Abstract
Osteoblastic and osteocytic cells are highly responsive to the lipid growth factor lysophosphatidic acid (LPA) but the mechanisms by which LPA alters bone cell functions are largely unknown. A major effect of LPA on osteocytic cells is the stimulation of dendrite membrane outgrowth, a process that we predicted to require changes in gene expression and protein distribution. We employed DNA microarrays for global transcriptional profiling of MLO-Y4 osteocytic cells grown for 6 and 24 hours in the presence or absence of LPA. We identified 932 transcripts that displayed statistically significant changes in abundance of at least 1.25-fold in response to LPA treatment. Gene ontology (GO) analysis revealed that the regulated gene products were linked to diverse cellular processes, including DNA repair, response to unfolded protein, ossification, protein-RNA complex assembly, and amine biosynthesis. Gene products associated with the regulation of actin microfilament dynamics displayed the most robust expression changes, and LPA-induced dendritogenesis in vitro was blocked by the stress fiber inhibitor cytochalasin D. Mass spectrometry-based proteomic analysis of MLO-Y4 cells revealed significant LPA-induced changes in the abundance of 284 proteins at 6 hours and 844 proteins at 24 hours. GO analysis of the proteomic data linked the effects of LPA to cell processes that control of protein distribution and membrane outgrowth, including protein localization, protein complex assembly, Golgi vesicle transport, cytoskeleton-dependent transport, and membrane invagination/endocytosis. Dendrites were isolated from LPA-treated MLO-Y4 cells and subjected to proteomic analysis to quantitatively assess the subcellular distribution of proteins. Sets of 129 and 36 proteins were enriched in the dendrite fraction as compared to whole cells after 6 hours and 24 hours of LPA exposure, respectively. Protein markers indicated that membranous organelles were largely excluded from the dendrites. Highly represented among the proteins with elevated abundances in dendrites were molecules that regulate cytoskeletal function, cell motility and membrane adhesion. Our combined transcriptomic/proteomic analysis of the response of MLO-Y4 osteocytic cells to LPA indicates that dendritogenesis is a membrane- and cytoskeleton-driven process with actin dynamics playing a particularly critical role.
Keywords: MLO-Y4 osteocyte-like cells, lysophosphatidic acid, gene expression, proteomics, dendrites
Introduction
Bone cell functions are modulated by a wide array of chemical and physical stimuli, and we and others have found that the bioactive lipid lysophosphatidic acid (LPA) elicits potent receptor-mediated regulatory effects on cultured osteoblastic and osteocytic cells. LPA induces acute signaling events in osteoblastic cells, such as elevations in cytosolic free Ca2+ and the activation of MAP kinase [1–5]. This lipid factor also triggered osteoblast mitogenesis and differentiation, and prolonged the survival of these cells when they were exposed to pro-apoptotic conditions [5–9]. LPA recently was reported to be a key autocrine mediator of nucleotide-coupled osteogenic activity in osteoblasts [10], and this lipid also has potential roles in the regulation of osteoclast function [11–13]. The role of LPA in the control of bone tissue function in vivo is not known, but mice lacking expression of the LPA1 receptor exhibited craniofacial malformations that might reflect effects on skeletal development [14, 15].
Platelets activated during early responses to tissue damage are the major source of LPA in vivo [16, 17], and the primary physiological roles for this lipid appear to relate to the stimulation of wound healing and angiogenesis [18]. It is likely that bone cells in the vicinity of skeletal damage are exposed to high levels of LPA released from hematomas. Pre-osteoblast migration is essential for proper fracture healing [19], and LPA has robust chemotactic effects on osteoblastic cells [1, 20, 21]. LPA induced membrane blebbing in primary cultured calvarial osteoblasts and stimulated the formation of membrane extensions in MC3T3-E1 pre-osteoblastic cells and MLO-Y4 osteocytic cells [21–23]. Osteocyte dendrites are critical for intercellular communication [24], and an enhancement of osteocyte membrane outgrowth in vivo would facilitate the re-establishment of the mechanosensory network in the newly-formed bone during fracture healing.
LPA exerts its effects on target cells through G protein-coupled receptors that subsequently are linked to signaling networks [25]. However, the mechanisms by which rapid signaling events elicit broader changes in bone cell function are less clear. We previously employed DNA microarray analysis to reveal that LPA treatment was linked to the regulation of over 500 gene products in MC3T3-E1 pre-osteoblastic cells [26]. The functions of many of these LPA-regulated transcripts were associated with cellular processes that control phenomena known to be important for skeletal healing, such as proliferation and migration. Thus, transcriptional profiling provided new insights into the mechanisms by which osteoblastic cells alter their function in response to lipid growth factors. We postulated that LPA would have similar effects on gene expression in osteocytic cells, particularly with respect to the ability of the lipid to stimulate dendrite outgrowth, and here we report the results of transcriptomic and proteomic profiling of LPA-treated MLO-Y4 cells.
Materials and methods
Materials
The bovine serum albumin (BSA) used in this study was essentially fatty acid-free (MP Biomedicals, Solon, OH). Ammonium bicarbonate and acetonitrile were purchased from Fisher Scientific (Fair Lawn, NJ), sequencing grade modified trypsin was purchased from Promega (Madison, WI), bicinchoninic acid (BCA) assay reagents and standards were purchased from Pierce (Rockford, IL). Unless otherwise noted, all other reagents were purchased from Sigma-Aldrich (St. Louis, MO).
Cell culture
MLO-Y4 osteocyte-like cells [27], a gift from Dr. Lynda Bonewald (University of Missouri-Kansas City), were grown on gelatin-coated plates in αMEM (Mediatech, Manassas, VA) containing 5% fetal bovine serum and 5% donor calf serum (both sera from Valley Biomedical, Winchester, VA) in a humidified 5% CO2/95% air atmosphere at 37°C. Where indicated, cells were serum-starved by incubation in αMEM containing 0.1% BSA (αMEM/BSA). LPA (1-oleoyl-2-hydroxy-sn-glycerol-3-phosphate; Biomol, Plymouth Meeting, PA) was added to cells from aqueous stock solutions.
DNA microarray analysis
Quadruplicate dishes of MLO-Y4 cells were serum-starved for 16 hours in αMEM/BSA and then incubated an additional 6 or 24 hours in the absence or presence of 1.0 µM LPA. Total RNA was extracted separately from each dish using the Qiagen RNeasy Mini kit (Qiagen, Valencia, CA). RNA quality was verified using an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). Biotin-labeled cRNA was synthesized and fragmented using Affymetrix One-Cycle Target Labeling reagents for hybridization to Mouse Genome 430A 2.0 GeneChips (Affymetrix, Santa Clara, CA). After hybridization, the arrays were washed and stained with streptavidin-phycoerythrin, and then scanned at a resolution of 2.5 microns using an Affymetrix GeneChip Scanner 3000. Quality control parameters were assessed throughout the experimental process to measure the efficiency of transcription, integrity of hybridization, and consistency of qualitative calls. The synthesis and fragmentation of cRNA were assessed using the Agilent 2100 Bioanalyzer. Spike-in control transcripts were monitored to verify hybridization integrity. The raw data files were normalized using the Robust Multi-Array Analysis [28], and significantly regulated genes were identified with multiple testing and false discovery rate (FDR) statistics at p < 0.01 [29] using GeneSpring software. Gene set enrichment for Gene Ontology biological process annotation was calculated using the DAVID web portal to identify the most significant cellular processes affected by LPA treatment [30]. The statistical scores in DAVID were calculated using a modified Fisher Exact test, where the p value (EASE score) was used to determine if the proportion of genes falling into a particular category was more than random chance compared to the background proportion of genes in that category for the whole microarray platform. Processes were chosen with p < 0.05 and at least 5 genes per process.
Real time RT-PCR
Quantitative assessment of mRNA expression was performed by real-time quantitative RT-PCR (qRT-PCR). Complementary DNA was synthesized from total RNA via reverse transcription using the Quantitect kit (Qiagen, Valencia, CA), which includes reagents for genomic DNA removal. To further ensure the specificity of the amplifications, all primer pairs (Table 1) spanned introns except for the destrin primers: the relatively simple intron-exon structure of the Dstn gene precluded the design of intron-spanning primers. PCR reactions were carried out using Roche FastStart DNA MasterPLUS SYBR Green I reagents according to the manufacturer's instructions in a Roche Lightcycler II (Roche Applied Science, Indianapolis, IN). Cycle parameters were: denaturation at 95°C for 10 seconds, annealing at 55°C for 5 seconds, and elongation at 72°C for 10 seconds for 45 cycles. Melting curve analyses were performed from 60°C to 95°C in 0.5°C increments. Quantitative RT-PCR data were normalized to the level of cyclophilin A transcript, the product of the mouse Ppia gene. Statistical analysis was performed using Student’s t-test, and results were considered to be significant if p < 0.05.
Table 1.
