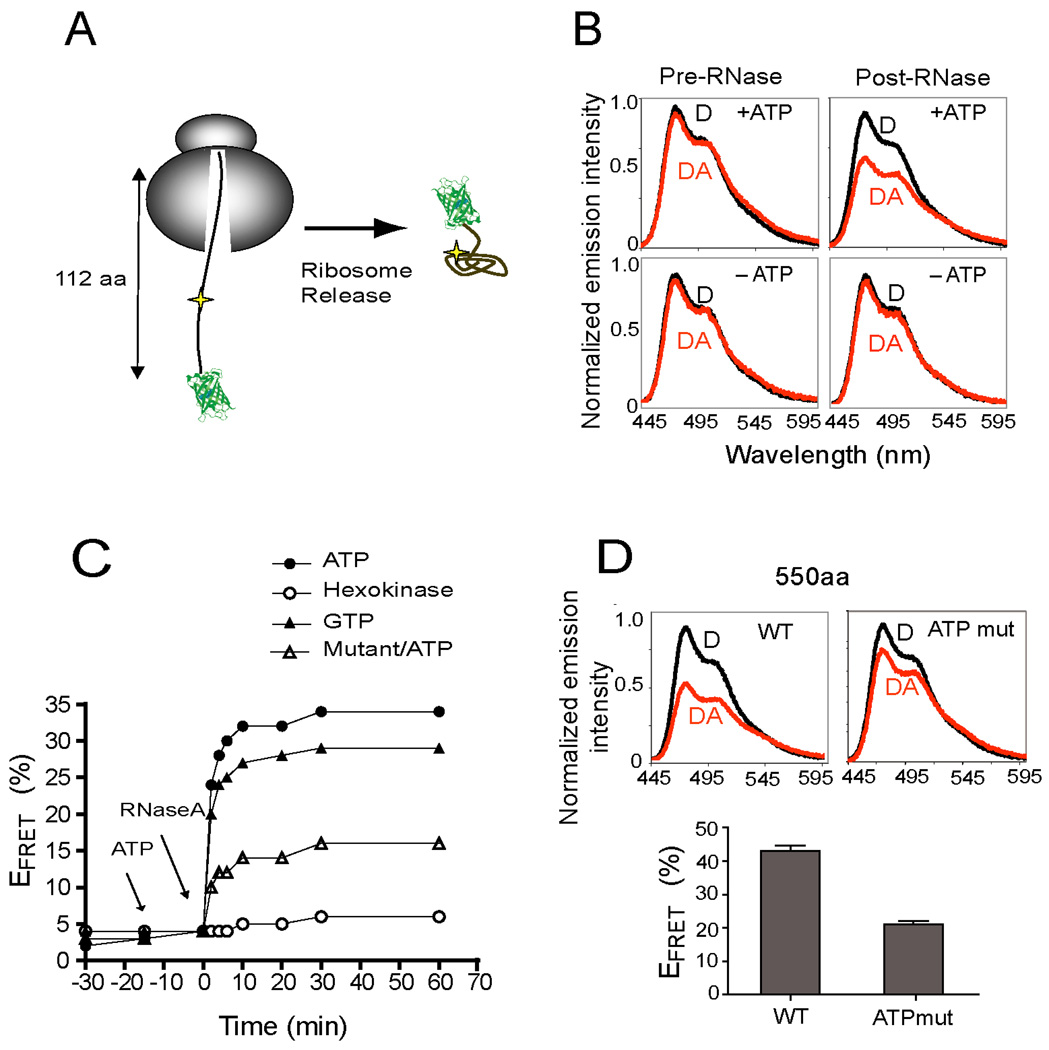
NBD1 residues 389–500 can acquire an ATP-binding competent conformation upon ribosome release (Figure 5). However, folding of ribosome-attached polypeptides occurred only after synthesis of residues 535–550 (Figure 4C). This suggests that during synthesis, N-terminus folding is transiently delayed by the ribosome. We therefore incorporated the acceptor dye at residue 450 and truncated NBD1 at residue 500. In these nascent chains the subdomain is kinetically trapped in an unfolded state by sequestration of critical C-terminal residues in the ribosome exit tunnel (Figure 6A). RNCs were then treated with RNase A, which rapidly cleaves the peptidyl-tRNA bond (in <60 sec, data not shown) and allows C-terminal residues to exit the ribosome, thereby mimicking the process of polypeptide elongation.
Figure 6.
ATP-dependent, subdomain folding. (A) Cartoon depicting ribosome release of CFP-NBD1 (truncation 500) with acceptor dye at residue 450. (B) Corrected CFP emission spectra for D (black) and DA (red) samples in the presence and absence of ATP (left) and 30 min after RNase treatment (right). (C) Real-time change in EFRET for CFP-NBD1 (truncation 500) following RNase treatment in the presence of ATP (closed circles), absence of ATP (open circles) and presence of GTP (closed triangles). Open triangles show results for ATP-binding mutant. Results show average of two independent experiments. (D) Corrected CFP emission spectra and calculated EFRET for ribosome attached wild type and ATP mutant NBD1 (truncation 550, n=3 +/− s.e.m.).
