Abstract
Interactions of the 8-kDa domain of the rat pol β and the intact enzyme with the ssDNA have been studied, using the quantitative fluorescence titration technique. The 8-kDa domain induces large topological changes in the bound DNA structure and engages much larger fragments of the DNA than when embedded in the intact enzyme. The DNA affinity of the domain is predominantly driven by entropy changes, dominated by the water release from the protein. The thermodynamic characteristics dramatically change when the domain is embedded in the intact polymerase, indicating the presence of significant communication between the 8-kDa domain and the catalytic 31-kDa domain. The diminished water release from the 31-kDa domain strongly contributes to its dramatically lower DNA affinity, as compared to the 8-kDa domain. Unlike the 8-kDa domain, the DNA binding of the intact pol β is driven by entropy changes, originating from the structural changes of the formed complexes.
Keywords: Fluorescence Titrations, Polymerases, DNA Repair, DNA Replication, Protein - ssDNA Interactions
INTRODUCTION
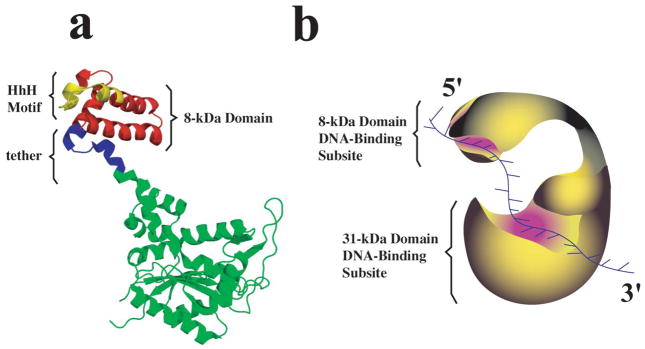
Polymerase β is one of several recognized DNA-directed polymerases of the eukaryotic nucleus (1–8). The enzyme plays a very specialized function in the DNA repair machinery in mammalian cells conducting “gap fillings” synthesis on gapped DNA. The protein is a single polypeptide of ~39 kDa whose crystal structure revealed a typical polymerase fold consisting of a thumb, palm, and fingers [9,10]. However, a characteristic feature of the pol β structure is the presence of an extra, small 8-kDa domain that is connected with the tip of the fingers through a tether of 14 amino acids, as depicted in Figure 1a [9,10].
Figure 1.
a. Three-generated using data from Brookhaven Protein Data Bank, under the code 1BPD, using PyMOL (DeLAno dimensional structure of the rat pol β based on crystallographic data [10]. The structure has been Scientific, San Carlos, CA). The 8-kDa domain is located at the tip of the finger domain and marked in red. Also, the location of the DNA-binding motif, HhH, is marked (yellow) in the structure. b. Schematic representation of the pol β molecule, which indicates the location of the DNA binding subsites on the 8-kDa and 31-kDa domain and their engagement in interactions with the ssDNA.
A fundamental feature of the pol β activities is that the enzyme must recognize a specific structure of the damaged DNA and the recognition process must precede the chemical step of the DNA synthesis. The complexity of the recognition process is reflected in the functional intricacy of the total DNA-binding site of the enzyme. It is built of two functionally different DNA-binding subsites located on the 8-kDa and the catalytic 31-kDa domains, respectively [11–18]. As a result, the enzyme binds the ssDNA in two binding modes which differ in the number of occluded nucleotides, the (pol β)16 and (pol β)5 binding modes [11–18]. In the (pol β)16 binding mode, both the 8-kDa and 31-kDa domains are involved in interactions with the ssDNA. In the (pol β)5 binding mode, only the 8-kDa domain is engaged in interactions with the nucleic acid.
Although both the 8-kDa and 31-kDa domains have DNA-binding capability, the DNA-affinity of the subsite on the 8-kDa domain is at least ~2 orders of magnitude higher than the affinity of the subsite on the 31-kDa domain [11,12,15]. The subsite on the 8-kDa domain is the primary DNA-binding area of the enzyme, which anchors the enzyme on the gapped DNA substrate [17,18]. The complex is initiated through the fast association of the 8-kDa domain with the DNA [17,18]. The 8-kDa domain recognizes the ss and/or dsDNA part of the substrate downstream from the primer. Association of the 31-kDa catalytic domain with the opposite dsDNA part of the substrate, which contains the primer, follows the initial recognition step.
Previous studies indicated very intricate characteristics of the isolated 8-kDa domain ssDNA interactions [15]. In complexes with polymer ssDNAs, the domain seems to occlude 13 ± 0.7 and 9 ± 0.6 nucleotides, in the absence and presence of Mg2+, respectively [15]. The stoichiometries of the isolated domain - polymer ssDNA complexes are significantly larger than the site-size of ~5 nucleotides of the (pol β)5 binding mode, indicating that the intact enzyme uses only part of the DNA-binding subsite of the domain in its interactions with the nucleic acid. The DNA-binding subsite of the domain has an energetically homogeneous structure, with crucial contacts for the DNA evenly distributed over the binding area.
Elucidation of the 8-kDa domain interactions with the ssDNA is one of the fundamental steps toward understanding the specificity of the molecular mechanism of the DNA recognition by pol β. Such analysis will also provide essential insights about the recognition mechanisms of DNA substrates by other DNA repair polymerases. Several puzzling questions are still unanswered. What is the major cause of such a large difference in the affinities between both DNA-binding subsites? To what extent are the interactions of the 8-kDa domain with the DNA independent from the interactions of the 31-kDa domain with the nucleic acid, i.e., is the 31-kDa domain imposing a specific orientation of the domain, which affects the site-size of the complex with the nucleic acid? In other words, what are the thermodynamic characteristics of these interactions, which would also indicate possible controls of their mechanisms?
In this communication, we examine the energetics of the interactions of the 8-kDa domain and the intact enzyme with the ssDNA. The 8-kDa domain engages much large fragments of the DNA than when the domain is embedded in the intact enzyme. The intrinsic affinity of the 8-kDa domain is predominantly driven by large entropy changes, generated by the water release from the domain. There is significant communication between the 8-kDa domain and the catalytic 31-kDa domain in the intact pol β, mediated through the 14 amino acid tether connecting the domains. Dramatically lower DNA affinity of the 31-kDa catalytic domain, as compared to the 8-kDa domain, results from the diminished release of water molecules from the catalytic subsite.
MATERIALS & METHODS
Reagents and Buffers
All chemicals were reagent grade. All solutions were made with distilled and deionized >18 MΩ(Milli -Q Plus) water. Buffer C is 10 mM sodium cacodylate adjusted to pH 7.0 with HCl, 100 mM NaCl, and 10% glycerol. The temperature and concentration of MgCl2 in the buffer are indicated in the text.
