Abstract
Background
Chronic tendon injuries, also known as tendinopathies, are common among professional and recreational athletes. These injuries result in a significant amount of morbidity and health care expenditure, yet little is known about the molecular mechanisms leading to tendinopathy.
Methods
We have used histological evaluation and molecular profiling to determine gene expression changes in 23 human patients undergoing surgical procedures for the treatment of chronic tendinopathy.
Results
Diseased tendons exhibit altered extracellular matrix, fiber disorientation, increased cellular content and vasculature, and the absence of inflammatory cells. Global gene expression profiling identified 983 transcripts with significantly different expression patterns in the diseased tendons. Global pathway analysis further suggested altered expression of extracellular matrix proteins and the lack of an appreciable inflammatory response.
Conclusions
Identification of the pathways and genes that are differentially regulated in tendinopathy samples will contribute to our understanding of the disease and the development of novel therapeutics.
Background
Chronic injuries to the Achilles, patellar, extensor carpi radialis brevis, and supraspinatus tendons remain a common problem for both elite and recreational athletes, as well as for individuals engaging in repetitive activities. These overuse type injuries account for 30-50% of all sports injuries and result in a significant amount of morbidity and health care expenditure [1].
Histologic studies have shown that the primary pathology is not inflammation as implied by the commonly used term "tendonitis." Instead, samples of diseased tendons show collagen degeneration, fiber disorientation, mucoid ground substance, hypercellularity, vascular ingrowth, and relative absence of inflammatory cells under light microscopy [2-4]. Tendinopathy (or tendinosis) is now the term most commonly used to describe the clinical entity and histologic findings. Interestingly, these findings are common to all tendinopathies, suggesting a similar etiology and pathophysiology.
The etiology of tendinopathy remains unclear, but most believe that a combination of extrinsic and intrinsic factors is responsible. The extrinsic theory suggests that direct mechanical contact leads to tendon fiber micro-damage and subsequent injury of the tendon that eventually results in weakness and pain. An example is impingement of the acromion on the supraspinatus tendon, which serves as the rationale behind acromioplasty surgery [5]. The intrinsic theory suggests that the tendon itself becomes inherently degenerative, probably as a result of microscopic fiber failure leading to accumulation of damage due to inability of the tendon to self-repair. Local ischemia may also be a contributory factor. Studies on the supraspinatus tendon have shown that its mid-portion is relatively hypovascular [6]. This lack of perfusion may result in the formation of oxygen free radicals or other molecules that initiate the pathological process.
Several observations have been made about the molecular mediators of tendinopathy. Tenocyte apoptosis or "programmed cell death" has been shown to occur at an increased frequency in tendinopathy specimens [7]. Free radicals as well as cyclic loading may induce the activation of molecules that lead to apoptosis [8,9]. In addition, animal studies have shown that various cytokines and matrix metalloproteinases (MMPs) may be disproportionately expressed in tendinopathy specimens. The application of cyclic strain has been shown to increase the production of prostaglandin E2 (PGE2), interleukin-6 (IL6), and IL1β [10,11]. IL1β in turn increases the production of MMP1, MMP3, and PGE2 [12]. Alfredson et al. studied samples from patients with Achilles tendinopathy and found downregulation of MMP3 mRNA and upregulation of MMP2 and vascular endothelial growth factor compared with control samples [13]. Riley et al. reported decreased MMP3 and MMP2 mRNA activity, with an increase in MMP14 [14]. These studies show that an imbalance in cytokines and MMPs exists in diseased tendons and probably contributes to the pathophysiology; however, inconsistencies in the expression of specific molecules in various studies indicate that more research needs to be done.
Currently, the only non-surgical therapies available to patients who suffer from chronic tendinopathies are physical therapy, activity modification, non-steroidal anti-inflammatory medications (NSAIDs), and steroid or platelet-rich plasma injections. These therapies offer unpredictable results, and in the case of steroids, can lead to serious side effects and more rapid degeneration of the tendon. By understanding the molecular mediators that lead to tendinopathy, novel therapeutic targets could potentially be identified for drug development. This will result in more effective treatments while minimizing side effects.
Microarray analysis has become a powerful tool in drug development. Microarrays allow researchers to screen samples of tissue for the expression of thousands of genes encoded in the human genome. This "shotgun" approach can identify which genes are active in a given sample by quantifying the production of specific mRNAs. Identification of the products of these mRNAs and their functions can serve to direct future research and development.
The purpose of this study was to investigate whether there is selective regulation of certain cytokines, matrix metalloproteinases, and protein kinases in human samples of tendinopathy compared with normal tendons, as evidenced by microarray analysis. Specifically, we were interested in determining which specific cytokines and degenerative enzymes are upregulated, and which are downregulated, in diseased samples compared with normal control tendons from the same patients. This information may contribute to the development of drugs that can selectively modulate the disease process.
Methods
Subjects
All protocols were approved by the Institutional Review Board at the Hospital for Special Surgery and informed consent was obtained from each participant. A prospective study was initiated to collect tissue from patients undergoing surgery as standard of care for tendinopathy. Biopsies (~3 mm3) of diseased tendons and a section of grossly normal appearing tendon were collected from 35 patients. Written informed consent was obtained from all patients prior to any study-related procedure. Information on patient demographics, diagnosis, prior treatments, current procedures, and operative findings was collected and is presented in Table 1. Patients with a diagnosis of inflammatory arthritis (such as rheumatoid arthritis or lupus) or those who had prior surgery on the involved tendon were excluded. Patients who received prior corticosteroid injection were included and this information was recorded.
Table 1.
Demographics and patient clinical diagnosis
| ID | Age | Sex | Diagnosis | Duration of Sx (months) | Num of Inject* | Pathology Sample | Control Sample |
|---|---|---|---|---|---|---|---|
| 5 | 45 | M | Distal Biceps Rupture | 0.3 | Distal Biceps | Brachialis | |
| 9 | 48 | M | ECRB tear | 12 | ECRB | EDC | |
| 10 | 41 | M | Patella Tendon Rupture | 0.1 | Patella | Quad tendon | |
| 12 | 62 | M | Flexor/Pronator | 18 | 1 | flexor/pronator | Distal Flexor/Pronator |
| 13 | 62 | F | RTC tear | 48 | 2 | Supraspinatus | Subscap |
| 15 | 32 | M | Patella Tendon Tear | 3 | Patella tendon | Nl Patellar Tendon | |
| 16 | 61 | F | RTC tear | 24 | 2 | Supraspinatus | Subscap |
| 17 | 59 | M | RTC tear | 6 | 1 | Supraspinatus | Subscap |
| 19 | 45 | M | Flexor/Pronator | 18 | 3 | flexor/pronator | Distal Flexor/Pronator |
| 20 | 63 | F | RTC tear | 9 | 1 | Supraspinatus | Subscap |
| 21 | 65 | F | RTC tear | 3 | 1 | Supraspinatus | Biceps |
| 23 | 64 | F | RTC tear | 8 | 1 | Supraspinatus | Subscap |
| 24 | 50 | M | RTC tear | 18 | 1 | Supraspinatus | Biceps |
| 26 | 55 | M | ECRB tear | 4 | ECRB | Distal ECRB | |
| 27 | 50 | M | RTC tear | 4 | Supraspinatus | Normal RTC | |
| 28 | 41 | M | Supra tear (PT), OA, Bankart | >24 | Supraspinatus | Biceps Tendon | |
| 29 | 46 | M | RTC tear, Adhesive Capsulitis | 4 | Supraspinatus | Biceps Tendon | |
| 30 | 52 | M | ECRB tear | 12 | ECRB | ECRL | |
| 31 | 66 | M | RTC tear | 6 | Supraspinatus | Subscap | |
| 32 | 46 | F | RTC tear | 2 | 1 | Supraspinatus | Teres minor |
| 33 | 44 | M | RTC tear | 6 | 1 | Supraspinatus | Subscap |
| 34 | 59 | F | RTC tear | 1 | 3 | Supraspinatus | Biceps |
| 35 | 49 | F | RTC tear | 24 | Supraspinatus | Subscap |
* Number of corticosteroids injections
ECRB Extensor Carpi Radialis Brevis
RTC Rotator Cuff
OA Osteoarthitis
EDC extensor digitorum comunis
RNA Preparation and Hybridization
RNA extraction and isolation were performed as previously described [15]. Fifty nanograms of RNA was labeled using the WT-Ovation FFPE System (Nugen, San Carlos, CA). Labeled cDNA was hybridized to GeneChip® Human Genome U133 2.0 arrays according to the manufacturer's protocol (Affymetrix, Santa Clara, CA). For each array, all probe sets were normalized to a mean signal intensity value of 100. The default GeneChip Operating Software statistical values were used for all analyses. The raw microarray data has been deposited in the Gene Expression Omnibus (GEO) database http://www.ncbi.nlm.nih.gov/projects/geo/ as accession number GSE26051.