Under these conditions, nascent chains remained in a stably unfolded conformation (EFRET = 5%) while attached to the ribosome, whereas EFRET increased to 35% upon RNAse digestion (Figure 6B). By monitoring the real-time increase in EFRET, we were able to measure de novo folding kinetics as the nascent chain exited from the ribosome tunnel. Remarkably, when RNase was added in the presence of ATP, FRET increased to 75% of the maximum value at the earliest measured (2 min) time point (Figure 6C). ATP addition had no effect on EFRET prior to ribosome release. In contrast, ATP depletion, by addition of hexokinase/2-deoxy D-glucose (Figure 6B,C) or apyrase (not shown), inhibited subdomain folding. These results demonstrate that as the N-terminus exits the ribosome, folding takes place on a time scale comparable to eukaryotic translation.
ATP serves as ligand that facilitates de novo NBD1 folding
To confirm that the rapid increase in EFRET was caused by direct ATP binding to the nascent polypeptide, nascent chains were released from ribosomes in the presence of GTP, which stabilizes full length NBD1 in solution (P. J. Thomas, personal communication), but does not release chaperones (e.g. Hsp70) from client proteins. Notably, GTP stimulated EFRET with similar kinetics and to approximately 80% of the level observed for ATP (Figure 6C). In addition, the ATP-binding mutant decreased EFRET by approximately 2.5 fold. Thus, multiple residues in the ATP binding site must be properly organized in the sub-domain for folding to occur. The ATP-binding mutant also exhibited reduced folding as demonstrated by a 50% reduction in EFRET for ribosome-attached nascent chains (truncation 550) even though ATP was present during translation (Figure 6D). Taken together, these results indicate that nucleotide stimulates rapid, cotranslational folding as the N-terminal subdomain exits the ribosome during the early stage of NBD1 synthesis.
The effect of F508del on N-terminal subdomain folding
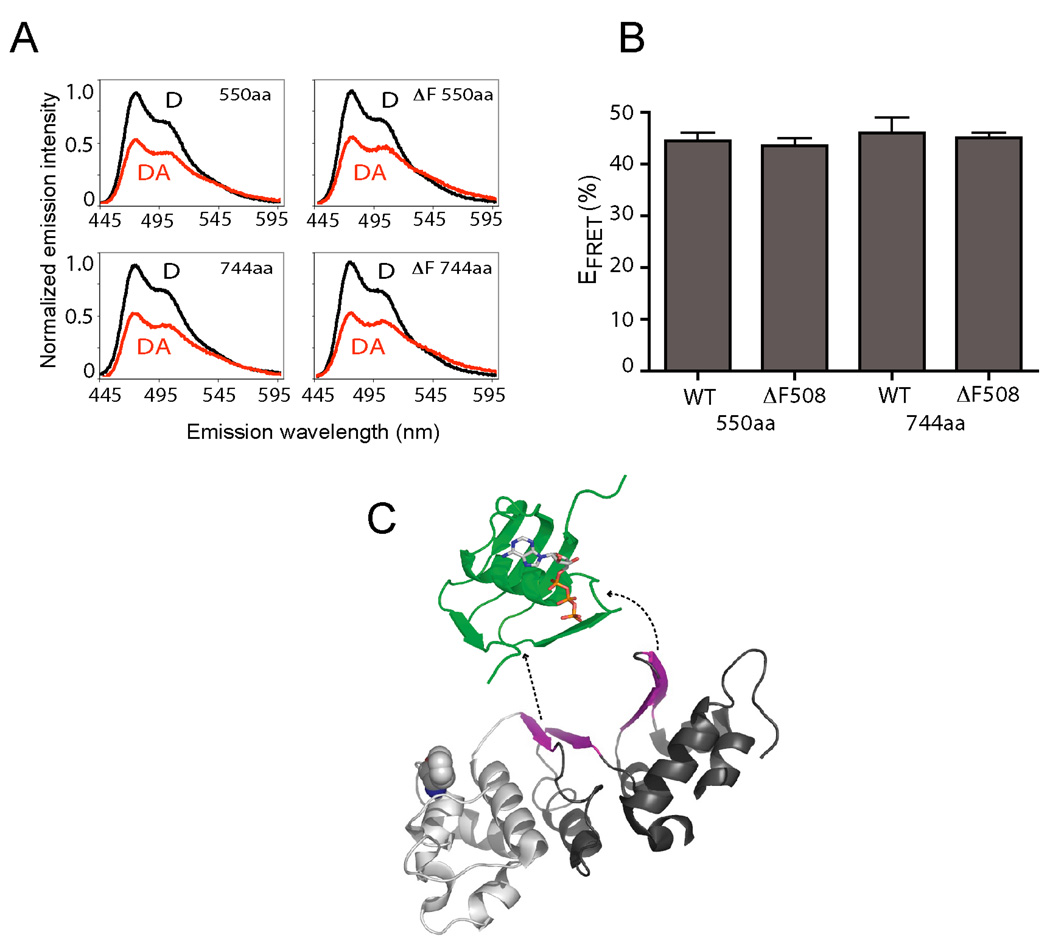
Finally, we tested the impact of Phe508 on NBD1 folding by comparing EFRET for wild type and F508del nascent chains. Results show that EFRET to residue 450 was indistinguishable for wild type and F508del constructs, both at the initial stage of folding (truncation 550) and after synthesis of the entire domain (truncation 744) (Figure 7A,B). This is consistent with our findings that residues 389–500 fold autonomously in the absence of the α-subdomain (Figure 5), and that Phe508 is likely still in the ribosome exit tunnel as residue 500 emerges and folding begins (Figure 4). Although we could not test whether F508del affected folding kinetics, because such experiments require truncation upstream of the α-subdomain, our results indicate that the N-terminal subdomain is capable of folding independent of Phe508, and that the F508del defect likely affects later events in the cotranslational folding pathway.