Primers for qRT-PCR. All sequences are shown in the 5′→3′ direction.
| Target | Forward primer | Reverse primer | Product (bp) | Accession no. |
|---|---|---|---|---|
| Acta1 | AATGAGCGTTTCCGTTGC | ATCCCCGCAGACTCCATAC | 68 | NM_009606 |
| Actg2 | AGGACTTCTCACACCCTTGG | GTGGTCTCTTCTTCACACATGG | 75 | NM_009610 |
| Actn1 | ATGAAGCCTGGACGGATG | GACAGGGTGGCTGTCTCATAA | 67 | NM_134156 |
| Cnn2 | GGAACATGACACAGGTGCAA | CAATGTCCACACCACTCTGC | 84 | NM_007725 |
| Ctnnd2 | GCGTATGCCCTACGTTCACT | GGGCAGGCCTTTCTCTTT | 96 | NM_008729 |
| Dstn | AGGAAGTGCCCTGTATCTGC | GACAGAAAGTCACTCTAAGCCATGT | 66 | NM_019771 |
| Flnb | TAGGAGCCCTGGTAGACAGC | GGTCCCAGGATTCCCAGT | 60 | NM_134080 |
| Hsbp1 | AGCTCACAGTGAAGACCAAGG | CATGTTCGTCCTGCCTTTCT | 72 | NM_013560 |
| IL33 | GACACATTGAGCATCCAAGG | AACAGATTGGTCATTGTATGTACTCAG | 78 | NM_133775 |
| Nrp1 | CCACACACAGTGGGCTTG | GGTCCAGCTGTAGGTGCTTC | 70 | NM_008737 |
| Ppia | GAGCTGTTTGCAGACAAAGTTC | CCCTGGCACATGAATCCTGG | 125 | NM_008907 |
| Ramp3 | TGTTGTTGCTGCTTTGTGGT | CTCTCCAGCATCCCTGTCTC | 67 | NM_019511 |
| sST2 | CCTCACGGCTCTGAGCTTAT | GGGTCCAGAAGAGAAATCACTG | 70 | NM_010743 |
| ST2L | AGACCTGTTACCTGGGCAAG | CACCTGTCTTCTGCTATTCTGG | 70 | NM_001025602 |
| Tagln | CCTTCCAGTCCACAAACGAC | GTAGGATGGACCCTTGTTGG | 64 | NM_011526 |
| Tnc | GGGCTATAGAACACCGATGC | CATTTAAGTTTCCAATTTCAGGTTC | 71 | NM_011607 |
| Tpm1 | AGCAAGCGGAGGCTGATA | TGACACCAGCTCATCTTCCA | 68 | NM_024427 |
| Tpm2 | AAAACCATTGATGATCTGGAAGA | TGATGTCATTGAGTGCGTTG | 94 | NM_009416 |
| Vcl | CCTCAGGAGCCTGACTTCC | AGCCAGCTCATCAGTTAGTCG | 69 | NM_009502 |
Measurement of dendrite outgrowth in vitro
Dendrite membrane extension was measured using a modification of the method we developed previously [22]. FluoroBlok cell culture inserts (1.0-µm pores; BD Biosciences, San Jose, CA) were coated on both sides by brief immersion in sterile 0.1% bovine gelatin at room temperature. The coated inserts were placed into wells of 24-well transwell “companion” plates containing 1 ml αMEM/BSA or αMEM/BSA/1.0 µM LPA. MLO-Y4 cells were trypsinized, counted with a Coulter counter, centrifuged and suspended in αMEM/BSA at 3.3 × 105 cells/ml. The cell suspension (300 µl) was added to the upper chambers, and the assembly was incubated in a humidified 5% CO2 atmosphere at 37°C for 4 hours. Where indicated, 10 µM cytochalasin D was added to the cell suspension prior to transfer into the transwells. The inserts were transferred to a 24-well plate containing 3 µg/ml calcein acetoxymethyl ester (Invitrogen Molecular Probes, Carlsbad, CA) in Hank’s buffered saline solution (HBSS; GIBCO-Invitrogen, Carlsbad, CA) and incubated for 30 min at 37°C in air. The inserts then were placed in a 24-well Sensoplate coverslip plate (Greiner Bio-One, Monroe, NC) containing HBSS, and images of dendrite outgrowth on the undersides of the microporous chambers were captured using a Nikon epifluorescence microscope and a 10X objective. Four fields from each of duplicate inserts were photographed using identical exposure conditions. Calcein fluorescence in the dendrites was quantified using Volocity image analysis software (Perkin Elmer, Waltham, MA). Data were evaluated statistically using Student’s two-tailed t-test and results were considered to be significant if p < 0.05.
Sample preparation for mass spectrometry(MS)-based proteomic analyses
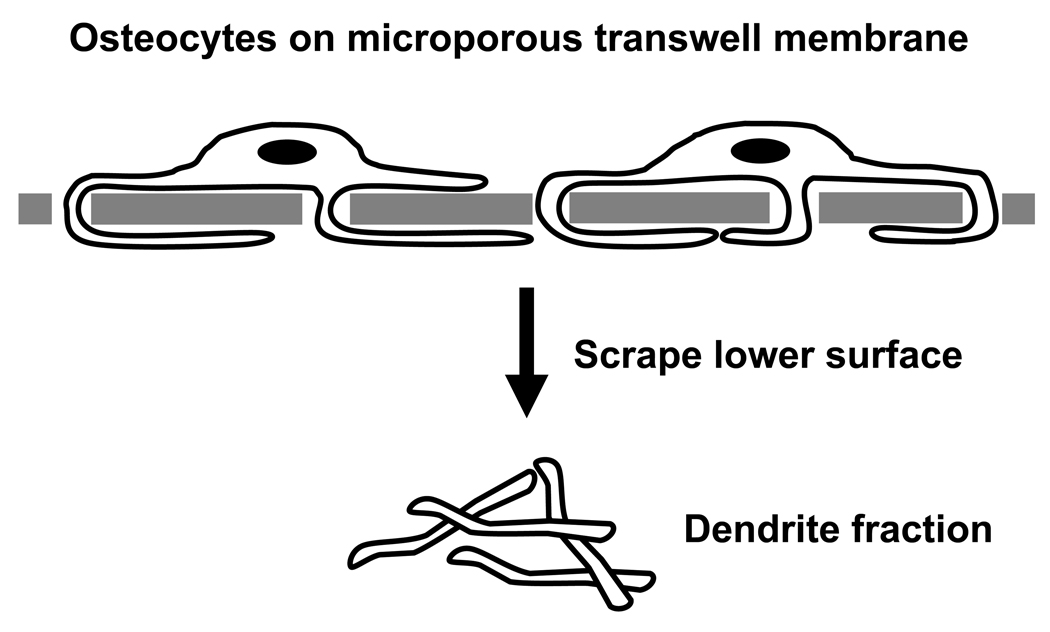
For the preparation of whole cell lysates, MLO-Y4 cells were grown to ~75% confluence in normal medium and then serum-starved for 16 hours in αMEM/BSA. The cells were then incubated an additional 6 or 24 hours in αMEM/BSA in the absence or presence of 1.0 µM LPA. The cells were rinsed three times with ice-cold HBSS after which they were scraped into ice-cold HBSS, collected by centrifugation and frozen as cell pellets at −80°C. For the proteomic analysis of isolated dendrites, 9.0 × 105 MLO-Y4 cells in 1.5 ml αMEM/BSA were seeded in the upper chambers of 6-well plate transwell inserts (1.0-µm pores; BD Biosciences, San Jose, CA) that were gelatin-coated on the upper surface. The cells were incubated for 6 or 24 hours in αMEM/BSA over lower chambers that contained 1.0 µM LPA in αMEM/BSA. The medium was aspirated and the inserts were rinsed twice by sequential immersion in ice-cold phosphate-buffered saline. The transwells were inverted and the dendrites on the lower surfaces of the inserts were scraped into 150-µl droplets of 25 mM NH4HCO3, pH 7.0 (Fig. 1). The yield was approximately 20 µg protein per insert and dendrites isolated from multiple inserts at each time point were pooled and frozen at −80°C to obtain sufficient material for MS analysis.
Fig. 1.
Isolation of dendrites from LPA-treated MLO-Y4 cells. Cells were seeded in the upper chambers of transwell chambers in αMEM/BSA medium and placed into 6-well plates containing 1.0 µM LPA in αMEM/BSA. Dendrites that penetrated the 1-µm pores of the transwell chamber were collected for MS/MS analysis.
Samples were incubated with 50% 2,2,2-trifluoroethanol at 60°C for 2 hours with constant shaking at 300 rpm. Lysis was performed in a sonication bath with ice for 2 minutes, after which protein concentrations were determined by the BCA protein assay (Pierce, Rockford, IL). The samples then were reduced with 2 mM DTT for 1 hour at 37°C with constant shaking at 300 rpm. Reduced samples were diluted 5-fold with 50 mM ammonium bicarbonate, pH 7.8, before tryptic digestion. Sequencing grade modified trypsin was prepared by adding 20 µl of 50 mM ammonium bicarbonate, pH 7.8, to a vial containing 20 µg lyophilized trypsin and incubated for 10 minutes at 37°C. Activated trypsin was added to the samples at a 1:50 (w/w) trypsin-to-protein ratio. Tryptic digestion was carried out at 37°C for 3 hours, followed by rapid freezing of the samples in liquid nitrogen. Samples were concentrated to 100 µl in a Speed-Vac SC 250 Express (Thermo Savant, Holbrook, NY). Concentrated samples were centrifuged at 13,000 rpm for 5 minutes and supernatants were placed into new tubes. A BCA protein assay was performed on samples to verify final concentration of peptides.