Identification of Differentially Expressed Genes
Expression values for all probe sets were subjected to locally weighted scatterplot smoothing (LOWESS) transformation. Correlation analysis and hierarchical clustering verified that all samples had similar patterns of expression. Only transcripts that were called as present in 65% of the samples and expressed greater than 35 signal units in either the normal or diseased samples were used for further analysis. Expression values were log2 transformed and transcripts were considered differentially regulated if the p-value based on a paired t-test analysis was < 0.01 and the average fold change was greater than 1.5. Extensive clinical information was collected for each patient. Analysis of variance models were constructed to determine whether any differences in expression could be explained by differences in histological disease score, disease duration, gender, steroid use, or NSAID use. Values presented are expressed as mean ± standard error of the mean. Differences were considered statistically significant for p < 0.01.
Identification of Significantly Regulated Gene Sets
Significantly regulated biological pathways were identified using a modified version of the sigPathway algorithm [16] incorporating a modified normalization routine and using gene sets collections C1 to C5 defined by the Molecular Signatures Database (MSigDB) [17]. A gene set was considered significant when q1 ≤0.05 and q2 ≤0.05, where q1 or q2 are the permutation-based false-discovery rates for the Q1 or Q2 hypotheses (see [18] for an explanation of Q1 and Q2). These cut-offs were met by1761 gene sets, which can be considered as differentially expressed.
Histology
Tissue samples were frozen and sectioned for histological analysis. Sections were processed using standard procedures, including Alcian blue (AB) staining. Sections were scored on four categories: intratendinous cellular proliferation, intratendinous vascular proliferation, intratendinous glycosaminoglycan (GAG) accumulation, and intratendinous fiber disorientation. Each category was scored based on a 5-point scale, from 0 for normal, 3 for moderate, and 5 for severe, by two independent histologists.
Results
Analysis of Tendinopathy Tissue Samples
Diseased and normal tendons were collected from 35 patients with rotator cuff tendinopathy, lateral epicondylitis (tennis elbow), patellar tendinopathy, or chronic Achilles tendinopathy who were undergoing surgery as standard of care treatment for tendinopathy. Tendinopathy was confirmed both macroscopically and microscopically. The diseased tendon appeared dull, yellow, and soft compared with normal tissue, which is shiny, white, and firm. Normal tendon tissue had relatively few cells, contained little vasculature, and had collagen fibers aligned in the direction of force. All diseased tendons had a similar loss of normal fibrillar structure and changes in cell orientation, which are histological characteristics of tendinopathy [2,19,20]. All diseased tissue had an increased number of fibroblasts, increased vasculature, and increased glycosaminoglycan staining (GAG content) although the degree of increase in each of the parameters was variable between patients (Figure 1, Table 2). There was little evidence of any inflammatory cells in either the tendon or the surrounding paratenon.
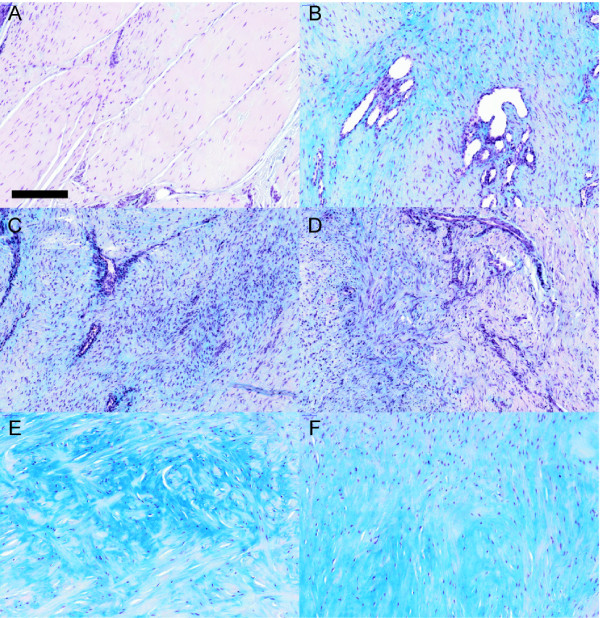
Figure 1.
Histological evaluation of human tendinopathy. Representative diseased tendons were stained with Alcian blue to visualize the proteoglycans in patient 12 (A), patient 10 (B), patient 15 (C), patient 24 (D), patient 25 (E), and patient 32 (F). Only a few bundles from patient 12 appear normal. The remaining sections show highly disorganized tissues with increased cell proliferation, increased vasculature, and/or increased GAG content. Scale bar is 200 μM.
Table 2.
Intratendinous tendinopathy scorea
| Patientb | Cellular Proliferation | Vascular Proliferation | GAG Accumulation | Fiber Disorientation | Total Score |
|---|---|---|---|---|---|
| 5 | 3.5 | 1.5 | 1.5 | 5 | 11.5 |
| 9 | 5 | 2 | 5 | 5 | 17 |
| 10 | 4.5 | 5 | 5 | 5 | 19.5 |
| 12 | 4 | 5 | 1 | 5 | 15 |
| 13 | 5 | 1 | 3.5 | 5 | 14.5 |
| 15 | 5 | 5 | 5 | 4.5 | 19.5 |
| 16 | 3.5 | 2.5 | 2 | 5 | 13 |
| 17 | 5 | 4.5 | 3 | 5 | 17.5 |
| 19 | 4.5 | 5 | 3 | 5 | 17.5 |
| 20 | 5 | 3 | 3 | 5 | 16 |
| 21 | 5 | 1 | 2 | 5 | 13 |
| 23 | 4.5 | 3 | 4 | 5 | 16.5 |
| 24 | 5 | 5 | 3 | 5 | 18 |
| 26 | 5 | 3 | 5 | 5 | 18 |
| 27 | 4.5 | 3 | 1 | 5 | 13.5 |
| 28 | 5 | 1.5 | 5 | 5 | 16.5 |
| 29 | 5 | 5 | 5 | 5 | 20 |
| 30 | 4 | 4.5 | 3 | 4.5 | 16 |
| 31 | 4.5 | 1.5 | 4 | 5 | 15 |
| 32 | 4.5 | 1.5 | 5 | 5 | 16 |
| 33 | 5 | 0.5 | 4 | 5 | 14.5 |
| 34 | 5 | 0.5 | 3 | 5 | 13.5 |
| 35 | 5 | 5 | 3 | 5 | 18 |
a Average of two independent histologist evaluation
b Normal tendon were scored 0 in all fields
Analysis of Gene Expression
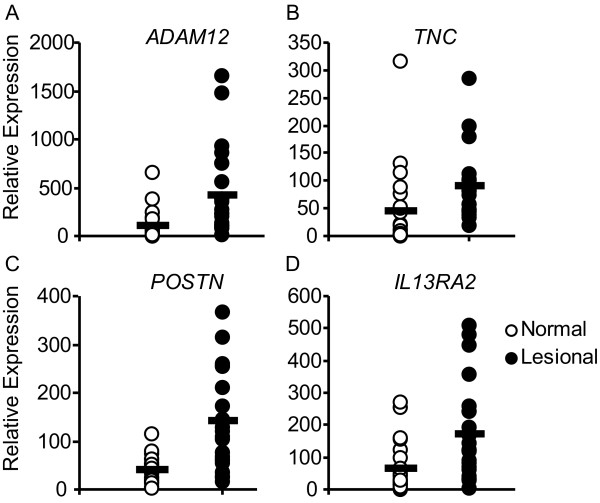
Global transcriptional analysis using Affymetrix genome-wide U133 2.0 Plus arrays identified 20055 transcripts expressed in 23 pairs of normal and diseased tendons that passed quality control filters. Of these, 983 transcripts were differentially regulated based on a paired t-test (p < 0.01, fold change ≥1.5. Among the top regulated genes were a disintegrin and metalloprotease 12 (ADAM12), tenascin C (TNC), periostin (POSTN), and interleukin 13 receptor alpha 2 (IL13RA2; Figure 2, Table 3). An analysis of covariance (ANCOVA) showed little relationship between the age of the patient and the level of gene expression for most transcripts. Furthermore, ANCOVA analysis suggested that < 1.2% of the transcripts showed any relationship between gene expression and histological disease score, disease duration, steroid use, NSAID use, gender, or duration of symptoms.