Figure 7.
Effects of F508del on cotranslational NBD1 folding. (A,B) Comparison of EFRET obtained for ribosome-bound wild type and F508del CFP-NBD1 nascent chains containing acceptor dye at residue 450 and truncated at residues 550 and 744 (n=3, +/− s.e.m). (C) Vectorial NBD1 topography (green-white-black in the order of synthesis) reflecting rapid folding of the ATP binding subdomain (green) and late formation of the 6-strand β-sheet F1-type ATPase core (magenta). (See also Figure S4.)
DISCUSSION
As proteins are synthesized in the cell, they must acquire native structure in a crowded environment while avoiding non-native conformations that lead to misfolding and/or aggregation. Little is known about the nature of these progressive de novo folding pathways. In this study, we developed a generally applicable FRET-based method using two translationally incorporated fluorophores that allow direct assessment of structural transitions in ribosome-bound nascent polypeptides. Our results define a multi-step cotranslational folding pathway for human CFTR NBD1 and provide a mechanistic description of how an ABC transporter acquires native tertiary structure in a cellular environment.
Specifically, we show that: i) NBD1 folding begins cotranslationally on the ribosome. These results support previous studies suggesting that CFTR domains fold cotranslationally in vitro (Kleizen et al., 2005) and in vivo (Du et al., 2005). ii) Folding is vectorial and initiated by local compaction of a small (~112 aa) N-terminal α/β-subdomain (helix H1, five β-strands (S1–S4, S6) and the RI) that forms a functional ATP binding site. iii) Subdomain folding is rapid and occurs coincident with synthesis. iv) Most remarkably, ATP, which stabilizes native NBD1 in solution (Lewis et al., 2004) and binds NBD1 peptide fragments (Thomas et al., 1991), also stimulates vectorial folding by inducing an energetically favored local (N-terminal) polypeptide conformation. One possibility is that ATP shifts the dynamic ensemble of nascent chains that are present at a critical stage of synthesis, thereby stabilizing a preexisting ATP-binding conformation. This would be consistent with the equilibrium-shift mechanism observed upon ligand binding to intact proteins (Henzler-Wildman and Kern, 2007). Alternatively, given that synthesis and folding are coupled in time, it is possible that ATP binding is initiated at a partially formed site (Thomas et al., 1991) that preferentially matures as additional residues are synthesized.
A unique aspect of our approach is the ability to characterize nascent protein folding using both conventional static snapshots obtained from truncated ribosome-tethered polypeptides (Cabrita et al., 2009; Eichmann et al., 2010; Woolhead et al., 2004), and dynamic conformational changes that take place as the nascent polypeptide exits the ribosome tunnel. By synchronously releasing kinetically trapped intermediates, we were able to measure real-time, de novo folding kinetics of a nascent polypeptide. Within the resolution of our study, N-terminal subdomain folding occurs on a time scale (seconds to minutes) comparable to eukaryotic translation (Netzer and Hartl, 1997). Although this resolution is modest by biophysical standards (Kubelka et al., 2004), when viewed from a vectorial perspective imposed by the rate of synthesis in cells (Hardesty and Kramer, 2001), N-terminus compaction occurs coincident with synthesis of the ABC α-subdomain and likely precedes formation of the F1-type β-sheet core (Lewis et al., 2004). Folding of N-terminal and C-terminal regions of NBD1 are therefore separated both spatially and temporally during synthesis. These data support a model predicted by in silico topological accessibility studies in which ancient α/β class proteins preferentially initiate directional protein folding via local N-terminal contacts (Ellis et al., 2010).
Given the preponderance of long-distance intra-chain interactions in native NBDs, (contact order = 24.2 (Plaxco et al., 1998), http://depts.washington.edu/bakerpg/contact_order/), it is reasonable to speculate that N-terminal subdomain folding provides a template or interface that contributes to the vectorial order of folding events (Figure 7C). Such an ordered folding pathway may function to minimize non-native interactions that predominate during in vitro refolding (Fedorov and Baldwin, 1999). This may be particularly important for the F1-type core subdomain, which contains the highly conserved Walker A and Walker B motifs, and requires intercalation of six β-strands (S3 and S6–S10) that are widely separated in primary sequence (Figure 7C and (Lewis et al., 2004)). If one considers N-terminal subdomain organization in the context of its vectorial folding pathway, then the first three β-strands, S1, S2 and S4, constitute the previously described ABC β-subdomain while S3, S6 and H1 are part of the previously described F1-type ATPase core (Figures 5A,B and 7C). The remaining four strands of the core β-sheet, S7–10, are synthesized in a discontinuous manner after the α-subdomain and additional intervening helices.