Liquid chromatography(LC)-strong cation exchange (SCX) separation
LC-SCX separation was performed with the flow rate of 0.2 ml/min using an Agilent 1100 HPLC System equipped with a quaternary pump, degasser, diode array detector, Peltier-cooled autosampler, and fraction collector (set at 4°C for all samples) (Agilent, Palo Alto, CA, USA) and a PolySulfoethyl A, 200 × 2.1 mm, 300-Å column with 10 × 2.1 mm guard column (PolyLC, Inc., Columbia, MD). Mobile phases were: solvent A = 10 mM ammonium formate, pH 3.0, 25% acetonitrile (ACN); solvent B = 500 mM ammonium formate, pH 6.8, 25% ACN. The injection amount for each separation run was 300 ug of tryptic peptides. Sample separation employed the following solvent gradient sequence: (a) 10 minutes of 100 % Solvent A; (b) 50 minutes changing from 100% Solvent A to 50% Solvent B; (c) 10 minutes increasing the concentration of Solvent B to 100%; and finally (d) 15 minutes of 100% Solvent B.
For fractionation of whole cell samples, 60 fractions were collected over 80 minutes. The first 20 fractions and last 5 fractions were not analyzed further; the remaining fractions were combined for LC MS/MS analysis. For fractionation of the MLO-Y4 cell dendrite samples 60 fractions were collected over 80 minutes. The first 20 fractions were not analyzed further; the remaining fractions were utilized for LC MS/MS analysis. Each fraction was vacuum-dried and stored at −20°C prior to LC MS/MS separation.
Reversed Phase LC Separation and MS/MS Analysis of Peptides
All fractions were subjected to the same LC-MS/MS analysis which included a constant pressure (5,000 psi) reversed phase capillary liquid chromatography system (75 µm i.d. × 65 cm capillary; Polymicro Technologies Inc., Phoenix, AZ) with a Finnigan LTQ ion trap mass spectrometer (ThermoFinnigan, San Jose, CA) and an electrospray ionization source manufactured in-house and which has been reported previously [31]. The instrument was operated in data-dependent mode with an m/z range of 400–2000. The 10 most abundant ions from MS analysis were selected for further MS/MS analysis using a normalized collision energy setting of 35%. A dynamic exclusion of 1 minute was applied to reduce repetitive analysis of the same abundant precursor ion.
LC-MS/MS Data Analysis
ExtractMSn (version 4.0) and SEQUEST analysis software (Version v.27, Rev 12; Thermo Fisher Scientific, Waltham MA) were used to match all MS/MS fragmentation spectra to sequences from the IPI mouse 2007 database (downloaded October 24, 2007), which contains a total of 51,489 protein entries. A search was performed using default parameters with no-enzyme rules within a +/− 1.5 Da parent mass window, +/− 0.5 fragment mass window, average parent mass, and monoisotopic fragment mass. The criteria selected for filtering were based on a method that utilizes a reverse database false positive model which gives a ~95% confidence level over the entire peptide dataset [32]. Specific filter criteria for this study to achieve this level of confidence included DelCN ≥0.1 coupled with Xcorr of ≥1.6 for full tryptic charge state +1, ≥2.4 charge state +2, and ≥3.2 charge state +3, and for partial tryptic peptides an Xcorr ≥4.3 for charge state +2 and ≥4.7 for charge state +3. Comparative protein quantification between control and LPA treatment utilized a tiered approach, described previously [33]. Briefly, proteins with greater than 10 peptide counts require a 1.6-fold change to be considered significant, proteins with more than 5 but less than 10 peptides require a 2.75-fold change to be considered significant, and proteins with 2–5 peptide identifications require a 5-fold difference to be considered significant. This process resulted in 284 significant proteins at 6 hours and 844 proteins at 24 hour after LPA treatment in whole cell lysates. Using the same process with the dendrite lysates, 59 proteins were significantly enriched in dendrites compared to LPA-treated whole cell lysates.
Integrative Data Analysis
Significant gene and protein lists were uploaded into the MetaCore software suite (GeneGo, Inc, St Joseph, MI) and mapped onto significant signaling networks using the shortest path and analyze network algorithms. Whole cell lysate and dendrite networks were merged in the software to identify common points of interaction between the respective signaling networks.
Results
Transcriptomic analysis of LPA-treated MLO-Y4 cells
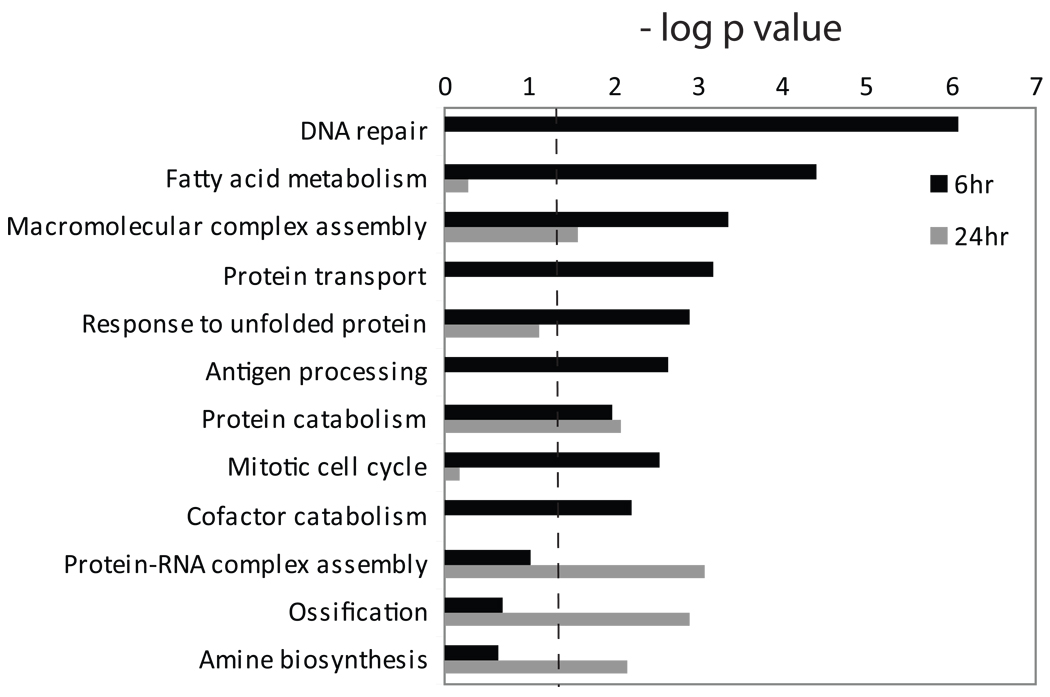
We postulated that the effects of LPA on osteocytic cells would include the regulation of gene expression, including the modulation of transcripts encoding proteins involved in cellular processes that control LPA-induced membrane outgrowth. MLO-Y4 cells were cultured for 6 and 24 hours with 1.0 µM LPA, a dose that we previously found to induce maximal dendrite outgrowth in vitro [22], after which global transcriptional profiling was performed using DNA microarrays. LPA induced statistically significant alterations in 2,976 probesets at a 1% FDR, with at least a 1.25-fold change in the levels of mRNAs detected by 952 probes (Table S1). Gene ontology (GO) analysis of the data revealed that LPA-regulated gene products were linked to a diverse group of cellular processes (Fig. 2). The early (6 hour) response included the regulation of gene products associated with GO processes that relate to inflammation and stress response pathways, such as DNA repair, response to unfolded protein, and antigen processing, as well as transcripts linked to cell proliferation (mitotic cell cycle). Gene products involved in ossification were regulated after 24 hours of LPA exposure, as were transcripts linked to protein-RNA complex assembly and amine biosynthesis. The GO analysis also indicated that genes that regulate protein transport were modulated in LPA-treated MLO-Y4 cells, a process that is important in membrane dynamics and includes elements that may regulate dendrite outgrowth.
Fig. 2.
Gene ontology analysis of LPA-regulated gene products. Biological process enrichment was calculated from the DNA microarray data using the DAVID web portal [30] which calculates a p value based on the probability that a process appears in the data set relative to that expected by random chance. Multifunctional gene products may be assigned to multiple biological processes. The dashed line indicates p = 0.05; all bars that extend to the right of that line were deemed statistically significant.