Figure 2.
Expression levels of genes that are differentially regulated in tendinopathy. Messenger RNA expression levels for (A) a disintegrin and metalloproteinase domain 12 (ADAM12), (B) tenascin C (TNC), (C) periostin (POSTN), (D) interleukin 13 receptor alpha 2 (IL13RA2) in tendons from patients with tendinopathy. Values represent the relative expression values as determined by microarray analysis.
Table 3.
Top regulated transcripts in human tendinopathy as determined by microarray analysis
| Gene Symbol | Description | p-value | q-value | Fold Change |
|---|---|---|---|---|
| ADAM12 | A disintegrin and metalloproteinase domain 12 | 1.57E-05 | 2.09E-04 | 5.6 |
| AP3M1 | Adaptor-related protein complex 3, mu 1 subunit | 1.24E-03 | 1.91E-03 | 1.7 |
| ARL7 | ADP-ribosylation factor-like 7 | 1.43E-03 | 2.11E-03 | 4.0 |
| ARPC5 | Actin related protein 2/3 complex, subunit 5 | 3.63E-05 | 2.10E-04 | 1.8 |
| ASXL1 | Additional sex combs like 1 (Drosophila) | 1.11E-03 | 1.78E-03 | 1.8 |
| ATP6V1A | ATPase, H+ transporting, lysosomal 70 kDa | 4.18E-05 | 2.10E-04 | 2.5 |
| BACE2 | Beta-site APP-cleaving enzyme 2 | 3.17E-06 | 8.01E-05 | 1.9 |
| BNC2 | Basonuclin 2 | 1.08E-04 | 3.15E-04 | 2.9 |
| CA12 | Carbonic anhydrase XII | 3.57E-05 | 2.10E-04 | 2.8 |
| CTHRC1 | Collagen triple helix repeat containing 1 | 5.20E-04 | 1.10E-03 | 4.4 |
| DDEF1 | Development and differentiation enhancing factor | 2.83E-05 | 2.10E-04 | 2.1 |
| DOCK10 | Dedicator of cytokinesis 10 | 7.91E-03 | 8.11E-03 | 3.3 |
| EDIL3 | EGF-like repeats and discoidin I-like domains 3 | 4.80E-03 | 5.34E-03 | 2.9 |
| FOXP1 | Forkhead box P1 | 4.67E-05 | 2.10E-04 | 1.7 |
| GPR161 | G protein-coupled receptor 161 | 2.76E-04 | 6.89E-04 | 1.9 |
| IGFBP3 | Insulin-like growth factor binding protein 3 | 7.29E-03 | 7.67E-03 | 3.5 |
| IL13RA2 | Interleukin 13 receptor, alpha 2 | 2.80E-03 | 3.50E-03 | 3.5 |
| ILK | Integrin-linked kinase | 1.00E-04 | 3.15E-04 | 1.6 |
| IQGAP1 | IQ motif containing GTPase activating protein 1 | 4.73E-05 | 2.10E-04 | 1.5 |
| ITGB1 | Integrin, beta 1 | 3.52E-04 | 8.29E-04 | 1.7 |
| ITPKB | Inositol 1,4,5-trisphosphate 3-kinase B | 7.37E-04 | 1.34E-03 | 2.5 |
| JAG1 | Jagged 1 (Alagille syndrome) | 1.90E-03 | 2.52E-03 | 3.1 |
| KCNE4 | Potassium voltage-gated channel 4 | 8.19E-03 | 8.19E-03 | 3.4 |
| LAMA4 | Laminin, alpha 4 | 6.73E-04 | 1.31E-03 | 3.4 |
| LRRC15 | Leucine rich repeat containing 15 | 3.50E-03 | 4.24E-03 | 3.2 |
| LRRC17 | Leucine rich repeat containing 17 | 1.74E-03 | 2.40E-03 | 2.8 |
| MARCKS | Myristoylated alanine-rich kinase C substrate | 3.93E-03 | 4.50E-03 | 2.8 |
| NOTCH3 | Notch homolog 3 (Drosophila) | 1.65E-03 | 2.36E-03 | 3.6 |
| PDIR | protein disulfide isomerase-related | 4.86E-04 | 1.08E-03 | 2.1 |
| POSTN | Periostin, osteoblast specific factor | 1.10E-04 | 3.15E-04 | 3.5 |
| RAP1GDS1 | RAP1, GTP-GDP dissociation stimulator 1 | 1.01E-03 | 1.68E-03 | 2.1 |
| S100A10 | S100 calcium binding protein A10 | 1.95E-03 | 2.52E-03 | 2.8 |
| SIN3A | SIN3 homolog A, transcription regulator (yeast) | 9.51E-05 | 3.15E-04 | 1.7 |
| SLC2A13 | Solute carrier family 2 13 | 5.41E-03 | 5.85E-03 | 2.8 |
| TAOK1 | TAO kinase 1 | 1.51E-04 | 4.03E-04 | 1.7 |
| TJP1 | Tight junction protein 1 (zona occludens 1) | 6.87E-04 | 1.31E-03 | 1.6 |
| TNC | Tenascin C (hexabrachion) | 9.20E-05 | 3.15E-04 | 5.3 |
| UBE2E3 | Ubiquitin-conjugating enzyme E2E 3 | 8.72E-04 | 1.52E-03 | 3.7 |
| WDR1 | WD repeat domain 1 | 4.01E-06 | 8.01E-05 | 1.6 |
| WISP1 | WNT1 inducible signaling pathway protein 1 | 3.93E-03 | 4.50E-03 | 3.4 |
Global pathway analysis, based on a modified sigPathway algorithm [16], identified increased expression of genes involved in the extracellular matrix, including genes involved in focal adhesion, integrin signaling, and collagen synthesis. In addition, expression of genes involved in cell cycle progression, TGFβ signaling, and NFκB signaling was increased (Table 4). This global pathway analysis did not suggest that cytokines as a whole were differentially regulated. However, of the 426 transcripts annotated by Gene Ontology as cytokines, cytokine signaling molecules, or related to cytokines, six were differentially regulated in diseased tendons, including chemokine-like factor super family 4 (CMTM4), interleukin 13 receptor alpha 2 (IL13RA2), interleukin 17D (IL17D), and interleukin 4 receptor (IL4R) (Table 5).