Intact Rat Pol β and the Rat Pol β8 -kDa Domain
Purification of the intact rat pol β and the 8 -kDa domain was performed as previously described [11–18]. The concentrations of the enzyme and the domain were spectrophotometrically determined using extinction coefficients ε280 = 2.1 × 104 cm−1 M−1, and ε280= 4.45 × 103 cm−1 M−1, respectively, obtained with the approach based on Edelhoch’s method [11–18,19,20].
Nucleic Acids
All nucleic acids were purchased from Midland Certified Reagents (Midland, Texas). The etheno-derivatives of the nucleic acids were obtained by modification with chloroacetaldehyde [11–15,21,22]. This modification goes to completion and provides a fluorescent derivative of the nucleic acid. The concentrations of the DNAs were spectrophotometrically determined using the extinction coefficient ε257= 3700 M−1 cm −1 (Nucleotide) [11–15,21,22]. The labeled ssDNA oligomer, dT(pT)19, contains a fluorescent marker, fluorescein (Fl), attached at the 5′ end through phosphoramidate chemistry, and/or coumarin derivative (CP), attached through the 6-carbon linker at the 3′ end. The labeled ssDNA oligomers are referred to as: 5′-Fl-dT(pT)19, dT(pT)19-CP-3′, and 5′-Fl-dT(pT)19-CP-3′.
Fluorescence Measurements
All steady -state fluorescence titrations were performed using the SLM-AMINCO 8100C spectrofluorometer. In order to avoid possible artifacts, due to the fluorescence anisotropy of the sample, polarizers were placed in excitation and emission channels and set at 90° and 55° (magic angle), respectively. The binding was followed by monitoring the fluorescence of the etheno-derivatives (λex = 325 nm, λem = 410 nm). Computer fits were performed using Mathematica (Wolfram, IL) and KaleidaGraph (Synergy Software, PA). The nucleic acid relative fluorescence increase, ΔF, upon binding the 8-kDa domain or the intact enzyme is defined as, ΔF = (Fi− Fo)/Fo, where Fi is the fluorescence of the DNA at a given titration point “i”, and Fo is the initial fluorescence value of the sample [11–15].
Quantitative Determination of Stoichiometries and Binding Isotherms of the Rat Pol β and the 8 -kDa Domain -ssDNA Complexes
In this work, we followed the binding of the rat pol β and the 8-kDa domain to the ssDNAs by monitoring the fluorescence increase, ΔF, of the nucleic acid etheno-derivatives upon the complex formation. The method of obtaining rigorous estimates of the total average degree of binding, ΣΘi, (number of domain molecules bound per oligomer) and the free protein concentration, PF, has been previously described in detail by us [23–25]. Briefly, the experimentally observed ΔF has a contribution from each of the different possible “i” complexes of the protein with the ssDNA. Thus, the observed fluorescence increase is functionally related to ΣΘi by
| (1) |
where Θi and ΔFimax are partial degree of binding and the molecular parameter characterizing the maximum fluorescence increase of the nucleic acid with the protein bound in complex “i”, respectively. The same value of ΔF, obtained at two different total nucleic acid concentrations, NT1 and NT2, indicates the same physical state of the nucleic acid, i.e., the total average degree of binding, ΣΘi, and the free protein concentration, PF, must be the same. The values of ΣΘi and PF are then related to the total protein concentrations, PT1 and PT2, and the total nucleic acid concentrations, NT1 and NT2, at the same value of ΔF, by
| (2) |
| (3) |
Fluorescence Energy Transfer Measurements
The Förster efficiency of the fluorescence energy transfer, E, from the coumarin donor, located at the 3′ end of the ssDNA oligomer, dT(pT)19, to the acceptor, fluorescein moiety, located at the 5′ end of the same oligomer, has been determined using two apparent fluorescence energy transfer efficiencies [26–29]. The energy transfer efficiency, ED, obtained from the quenching of the donor fluorescence is defined as [16,29]
| (4) |
where FD and F DA are the fluorescence of the donor in the absence and presence of the acceptor, respectively, νD is the fraction of the donor in the complex with the acceptor [16,29].
The apparent fluorescence transfer efficiency, EA, has been determined by measuring the fluorescence intensity of the acceptor (fluorescein at the 5′ end of the ssDNA oligomer), excited at a wavelength where a donor (coumarin at the 3′ end of the ssDNA oligomer) predominantly absorbs, in the absence and presence of the donor. The value of EA is defined as [16,29]
| (5) |
where The FA, and FAD, are fluorescence intensities of the acceptor in the absence and presence of the donor, CAT and C DT are the total concentrations of the acceptor and the donor, νA is the fraction of acceptors in the complex with donors, εA and εD are the molar absorption coefficients of the acceptor and the donor at the excitation wavelength, respectively, and are the quantum yields of the free acceptor and the acceptor in the presence of the donor. All quantities in eq. 5 can be experimentally determined. The Förster energy transfer efficiency, E, is related to ED and EA, by [30,31]
| (6) |
The fluorescence energy transfer efficiency between the donor and the acceptor dipoles, E, is related to the average distance, R, separating the absorption and emission dipoles by [26–29,30,31]
| (7) |
where, Ro = 9790(κ2n−4φd J)1/6, is the so called Förster critical distance (in angstroms), the distance at which the transfer efficiency is 50%, κ2 is the orientation factor, φd is the donor quantum yield in the absence of the acceptor, and n is the refractive index of the medium (n = 1.4), the overlap integral, J, characterizes the resonance between the donor and acceptor dipoles. The Förster critical distance, Ro = 52 Å, for fluorescein and coumarin, attached to the 5′ and 3′ end of dT(pT)19, respectively, has been previously determined by us [29].
RESULTS
The Site-Sizes of the Pol β8 -kDa Domain - ssDNA Complexes In the Presence and Absence of Magnesium
Studies with the polymer ssDNA indicated that the isolated rat pol β8 -kDa domain changes its site-size in the complex with the nucleic acid, depending on the presence of the magnesium cations in solution (see above) [15]. However, the presence of positive cooperative interactions, characterizing the binding process has made these analyses rather difficult [15]. To quantitatively and systematically address the issue of the site-size of the domain - ssDNA complex, we examined interactions of the isolated 8-kDa domain with the ssDNA oligomers, differing in the number of nucleotides in the presence and absence of Mg2+, using a series of ssDNA oligomers, containing different numbers of nucleotides. Moreover, the selected oligomers accommodate only a single molecule of the domain, as tested by the quantitative method outlined in Materials and Methods [23–25]. We applied an analogous strategy in our previous studies of various protein -DNA systems, including E. coli DnaB and PriA helicases, PriB protein, plasmid 1010 RepA helicase, and African Swine Fever virus polymerase X [32–39].