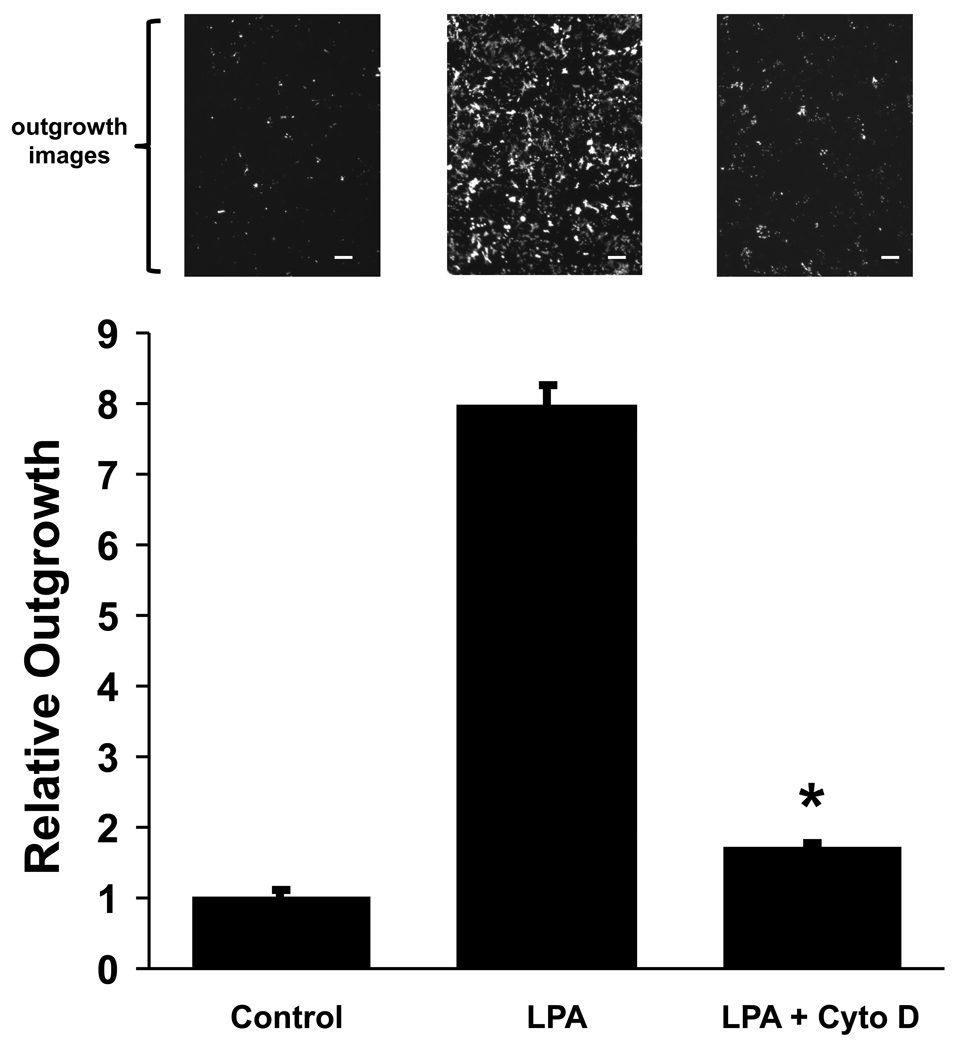
While there were many statistically significant LPA-induced changes in osteocytic cell gene expression relatively few transcripts exhibited changes of 1.5-fold or greater, and the most substantial effects were seen in cells treated for the shorter time period. A heat map profile of the gene expression changes is shown in Figure S1. After 6 hours of LPA exposure 179 gene products exhibited ≥ 1.5-fold increases (109 mRNAs) or decreases (70 mRNAs) in abundance, and 23 transcripts were up-regulated (18 mRNAs) or down-regulated (7 mRNAs) by at least 2.0-fold (Table S1). Genes that encode regulators of actin microfilament function were highly represented in the group most strongly modulated by a 6-hour exposure to the growth factor, a result we verified by qRT-PCR analysis (Table 2). Osteocyte dendrites contain many actin microfilaments that have been implicated the control of cell morphology [22, 34, 35], and the mRNA expression data pointed to the control of actin cytoskeletal dynamics as a potential mechanism by which LPA induces dendrite extension. Therefore, we measured the effect of cytochalasin D, an inhibitor of actin polymerization, on LPA-induced dendrite outgrowth in MLO-Y4 cells. Our results showed that perturbing the actin cytoskeleton profoundly inhibited the ability of LPA to induce dendritogenesis in vitro (Fig. 3).
Table 2.
Relative abundance of LPA-regulated mRNAs encoding proteins related to the cytoskeleton, cell adhesion and cell motility.
| Gene Product | Gene Name | DNA arrays† | qRT-PCR† |
|---|---|---|---|
| Smooth muscle actin γ2 | Actg2 | 5.0 | 30.6 ± 3.7 * |
| Transgelin | Tagln | 4.2 | 14.5 ± 0.9 * |
| Catenin δ2 | Ctnnd2 | 3.0 | 5.5 ± 0.7 * |
| Tenascin C | Tnc | 2.6 | 5.1 ± 0.8 ** |
| Tropomyosin 1α | Tpm1 | 2.5 | 4.9 ± 0.3 * |
| Skeletal muscle actin α1 | Acta1 | 12.5 | 3.0 ± 1.4 |
| Tropomyosin 2β | Tpm2 | 1.6 | 2.4 ± 0.4 * |
| Vinculin | Vcl | 2.3 | 2.3 ± 0.2 *** |
| Neuropilin 1 | Nrp1 | 2.1 | 2.2 ± 0.1 * |
| α-Actinin | Actn1 | 1.7 | 1.7 ± 0.1 * |
| Calponin 2 | Cnn2 | 1.5 | 1.6 ± 0.1 * |
| Destrin | Dstn | 1.5 | 1.6 ± 0.2 *** |
| Filamin β | Flnb | 1.6 | 1.6 ± 0.1 *** |
Data indicate fold-changes relative to control cells after a 6-hr treatment. qRT-PCR results are indicated ± S.E.
p < 0.001;
p < 0.002;
p < 0.05.
Fig. 3.
Effect of actin cytoskeletal disruption on LPA-induced dendrite outgrowth. Dendritogenesis in MLO-Y4 cells was measured using a transwell assay. The data show the quantification of membrane extension where the αMEM/BSA medium in the lower chamber contained no additives (Control), 1.0 µM LPA (LPA), or 1.0 µM LPA + 10 µM cytochalasin D (LPA + Cyto D). Outgrowth was normalized to Control values. The error bars represent S.E. * p < 0.0001 for LPA + Cyto D vs. LPA. Representative micrographs above each column show calcein-loaded dendrite membranes on the undersides of the transwell chambers under each treatment condition. Scale bars: 50 µm.
The DNA microarray and qRT-PCR data revealed that LPA treatment was coupled to changes in the levels of transcripts encoding proteins involved in the control of inflammation: ST2L and sST2 (alternative splicing products of the interleukin 1 receptor-like gene 1, Il1rl); heat shock protein 25 (Hsp1b); interleukin 33, the natural ligand for ST2L [36] (Table 3). LPA also stimulated the expression of RAMP3 mRNA (Table 3), which encodes a receptor for the bone anabolic agents adrenomedullin and amylin [37, 38].
Table 3.
Relative abundance of LPA-regulated mRNAs encoding proteins related to receptor function and inflammation.
| Gene Product | Gene Name | DNA arrays† | qRT-PCR† |
|---|---|---|---|
| Receptor activity modifying protein 3 |
Ramp3 | 5.1 | 5.7 ± 0.5 * |
| Interleukin 1 receptor-like membrane protein (ST2L) |
Il1rl1 | 2.9 | 3.1 ± 0.3 * |
| Interleukin 1 receptor-like soluble protein (sST2) |
Il1rl1 | 2.8 | 2.9 ± 0.1 * |
| Heat-shock protein 25 | Hsbp1 | 1.6 | 1.6 ± 0.1 * |
| Interleukin 33 | Il33 | 2.4 | 1.6 ± 0.1 ** |
Data indicate fold-changes relative to control cells after a 6-hr treatment. qRT-PCR results are indicated ± S.E.
p < 0.001;
p < 0.01.