Table 4.
Genesets regulated in human tendinopathy as determined by sigpathway
| NTk* | |
|---|---|
| Up regulated | |
| HSA04110_CELL_CYCLE | 6.52 |
| RNA_PROCESSING | 6.44 |
| SPINDLE | 6.25 |
| HSA04510_FOCAL_ADHESION | 6.10 |
| TGFBETA_LATE_UP | 6.02 |
| CELL_CYCLE_KEGG | 6.00 |
| CELL_CYCLE | 5.94 |
| ST_INTEGRIN_SIGNALING_PATHWAY | 5.40 |
| COLLAGEN | 5.32 |
| RUIZ_TENASCIN_TARGETS | 5.24 |
| I_KAPPAB_KINASE_NF_KAPPAB_CASCADE | 5.23 |
| MITOSIS | 5.03 |
| INOS_ALL_UP | 4.89 |
| Down Regulated | |
| ION_TRANSMEMBRANE_TRANSPORTER_ACTIVITY | -4.99 |
| G_PROTEIN_COUPLED_RECEPTOR_ACTIVITY | -5.01 |
| ELECTRON_TRANSPORT_CHAIN | -5.21 |
| MOOTHA_VOXPHOS | -6.01 |
| ION_CHANNEL_ACTIVITY | -6.20 |
| SUBSTRATE_SPECIFIC_CHANNEL_ACTIVITY | -6.27 |
* As defined in Tian et. al [18]
Table 5.
Chemokines regulated in human tendinopathy
| Gene Symbol | Description | p-value | q-value | Fold Change |
|---|---|---|---|---|
| BMP1 | Bone morphogenetic protein 1 | 0.000 | 0.005 | 1.69 |
| BMP8B | Bone morphogenetic protein 8b | 0.002 | 0.011 | 1.79 |
| CCL2 | Chemokine (C-C motif) ligand 2 | 0.005 | 0.014 | 1.74 |
| CMTM4 | Chemokine-like factor super family 4 | 0.008 | 0.017 | 1.98 |
| CMTM6 | Chemokine-like factor super family 6 | 0.014 | 0.023 | 1.37 |
| IFNGR1 | Interferon gamma receptor 1 | 0.027 | 0.034 | 1.42 |
| IL13RA2 | Interleukin 13 receptor, alpha 2 | 0.003 | 0.011 | 3.48 |
| IL17D | Interleukin 17D | 0.008 | 0.017 | 0.54 |
| IL6R | Interleukin 6 receptor | 0.026 | 0.034 | 0.73 |
| LEPR | Leptin receptor | 0.028 | 0.034 | 0.49 |
| OSMR | Oncostatin M receptor | 0.038 | 0.043 | 1.81 |
| SP110 | SP110 nuclear body protein | 0.010 | 0.019 | 1.51 |
| STAT2 | Signal transducer and activator of transcription 2 | 0.051 | 0.054 | 1.4 |
| STAT3 | Signal transducer and activator of transcription 3 | 0.004 | 0.013 | 1.87 |
| TXLNA | Taxilin | 0.001 | 0.006 | 1.39 |
Consistent with a role in increased matrix turnover, tendonopathy tissue exhibited increased expression of genes encoding MMPs (Table 6) and genes encoding collagens (Table 7). The majority of collagens interrogated showed a modest 2-fold increase in expression in diseased tendons. Diseased tendons also showed increased expression of several enzymes that mediate collagen breakdown, including MMP2, MMP9, MMP14, MMP13, and MMP19. While many MMP genes were induced in diseased tendons, not all transcripts encoding MMPs were upregulated; in fact, MMP3 and MMP24 were significantly downregulated in diseased tissue. Of the four known tissue inhibitors of metalloproteases (TIMPs), only expression of TIMP1 was increased. No ADAMTS (a disintegrin and metalloprotease with thrombospondin motifs) family members were differentially regulated based on our cut-offs.
Table 6.
Change in matrix metallopeptidase gene expression in human tendinopathy
| Gene Symbol | Description | p-value | q-value | Fold Change | |
|---|---|---|---|---|---|
| Regulated | |||||
| MMP2 | Matrix metallopeptidase 2 | Expressed | 7.42E-03 | 4.82E-02 | 1.8 |
| MMP3 | Matrix metallopeptidase 3 | Expressed | 5.50E-02 | 2.38E-01 | 0.3 |
| MMP14 | Matrix metallopeptidase 14 | Expressed | 1.79E-03 | 1.66E-02 | 1.9 |
| MMP19 | Matrix metallopeptidase 19 | Expressed | 1.91E-03 | 1.66E-02 | 3.4 |
| Not Regulated | |||||
| MMP28 | Matrix metallopeptidase 28 | Expressed | 0.09 | 0.30 | 1.4 |
| TIMP1 | TIMP metallopeptidase inhibitor 1 | Expressed | 0.10 | 0.30 | 1.6 |
| TIMP2 | TIMP metallopeptidase inhibitor 2 | Expressed | 0.37 | 0.50 | 1.1 |
| TIMP3 | TIMP metallopeptidase inhibitor 3 | Expressed | 0.26 | 0.49 | 0.9 |
| TIMP4 | TIMP metallopeptidase inhibitor 4 | Expressed | 0.71 | 0.80 | 0.9 |
| Expressed below limited of detection | |||||
| MMP1 | Matrix metallopeptidase 1 | - | 0.96 | 0.96 | 1.0 |
| MMP7 | Matrix metallopeptidase 7 | - | 0.34 | 0.49 | 0.8 |
| MMP8 | Matrix metallopeptidase 8 | - | 0.81 | 0.84 | 0.8 |
| MMP9 | Matrix metallopeptidase 9 | - | 0.00 | 0.02 | 3.2 |
| MMP10 | Matrix metallopeptidase 10 | - | 0.08 | 0.30 | 0.8 |
| MMP12 | Matrix metallopeptidase 12 | - | 0.79 | 0.84 | 0.9 |
| MMP13 | Matrix metallopeptidase 13 | - | 0.03 | 0.14 | 2.6 |
| MMP15 | Matrix metallopeptidase 15 | - | 0.15 | 0.35 | 0.5 |
| MMP16 | Matrix metallopeptidase 16 | - | 0.57 | 0.67 | 0.6 |
| MMP17 | Matrix metallopeptidase 17 | - | 0.56 | 0.67 | 0.8 |
| MMP20 | Matrix metallopeptidase 20 | - | 0.53 | 0.67 | 0.7 |
| MMP21 | Matrix metallopeptidase 21 | - | 0.13 | 0.33 | 1.5 |
| MMP23A/B | Matrix metallopeptidase 23A/B | - | 0.34 | 0.49 | 0.8 |
| MMP24 | Matrix metallopeptidase 2 | - | 0.29 | 0.49 | 0.6 |
| MMP25 | Matrix metallopeptidase 25 | - | 0.18 | 0.40 | 0.7 |
| MMP26 | Matrix metallopeptidase 26 | - | 0.34 | 0.49 | 0.8 |
| MMP27 | Matrix metallopeptidase 27 | - | 0.33 | 0.49 | 0.8 |
Table 7.