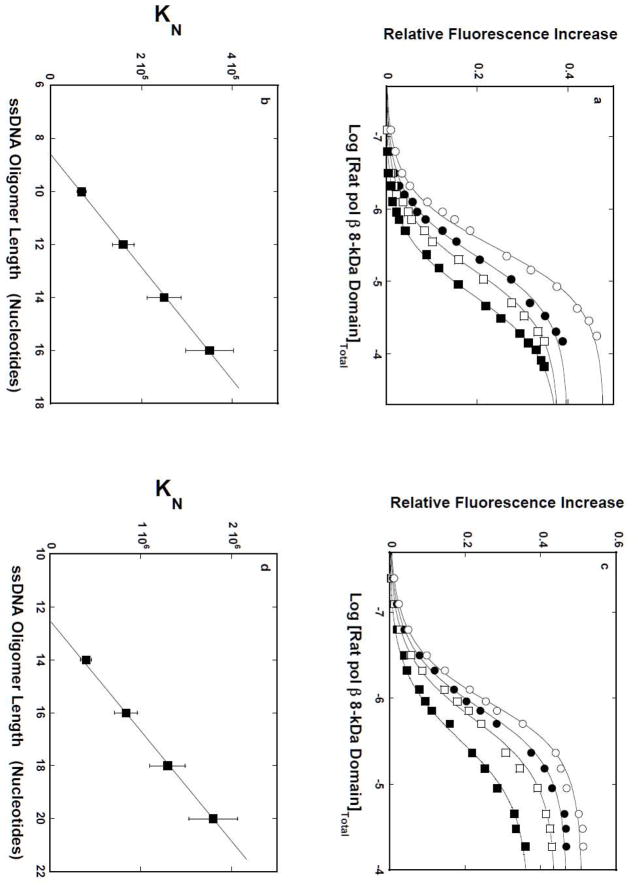
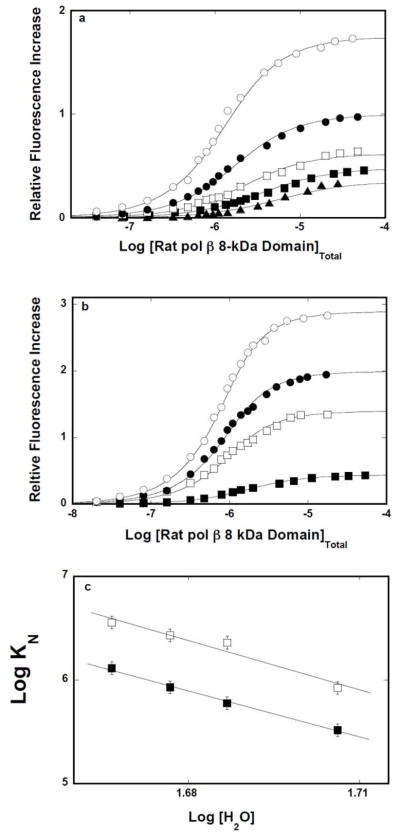
Fluorescence titrations of the 10-, 12-, 14-, 16-mers, dεA(pεA)9, dεA(pεA)11, dεA(pεA)13, and dεA(pεA)15, with the rat pol β8 -kDa domain in buffer C, containing 1 mM MgCl2, are shown in Figure 2a. The mid point of the titration curves shifts toward the lower concentrations of the domain as the length of the ssDNA increases, indicating an increased macroscopic affinity of the domain [23–25]. Also, binding of the domain induces a strong increase of the nucleic acid fluorescence, indicating a significant change in the structure of the bound etheno-derivatives of the ssDNAs [22,23,32,33]. The solid lines in Figures 2a are the nonlinear least-squares fits of the experimental titration curves to the single site-binding isotherm, defined by eq. 8
Figure 2.
a. The rat pol β8 -kDa domain -ssDNA site -size in the presence of magnesium. Fluorescence titrations of the 10-, 12-, 14-, and 16-mer, dεA(pεA)9, dεA(pεA)11, dεA(pεA)13, and dεA(pεA)15(λex = 325 nm, λe m= 410 nm) with the rat pol β8 -kDa domain, in buffer C (pH 7.0, 10°C), containing 1 mM MgCl2. Concentrations of all ssDNA oligomers are 1 × 10−6 M; (■), K = 6.8 × 105 M−1, ΔFmax = 0.38; (❑), K = 1.6 × 105 M−1, ΔFmax = 0.38; (●), K = 2.5 × 105 M−1, ΔFmax = 0.40; (❍), K = 3.5 × 105 M−1, ΔFmax = 0.48. The solid lines are nonlinear least-squares fits using the single-site binding isotherm (eq. 8). b. The dependence of the macroscopic binding constant KN, upon the length of the ssDNA. The solid line is a linear least-squares fit to eq. 10. Extrapolation of the line to KN = 0 intercepts the DNA length axis at Np= 8.6 ± 0.7. c. Fluorescence titrations of the 14-, 16-, 18-, and 20-mer, dεA(pεA)13, dεA(pεA)15, dεA(pεA)17, and dεA(pεA)19(λex = 325 nm, λe m= 410 nm) with the rat pol β8 -kDa domain, in buffer C (pH 7.0, 10°C). Concentrations of all ssDNA oligomers are 1 × 10−6 M; (■) K = 4.0 × 105 M−1, ΔFmax = 0.37; (❑), K = 8.4 × 105 M−1, ΔFmax = 0.44; (●), K = 1.3 × 106 M−1, ΔFmax = 0.47; (❍), K = 1.8 × 106 M−1, ΔFmax = 0.51. The solid lines are nonlinear least-squares fits using the single-site binding isotherm (eq. 8). d. The dependence of the macroscopic binding constant KN, upon the length of the ssDNA. The solid line is a linear least square fit to eq. 10. Extrapolation of the line to KN = 0 intercepts the DNA length axis at Np = 12.5 ± 0.8 (details in text).
| (8) |
where KN is the macroscopic binding constant characterizing the affinity for a given ssDNA oligomer, containing N nucleotides and ΔFmax is the maximum induced fluorescence increase of the nucleic acid. The analysis includes both KN and ΔFmax as fitting parameters.
The dependence of KN, upon the length of the ssDNA oligomer, is shown in Figure 2b. Within experimental accuracy, the plot is linear. The simplest explanation of the linear character of the plot is that the DNA-binding subsite of the 8-kDa domain, which occludes the p number of nucleotides and binds with the intrinsic binding constant, Kp, experiences the presence of several potential binding sites on the ssDNA oligomers [40–42]. Therefore, the value of KN contains a statistical factor that can analytically be defined in terms of p and Kp, as
| (9) |
and
| (10) |
Extrapolation of the plot in Figure 2b to the zero value of KN intercepts the abscissa at the DNA length, Np= p − 1, corresponding to the length of the ssDNA oligomer, which is too short to form a complex with the domain. The plot in Figure 2b gives p = 9.6 ± 0.7, which is in excellent agreement with the value of 9 ± 0.6, previously obtained using the polymer ssDNA [15]. The intrinsic binding constant, Kp, is determined by the slope of the linear region in Figure 2b (eq. 10), which provides the value of (4.7 ± 0.8) × 104 M−1(see Discussion).