Proteomic analysis of LPA-treated MLO-Y4 cells
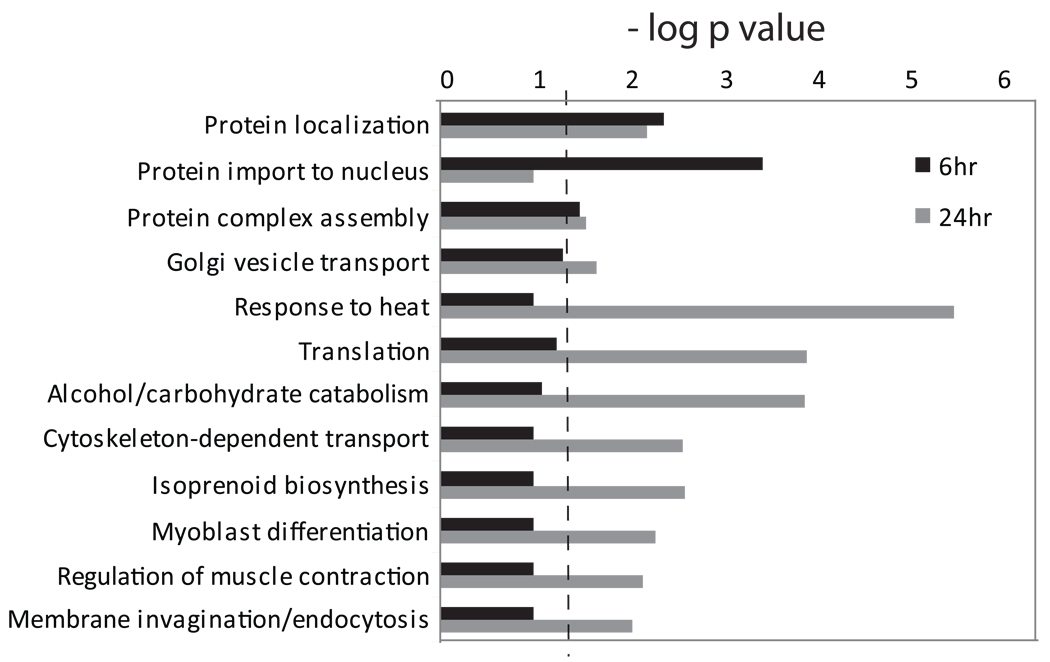
The effects of LPA on gene expression in MLO-Y4 cells were relatively modest in terms of the magnitude of transcript level modulation. This suggested that post-transcriptional regulation might have a substantial role in mediating the ability of this growth factor to alter osteocytic cell function and morphology. We prepared lysates from control and LPA-treated MLO-Y4 cells for tandem mass spectrometry (MS/MS) analysis to identify proteins that exhibit changes in abundance in response to the lipid factor. MS/MS analysis of lysates from whole cells treated with LPA for 6 hours and 24 hours revealed peptides corresponding to 5,565 proteins and 6,122 proteins, respectively; a total of 7,123 proteins with at least 2 peptide hits per protein were identified when both treatment times were compared (Table S2). Statistically significant changes in the levels of 284 proteins were measured after 6 hours of LPA treatment, and 844 proteins exhibited abundance changes after exposure of osteocytic cells to LPA for 24 hours (Table S3). The results of global proteomic profiling linked the effects of LPA on MLO-Y4 cells to several GO categories that have high relevance to the control of protein distribution and membrane outgrowth, including protein localization, protein complex assembly, Golgi vesicle transport, cytoskeleton-dependent transport, and membrane invagination/endocytosis (Figure 4). LPA-induced alterations in proteins associated with the GO categories of regulation of muscle contraction and myoblast differentiation further reflected the extent to which molecules that regulate cytoskeletal dynamics were modulated in response to this lipid factor.
Fig. 4.
Gene ontology analysis of LPA-regulated protein expression. Biological process enrichment was calculated from the MS/MS analysis of MLO-Y4 cells as in Figure 2. The dashed line indicates p = 0.05; all bars that extend to the right of that line were considered to be statistically significant.
We also postulated that a redistribution of specific proteins plays a major role in the outgrowth of membrane processes, and MS-based proteomics was employed to identify proteins that were enriched in LPA-induced dendritic membranes. We isolated membranous extensions from MLO-Y4 cells using a modification of our in vitro dendrite outgrowth assay [22] (Fig. 1). Cells were treated with LPA for either 6 or 24 hours prior to the isolation of membranes from the lower surfaces of transwell chambers. The protein composition of dendrite fractions was compared to the proteomic profiles of whole cell lysates that were prepared from parallel dishes of LPA-treated cells. The dendrite proteome consisted of 900 and 632 proteins (represented by at least 3 peptides each) after exposure of osteocytic cells to LPA for 6 hours and 24 hours, respectively. When the abundance of dendrite proteins was calculated relative to the protein levels in whole cells, 129 proteins showed dendrite enrichment of at least 1.5-fold in cells treated with LPA for 6 hours, and 36 proteins exhibited this level of enrichment after 24 hours of treatment with the lipid factor (Table S2).
Proteins that modulate actin cytoskeletal dynamics were among the peptides found to be enriched in isolated dendrites (Table 4). Thymosin β10, a small G-actin sequestering protein [39], showed particularly large dendrite:whole cell distribution ratios at the two LPA exposure times. Other proteins that regulate actin polymerization also were more abundant in dendrites: actin-related protein 2/3, F-actin-capping protein, phosphatase and actin regulator 4, transgelin-3, LIM domain-containing protein 1, cofilin isoforms, and members of the protein S100 family. Additional cytoskeleton-associated proteins with functions related to cell motility (myotrophin, myosin light polypeptide 6B, tropomyosin family members, radixin) and membrane adhesion (galectin-1) had higher relative abundances in dendrites compared to whole cells (Table 4). LPA treatment led to the redistribution to dendrites of three septin proteins which control membrane architecture via the modulation of microfilament and microtubule organization [40]. Isolated dendrites also were enriched for several proteins associated with microtubule function: tubulin folding cofactor B, kinesin heavy chain isoform 5A, stathmin-2, tubulin-specific chaperone A, microtubule-associated protein 4, and tubulin β2A chain (Table S2).
Table 4. Relative enrichment in LPA-induced osteocyte dendrites of proteins that regulate actin cytoskeletal dynamics.
The data are expressed as the ratio of peptide abundance in isolated dendrites relative to whole osteocytes.
| Protein | 6 hr | 24 hr |
|---|---|---|
| Thymosin β10 | 12.7 | 25.0 |
| Myotrophin | 7.3 | 1.1 |
| Cofilin 1, non-muscle homolog | 5.3 | 1.2 |
| Phosphatase and actin regulator 4 | 5.0 | 0.7 |
| Actin, cytoplasmic type 5 homolog | 4.0 | 7.0 |
| Cofilin, isoform 1 | 4.0 | 0.9 |
| LIM domain-containing protein 1 | 4.0 | 1.0 |
| Galectin-1 | 3.5 | 1.0 |
| Radixin isoform a | 3.0 | 0.5 |
| Tropomyosin 1α chain, isoform 1 | 2.6 | 0.9 |
| Tropomyosin 3α chain, isoform 2 | 2.5 | 1.0 |
| Tropomyosin 3γ | 2.5 | 1.3 |
| Actin-related protein 2/3 complex subunit 3 | 2.3 | 0.8 |
| F-actin-capping protein subunit β | 2.1 | 0.6 |
| Protein S100-A4 | 2.1 | 3.2 |
| Septin 11 | 2.0 | 0.9 |
| Septin 6, isoform V | 2.0 | 0.5 |
| Myosin light polypeptide 6B | 1.7 | 1.0 |
| Transgelin-3 | 1.5 | 0.8 |
| Septin 5 | 1.5 | 0 |
| Protein S100-A10 | 1.5 | 0.2 |
In addition to molecules that regulate cytoskeletal dynamics, isolated dendrites exhibited elevated levels of proteins involved in intermediary metabolism and the control of intracellular redox homeostasis (Table S2): fructose-bisphosphate aldolase C, phosphoglycerate kinase 1, glyoxylate reductase/hydroxypyruvate reductase, glyceraldehyde-3-phosphate dehydrogenase isoform 1, phosphoglycerate mutase 1 and 2, aldehyde dehydrogenase, fructose-bisphosphate aldolase, superoxide dismutase, sulfiredoxin 1 homolog, glutathione S-transferase A2, and thioredoxin domain-containing protein 17. Several proteins linked to receptor-mediated signaling pathways also segregated preferentially to LPA-induced dendrite domains, such as histidine triad nucleotide-binding protein 1, calcium-regulated heat stable protein 1, fibulin-2, Ran-specific GTPase-activating protein, and calcium-binding protein p22. Dendrite membranes were enriched in macrophage migration inhibitory factor and osteoclast-stimulating factor 1, which have been linked to the regulation of bone resorption [41, 42].
Proteins associated with organelles such as mitochondria, (succinate dehydrogenase, phosphoenolpyruvate carboxykinase 2, fumerate hydratase, ATP synthase subunit O), endolysosomes (lysosome membrane protein 2, cathepsin B, vesicle-fusing ATPase, vacuolar protein sorting-associated protein 35, vacuolar ATP synthase subunits), and the endoplasmic reticulum-Golgi complex (calumenin, inositol 1,4,5-trisphosphate receptor type 2, endoplasmin, calreticulin, mannose-6-phosphate receptor, dolichyl-diphospho-oligosaccharide-protein glycosyltransferase, Golgi phosphoprotein 3-like), were largely excluded from the dendritic extensions (Table S2). Several plasma membrane proteins also were de-enriched in the isolated dendrites (integrins αV, α5, β1 and β5, clathrin heavy chain, sodium-potassium ATPase subunits α3 and β3, ectonucleoside triphosphate diphosphohydrolase 5, ectonucleotide pyrophosphatase-phosphodiesterase family member 3) (Table S2).
We employed the MetaCore knowledgebase and software suite to integrate the dendrite proteome with the whole-cell proteomic data and transcriptomic profiling to reveal signaling complexes that are predicted to modulate osteocytic cell functions, particularly networks that may control LPA-induced dendrite outgrowth. LPA-coupled signaling networks were found with src, jun and p53 as major regulatory nodes, proteins which control a wide range of cellular functions (data not shown). Another LPA-coupled network was predicted with the cytoskeletal regulator radixin as the node connecting the dendrite proteome network to whole cell protein and gene expression network (Fig. S2).