Change in collagen gene expression in human tendinopathy
| Gene Symbol | Description | p-value | q-value | Fold Change | |
|---|---|---|---|---|---|
| Regulated | |||||
| COL1A1 | Collagen, type I, alpha 1 | Expressed | 9.37E-03 | 3.96E-02 | 1.7 |
| COL1A2 | Collagen, type I, alpha 2 | Expressed | 4.45E-03 | 3.16E-02 | 1.6 |
| COL3A1 | Collagen, type III, alpha 1 | Expressed | 1.50E-02 | 4.28E-02 | 2.0 |
| COL4A1 | Collagen, type IV, alpha 1 | Expressed | 1.91E-02 | 4.53E-02 | 2.7 |
| COL4A2 | Collagen, type IV, alpha 2 | Expressed | 6.09E-03 | 3.16E-02 | 2.3 |
| COL5A1 | Collagen, type V, alpha 1 | Expressed | 1.20E-02 | 4.28E-02 | 2.0 |
| COL5A2 | Collagen, type V, alpha 2 | Expressed | 3.99E-03 | 3.16E-02 | 2.1 |
| COL5A3 | Collagen, type V, alpha 3 | Expressed | 3.02E-03 | 3.16E-02 | 2.0 |
| COL6A1 | Collagen, type VI, alpha 1 | Expressed | 2.93E-02 | 6.18E-02 | 1.8 |
| COL6A2 | Collagen, type VI, alpha 2 | Expressed | 3.62E-02 | 6.89E-02 | 1.8 |
| COL6A3 | Collagen, type VI, alpha 3 | Expressed | 5.45E-03 | 3.16E-02 | 1.7 |
| COL8A1 | Collagen, type VIII, alpha 1 | Expressed | 2.49E-02 | 5.57E-02 | 2.0 |
| COL8A2 | Collagen, type VIII, alpha 2 | Expressed | 1.48E-02 | 4.28E-02 | 1.8 |
| COL10A1 | Collagen, type X, alpha 1 | Expressed | 1.69E-02 | 4.28E-02 | 2.1 |
| COL12A1 | Collagen, type XII, alpha 1 | Expressed | 4.10E-03 | 3.16E-02 | 2.1 |
| COL18A1 | Collagen, type XVIII, alpha 1 | Expressed | 6.65E-03 | 3.16E-02 | 2.2 |
| COL27A1 | Collagen, type XXVII, alpha 1 | Expressed | 1.20E-03 | 3.16E-02 | 2.5 |
| Not Regulated | |||||
| COL7A1 | Collagen, type VII, alpha 1 | Expressed | 0.96 | 0.96 | 1.0 |
| COL11A1 | Collagen, type XI, alpha 1 | Expressed | 0.58 | 0.71 | 1.2 |
| COL15A1 | Collagen, type XV, alpha 1 | Expressed | 0.51 | 0.64 | 1.2 |
| COL16A1 | Collagen, type XVI, alpha 1 | Expressed | 0.12 | 0.19 | 1.4 |
| COL21A1 | Collagen, type XXI, alpha 1 | Expressed | 0.34 | 0.45 | 1.4 |
| Expressed below limited of detection | |||||
| COL2A1 | Collagen, type II, alpha 1 | - | 0.21 | 0.31 | 0.5 |
| COL4A3 | Collagen, type IV, alpha 3 | - | 0.15 | 0.23 | 0.6 |
| COL4A4 | Collagen, type IV, alpha 4 | - | 0.90 | 0.96 | 1.0 |
| COL4A5 | Collagen, type IV, alpha 5 | - | 0.71 | 0.84 | 0.9 |
| COL4A6 | Collagen, type IV, alpha 6 | - | 0.94 | 0.96 | 1.0 |
| COL9A1 | Collagen, type IX, alpha 1 | - | 0.94 | 0.96 | 1.0 |
| COL9A2 | Collagen, type IX, alpha 2 | - | 0.05 | 0.08 | 1.9 |
| COL11A2 | Collagen, type XI, alpha 2 | - | 0.12 | 0.19 | 0.7 |
| COL13A1 | Collagen, type XIII, alpha 1 | - | 0.08 | 0.13 | 2.0 |
| COL14A1 | Collagen, type XIV, alpha 1 | - | 0.02 | 0.04 | 1.8 |
| COL17A1 | Collagen, type XVII, alpha 1 | - | 0.33 | 0.45 | 1.3 |
| COL19A1 | Collagen, type XIX, alpha 1 | - | 0.02 | 0.04 | 0.5 |
| COL22A1 | Collagen, type XXII, alpha 1 | - | 0.03 | 0.07 | 0.5 |
| COL23A1 | Collagen, type XXIII, alpha 1 | - | 0.75 | 0.84 | 1.0 |
| COL24A1 | Collagen, type XXIV, alpha 1 | - | 0.33 | 0.45 | 1.3 |
| COL25A1 | Collagen, type XXV, alpha 1 | - | 0.76 | 0.84 | 0.9 |
Diseased tendons showed increased expression of a number of relevant and important signaling pathways; for example the fibroblast growth factors FGFR1 (1.7-fold) and FGFR2 (2.7-fold), and genes in the Notch signaling pathway, including JAG1 (3.1-fold) and Notch3 (3.6-fold). There was also evidence for increased WNT signaling. Among the highest regulated transcripts, four are involved in WNT signaling: WISP1 (4.2-fold), DKK3 (3.6-fold), WNT3 (3.6-fold), and LEF1 (4.1-fold). Additionally, tendinopathy tissue showed differential expression of other WNT pathway genes, including increased expression of CTNNB (1.5-fold), WNT5a (2.7-fold), and FZD1 (1.7-fold) and decreased expression of LRP5 (0.59-fold) and FRZB (0.35-fold).
We found little evidence for endochondral ossification even though this has been reported in Achilles and patella tendinopathy [21]. Histological evaluation failed to identify any gross areas of ossification in any of the patients and RNA analysis did not show increased expression of the bone markers osteocalcin or osterix. However, expression of CBFA1, a transcription factor associated with osteoblast differentiation, was increased 1.8-fold and that of RANKL, a key factor for osteoclast differentiation and activation, was increased 4.0-fold in diseased tendons.
In addition to the global pathway analysis, we specifically investigated the regulation of kinases in diseased tendons (Table 8). The most statistically significant regulated kinase is diseased tendons is the thousand and one amino acid kinase (TAOK1), which showed a 1.7-fold increase in expression. TAOK1 activates the p38 MAP kinase pathway and tendonopathy tissue also showed increased expression of multiple MAP kinases, including MAP3K2 (1.8-fold), MAP4K5 (1.9-fold), and MAPK8 (2.0-fold).
Table 8.