Our data therefore suggest a modification to the canonical NBD subdomain classification, which was originally based on alignments with adenylate kinase (Hyde et al., 1990; Mimura et al., 1991) and the F0, F1 ATPase (Annereau et al., 1997). Notably, synthesis of the entire F1-type ATPase core domain is not a prerequisite for ATP binding. An alternative classification might therefore include: i) an N-terminal α/β subdomain (residues 389 to ~495), which comprises the minimal ATP-binding site, and ii) the ABC α-subdomain (residues 495–567), which contains the LSGGQ signature motif and forms key structural contacts with transmembrane domains (Serohijos et al., 2008). Accordingly, the β-sheet core represents a distinct folding module that is likely formed during the final assembly of the domain. Because this vectorial topography is highly conserved among ABC transporter NBDs, an important future question is whether key elements of the cotranslational folding pathway identified here are specific to CFTR or represent general features of this transporter superfamily (Figure S4).
Interestingly, Phe508 is not required for ATP-binding and is still in the ribosome tunnel when the N-terminal subdomain begins to fold. Based on the EFRET to residue 450, the F508del mutation had no effect on cotranslational folding of either truncated or full-length NBD1. The simplest explanation for these findings is that F508del impacts the efficiency and/or kinetics during a late stage(s) of NBD1 folding that involves the α-subdomain and/or formation of the β-sheet core. This may explain why second-site suppressors of F508del misfolding frequently reside within the α-subdomain, or at the N-terminal subdomain interface (He et al., 2010; Pissara et al., 2008; Teem et al., 1993). Interestingly though, removal of the RI within the N-terminal subdomain partially restores full length F508del CFTR trafficking and function in mammalian cells (Aleksandrov et al., 2010). Thus, it remains possible that some aspects of N-terminus folding and/or structure, particularly involving the RI, may be impacted by Phe508 through allosteric interactions through the intact domain.
From a mechanistic standpoint, the extensive surface contacts between NBD1 subdomains, and in particular the final inter-digitations of β-strands in the domain core, likely require maintenance of a folding-competent conformation until after most or all of the domain has been synthesized. How is this achieved in the crowded environment of the cell? While we did not observe a direct role for chaperones in N-terminal subdomain folding in this study, Hsp70 and its co-chaperones have been shown to stabilize NBD1 in vitro (Strickland et al., 1997), and interact with NBD1 in vivo (Meacham et al., 1999; Rosser et al., 2008). It is possible that transient chaperone binding to the N-terminus may occur in the cytosol during synthesis and prior to RNC isolation. In addition, peptide binding studies have identified a putative Hsp70 binding site (residues 545–561) that resides near the interface of the α-subdomain and β-sheet core (Strickland et al., 1997). Hsp/c70 also binds to full length wild type CFTR and exhibits prolonged binding to F508del CFTR (Yang et al., 1993). Thus, cellular chaperones almost certainly play a role in the late stages of α-subdomain folding and/or core formation, which likely represent the rate limiting, and possibly the pathologic step of NBD1 folding in CF patients.
EXPERIMENTAL PROCEDURES
Plasmids
CFP-NBD1 fusion constructs were generated by removing the last 5 C-terminal residues from eCFP (Clontech), inserting a gly residue, and ligating to the first residue (Thr389) of the CFTR NBD1 domain (wild type or F508del) in a pSP64 vector (Promega). TAG stop codons were engineered at indicated sites by PCR overlap extension, and truncated cDNA for in vitro transcription was generated by PCR amplification using appropriate antisense oligonucleotides. The CFP-NBD1 ATP-binding mutant contained W401A, A462F, and K464A mutations in the ATP binding site as predicted by the crystal structure (2BBO). All cloned PCR fragments were verified by sequencing.
In Vitro Transcription/Translation
Translation was carried out in RRL using in vitro transcribed mRNA templates for 72 min at 24°C. Four 250 µl reactions were translated in parallel in the presence of 0.8–1 µM [14C]Lys-tRNAamb or ε-NBD-[14C]Lys-tRNAamb to generate samples containing: Blank with no Donor or Acceptor dye (B), Donor only (D), Acceptor only (A), and Donor + Acceptor (DA). Ribosome-nascent chain complexes were isolated at 4°C by size exclusion chromatography. For Figure 2, translation was carried out in the presence of [35S]Met and products were analyzed by SDS-PAGE before or after pelleting RNCs through 35% sucrose, 40mM HEPES-pH 7.6, 100mM KOAc, and 15mM MgOAc at 395,000 × g for 1hr
Steady State Fluorescence Measurements
CFP fluorescence emission spectra (λex = 430 nm, λem = 445–600 nm) were obtained using a Fluorolog 3–22 (HORIBA Jobin Yvon) fluorimeter. Measurements were performed at 4°C unless otherwise stated. FRET efficiency (EFRET) was determined from the acceptor-induced decrease in donor fluorescence emission intensity (at λmax = 475 nm) using E = 1 - [F(DA - A)/FD] after (i) subtraction of the background signal, and (ii) normalization of the net FDA, FD, and FA emission intensities to the same donor dye concentration as determined by scintillation counting of 14C-labeled polypeptides in each sample. Because FA was undetectable at λem = 475 nm, FDA-FA = FDA and the equation E = 1-FDA/FD was also used (See Figures 1, S2, and Supplementary Experimental Procedures). Folding kinetics of ribosome released nascent chains were determined at 25°C from the CFP emission intensity of each sample (λex = 430 nm and λem = 475 nm) measured at indicated times before and after addition of RNase A (0.2 mg/ml) following pretreatment with Mg/ATP (3 mM), Mg/GTP (3 mM), or hexokinase/2-deoxy-D-glucose.