Discussion
The exposure of osteocytic cells to a physiological concentration of LPA led to statistically significant changes in the levels of many gene transcripts. GO analysis revealed distinct biological processes to which the functions of the encoded proteins are associated. As expected for the response of cells to a pleiotropic growth factor, a variety of diverse biological processes appeared to be modulated in LPA-treated MLO-Y4 cells. For example, LPA treatment led to elevated levels of transcripts encoding RAMP3 and proteins linked to the regulation of inflammation: ST2L, sST2, heat shock protein 25 (Hsp1b), and interleukin 33. Each of these gene products was among the LPA-induced transcripts in MC3T3-E1 osteoblastic cells [26], but this does not appear to be a generic action of LPA on all cells because Ramp3, Hsp1b and Il33 were not among the LPA-regulated gene products in mouse embryonic fibroblasts [43].
Relatively few large magnitude changes in gene expression were observed in MLO-Y4 cells in response to LPA treatment, in contrast to the response of MC3T3-E1 pre-osteoblastic cells to LPA in which the levels of over 500 gene products were altered by at least two-fold [26]. This difference occurs despite the fact that the two cell types express the same LPA receptor forms [21, 22], which suggests that the osteoblast-to-osteocyte transition is accompanied by a significant change in the intracellular signaling pathways that are coupled to LPA receptor occupancy, particularly those networks that control gene expression. The effects of LPA on gene expression in MLO-Y4 cells were much more pronounced after 6 hours of treatment than after 24 hours of exposure to the growth factor. We observed similar kinetics of gene alterations in LPA-treated MC3T3-E1 cells [26], and it is possible that bone cell LPA receptors exhibit homologous desensitization over time, a phenomenon proposed to the be the major mechanism by which LPA responsiveness is negatively regulated [18, 44]. Alternatively, LPA can be inactivated by dephosphorylation as a result of the activity of lipid phosphate phosphatases expressed on the plasma membrane of many cell types [45], and enzymes present on bone cell surfaces may contribute to a progressive loss of LPA activity: LPA phosphatase type 6 was among the proteins found in our proteomic analysis (Table S2).
LPA is a potent stimulator of osteocytic cell membrane outgrowth and chemotaxis [22]. Many of the transcripts up-regulated most robustly by LPA encode proteins that control cytoskeletal dynamics, cell motility, and cell contact phenomena, which suggest mechanisms by which this lipid regulates bone cell membrane behavior. Gene products with functions potentially relating to dendritogenesis that were up-regulated by LPA included catenin δ2 and neuropilin 1, which in neuronal cells have been linked to the outgrowth of dendritic membrane extensions via an F-actin-mediated process [46, 47]. LPA increased the abundance of mRNA encoding tropomyosins 1 and 2, molecules that mediate actin-myosin interactions in the control of cytoskeletal contraction and membrane structural dynamics [48]. Vinculin is an actin-binding protein associated with cell attachment and focal adhesions, and α-actinin may recruit vinculin to actin fibers to regulate cell morphology during mechanosensation [48]. Filamin β and α-actinin are actin binding proteins that function to crosslink and bundle F-actin fibers, and destrin (actin-depolymerizing factor; ADF) is involved in actin treadmilling during lamellipodia formation [48]. Tenascin C is an extracellular matrix protein transiently expressed during soft tissue wound healing that appears to promote a migratory phenotype in fibroblasts in vitro [49]. Transgelin is a member of calponin family and was found to associate with actin bundles in podosomes, which links this protein to the control of migration and membrane outgrowth [50]. This protein may have other important LPA-modulated functions in osteocyte, such as a role in cell differentiation: transgelin expression was up-regulated during the osteogenic commitment of dental follicle progenitor cells [51].
While LPA-induced changes in MLO-Y4 cell transcript levels were larger after 6 hours of lipid exposure than after 24 hours, protein abundance changes were more pronounced at the later treatment period. Proteins that exhibited LPA-induced changes in expression levels were found by GO analysis to be associated with protein localization, protein complex assembly, Golgi vesicle transport, cytoskeleton-dependent transport, and membrane invagination/endocytosis, cellular processes that are likely to play key roles in the outgrowth of dendritic membrane extensions. Similarly, LPA-treated MLO-Y4 cells displayed changes in the expression of mRNAs associated with the GO category protein transport. Only 60 genes/proteins were found to overlap when the statistically significant transcriptomic and proteomic datasets were compared (data not shown). Notably with respect to LPA-induced dendritogenesis, this list included mRNAs and proteins directly related to cytoskeletal dynamics, such as tropomyosins 1 and 2, calponin 2, destrin, filamin β, and transgelin 2. While the correlation between changes in the levels of mRNA and their encoded proteins typically is less than 50% in mammalian cells [52], the rather small number of common elements between these two screening platforms most likely reflects the comparison of only two widely-spaced time points of LPA treatment.
MS/MS analysis also revealed that proteins linked to the regulation of actin microfilament functions were enriched in dendrites isolated from LPA-treated MLO-Y4 cells. This was consistent with the results of the transcriptomic profiling and with the observation that LPA-induced dendritogenesis in vitro was abrogated by cytochalasin D. Twenty-three proteins with functions associated with the control of actin dynamics were enriched in the isolated dendrites. Thymosin β10, which exhibited the largest enrichment of all proteins found in this subcellular fraction, is a 5 kDa peptide that regulates microfilament polymerization by buffering monomeric G-actin [39]. A moderate overexpression of thymosin β10 in NIH3T3 cells led to increased motility and spreading, and elevated thymosin β10 levels were observed in migrating breast cancer cells [53, 54]. A role for thymosin β10 in osteocyte membrane outgrowth would support our previous proposal that common mechanisms govern aspects of dendritogenesis and cell motility [22]. Other dendrite proteins included molecules that participate in actin filament capping and elongation, including many that have roles in the regulation of cell surface architecture and adhesion. It will be important for future studies to reveal the contribution of these individual peptides to the control of osteocyte morphology.
LPA-induced dendrites contained several proteins with functions linked to tubulin dynamics. However, Murshid et al. observed microtubules extending only partially into the dendrites of chick osteocytes, and exposure of these cells to the microtubule depolymerizing agent nocodazole had no effect on their morphology [34]. Similarly, we treated MLO-Y4 cells with nocodazole and found that LPA-stimulated dendrite outgrowth in vitro was not inhibited (data not shown). Thus, the contribution of the microtubule-associated protein factors in the dendritic membrane processes to osteocyte function remains unknown.
The lack of protein markers for endoplasmic reticulum, Golgi apparatus, endolysosomes and mitochondria in the isolated dendrite fraction points to an apparent exclusion of most membranous organelles from the dendrite region. Several plasma membrane proteins in the dendrite fraction were de-enriched relative to whole cells, including clathrin, galectin-3, adhesion molecules (integrins), ecto-phosphatases, and sodium-potassium ATPase subunits, which implies that dendritogenesis is linked to the creation of specialized cell surface domains.
Integration of proteomic and transcriptomic data onto LPA-induced signaling pathways revealed a network containing important regulators of actin cytoskeletal dynamics with radixin serving as the connecting node (Fig. S2). Radixin is a key member of the ezrin-radixin-moesin (ERM) family of proteins that modulate cytoskeleton-membrane interactions to control the morphology of a variety of cell types. ERMs are particularly important in the organization of specialized membrane domains, such as epithelial cell microvilli [55], and radixin may have a significant role in the regulation of LPA-induced dendritogenesis. This hypothesis will be the focus of future studies.
In conclusion, our combined transcriptomic-proteomic analysis of the response of MLO-Y4 osteocytic cells to the lipid growth factor LPA indicates that dendritogenesis is a cytoskeleton-driven process with actin dynamics playing a particularly critical role. Osteocyte dendrites are essential for cell-cell communication during bone mechanotransduction [56], and decreases in osteocyte connectivity are linked to several bone pathologies [24]. Thus, therapies that foster osteocyte dendrite formation and stability may be useful for the treatment of bone wasting diseases and skeletal damage. The design of such interventions will require a thorough understanding of dendritogenesis, and the data reported here provide new insights into potential mechanisms by which osteocyte morphology is controlled.
Supplementary Material
Heat map of LPA-induced changes in the levels of MLO-Y4 cell gene products. The map depicts the 932 Affymetrix probes that exhibited quantitative modulation of at least 25%. The scale bar indicates the color ranges for transcript levels (log base 2 of fold change).
Integrative analysis of transcriptomic and proteomic data linked to actin cytoskeletal dynamics. Global gene expression data (Affymetrix arrays) and MS-based proteomic data from whole osteocytes and isolated dendritic processes were subjected to network analysis using MetaCore software. Pathways resulting from transcriptomic profiling and whole-cell proteomics are connected by green lines; pathways generated from the dendrite proteome are indicated by green connections.