Analogous fluorescence titrations of the 14-, 16-, 18-, 20-mers, dεA(pεA)13, dεA(pεA)15, dεA(pεA)17, and dεA(pεA)19, with the rat pol β8 -kDa domain in buffer C, in the absence of MgCl2, are shown in Figure 2c. The solid lines in Figures 2c are the nonlinear least-squares fits of the experimental titration curves to the single site-binding isotherm, defined by eq. 8. The dependence of the macroscopic binding constant, KN, upon the length of the ssDNA oligomer, is shown in Figure 2d. As observed for the solutions containing magnesium (Figure 2b), the plot is linear, indicating that the DNA-binding subsite of the 8-kDa domain experiences the presence of several potential binding sites on the ssDNA oligomers [40–42]. The values of KN, in terms of p and Kp, are defined by eqs. 9 and 10. Extrapolation of the plot in Figure 2d to the zero value of KN intercepts the abscissa at the DNA length, Np= p − 1, which is clearly larger than that observed for the samples containing Mg2+ (Figure 2b). The plot in Figure 2d gives p = 13.5 ± 0.8, as compared to 13 ± 0.7 obtained for the polymer ssDNA [15]. The slope of the plot in Figure 2d provides the intrinsic binding constant, Kp= (2.3 ± 0.6) × 105 M−1 (eq. 10) (see Discussion).
Topology of the ssDNA Associated With the Rat Pol β8 -kDa Domain
The large site-size of the 8-kDa domain -ssDNA, particularly in the absence of magnesium, indicates that the domain engages an additional binding area in interactions with the nucleic acid, as compared to the (pol β)5 binding mode, where the intact enzyme also engages only the 8 -kDa domain in interactions with the DNA [11–14]. The candidates for such binding areas are two patches containing lysine and arginine residues on both sides of the HhH DNA-binding motif in the domain (Figure 1b) (see Discussion). Because of the small size of the domain and the locations of the two positively charged patches, such an engagement would require significant bending of the nucleic acid, which can be detected using the fluorescence resonance energy transfer (FRET) method [26–29]. To address the topology of the 8-kDa domain - ssDNA complex, we performed FRET measurements of the protein complex with the ssDNA 20-mer, 5′-Fl-dT(pT)19-CP-3′, in the absence of magnesium (Materials and Methods). The fluorescein (Fl) at the 5′ end, and coumarin derivative (CP) at the 3′ end of the oligomer serve as the fluorescence energy transfer acceptor, and donor, respectively [29]. With a site-size of ~13 nucleotides, only a single domain molecule associates with the nucleic acid and the affinity for the labeled oligomer is indistinguishable from the affinity determined for the unmodified DNA (data not shown). The values of the limiting anisotropy for the free ssDNA oligomer and its complexes with the enzyme do not exceed ~0.19 (data not shown), an indication that the orientation factor, κ2, does not significantly affect the FRET measurements [29,30].
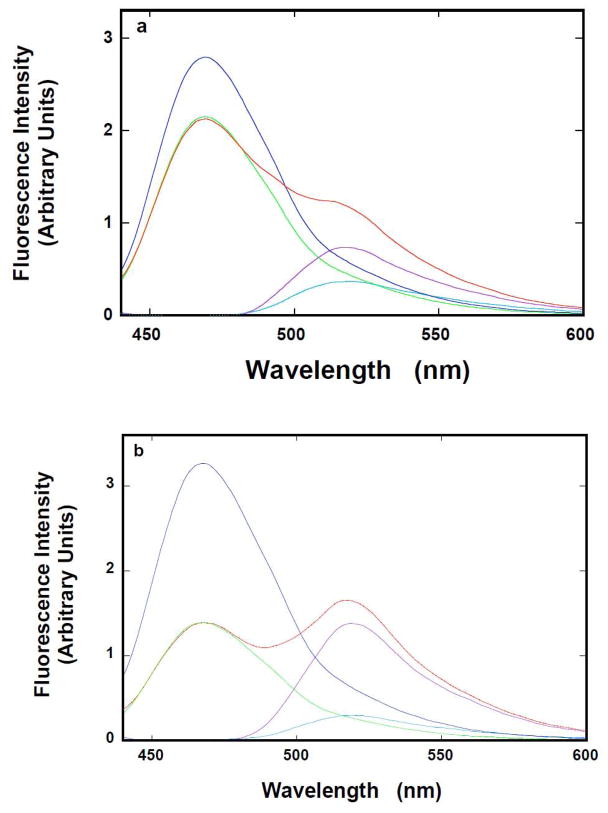
Fluorescence emission spectra (λex = 425 nm) of the ssDNA 20 -mer, dT(pT)19-CP-3′, containing only the donor, 5′-Fl-dT(pT)19, containing only the acceptor, and 5′-Fl-dT(pT)19-CP-3′, containing both the donor and the acceptor, in the absence of the 8-kDa domain, are shown in Figure 3a. There is significant quenching of the donor fluorescence, accompanied by an increase of the acceptor fluorescence emission in the presence of the donor, indicating that an efficient fluorescence energy transfer process occurs in the system [26–30]. The emission spectrum of the CP donor, in the complex with the Fl acceptor, has been obtained by normalizing the peak of the donor emission spectrum, recorded in the absence of the acceptor, to the emission intensity of the donor recorded in the presence of the acceptor. Subsequently, the emission spectrum of the acceptor, in the complex with the donor, has been obtained by subtracting the normalized spectrum of the donor from the spectrum of 5′-Fl-dT(pT)19-CP-3′. The normalized emission spectrum of the CP donor in the presence of the acceptor and the emission spectrum of the acceptor in the presence of the donor are included in Figure 3a.
Figure 3.
a. Topology of the ssDNA bound to the rat pol β8 -kDa domain. Fluorescence emission spectrum (λex = 425 nm) of dT(pT) 19-CP-3′ (——), 5′ Fl-dT(pT)19(——), and 5′-Fl-dT(pT)19-CP-3′ (——) in buffer C (pH 7.0, 10°C) (Materials and Methods); the normalized emission spectrum of dT(pT)19-CP-3′ to the maximum of CP emission in 5′-Fl-dT(pT)19-CP-3′ (——), the sensitized emission spectrum of 5′-Fl-dT(pT)19-CP-3′ (——). b. Fluorescence emission spectrum, recorded in the presence of the rat pol β8 -kDa domain (λex = 425 nm), of dT(pT)19-CP-3′ (——), 5′-Fl-dT(pT)19(blue), and 5′-Fl-dT(pT)19-CP-3′ (——) in buffer C (10°C); the normalized emission spectrum of dT(pT)19-CP-3′ to the maximum of CP emission in 5′-Fl-dT(pT)19-CP-3′ (——), the sensitized emission spectrum of 5′-Fl-dT(pT)19-CP-3′ (——). Concentrations of the ssDNA oligomers and the 8-kDa domain are: 5 × 10−8 M (oligomer) and 2 × 10−5 M (details in text).