ATP binding Assay
CFP-NBD1 constructs were translated in RRL the presence of [35S]Met. Nascent chains were released from ribosomes by RNase digestion, and lysates were gel filtered (Sephadex G-25) and incubated with γ-ATP-conjugated polyacrylamide beads as per manufacturer’s recommendations (ProteoEnrich™ ATP-Binders™ Kit, EMD Chemicals Inc) at 4°C. Beads were washed and eluted with 1% SDS or elution buffer containing 20 mM ATP. Eluate was concentrated and analyzed by SDS-PAGE and quantified by phosphorimaging. For ATP competition, binding was performed in the presence of 35 mM ATP. Where indicated, translations were performed in the presence of 14 µM recombinant c-Bag protein.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Drs. Volodmir Zeeneko for technical support, Devaki Kelkar for sharing experimental information, Jason Young for providing Bag1 cDNA and Yoshihiro Matsumura for purified c-Bag protein, Elisa Cooper for early development of CFP-CFTR fusion proteins, David Farrens and Ujwal Shinde for critical reading of manuscript, and Mr. Karl Rusterholtz for technical assistance. This work was supported by research grants from Cystic Fibrosis Foundation Therapeutics Inc. (CFFTI) and NIH GM53457, DK51818 (W.R.S), and also NIH GM26494, NSF EF-0623664, and Robert A. Welch Foundation Chair grant BE-0017 (A.E.J).
Abbreviations
- ABC transporter
ATP Binding Cassette transporter
- CFP
cyan fluorescent protein
- C.O.
contact order
- CFTR
Cystic fibrosis transmembrane conductance regulator
- FRET
Förster resonance energy transfer
- EFRET
FRET efficiency
- CF
cystic fibrosis
- NBD1
nucleotide binding domain 1
- RI
regulatory insertion
- RNC
ribosome nascent chain complex
- RRL
rabbit reticulocyte lysate
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental information includes Supplementary Data: Figures S1, S2, S3, S4 and Tables S1, S2. Additional experimental details and theoretical considerations used for FRET calculations are included in Supplementary Experimental Procedures.
REFERENCES
- Aleksandrov A, Kota P, Aleksandrov L, He L, Jenson T, Cui L, Gentzsch M, Dokholyan N, Riordan J. Regulatory insertion removal restores maturation, stability and function of deltaF508 CFTR. J. Mol. Biol. 2010;401:194–210. doi: 10.1016/j.jmb.2010.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annereau J, Wulbrand U, Vankeerberghen A, Cuppens H, Bontems F, Tümmler B, Cassiman J, Stoven V. A novel model for the first nucleotide binding domain of the cystic fibrosis transmembrane conductance regulator. FEBS lett. 1997;407:303–308. doi: 10.1016/s0014-5793(97)00363-3. [DOI] [PubMed] [Google Scholar]
- Balch W, Morimoto R, Dillin A, Kellly J. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- Barral JM, Broadley SA, Schaffar G, Hartl FU. Roles of molecular chaperones in protein misfolding diseases. Semin Cell Dev Biol. 2004;15:17–29. doi: 10.1016/j.semcdb.2003.12.010. [DOI] [PubMed] [Google Scholar]
- Becker T, Bhushan S, Jarasch A, Armache J, Funes S, Jossinet F, Gumbart J, Mielke T, Berminghausen O, Schulten K, et al. Structure of momomeric yeast and mammalian Sec671 complexes interacting with the translating ribosome. Science. 2009;326:1369–1373. doi: 10.1126/science.1178535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger AL, Ikuma M, Welsh MJ. Normal gating of CFTR requires ATP binding to both nucleotide-binding domains and hydrolysis at the second nucleotide-binding domain. Proc Natl Acad Sci U S A. 2005;102:455–460. doi: 10.1073/pnas.0408575102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrita LD, Dobson CM, Christodoulou J. Protein folding on the ribosome. Curr Opin Struct Biol. 2010;20:33–45. doi: 10.1016/j.sbi.2010.01.005. [DOI] [PubMed] [Google Scholar]
- Cabrita LD, Hsu ST, Launay H, Dobson CM, Christodoulou J. Probing ribosome-nascent chain complexes produced in vivo by NMR spectroscopy. Proc Natl Acad Sci U S A. 2009;106:22239–22244. doi: 10.1073/pnas.0903750106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carveth K, Buck T, Anthony V, Skach WR. Cooperativity and flexibility of cystic fibrosis transmembrane conductance regulator transmembrane segments participate in membrane localization of a charged residue. J Biol Chem. 2002;277:39507–39514. doi: 10.1074/jbc.M205759200. [DOI] [PubMed] [Google Scholar]