Acknowledgements
This work was supported by National Institutes of Health grant AR055192 (N.J.K.) and the Laboratory-Directed Research and Development Program at the Pacific Northwest National Laboratory, operated by Battelle for the U.S. Department of Energy under contract DE-AC06-76RLO1830. A portion of this research was performed using the Environmental Molecular Sciences Laboratory (EMSL), a national scientific user facility sponsored by the Department of Energy's Office of Biological and Environmental Research and located at the Pacific Northwest National Laboratory.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Karagiosis SA, Chrisler WB, Bollinger N, Karin NJ. Lysophosphatidic acid-induced ERK activation and chemotaxis in MC3T3-E1 preosteoblasts are independent of EGF receptor transactivation. J Cell Physiol. 2009;219:716–723. doi: 10.1002/jcp.21720. [DOI] [PubMed] [Google Scholar]
- 2.Lyons JM, Karin NJ. A role for G protein-coupled lysophospholipid receptors in sphingolipid-induced Ca2+ signaling in MC3T3-E1 osteoblastic cells. J Bone Miner Res. 2001;16:2035–2042. doi: 10.1359/jbmr.2001.16.11.2035. [DOI] [PubMed] [Google Scholar]
- 3.Liu R, Farach-Carson MC, Karin NJ. Effects of sphingosine derivatives on MC3T3-E1 pre-osteoblasts: psychosine elicits release of calcium from intracellular stores. Biochem Biophys Res Commun. 1995;214:676–684. doi: 10.1006/bbrc.1995.2339. [DOI] [PubMed] [Google Scholar]
- 4.Carpio LC, Stephan E, Kamer A, Dziak R. Sphingolipids stimulate cell growth via MAP kinase activation in osteoblastic cells. Prostaglandins Leukot Essent Fatty Acids. 1999;61:267–273. doi: 10.1054/plef.1999.0100. [DOI] [PubMed] [Google Scholar]
- 5.Caverzasio J, Palmer G, Suzuki A, Bonjour JP. Evidence for the involvement of two pathways in activation of extracellular signal-regulated kinase (Erk) and cell proliferation by Gi and Gq protein-coupled receptors in osteoblast-like cells. J Bone Miner Res. 2000;15:1697–1706. doi: 10.1359/jbmr.2000.15.9.1697. [DOI] [PubMed] [Google Scholar]
- 6.Dziak R, Yang BM, Leung BW, Li S, Marzec N, Margarone J, Bobek L. Effects of sphingosine-1-phosphate and lysophosphatidic acid on human osteoblastic cells. Prostaglandins Leukot Essent Fatty Acids. 2003;68:239–249. doi: 10.1016/s0952-3278(02)00277-6. [DOI] [PubMed] [Google Scholar]
- 7.Grey A, Banovic T, Naot D, Hill B, Callon K, Reid I, Cornish J. Lysophosphatidic acid is an osteoblast mitogen whose proliferative actions involve Gi proteins and protein kinase C, but not P42/44 mitogen-activated protein kinases. Endocrinology. 2001;142:1098–1106. doi: 10.1210/endo.142.3.8011. [DOI] [PubMed] [Google Scholar]
- 8.Grey A, Chen Q, Callon K, Xu X, Reid IR, Cornish J. The phospholipids sphingosine-1- phosphate and lysophosphatidic acid prevent apoptosis in osteoblastic cells via a signaling pathway involving Gi proteins and phosphatidylinositol-3 kinase. Endocrinology. 2002;143:4755–4763. doi: 10.1210/en.2002-220347. [DOI] [PubMed] [Google Scholar]
- 9.Liu YB, Kharode Y, Bodine PV, Yaworsky PJ, Robinson JA, Billiard J. LPA induces osteoblast differentiation through interplay of two receptors: LPA1 and LPA4. J Cell Biochem. 2010;109:794–800. doi: 10.1002/jcb.22471. [DOI] [PubMed] [Google Scholar]
- 10.Panupinthu N, Rogers JT, Zhao L, Solano-Flores LP, Possmayer F, Sims SM, Dixon SJ. P2×7 receptors on osteoblasts couple to production of lysophosphatidic acid: a signaling axis promoting osteogenesis. J Cell Biol. 2008;181:859–871. doi: 10.1083/jcb.200708037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boucharaba A, Serre CM, Guglielmi J, Bordet JC, Clezardin P, Peyruchaud O. The type 1 lysophosphatidic acid receptor is a target for therapy in bone metastases. Proc Natl Acad Sci U S A. 2006;103:9643–9648. doi: 10.1073/pnas.0600979103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aki Y, Kondo A, Nakamura H, Togari A. Lysophosphatidic acid-stimulated interleukin-6 and −8 synthesis through LPA1 receptors on human osteoblasts. Arch Oral Biol. 2008;53:207–213. doi: 10.1016/j.archoralbio.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 13.Lapierre DM, Tanabe N, Pereverzev A, Spencer M, Shugg RP, Dixon SJ, Sims SM. Lysophosphatidic acid signals through multiple receptors in osteoclasts to elevate cytosolic calcium concentration, evoke retraction and promote cell survival. J Biol Chem. 2010;285:25792–25801. doi: 10.1074/jbc.M110.109322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Contos JJ, Fukushima N, Weiner JA, Kaushal D, Chun J. Requirement for the lpA1 lysophosphatidic acid receptor gene in normal suckling behavior. Proc Natl Acad Sci U S A. 2000;97:13384–13389. doi: 10.1073/pnas.97.24.13384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harrison SM, Reavill C, Brown G, Brown JT, Cluderay JE, Crook B, Davies CH, Dawson LA, Grau E, Heidbreder C, Hemmati P, Hervieu G, Howarth A, Hughes ZA, Hunter AJ, Latcham J, Pickering S, Pugh P, Rogers DC, Shilliam CS, Maycox PR. LPA1 receptor-deficient mice have phenotypic changes observed in psychiatric disease. Mol Cell Neurosci. 2003;24:1170–1179. doi: 10.1016/j.mcn.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 16.Eichholtz T, Jalink K, Fahrenfort I, Moolenaar WH. The bioactive phospholipid lysophosphatidic acid is released from activated platelets. Biochem J. 1993;291(Pt 3):677–680. doi: 10.1042/bj2910677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sano T, Baker D, Virag T, Wada A, Yatomi Y, Kobayashi T, Igarashi Y, Tigyi G. Multiple mechanisms linked to platelet activation result in lysophosphatidic acid and sphingosine 1-phosphate generation in blood. J Biol Chem. 2002;277:21197–21206. doi: 10.1074/jbc.M201289200. [DOI] [PubMed] [Google Scholar]
- 18.Moolenaar WH, van Meeteren LA, Giepmans BN. The ins and outs of lysophosphatidic acid signaling. Bioessays. 2004;26:870–881. doi: 10.1002/bies.20081. [DOI] [PubMed] [Google Scholar]
- 19.Einhorn TA. The cell and molecular biology of fracture healing. Clin Orthop Relat Res. 1998;355 Suppl:S7–S21. doi: 10.1097/00003086-199810001-00003. [DOI] [PubMed] [Google Scholar]
- 20.Fotos JS, Patel VP, Karin NJ, Koh JT, Temburni MK, Galileo DS. Automated time-lapse microscopy and high-resolution tracking of cell migration. Cytotechnology. 2006;51:7–19. doi: 10.1007/s10616-006-9006-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masiello LM, Fotos JS, Galileo DS, Karin NJ. Lysophosphatidic acid induces chemotaxis in MC3T3-E1 osteoblastic cells. Bone. 2006;39:72–82. doi: 10.1016/j.bone.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 22.Karagiosis SA, Karin NJ. Lysophosphatidic acid induces osteocyte dendrite outgrowth. Biochem Biophys Res Commun. 2007;357:194–199. doi: 10.1016/j.bbrc.2007.03.121. [DOI] [PubMed] [Google Scholar]
- 23.Panupinthu N, Zhao L, Possmayer F, Ke HZ, Sims SM, Dixon SJ. P2×7 nucleotide receptors mediate blebbing in osteoblasts through a pathway involving lysophosphatidic acid. J Biol Chem. 2007;282:3403–3412. doi: 10.1074/jbc.M605620200. [DOI] [PubMed] [Google Scholar]
- 24.Knothe Tate ML, Adamson JR, Tami AE, Bauer TW. The osteocyte. Int J Biochem Cell Biol. 2004;36:1–8. doi: 10.1016/s1357-2725(03)00241-3. [DOI] [PubMed] [Google Scholar]
- 25.Lin ME, Herr DR, Chun J. Lysophosphatidic acid (LPA) receptors: signaling properties and disease relevance. Prostaglandins Other Lipid Mediat. 2010;91:130–138. doi: 10.1016/j.prostaglandins.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Waters KM, Tan R, Genetos DC, Verma S, Yellowley CE, Karin NJ. DNA microarray analysis reveals a role for lysophosphatidic acid in the regulation of anti-inflammatory genes in MC3T3-E1 cells. Bone. 2007;41:833–841. doi: 10.1016/j.bone.2007.06.029. [DOI] [PubMed] [Google Scholar]
- 27.Kato Y, Windle JJ, Koop BA, Mundy GR, Bonewald LF. Establishment of an osteocyte-like cell line, MLO-Y4. J Bone Miner Res. 1997;12:2014–2023. doi: 10.1359/jbmr.1997.12.12.2014. [DOI] [PubMed] [Google Scholar]