Kinases regulated in human tendinopathy
| Gene Symbol | Description | pValue | q-value | Fold Change |
|---|---|---|---|---|
| ABL2 | v-abl Abelson leukemia viral oncogene homolog 2 | 4.79E-03 | 7.83E-03 | 31.0 |
| BMPR2 | Bone morphogenetic protein receptor, type II | 4.10E-03 | 7.83E-03 | 27.0 |
| CAV2 | Caveolin 2 | 1.22E-02 | 1.44E-02 | 43.0 |
| CDK6 | Cyclin-dependent kinase 6 | 1.37E-02 | 1.53E-02 | 24.0 |
| CSNK1D | Casein kinase 1, delta | 5.21E-03 | 8.13E-03 | 19.0 |
| CSNK1G1 | Casein kinase 1, gamma 1 | 8.18E-03 | 1.03E-02 | 40.0 |
| DCLK1 | Doublecortin-like kinase 1 | 3.78E-03 | 7.83E-03 | 28.0 |
| EML4 | Echinoderm microtubule associated protein like 4 | 9.69E-04 | 3.15E-03 | 41.0 |
| EPS8 | Epidermal growth factor receptor substrate 8 | 1.96E-04 | 1.67E-03 | 9.0 |
| FGFR1 | Fibroblast growth factor receptor 1 | 5.64E-03 | 8.46E-03 | 1.7 |
| ILK | Integrin-linked kinase | 1.00E-04 | 1.67E-03 | 1.6 |
| JAK3 | Janus kinase 3 | 4.62E-03 | 7.83E-03 | 1.8 |
| KIAA0220 | PI-3-kinase-related kinase SMG-1 | 2.70E-03 | 7.02E-03 | 1.6 |
| KIAA0999 | KIAA0999 protein | 1.62E-03 | 4.86E-03 | 2.1 |
| MAP3K2 | Mitogen-activated protein kinase kinase kinase 2 | 4.82E-03 | 7.83E-03 | 1.8 |
| MAP4K5 | Mitogen-activated protein kinase kinase kinase 5 | 1.31E-02 | 1.50E-02 | 1.9 |
| MAPK8 | Mitogen-activated protein kinase 8 | 1.42E-02 | 1.54E-02 | 2.0 |
| NAGK | N-acetylglucosamine kinase | 4.16E-03 | 7.83E-03 | 1.8 |
| PALM2-AKAP2 | A kinase (PRKA) anchor protein 2 | 2.31E-02 | 2.37E-02 | 1.9 |
| PANK2 | Pantothenate kinase 2 | 6.39E-04 | 2.67E-03 | 1.6 |
| PAPSS1 | 3'-phosphoadenosine 5'-phosphosulfate synthase 1 | 2.35E-04 | 1.67E-03 | 1.8 |
| PDLIM5 | PDZ and LIM domain 5 | 8.94E-03 | 1.09E-02 | 1.9 |
| PFTK1 | PFTAIRE protein kinase 1 | 4.74E-03 | 7.83E-03 | 1.6 |
| PGAM1 | Phosphoglycerate mutase 1 (brain) | 4.81E-04 | 2.35E-03 | 1.6 |
| PI4K2B | Phosphatidylinositol 4-kinase type 2 beta | 7.80E-03 | 1.02E-02 | 1.7 |
| PIK3C2A | Phosphoinositide-3-kinase, class 2, alpha | 2.28E-02 | 2.37E-02 | 1.5 |
| PIP4K2A | Phosphatidylinositol-5-phosphate 4-kinase, Iia | 7.52E-04 | 2.67E-03 | 1.5 |
| PRKAA1 | Protein kinase, AMP-activated, alpha 1 | 5.32E-02 | 5.32E-02 | 1.7 |
| PRKD1 | Protein kinase D1 | 7.86E-03 | 1.02E-02 | 1.8 |
| PTK9 | Twinfilin, actin-binding protein, homolog 1 | 1.62E-04 | 1.67E-03 | 1.7 |
| RIPK2 | Receptor-interacting serine-threonine kinase 2 | 1.84E-03 | 5.11E-03 | 1.6 |
| RIPK5 | Receptor interacting protein kinase 5 | 5.91E-03 | 8.53E-03 | 1.7 |
| SGMS2 | Sphingomyelin synthase 2 | 3.38E-03 | 7.76E-03 | 2.3 |
| SHC1 | Src homology 2 domain containing protein 1 | 2.99E-04 | 1.67E-03 | 1.7 |
| SOCS3 | Suppressor of cytokine signaling 3 | 3.07E-03 | 7.48E-03 | 1.7 |
| SSA2 | TROVE domain family, member 2 | 2.72E-04 | 1.67E-03 | 1.6 |
| TAOK1 | TAO kinase 1 | 1.51E-04 | 1.67E-03 | 1.7 |
| TRIM27 | Tripartite motif-containing 27 | 6.94E-04 | 2.67E-03 | 1.6 |
| TTN | Titin | 6.67E-03 | 9.29E-03 | 0.2 |
Discussion
We have evaluated gene expression and histological changes associated with human tendinopathy. While gene expression changes in human tendinopathy have previously been described [13,22,23], this study represents the largest and most complete study to date. However, our study has several limitations. First, control tendons were taken from the same joints as the diseased tendons. These control tendons appeared normal macroscopically but their proximity to the diseased tendon may have some effect on the control tendon. Furthermore, the control tendon was an anatomically distinct tendon exposed to different loads and strains and it is not known how this may affect the gene expression profile. Moreover, the tendon specimens were collected from a heterogeneous population with differences in age, gender, symptoms, duration of disease, and physical activity (Table 1). While changes in gene expression were not related to any of these differences, they all contribute to variations in the data. Given the vast variation in the human population, we reasoned that control samples from the same patients would be a better control for interpatient variability than tendons from control patients without any tendon symptoms.
Progression of tendinopathy is dependent on extracellular matrix integrity and remodeling of the tendon is a common feature of tendinopathy [24]. The extracellular matrix of normal tendons consists of many of structural proteins (collagens) and proteoglycans. Collagen is the main component of tendons and type I collagens account for 65% to 80% of the total tendon mass [25]. mRNA levels for type I and type III collagens (COL1A1, COL1A2, and COL3A1) were increased in the diseased tendons. Tendons also contain several proteoglycans, although none of the proteoglycans analyzed (including aggrecan, versican, decorin, biglycan, fibromodulin, and lumican) showed increased expression in diseased tendons. In contrast, expression of the non-collagen glycoproteins fibronectin, tenascin C, fibrillin, and laminin was increased in diseased tendons. Consistent with active remodeling, increased MMP activity, particularly of MMP2 and MMP9, has been associated with ruptured Achilles tendon [26]. Diseased tendons showed increased expression of several matrix associated proteins including MMP19, MMP9, MMP13, MMP14, and MMP2. In contrast, MMP3 expression was decreased in tendinopathy in both our study and in others [13,22].
We can begin to speculate how MMP activity is regulated in tendinopathy tissue. Diseased tissues showed increased expression of components of the JAK/STAT pathway (STAT3, 1.9-fold, and JAK3, 1.8-fold). The JAK/STAT pathway is known to activate IL4R [27], which induces expression of IL13RA2 [28]. Expression of both IL4R and IL13RA2 was elevated in diseased tissue (3.2- and 3.5-fold, respectively). IL13, secreted by T helper type 2 cells, has anti-inflammatory properties and through interaction with its receptor IL13RA2 induces certain MMPs [29] including MMP9 and MMP14 [30]. Together, these findings suggest that activation of the JAK/STAT pathway may lead to increased MMP activation.
Chard et al. previously suggested that tendon pathology is linked to formation of excessive fibrocartilage [31]; however, other prior studies [22] and our current study found little evidence of chondrocyte-like cells. Tendinopathy tissue showed decreased expression of a number of cartilage markers including COMP, but expression of other cartilage markers such as aggrecan was not altered. In contrast, expression of type I collagen was increased, further supporting the absence of increased chondrocytic cells in diseased tendons. This is in contrast to an overuse animal model of tendinopathy [32,33] that suggested increased formation of fibrocartilage associated with overuse. This discrepancy can possibly be explained by differences in disease severity. The human samples were collected during surgery following tendon tear or rupture and represent late stage disease in which surgical intervention became necessary, while the animal models represent earlier development of disease. Furthermore, the human tendon samples represent small biopsies, mostly from the midsubstance. One might expect any increase in fibrocartilage to occur near the bone insertion site, an area from which samples were not collected.
Global pathway analysis identified novel pathways that are differentially regulated in human tendinopathy including regulation of genes involved in WNT signaling. Tendinopathy is characterized by increased cellular infiltration and proliferation. Increases in WNT signaling can promote proliferation and maintain cells in an undifferentiated state [34] and the regulation of multiple WNT signaling genes suggests that this pathway is activated in tendinopathy. Pathway analysis also suggested an increased regulation of genes involved in integrin signaling. Integrins are receptors involved in cell adhesion to the extracellular matrix and transmit extracellular signals into the cell to regulate gene expression. While many of the genes encoding integrins were only slightly upregulated, expression of integrin beta 1 (fibronectin receptor) was significantly increased in tendinopathy tissue.