- Cheng SH, Gregory RJ, Marshall J, Paul S, Souza DW, White GA, O'Riordan CR, Smith AE. Defective intracellular transport and processing of CFTR is the molecular basis of most cystic fibrosis. Cell. 1990;63:827–834. doi: 10.1016/0092-8674(90)90148-8. [DOI] [PubMed] [Google Scholar]
- Clark PL. Protein folding in the cell: reshaping the folding funnel. Trends Biochem Sci. 2004;29:527–534. doi: 10.1016/j.tibs.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Clark PL, King J. A newly synthesized, ribosome-bound polypeptide chain adopts conformations dissimilar from early in vitro refolding intermediates. J Biol Chem. 2001;276:25411–25420. doi: 10.1074/jbc.M008490200. [DOI] [PubMed] [Google Scholar]
- Dawson RJ, Locher KP. Structure of a bacterial multidrug ABC transporter. Nature. 2006;443:180–185. doi: 10.1038/nature05155. [DOI] [PubMed] [Google Scholar]
- Du K, Sharma M, Lukacs GL. The DeltaF508 cystic fibrosis mutation impairs domain-domain interactions and arrests post-translational folding of CFTR. Nat Struct Mol Biol. 2005;12:17–25. doi: 10.1038/nsmb882. [DOI] [PubMed] [Google Scholar]
- Eichmann C, Preissler S, Reik R, Deuerling E. Cotranslational structure acquisition of nascent polypeptides monitored by NMR spectroscopy. Proc Nat Acad Sci, USA. 2010;107:9111–9116. doi: 10.1073/pnas.0914300107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis J, Huard F, Deane C, Srivastava S, Wood G. Directionality in protein folding prediction. BMC Bioinformatics. 2010;11:172. doi: 10.1186/1471-2105-11-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis RJ. Macromolecular crowding: obvious but underappreciated. Trends Biochem Sci. 2001;26:597–604. doi: 10.1016/s0968-0004(01)01938-7. [DOI] [PubMed] [Google Scholar]
- Ellis RJ. Protein misassembly: macromolecular crowding and molecular chaperones. Adv Exp Med Biol. 2007;594:1–13. doi: 10.1007/978-0-387-39975-1_1. [DOI] [PubMed] [Google Scholar]
- Evans MS, Sander IM, Clark PL. Cotranslational folding promotes beta-helix formation and avoids aggregation in vivo. J Mol Biol. 2008;383:683–692. doi: 10.1016/j.jmb.2008.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorov AN, Baldwin TO. Cotranslational protein folding. J Biol Chem. 1997;272:32715–32718. doi: 10.1074/jbc.272.52.32715. [DOI] [PubMed] [Google Scholar]
- Fedorov AN, Baldwin TO. Process of biosynthetic protein folding determines the rapid formation of native structure. J Mol Biol. 1999;294:579–586. doi: 10.1006/jmbi.1999.3281. [DOI] [PubMed] [Google Scholar]
- Frydman J. Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu Rev Biochem. 2001;70:603–647. doi: 10.1146/annurev.biochem.70.1.603. [DOI] [PubMed] [Google Scholar]
- Hardesty B, Kramer G. Folding of a nascent peptide on the ribosome. Prog Nucleic Acid Res Mol Biol. 2001;66:41–66. doi: 10.1016/s0079-6603(00)66026-9. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M. Converging concepts of protein folding in vitro and in vivo. Nat Struct Mol Biol. 2009;16:574–581. doi: 10.1038/nsmb.1591. [DOI] [PubMed] [Google Scholar]
- He L, Aleskandrov L, Cui L, Jension T, Nesbitt K, Riordan J. Restoration of domain folding and interdomain assembly by second-site suppressors of the deltaF508 mutation in CFTR. FASEB J. 2010;24:3103–3112. doi: 10.1096/fj.09-141788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henzler-Wildman K, Kern D. Dynamic personalities of proteins. Nature. 2007;450:964–972. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- Hsu ST, Fucini P, Cabrita LD, Launay H, Dobson CM, Christodoulou J. Structure and dynamics of a ribosome-bound nascent chain by NMR spectroscopy. Proc Natl Acad Sci U S A. 2007;104:16516–16521. doi: 10.1073/pnas.0704664104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde S, Emsley P, Hartshorn M, Mimmack M, Gileadi U, Pearce S, Gallaghe M, Gill D, Hubbard R, Higgens C. Structural model of ATP-binding proteins associated with cystic fibrosis, multidrug resistance and bacterial transport. Nature. 1990;346:312–313. doi: 10.1038/346362a0. [DOI] [PubMed] [Google Scholar]
- Johnson AE. The co-translational folding and interactions of nascent protein chains: a new approach using fluorescence resonance energy transfer. FEBS Lett. 2005;579:916–920. doi: 10.1016/j.febslet.2004.11.046. [DOI] [PubMed] [Google Scholar]