- 28.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 29.Dudoit S, van der Laan MJ, Pollard KS. Multiple testing. Part I. Single-step procedures for control of general type I error rates. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1040. Article13. [DOI] [PubMed] [Google Scholar]
- 30.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 31.Shen Y, Zhao R, Belov ME, Conrads TP, Anderson GA, Tang K, Pasa-Tolic L, Veenstra TD, Lipton MS, Udseth HR, Smith RD. Packed capillary reversed-phase liquid chromatography with high-performance electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry for proteomics. Anal Chem. 2001;73:1766–1775. doi: 10.1021/ac0011336. [DOI] [PubMed] [Google Scholar]
- 32.Qian WJ, Monroe ME, Liu T, Jacobs JM, Anderson GA, Shen Y, Moore RJ, Anderson DJ, Zhang R, Calvano SE, Lowry SF, Xiao W, Moldawer LL, Davis RW, Tompkins RG, Camp DG, 2nd, Smith RD. Quantitative proteome analysis of human plasma following in vivo lipopolysaccharide administration using 16O/18O labeling and the accurate mass and time tag approach. Mol Cell Proteomics. 2005;4:700–709. doi: 10.1074/mcp.M500045-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qian WJ, Jacobs JM, Camp DG, 2nd, Monroe ME, Moore RJ, Gritsenko MA, Calvano SE, Lowry SF, Xiao W, Moldawer LL, Davis RW, Tompkins RG, Smith RD. Comparative proteome analyses of human plasma following in vivo lipopolysaccharide administration using multidimensional separations coupled with tandem mass spectrometry. Proteomics. 2005;5:572–584. doi: 10.1002/pmic.200400942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murshid SA, Kamioka H, Ishihara Y, Ando R, Sugawara Y, Takano-Yamamoto T. Actin and microtubule cytoskeletons of the processes of 3D–cultured MC3T3-E1 cells and osteocytes. J Bone Miner Metab. 2007;25:151–158. doi: 10.1007/s00774-006-0745-5. [DOI] [PubMed] [Google Scholar]
- 35.Tanaka-Kamioka K, Kamioka H, Ris H, Lim SS. Osteocyte shape is dependent on actin filaments and osteocyte processes are unique actin-rich projections. J Bone Miner Res. 1998;13:1555–1568. doi: 10.1359/jbmr.1998.13.10.1555. [DOI] [PubMed] [Google Scholar]
- 36.Schmitz J, Owyang A, Oldham E, Song Y, Murphy E, McClanahan TK, Zurawski G, Moshrefi M, Qin J, Li X, Gorman DM, Bazan JF, Kastelein RA. IL-33, an interleukin-1-like cytokine that signals via the IL-1 receptor-related protein ST2 and induces T helper type 2-associated cytokines. Immunity. 2005;23:479–490. doi: 10.1016/j.immuni.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 37.Cornish J, Grey A, Callon KE, Naot D, Hill BL, Lin CQ, Balchin LM, Reid IR. Shared pathways of osteoblast mitogenesis induced by amylin, adrenomedullin, and IGF-1. Biochem Biophys Res Commun. 2004;318:240–246. doi: 10.1016/j.bbrc.2004.04.020. [DOI] [PubMed] [Google Scholar]
- 38.McLatchie LM, Fraser NJ, Main MJ, Wise A, Brown J, Thompson N, Solari R, Lee MG, Foord SM. RAMPs regulate the transport and ligand specificity of the calcitonin-receptor-like receptor. Nature. 1998;393:333–339. doi: 10.1038/30666. [DOI] [PubMed] [Google Scholar]
- 39.Huff T, Muller CS, Otto AM, Netzker R, Hannappel E. β-Thymosins, small acidic peptides with multiple functions. Int J Biochem Cell Biol. 2001;33:205–220. doi: 10.1016/s1357-2725(00)00087-x. [DOI] [PubMed] [Google Scholar]
- 40.Spiliotis ET, Nelson WJ. Here come the septins: novel polymers that coordinate intracellular functions and organization. J Cell Sci. 2006;119:4–10. doi: 10.1242/jcs.02746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jacquin C, Koczon-Jaremko B, Aguila HL, Leng L, Bucala R, Kuchel GA, Lee SK. Macrophage migration inhibitory factor inhibits osteoclastogenesis. Bone. 2009;45:640–649. doi: 10.1016/j.bone.2009.06.028. [DOI] [PubMed] [Google Scholar]
- 42.Reddy S, Devlin R, Menaa C, Nishimura R, Choi SJ, Dallas M, Yoneda T, Roodman GD. Isolation and characterization of a cDNA clone encoding a novel peptide (OSF) that enhances osteoclast formation and bone resorption. J Cell Physiol. 1998;177:636–645. doi: 10.1002/(SICI)1097-4652(199812)177:4<636::AID-JCP14>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 43.Stortelers C, Kerkhoven R, Moolenaar WH. Multiple actions of lysophosphatidic acid on fibroblasts revealed by transcriptional profiling. BMC Genomics. 2008;9:387. doi: 10.1186/1471-2164-9-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jalink K, van Corven EJ, Moolenaar WH. Lysophosphatidic acid, but not phosphatidic acid, is a potent Ca2+-mobilizing stimulus for fibroblasts. Evidence for an extracellular site of action. J Biol Chem. 1990;265:12232–12239. [PubMed] [Google Scholar]
- 45.Brindley DN, Pilquil C. Lipid phosphate phosphatases and signaling. J Lipid Res. 2009;50 Suppl:S225–S230. doi: 10.1194/jlr.R800055-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim K, Sirota A, Chen Yh YH, Jones SB, Dudek R, Lanford GW, Thakore C, Lu Q. Dendrite-like process formation and cytoskeletal remodeling regulated by δ-catenin expression. Exp Cell Res. 2002;275:171–184. doi: 10.1006/excr.2002.5503. [DOI] [PubMed] [Google Scholar]
- 47.Kim S, Chiba A. Dendritic guidance. Trends Neurosci. 2004;27:194–202. doi: 10.1016/j.tins.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 48.Le Clainche C, Carlier MF. Regulation of actin assembly associated with protrusion and adhesion in cell migration. Physiol Rev. 2008;88:489–513. doi: 10.1152/physrev.00021.2007. [DOI] [PubMed] [Google Scholar]
- 49.Trebaul A, Chan EK, Midwood KS. Regulation of fibroblast migration by tenascin-C. Biochem Soc Trans. 2007;35:695–697. doi: 10.1042/BST0350695. [DOI] [PubMed] [Google Scholar]
- 50.Assinder SJ, Stanton JA, Prasad PD. Transgelin: an actin-binding protein and tumour suppressor. Int J Biochem Cell Biol. 2009;41:482–486. doi: 10.1016/j.biocel.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 51.Morsczeck C, Petersen J, Vollner F, Driemel O, Reichert T, Beck HC. Proteomic analysis of osteogenic differentiation of dental follicle precursor cells. Electrophoresis. 2009;30:1175–1184. doi: 10.1002/elps.200800796. [DOI] [PubMed] [Google Scholar]
- 52.Waters KM, Pounds JG, Thrall BD. Data merging for integrated microarray and proteomic analysis. Brief Funct Genomic Proteomic. 2006;5:261–272. doi: 10.1093/bfgp/ell019. [DOI] [PubMed] [Google Scholar]
- 53.Maelan AE, Rasmussen TK, Larsson LI. Localization of thymosin β10 in breast cancer cells: relationship to actin cytoskeletal remodeling and cell motility. Histochem Cell Biol. 2007;127:109–113. doi: 10.1007/s00418-006-0208-z. [DOI] [PubMed] [Google Scholar]
- 54.Sun HQ, Kwiatkowska K, Yin HL. β-Thymosins are not simple actin monomer buffering proteins. Insights from overexpression studies. J Biol Chem. 1996;271:9223–9230. [PubMed] [Google Scholar]
- 55.Fehon RG, McClatchey AI, Bretscher A. Organizing the cell cortex: the role of ERM proteins. Nat Rev Mol Cell Biol. 2010;11:276–287. doi: 10.1038/nrm2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Burger EH, Klein-Nulend J. Mechanotransduction in bone – role of the lacuno- canalicular network. FASEB J. 1999;13 Suppl:S101–S112. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Heat map of LPA-induced changes in the levels of MLO-Y4 cell gene products. The map depicts the 932 Affymetrix probes that exhibited quantitative modulation of at least 25%. The scale bar indicates the color ranges for transcript levels (log base 2 of fold change).
Integrative analysis of transcriptomic and proteomic data linked to actin cytoskeletal dynamics. Global gene expression data (Affymetrix arrays) and MS-based proteomic data from whole osteocytes and isolated dendritic processes were subjected to network analysis using MetaCore software. Pathways resulting from transcriptomic profiling and whole-cell proteomics are connected by green lines; pathways generated from the dendrite proteome are indicated by green connections.