We found little direct evidence to support an inflammatory response in established human tendinopathy. Histological evaluation did not identify a significant number of inflammatory cells and analysis of the gene expression of inflammatory cytokines identified only a handful of cytokine genes that were differentially regulated in diseased tendons, although a number of these are implicated in tendon function and tendon healing. Expression of the proinflammatory cytokine IL17D was reduced in diseased tendons. IL17 increases the turnover of type I collagen through both inhibiting its synthesis and promoting its breakdown [35], and members of the IL17 cytokine family members are inhibitors of human hematopoietic progenitor proliferation [36]. The oncostatin M receptor was significantly upregulated in diseased tendons. Oncostatin M contributes to the release of proteoglycans and the breakdown of collagens [37]. IL6 is a pro-inflammatory cytokine important for tendon healing; lack of IL6 prevents proper tendon healing [38]. Evidence for aberrant regulation of the IL6 pathway in damaged tendons includes decreased expression of the IL6 receptor. IL6 can also signal through STAT3, which was upregulated in diseased tendons. STAT3 expression has also been identified in ruptured rotator cuff. However, activation of STAT3 is mainly induced by proliferating vessels [39], and since diseased tendons have increased vasculature, many of the observed changes in cytokine expression may simply be due to this change in vasculature. Therefore, whether these cytokines play a direct role in tendonopathy requires further study. In addition to the lack of evidence for direct regulation of many pro-inflammatory cytokines, there is also indirect evidence for the absence of inflammatory cytokine activity. The lack of expression of MMP1 and MMP13, which are known to be induced by inflammatory cytokines [40], further supports the proposal that pro-inflammatory cytokines do not play a major role in tendinopathy at this late stage of disease.
Conclusions
In this paper we describe the global transcriptome of human tendinopathy. Although we have identified a number of genes that are differentially regulated, the ultimate roles of these genes and pathways in tendon disease are yet to be determined. However, we have provided a resource that we and other investigators can use to explore the molecular changes associated with tendinopathy.
Competing interests
Howard Seeherman, Jian Li, Joanne Archambault and Scott Jelinsky were employed by Wyeth Research/Pfizer at the time of this research. Scott A Rodeo received research funding from Wyeth at the time of this study.
Authors' contributions
HJS, JMA and SAR worked on the study design. SAR recruited patients and collected the samples. LVG wrote the IRB and informed consents and help collect the samples. JL performed the histology analysis. SAJ performed the microarray analysis and drafted the manuscript. All authors have read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Contributor Information
Scott A Jelinsky, Email: Scott.Jelinsky@pfizer.com.
Scott A Rodeo, Email: rodeos@hss.edu.
Jian Li, Email: Jian.Li@pfizer.com.
Lawrence V Gulotta, Email: GulottaL@hss.edu.
Joanne M Archambault, Email: joarch@comcast.net.
Howard J Seeherman, Email: Howard.Seeherman@pfizer.com.
Acknowledgements
Some authors were employed by Wyeth Research at the time of the research. The authors wish to thank Christine Huard and Ying Zhang for microarray processing and Katy Wallace and Donna Gavin for histology processing.
References
- Maffulli N, Wong J, Almekinders LC. Types and epidemiology of tendinopathy. Clin Sports Med. 2003;22(4):675–692. doi: 10.1016/S0278-5919(03)00004-8. [DOI] [PubMed] [Google Scholar]
- Astrom M, Rausing A. Chronic Achilles tendinopathy. A survey of surgical and histopathologic findings. Clin Orthop Relat Res. 1995. pp. 151–164. [PubMed]
- Khan KM, Maffulli N. Tendinopathy: an Achilles' heel for athletes and clinicians. Clin J Sport Med. 1998;8(3):151–154. doi: 10.1097/00042752-199807000-00001. [DOI] [PubMed] [Google Scholar]
- Movin T, Gad A, Reinholt FP, Rolf C. Tendon pathology in long-standing achillodynia. Biopsy findings in 40 patients. Acta Orthop Scand. 1997;68(2):170–175. doi: 10.3109/17453679709004002. [DOI] [PubMed] [Google Scholar]
- Bigliani L, Morisson D, April E. Morphology of the acromion and its relationship to rotator cuff tears. Orthop Trans. 1986;10:459–460. [Google Scholar]
- Rathbun JB, Macnab I. The microvascular pattern of the rotator cuff. J Bone Joint Surg Br. 1970;52(3):540–553. [PubMed] [Google Scholar]
- Yuan J, Murrell GA, Wei AQ, Wang MX. Apoptosis in rotator cuff tendonopathy. J Orthop Res. 2002;20(6):1372–1379. doi: 10.1016/S0736-0266(02)00075-X. [DOI] [PubMed] [Google Scholar]
- Arnoczky SP, Tian T, Lavagnino M, Gardner K, Schuler P, Morse P. Activation of stress-activated protein kinases (SAPK) in tendon cells following cyclic strain: the effects of strain frequency, strain magnitude, and cytosolic calcium. J Orthop Res. 2002;20(5):947–952. doi: 10.1016/S0736-0266(02)00038-4. [DOI] [PubMed] [Google Scholar]
- Yuan J, Murrell GA, Trickett A, Wang MX. Involvement of cytochrome c release and caspase-3 activation in the oxidative stress-induced apoptosis in human tendon fibroblasts. Biochim Biophys Acta. 2003;1641(1):35–41. doi: 10.1016/S0167-4889(03)00047-8. [DOI] [PubMed] [Google Scholar]
- Skutek M, van Griensven M, Zeichen J, Brauer N, Bosch U. Cyclic mechanical stretching enhances secretion of Interleukin 6 in human tendon fibroblasts. Knee Surg Sports Traumatol Arthrosc. 2001;9(5):322–326. doi: 10.1007/s001670100217. [DOI] [PubMed] [Google Scholar]
- Tsuzaki M, Bynum D, Almekinders L, Yang X, Faber J, Banes AJ. ATP modulates load-inducible IL-1beta, COX 2, and MMP-3 gene expression in human tendon cells. J Cell Biochem. 2003;89(3):556–562. doi: 10.1002/jcb.10534. [DOI] [PubMed] [Google Scholar]
- Tsuzaki M, Guyton G, Garrett W, Archambault JM, Herzog W, Almekinders L, Bynum D, Yang X, Banes AJ. IL-1 beta induces COX2, MMP-1, -3 and -13, ADAMTS-4, IL-1 beta and IL-6 in human tendon cells. J Orthop Res. 2003;21(2):256–264. doi: 10.1016/S0736-0266(02)00141-9. [DOI] [PubMed] [Google Scholar]
- Alfredson H, Lorentzon M, Backman S, Backman A, Lerner UH. cDNA-arrays and real-time quantitative PCR techniques in the investigation of chronic Achilles tendinosis. J Orthop Res. 2003;21(6):970–975. doi: 10.1016/S0736-0266(03)00107-4. [DOI] [PubMed] [Google Scholar]
- Riley GP, Curry V, DeGroot J, van El B, Verzijl N, Hazleman BL, Bank RA. Matrix metalloproteinase activities and their relationship with collagen remodelling in tendon pathology. Matrix Biol. 2002;21(2):185–195. doi: 10.1016/S0945-053X(01)00196-2. [DOI] [PubMed] [Google Scholar]