Corresponding fluorescence emission spectra of the ssDNA 20-mer, dT(pT)19-CP-3′, containing only the donor, 5′-Fl-dT(pT)19, containing only the acceptor, and 5′-Fl-dT(pT)19-CP-3′, containing both the donor and the acceptor, saturated with the 8-kDa domain, are shown in Figure 3b. Both the quenching of the donor fluorescence and the increase of the acceptor fluorescence emission are significantly more pronounced in the presence of the protein, than observed for the ssDNA oligomer alone, indicating a considerable increase of the fluorescence energy transfer efficiency in the 8-kDa domain -ssDNA 20 -mer complex. Analysis of the spectra has been performed as described above (Figure 3a). The normalized emission spectrum of the donor in the presence of the acceptor and the emission spectrum of the acceptor in the presence of the donor, in the 8-kDa domain -20 -mer complex, are included in Figure 3b.
The apparent fluorescence energy transfer efficiencies, ED and EA, Förster fluorescence energy transfer efficiency, E, and the average distance between the donor and acceptor, R, have been calculated using eqs. 4–7 [26–30]. In the absence of the domain, ED= 0.23 ± 0.03, EA = 0.46 ± 0.05, E = 0.37 ± 0.05, and R = 56.7 ± 3 Å. The value of R is shorter than the distance (~70 Å) between the 5′ and 3′ ends of the nucleic acid in the dsDNA B structure, indicating that dT(pT)19 is folded in solution, due to the low persistent length of the oligomer [43]. In the presence of the domain, ED= 0.57 ± 0.03, EA = 0.99 ± 0.03, E = 0.70 ± 0.03, and R = 45.2 ± 3 Å. These much larger values of the FRET efficiencies and the value of the distance between the acceptor and the donor, which is ~11.5 Å shorter than determined for the free oligomer, strongly indicate that the nucleic acid is profoundly bent in the complex with the 8-kDa domain. Analogous FRET studies of the 8-kDa domain -20-mer complex with the 1:1 stoichiometry, in the presence of magnesium, provide, ED= 0.46 ± 0.03, EA = 0.76 ± 0.03, E = 0.59 ± 0.03, and R = 49.1 ± 3 Å, for the 8-kDa domain -20 -mer complex (data not shown). The values of all the FRET efficiencies are lower than in the absence of Mg2+ cations and the value of the distance between the acceptor and the donor is ~7.6 Å shorter than determined for the free oligomer. The data indicate that, in the presence of magnesium, the bending of the nucleic acid in the complex is less pronounced than that when Mg2+cations are present in solution (see Discussion).
Temperature Effect On the Rat Pol β and the Rat Pol β8 -kDa Domain Interactions With the ssDNA In the Presence and Absence of Magnesium
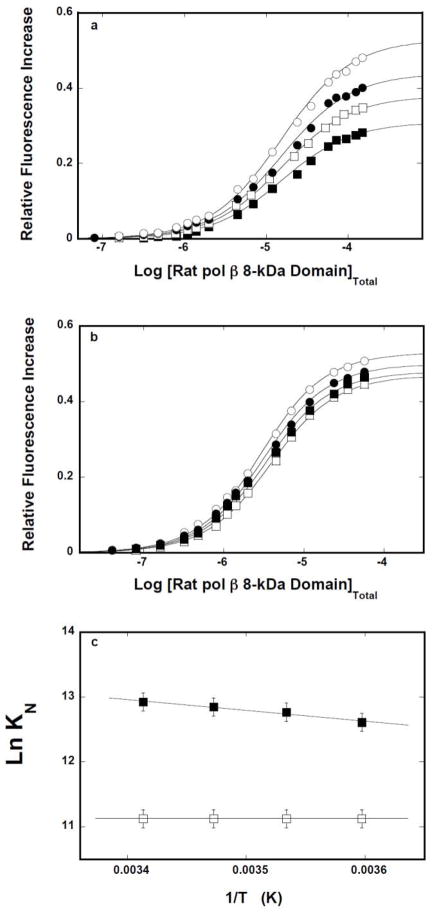
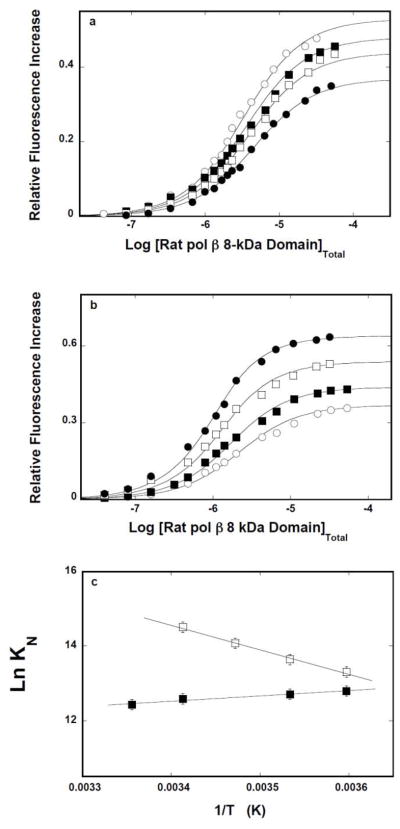
The thermodynamic characteristics of the 8-kDa domain interactions with the ssDNA have been addressed by examining the temperature effect on the protein binding to the ssDNA oligomers, in the presence and absence of magnesium. Fluorescence titrations of 10-mer dεA(pεA)9 with the 8 -kDa domain, performed in buffer C containing 1 mM MgCl2 and at different temperatures, are shown in Figure 4a. Analogous titrations of the 16-mer, dεA(pεA)15, are shown in Figure 4b. The solid lines in Figures 4a and 4b are nonlinear least-squares fits of the titration curves to a single-site isotherm (eq. 8), with KN and ΔFmax as fitting parameters. Although the increase of the temperature affects the observe fluorescence change, it has no effect on the macroscopic affinity of the protein -10 -mer complex. In the case of the 16-mer, the value of KN slightly increases with the temperature.