- Kaiser CM, Chang HC, Agashe VR, Lakshmipathy SK, Etchells SA, Hayer-Hartl M, Hartl FU, Barral JM. Real-time observation of trigger factor function on translating ribosomes. Nature. 2006;444:455–460. doi: 10.1038/nature05225. [DOI] [PubMed] [Google Scholar]
- Kajihara D, Abe R, Iijima I, Komiyama C, Sisido M, Hohsaka T. FRET analysis of protein conformational change through position-specific incorporation of fluorescent amino acids. Nat Methods. 2006;3:923–929. doi: 10.1038/nmeth945. [DOI] [PubMed] [Google Scholar]
- Kleizen B, van Vlijmen T, de Jonge HR, Braakman I. Folding of CFTR is predominantly cotranslational. Mol Cell. 2005;20:277–287. doi: 10.1016/j.molcel.2005.09.007. [DOI] [PubMed] [Google Scholar]
- Komar AA. A pause for thought along the co-translational folding pathway. Trends Biochem Sci. 2009;34:16–24. doi: 10.1016/j.tibs.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Kubelka J, Hofrichter J, Eaton W. The protein folding "Speed limit". Curr. Opin. Stuct. Biol. 2004;14:76–88. doi: 10.1016/j.sbi.2004.01.013. [DOI] [PubMed] [Google Scholar]
- Lewis HA, Buchanan SG, Burley SK, Conners K, Dickey M, Dorwart M, Fowler R, Gao X, Guggino WB, Hendrickson WA, et al. Structure of nucleotide-binding domain 1 of the cystic fibrosis transmembrane conductance regulator. EMBO J. 2004;23:282–293. doi: 10.1038/sj.emboj.7600040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis HA, Wang C, Zhao X, Hamuro Y, Conners K, Kearins MC, Lu F, Sauder JM, Molnar KS, Coales SJ, et al. Structure and dynamics of NBD1 from CFTR characterized using crystallography and hydrogen/deuterium exchange mass spectrometry. J Mol Biol. 2010;396:406–430. doi: 10.1016/j.jmb.2009.11.051. [DOI] [PubMed] [Google Scholar]
- Lewis HA, Zhao X, Wang C, Sauder JM, Rooney I, Noland BW, Lorimer D, Kearins MC, Conners K, Condon B, et al. Impact of the deltaF508 mutation in first nucleotide-binding domain of human cystic fibrosis transmembrane conductance regulator on domain folding and structure. J Biol Chem. 2005;280:1346–1353. doi: 10.1074/jbc.M410968200. [DOI] [PubMed] [Google Scholar]
- Lu J, Deutsch C. Folding zones inside the ribosomal exit tunnel. Nat. Struct. Mol. Biol. 2005;12:1123–1129. doi: 10.1038/nsmb1021. [DOI] [PubMed] [Google Scholar]
- Luheshi LM, Crowther DC, Dobson CM. Protein misfolding and disease: from the test tube to the organism. Curr Opin Chem Biol. 2008;12:25–31. doi: 10.1016/j.cbpa.2008.02.011. [DOI] [PubMed] [Google Scholar]
- Meacham GC, Lu Z, King S, Sorscher E, Tousson A, Cyr DM. The Hdj-2/Hsc70 chaperone pair facilitates early steps in CFTR biogenesis. EMBO J. 1999;18:1492–1505. doi: 10.1093/emboj/18.6.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza JL, Thomas PJ. Building an understanding of cystic fibrosis on the foundation of ABC transporter structures. J Bioenerg Biomembr. 2007;39:499–505. doi: 10.1007/s10863-007-9117-7. [DOI] [PubMed] [Google Scholar]
- Mimura C, Holbrook S, Ames G. Structural model of the nucleotide-binding conserved component of periplasmic permeases. Proc Nat Acad Sci, USA. 1991;88:84–88. doi: 10.1073/pnas.88.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake-Stoner SJ, Miller AM, Hammill JT, Peeler JC, Hess KR, Mehl RA, Brewer SH. Probing protein folding using site-specifically encoded unnatural amino acids as FRET donors with tryptophan. Biochemistry. 2009;48:5953–5962. doi: 10.1021/bi900426d. [DOI] [PubMed] [Google Scholar]
- Netzer WJ, Hartl FU. Recombination of protein domains facilitated by co-translational folding in eukaryotes. Nature. 1997;388:343–349. doi: 10.1038/41024. [DOI] [PubMed] [Google Scholar]
- Pissara LS, Farinha CM, Xu Z, Schmidt A, Thibodeau PH, Cai Z, Thomas PJ, Sheppard DN, Amaral MD. Solubilizing mutations used to crystalize one CFTR domain attenuate the trafficking and channel defects caused by the major cystic fibrosis mutation. Chem Biol. 2008;1:62–69. doi: 10.1016/j.chembiol.2007.11.012. [DOI] [PubMed] [Google Scholar]
- Plaxco KW, Simons KT, Baker D. Contact order, transition state placement and the refolding rates of single domain proteins. J Mol Biol. 1998;277:985–994. doi: 10.1006/jmbi.1998.1645. [DOI] [PubMed] [Google Scholar]
- Qu BH, Strickland EH, Thomas PJ. Localization and suppression of a kinetic defect in cystic fibrosis transmembrane conductance regulator folding. J Biol Chem. 1997;272:15739–15744. doi: 10.1074/jbc.272.25.15739. [DOI] [PubMed] [Google Scholar]