- Jelinsky SA, Archambault J, Li L, Seeherman H. Tendon-selective genes identified from rat and human musculoskeletal tissues. J Orthop Res. 2009. [DOI] [PubMed]
- Crabtree JS, Jelinsky SA, Harris HA, Choe SE, Cotreau MM, Kimberland ML, Wilson E, Saraf KA, Liu W, McCampbell AS. et al. Comparison of Human and Rat Uterine Leiomyomata: Identification of a Dysregulated Mammalian Target of Rapamycin Pathway. Cancer Research. 2009;69(15):6171–6178. doi: 10.1158/0008-5472.CAN-08-4471. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Kuehn H, Gould J, Tamayo P, Mesirov JP. GSEA-P: a desktop application for Gene Set Enrichment Analysis. Bioinformatics. 2007;23(23):3251–3253. doi: 10.1093/bioinformatics/btm369. [DOI] [PubMed] [Google Scholar]
- Tian L, Greenberg SA, Kong SW, Altschuler J, Kohane IS, Park PJ. Discovering statistically significant pathways in expression profiling studies. Proc Natl Acad Sci USA. 2005;102(38):13544–13549. doi: 10.1073/pnas.0506577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvinen M, Jozsa L, Kannus P, Jarvinen TL, Kvist M, Leadbetter W. Histopathological findings in chronic tendon disorders. Scand J Med Sci Sports. 1997;7(2):86–95. doi: 10.1111/j.1600-0838.1997.tb00124.x. [DOI] [PubMed] [Google Scholar]
- Kannus P, Jozsa L. Histopathological changes preceding spontaneous rupture of a tendon. A controlled study of 891 patients. J Bone Joint Surg Am. 1991;73(10):1507–1525. [PubMed] [Google Scholar]
- Fenwick S, Harrall R, Hackney R, Bord S, Horner A, Hazleman B, Riley G. Endochondral ossification in Achilles and patella tendinopathy. Rheumatology (Oxford) 2002;41(4):474–476. doi: 10.1093/rheumatology/41.4.474. [DOI] [PubMed] [Google Scholar]
- Ireland D, Harrall R, Curry V, Holloway G, Hackney R, Hazleman B, Riley G. Multiple changes in gene expression in chronic human Achilles tendinopathy. Matrix Biol. 2001;20(3):159–169. doi: 10.1016/S0945-053X(01)00128-7. [DOI] [PubMed] [Google Scholar]
- Jones GC, Corps AN, Pennington CJ, Clark IM, Edwards DR, Bradley MM, Hazleman BL, Riley GP. Expression profiling of metalloproteinases and tissue inhibitors of metalloproteinases in normal and degenerate human achilles tendon. Arthritis Rheum. 2006;54(3):832–842. doi: 10.1002/art.21672. [DOI] [PubMed] [Google Scholar]
- Eriksen HA, Pajala A, Leppilahti J, Risteli J. Increased content of type III collagen at the rupture site of human Achilles tendon. J Orthop Res. 2002;20(6):1352–1357. doi: 10.1016/S0736-0266(02)00064-5. [DOI] [PubMed] [Google Scholar]
- Kannus P. Structure of the tendon connective tissue. Scand J Med Sci Sports. 2000;10(6):312–320. doi: 10.1034/j.1600-0838.2000.010006312.x. [DOI] [PubMed] [Google Scholar]
- Karousou E, Ronga M, Vigetti D, Passi A, Maffulli N. Collagens, proteoglycans, MMP-2, MMP-9 and TIMPs in human achilles tendon rupture. Clin Orthop Relat Res. 2008;466(7):1577–1582. doi: 10.1007/s11999-008-0255-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang R, Patel D, Morris JJ, Rutschman RL, Murray PJ. Shaping gene expression in activated and resting primary macrophages by IL-10. J Immunol. 2002;169(5):2253–2263. doi: 10.4049/jimmunol.169.5.2253. [DOI] [PubMed] [Google Scholar]
- Khodoun M, Lewis CC, Yang JQ, Orekov T, Potter C, Wynn T, Mentink-Kane M, Hershey GK, Wills-Karp M, Finkelman FD. Differences in expression, affinity, and function of soluble (s)IL-4Ralpha and sIL-13Ralpha2 suggest opposite effects on allergic responses. J Immunol. 2007;179(10):6429–6438. doi: 10.4049/jimmunol.179.10.6429. [DOI] [PubMed] [Google Scholar]
- Zheng T, Zhu Z, Wang Z, Homer RJ, Ma B, Riese RJ Jr, Chapman HA Jr, Shapiro SD, Elias JA. Inducible targeting of IL-13 to the adult lung causes matrix metalloproteinase- and cathepsin-dependent emphysema. J Clin Invest. 2000;106(9):1081–1093. doi: 10.1172/JCI10458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa T, Joshi B, Nakajima A, Puri RK. A novel role of interleukin-13 receptor alpha2 in pancreatic cancer invasion and metastasis. Cancer Res. 2009;69(22):8678–8685. doi: 10.1158/0008-5472.CAN-09-2100. [DOI] [PubMed] [Google Scholar]
- Chard MD, Cawston TE, Riley GP, Gresham GA, Hazleman BL. Rotator cuff degeneration and lateral epicondylitis: a comparative histological study. Ann Rheum Dis. 1994;53(1):30–34. doi: 10.1136/ard.53.1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archambault JM, Jelinsky SA, Lake SP, Hill AA, Glaser DL, Soslowsky LJ. Rat supraspinatus tendon expresses cartilage markers with overuse. J Orthop Res. 2007;25(5):617–624. doi: 10.1002/jor.20347. [DOI] [PubMed] [Google Scholar]
- Jelinsky SA, Lake SP, Archambault JM, Soslowsky LJ. Gene expression in rat supraspinatus tendon recovers from overuse with rest. Clin Orthop Relat Res. 2008;466(7):1612–1617. doi: 10.1007/s11999-008-0270-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Berge D, Brugmann SA, Helms JA, Nusse R. Wnt and FGF signals interact to coordinate growth with cell fate specification during limb development. Development. 2008;135(19):3247–3257. doi: 10.1242/dev.023176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabaud M, Lubberts E, Joosten L, van Den Berg W, Miossec P. IL-17 derived from juxta-articular bone and synovium contributes to joint degradation in rheumatoid arthritis. Arthritis Res. 2001;3(3):168–177. doi: 10.1186/ar294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broxmeyer HE, Starnes T, Ramsey H, Cooper S, Dahl R, Williamson E, Hromas R. The IL-17 cytokine family members are inhibitors of human hematopoietic progenitor proliferation. Blood. 2006;108(2):770.. doi: 10.1182/blood-2006-01-0292. [DOI] [PubMed] [Google Scholar]
- Cawston TE, Curry VA, Summers CA, Clark IM, Riley GP, Life PF, Spaull JR, Goldring MB, Koshy PJ, Rowan AD. et al. The role of oncostatin M in animal and human connective tissue collagen turnover and its localization within the rheumatoid joint. Arthritis Rheum. 1998;41(10):1760–1771. doi: 10.1002/1529-0131(199810)41:10<1760::AID-ART8>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Lin TW, Cardenas L, Glaser DL, Soslowsky LJ. Tendon healing in interleukin-4 and interleukin-6 knockout mice. J Biomech. 2006;39(1):61–69. doi: 10.1016/j.jbiomech.2004.11.009. [DOI] [PubMed] [Google Scholar]
- Nakama K, Gotoh M, Yamada T, Mitsui Y, Yasukawa H, Imaizumi T, Higuchi F, Nagata K. Interleukin-6-induced activation of signal transducer and activator of transcription-3 in ruptured rotator cuff tendon. J Int Med Res. 2006;34(6):624–631. doi: 10.1177/147323000603400607. [DOI] [PubMed] [Google Scholar]
- Vincenti MP, Brinckerhoff CE. Transcriptional regulation of collagenase (MMP-1, MMP-13) genes in arthritis: integration of complex signaling pathways for the recruitment of gene-specific transcription factors. Arthritis Res. 2002;4(3):157–164. doi: 10.1186/ar401. [DOI] [PMC free article] [PubMed] [Google Scholar]