Figure 4.
a. Temperature effect on the 8-kDa domain- ssDNA interactions in the presence of magnesium. Fluorescence titrations of the ssDNA 10-mer, dεA(pεA)9, with the rat pol β8 -kDa domain (λex = 325 nm, λem = 410 nm) in buffer C (pH 7.0), containing 1 mM MgCl2, at different temperatures: (■) 5°C, (❑)10°C, (●),15°C, (❍) 20°C. The concentration of the ssDNA 10-mer is 1 × 10−6 M. The solid line is the nonlinear least squares fits of the titration curves, using the single-site binding isotherm defined by eq. 8. b. Fluorescence titrations of the ssDNA 16-mer, dεA(pεA)15, with the rat pol β8 -kDa domain(λex = 325 nm, λem = 410 nm) in buffer C (pH 7.0), containing 1 mM MgCl2, at different temperatures: (■) 5°C,(❑)10°C, (●), 15°C, (❍) 20°C. The concentration of the ssDNA 10-mer is 1 × 10−6 M. The solid line is the nonlinear least-squares fits of the titration curves, using the single-site binding isotherm defined by eq. 8. c. The dependence of the natural logarithm of the macroscopic binding constants, K10 (❑) and K16 (■), upon the reciprocal of the temperature (Kelvin) (van’t Hoff plot). The apparent enthalpies and entropies of the examined binding processes are included in Table 1.
Figure 4c shows the dependence of the natural logarithm of the binding constant, KN, upon the reciprocal of the temperature (Kelvin) (van’t Hoff plot) for the 10-and 16-mer [44]. Within experimental accuracy, the plots are linear. The slopes of the plots are related to the apparent enthalpy of the corresponding binding process by
| (11) |
The obtained values of the apparent enthalpy, ΔHo, for both ssDNA oligomers are included in Table 1. Using the standard thermodynamic formulas, ΔGo = −RTLnKN, and ΔSo= (−ΔGo +ΔHo)/T, one calculates the values of the apparent entropy, ΔSo, which are also included in Table 1. The data indicate that interactions of the isolated 8-kDa domain with the ssDNA are characterized by the apparent enthalpy changes of ~0, or slightly positive, in the case of the 10-and 16-mer, respectively. Nevertheless, interactions of the domain with the ssDNA, in the presence of Mg2+, are completely driven by the apparent entropy changes, ΔSo (see Discussion).
Table 1.
Thermodynamic parameters of the binding of the rat pol β 8-kDa domain to the ssDNA oligomers dεA(pεA)9 and dεA(pεA)15, in buffer C (pH 7.0, 10°C), in the absence and presence of 1 mM MgCl2(details in text)*.
| ssDNA Oligomer | MgCl2 | Stoichiometry | Site-size | **∂LogKN/∂Log[H2O] | ***∂LogKN/∂Log[H2O] | ΔHo kcal/mol | ****ΔSo cal/(mol deg) |
|---|---|---|---|---|---|---|---|
| dεA(pεA)9 | - | 1 | 13 ± 1 | −14.9 ± 2.3 | −14.2 ± 2.3 | −2.8 ± 0.9 | 15 ± 5.0 |
| 1 mM | 1 | 9 ± 1 | −19.1 ± 2.5 | −18.3 ± 2.5 | 0.0 ± 0.8 | 22.0 ± 7.0 | |
| dεA(pεA)15 | - | 1 | 13 ± 1 | −15.9 ± 2.3 | −15.4 ± 2.3 | 13.1 ± 2.8 | 73.0 ± 21.0 |
| 1 mM | 1 | 9 ± 1 | −19.4 ± 2.5 | −16.2 ± 2.5 | 3.3 ± 1.1 | 33.0 ± 9.0 |
Errors are standard deviations determined using 3 – 4 independent experiments.
Determined in the presence of glycerol.
Determined in the presence of DMSO.
Calculated using the intrinsic binding constant obtained at 10°C.
Fluorescence titrations of the same 10-and 16-mer with the 8-kDa domain, performed in buffer C, in the absence of magnesium and at different temperatures, are shown in Figure 5a and 5b, respectively. The solid lines in Figures 5a and 5b are nonlinear least-squares fits of the titration curves to a single -site isotherm (eq. 8), with KN and ΔFmax as fitting parameters. The dependence of the natural logarithm of the binding constant, KN, upon the reciprocal of the temperature (Kelvin) (van’t Hoff plot) for the 10-and 16 -mer is shown in Figure 5c [44]. Within experimental accuracy, the plots are linear. The obtained values of the apparent enthalpy, ΔHo and ΔSo, for both ssDNA oligomers are included in Table 1. Unlike the data obtained in the presence of magnesium (Figures 4a, 4b, and 4c), there is a significant difference in thermodynamic parameters characterizing the binding of the 10- and 16-mer. The 10-mer binds with the apparent negative enthalpy changes of ~−2.8 kcal/mol, while ΔHo ~13 kcal/mol is strongly positive for the 16-mer. Nevertheless, the interactions in the absence of magnesium are still dominated by the apparent positive entropy changes, ΔSo (see Discussion).
Figure 5.
a. Temperature effect on the 8-kDa domain - ssDNA interactions in the absence of magnesium. Fluorescence titrations of the ssDNA 10-mer, dεA(pεA)9, with the rat pol β8 -kDa domain (λex = 325 nm, λem = 410 nm) in buffer C (pH 7.0) at different temperatures: (■) 5°C, (❑)10°C, (●),20°C, (❍) 25°C. The concentration of the ssDNA 10 -mer is 1 × 10−6 M. The solid line is the nonlinear least-squares fits of the titration curves, using the single-site binding isotherm defined by eq. 8. b. Fluorescence titrations of the ssDNA 16-mer, dεA(pεA)15, with the rat pol β8 -kDa domain (λex = 325 nm, λem = 410 nm) in buffer C (pH 7.0) at different temperatures: (■) 5°C, (❑)10°C, (●),15°C. (❍) 20°C. The concentration of the ssDNA 10 -mer is 1 × 10−6M. The solid line is the nonlinear lea st-squares fits of the titration curves, using the single-site binding isotherm defined by eq. 8. c. The dependence of the natural logarithm of the macroscopic binding constants, K10 (❑) and K16 (■), upon the reciprocal of the temperature (Kelvin) (van’t Hoff plot). The apparent enthalpies and entropies of the examined binding processes are included in Table 1.
Analogous thermodynamic analyses have been performed for the binding of the intact rat pol β to the ssDNA 14 -mer and 20-mer, which exclusively engage the 8-kDa domain, forming the (pol β)5 binding mode and the total DNA -binding site, forming the (pol β)16 binding mode, respectively (data not shown). The obtained apparent enthalpies and entropies, ΔHo and ΔSo, are included in Table 2. In general, in the presence and absence of magnesium, binding of the intact enzyme to the 14- and the 20-mer is characterized by apparent positive enthalpy changes. With the exception of the formation of the (pol β)16 binding mode in the absence of magnesium (Table 2), the values of ΔHo are significantly more positive than observed for the isolated 8-kDa domain (Table 1). Nonetheless, association of the intact enzyme is predominantly driven by large entropy changes (see Discussion).