- Riordan J. CFTR function and prospects for therapy. Annu Rev Biochem. 2008;77:701–726. doi: 10.1146/annurev.biochem.75.103004.142532. [DOI] [PubMed] [Google Scholar]
- Rosser MF, Grove DE, Chen L, Cyr DM. Assembly and misassembly of cystic fibrosis transmembrane conductance regulator: folding defects caused by deletion of F508 occur before and after the calnexin-dependent association of membrane spanning domain (MSD) 1 and MSD2. Mol Biol Cell. 2008;19:4570–4579. doi: 10.1091/mbc.E08-04-0357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serohijos AW, Hegedus T, Aleksandrov AA, He L, Cui L, Dokholyan NV, Riordan JR. Phenylalanine-508 mediates a cytoplasmic-membrane domain contact in the CFTR 3D structure crucial to assembly and channel function. Proc Natl Acad Sci U S A. 2008;105:3256–3261. doi: 10.1073/pnas.0800254105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siller E, DeZwaan DC, Anderson JF, Freeman BC, Barral JM. Slowing bacterial translation speed enhances eukaryotic protein folding efficiency. J Mol Biol. 2010;396:1310–1318. doi: 10.1016/j.jmb.2009.12.042. [DOI] [PubMed] [Google Scholar]
- Strickland E, Qu BH, Millen L, Thomas PJ. The molecular chaperone Hsc70 assists the in vitro folding of the N-terminal nucleotide-binding domain of the cystic fibrosis transmembrane conductance regulator. J Biol Chem. 1997;272:25421–25424. doi: 10.1074/jbc.272.41.25421. [DOI] [PubMed] [Google Scholar]
- Stryer L. Fluorescence energy transfer as a spectroscopic ruler. Annu Rev Biochem. 1978;47:819–846. doi: 10.1146/annurev.bi.47.070178.004131. [DOI] [PubMed] [Google Scholar]
- Taraska JW, Zagotta WN. Structural dynamics in the gating ring of cyclic nucleotide-gated ion channels. Nat Struct Mol Biol. 2007;14:854–860. doi: 10.1038/nsmb1281. [DOI] [PubMed] [Google Scholar]
- Teem J, Berger H, Ostedgaard LDR, Tsui LC, Welsh M. Identification of revertants for the cystic fibrosis delta F508 mutation using STE6-CFTR chimeras in yeast. Cell. 1993;73:335–346. doi: 10.1016/0092-8674(93)90233-g. [DOI] [PubMed] [Google Scholar]
- Thibodeau P, Richardson J, Wang W, Millen L, Watson J, Mendoza J, Du K, Fischman S, Senderowitz H, Lukacs G, et al. The cystic fibrosis-causing mutation ΔF508 affects multiple steps in cystic fibrosis transmembrane conductance regulator biogenesis. J. Biol. Chem. 2010;285:35825–35835. doi: 10.1074/jbc.M110.131623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibodeau PH, Brautigam CA, Machius M, Thomas PJ. Side chain and backbone contributions of Phe508 to CFTR folding. Nat Struct Mol Biol. 2005;12:10–16. doi: 10.1038/nsmb881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PJ, Shenbagamurthi P, Ysern X, Pedersen PL. Cystic fibrosis transmembrane conductance regulator: nucleotide binding to a synthetic peptide. Science. 1991;251:555–557. doi: 10.1126/science.1703660. [DOI] [PubMed] [Google Scholar]
- Voss NR, Gerstein M, Steitz TA, Moore PB. The geometry of the ribosomal polypeptide exit tunnel. J Mol Biol. 2006;360:893–906. doi: 10.1016/j.jmb.2006.05.023. [DOI] [PubMed] [Google Scholar]
- Wang C, Protasevich I, Yang Z, Seehausen D, Skalak T, Zhao X, Atwell S, Emtage J, Wetmore D, Brouillette C, Hunt J. Integrated biophysical studies implicate partial unfolding of NBD1 of CFTR in the molecular pathogenesis of F508del cystic fibrosis. Pro. Sci. 2010 doi: 10.1002/pro.480. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolhead CA, McCormick PJ, Johnson AE. Nascent membrane and secretory proteins differ in FRET-detected folding far inside the ribosome and in their exposure to ribosomal proteins. Cell. 2004;116:725–736. doi: 10.1016/s0092-8674(04)00169-2. [DOI] [PubMed] [Google Scholar]
- Yang Y, Janich S, Cohn J, Wilson J. The common variant of cystic fibrosis transmembrane conductance regulator is recognized by hsp70 and degraded in a pre-Golgi nonlysosomal compartment. Proc. Natl. Acad. Sci. USA. 1993;90:9480–9484. doi: 10.1073/pnas.90.20.9480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JC, Hoogenraad NJ, Hartl FU. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell. 2003;112:41–50. doi: 10.1016/s0092-8674(02)01250-3. [DOI] [PubMed] [Google Scholar]
- Ziv G, Haran G, Thirumalai D. Ribosome exit tunnel can entropically stabilize alpha-helices. Proc Natl Acad Sci U S A. 2005;102:18956–18961. doi: 10.1073/pnas.0508234102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.