Table 2.
Thermodynamic parameters of the binding of the rat pol β to the ssDNA oligomers dεA(pεA)13 and dεA(pεA)19, in buffer C (pH 7.0, 10°C), in the absence and presence of 1 mM MgCl2(details in text)*.
| ssDNA Oligomer | MgCl2 | Stoichiometry | Site-size | ** dLogKN/dLog[H2O] | *** dLogKN/dLog[H2O] | ΔH° kcal/mol | ****ΔS° cal/mol deg |
|---|---|---|---|---|---|---|---|
| dεA(pεA)13 | - | 1 | 5 ± 1 | −7.8 ± 1.5 | −8.4 ± 1.7 | 2.5 ± 8.9 | 31 ± 10 |
| 1 mM | 1 | 5 ± 1 | −6.7 ± 1.3 | −7.3 ± 1.5 | 14.4 ± 2.8 | 73 ± 21 | |
| dεA(pεA)19 | - | 1 | 16 ± 1 | −5.4 ± 1.1 | −5.9 ± 1.2 | 1.7 ± 0.5 | 32 ± 10 |
| 1 mM | 1 | 16 ± 1 | −9.8 ± 1.9 | −9.3 ± 1.8 | 9.9 ± 2.1 | 56 ± 16 |
Errors are standard deviations determined using 3 – 4 independent experiments.
Determined in the presence of glycerol.
Determined in the presence of DMSO.
Calculated using the intrinsic binding constant obtained at 10°C.
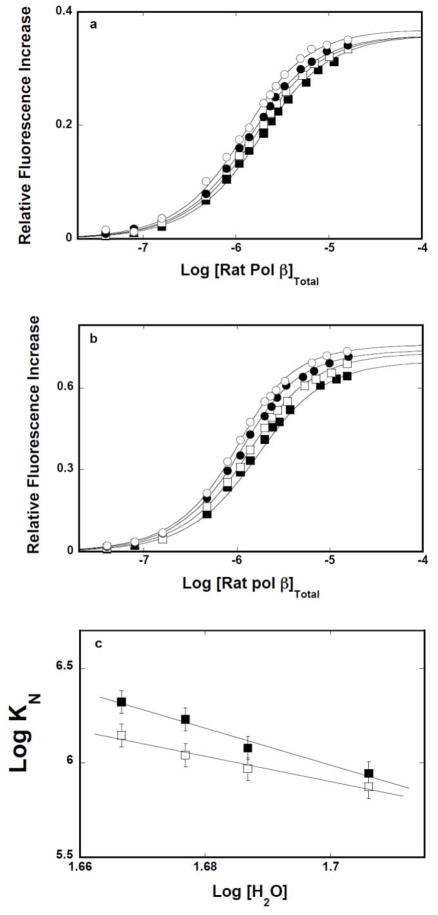
Solvent Effect On Interactions of the Rat Pol β8 -kDa Domain and the Intact Rat Pol β With the ssDNA, In the Presence and Absence of Magnesium
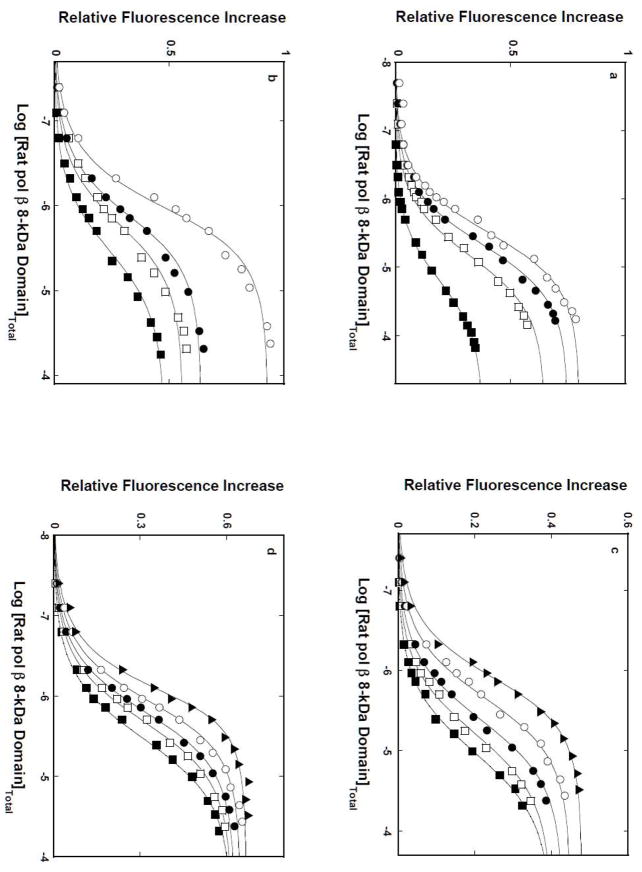
The nature of interactions of the isolated 8-kDa domain and the intact pol β with the ssDNA has been further analyzed by examining the solvent effect on the protein binding to selected ssDNA oligomers. Fluorescence titrations of the 10-mer, dεA(pεA)9, and 16-mer, dεA(pεA)15, with the 8-kDa domain in buffer C, containing 1 mM MgCl2 and different concentrations of the neutral solute, glycerol (w/v %), are shown in Figures 6a and 6b, respectively. The effect of glycerol on the domain -ssDNA interactions is striking. The titration curves are significantly shifted toward lower total protein concentrations, indicating a strong increase of the macroscopic affinity of the protein - nucleic acid associations, for both the 10-and the 16 -mer. Moreover, in both cases, the values of ΔFmax accompanying the binding are greatly increased at higher glycerol concentrations, indicating a profound change of the nucleic acid structure in the complex [11,15,21,22].
Figure 6.
a. Glycerol and DMSO effects on the 8-kDa domain - ssDNA interactions in the presence of magnesium. Fluorescence titrations of the ssDNA 10-mer, dεA(pεA)9, with the rat pol β8 -kDa domain (λex = 325 nm,λ em = 410 nm) in buffer C (pH 7.0, 10°C), containing 1 mM NgCl2, and different glycerol concentrations (w/v): 10 % (■);15% (❑); 17.5% (●); 20%(❍). b. Fluorescence titrations of the ssDNA 16-mer, dεA(pεA)15, with the rat pol β8 -kDa domain in buffer C (pH 7.0, 10°C), 1 mM MgCl2, and different glycerol concentrations (w/v): 10 % (■), 15% (❑), 17.5% (●), 20%(❍). The concentration of the ssDNA 10-and the 16 -mer is 1 × 10−6 M. The solid lines are nonlinear least-squares fits of the titration curves, using the single-site binding isotherm defined by eq 8. c. Fluorescence titrations of the ssDNA 10-mer, dεA(pεA)9, with the rat pol β8 -kDa domain (λex = 325 nm,λ em = 410 nm) in buffer C (pH 7.0, 10°C), containing 1 mM NgCl2, and different DMSO concentrations (w/v): 10 % (■),15% (❑), 17.5% (●), 20% (❍), 25%(▲). b. Analogous fluorescence titrations of the ssDNA 16-mer, dεA(pεA)15, with the rat pol β8 -kDa domain in the presence of DMSO. The concentration of the ssDNA 10- and the 16-mer is 1 × 10−6M. The solid lines are nonlinear -least squares fits of the titration curves, using the single-site binding isotherm defined by eq 8.
Parallel titrations have been performed in the presence of another neutral solute, DMSO, in order to address the question, as to what extent the dramatic glycerol effect could be specific for the solute. Fluorescence titrations of the ssDNA 10-mer and the 16-mer with the 8-kDa domain at different concentrations of DMSO (w/v %) are shown in Figures 6c and 6d. Similar to the effect of glycerol, the presence of DMSO dramatically increases the macroscopic affinity of the examined 8-kDa domain - DNA complexes. Nevertheless, although considerable, the accompanying increases of ΔFmax are less pronounced than in the presence of glycerol. The solid lines in figures 6a, 6b, 6c, and 6d are nonlinear least-squares fits of the experimental titration curves to eq. 8 with KN and ΔFmax as fitting parameters.
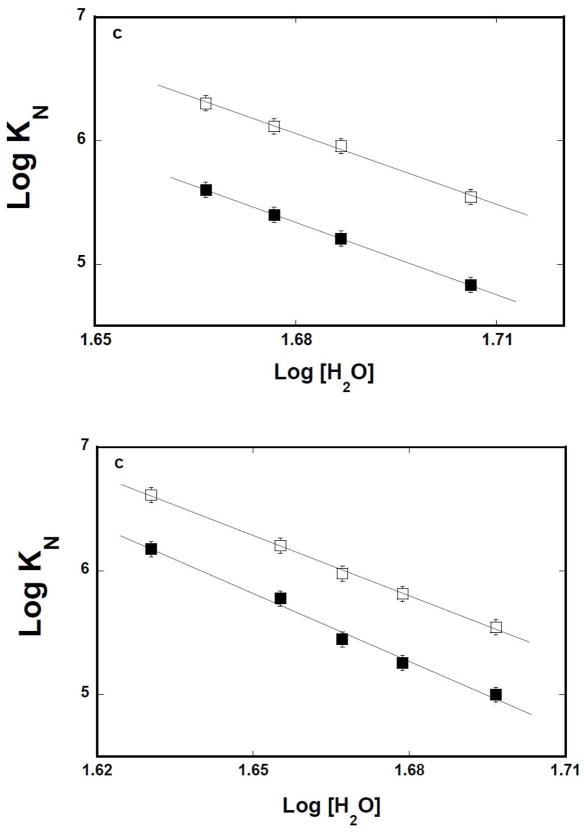
The simplest thermodynamic model of the observed effect of both solutes is that water molecules participate in the association and the process is affected by the changes of the water activity in sample [45–47]. It should be pointed out that the effect of the “neutral” solute on the macromolecular interactions could be an intricate process, involving the presence of preferential interactions [45]. However, the very similar effect of two different solutes on the same binding process strongly suggests that the preferential interactions do not dominate the observed effects [45–47]. The dependences of the logarithm of the macroscopic binding constants, K10 and K 16 upon the logarithm of [H2O] (log-log plot), determined in the presence of glycerol, or DMSO, are shown in Figures 7a and 7b, respectively [48–51]. The plots are linear in examined solute concentration ranges. In the presence of glycerol, the slopes of the plots are ∂logK10/∂log[H2O] = −19.1 ± 2.5 and ∂logK16/∂log[H2O] = −19.4 ± 2.5, respectively. The values of the slopes of the log-log plots, obtained in the presence of DMSO, are ∂logK10/∂log[H2O] = −18.3 ± 2.5 and ∂logK16/∂log[H2O] = −16.2 ± 2.5, respectively (Table 1). The slopes of the log -log plots, obtained for two different solutes, are, within experimental accuracy, very similar, indicating that the water activity is indeed the dominant factor in the observed solute effects. The data indicate that association of the isolated 8-kDa domain with the ssDNA, in the presence of Mg2+, is accompanied by a net release of ~18 water molecules [48–51] (see Discussion).
Figure 7.
a. Log-log plots of the net release of water molecules from the 8-kDa domain – ssDNA complexes in the presence of glycerol or DMSO and magnesium. The dependence of the logarithm of the binding constants, K10(■) and K16(❑), upon the logarithm of water concentration, in buffer C (pH 7.0, 10°C), containing 1 mM NgCl 2, in the presence of glycerol. The solid lines are the linear least-squares fits, which provides the slopes, ∂LogK10/∂Log[H2O] = −19.1 ± 2.5 and ∂LogK16/∂Log[H2O] = −19.4 ± 2.5, for the 10-and 16 -mer, respectively (details in text). b. The dependence of the logarithm of the binding constants, K10(■) and K16(❑), upon the logarithm of water concentration, in buffer C (pH 7.0, 10°C), containing 1 mM NgCl2, in the presence of DMSO. The solid lines are the linear-least squares fits, which provides the slopes, ∂LogK10/∂Log[H2O] = −16.2 ± 2.5 and ∂LogK16/∂Log[H2O] = −18.3 ± 2.5, for the 10-and 16 -mer, respectively (details in text).
Corresponding fluorescence titrations of dεA(pεA)9 and dεA(pεA)15 with the 8 -kDa domain in buffer C, containing different concentrations of glycerol, in the absence of magnesium, are shown in Figures 8a and 8b. Both the macroscopic affinity of the protein - ssDNA oligomer associations and the values of ΔFmax accompanying the binding are clearly increased at higher glycerol concentrations, as indicated by the strong shift of the titration curves toward lower total protein concentrations and large observed fluorescence changes. The solid lines in figures 8a, and 8b are nonlinear least-squares fits of the experimental titration curves to eq. 8 with K N and ΔFmax as fitting parameters. The dependences of the logarithm of the macroscopic binding constants, K10 and K16 upon the logarithm of [H 2O] (log-log plot), determined in the presence of glycerol (see above), are shown in Figure 8c [48–51]. The plots are linear in examined solute concentration ranges. The slopes of the plots are ∂logK10/∂log[H2O] = −14.9 ± 2.3 and ∂logK16/∂log[H2O] = −15.9 ± 2.5, respectively. Similar values of the slopes were obtained in the presence of DMSO (data not shown) (Table 1). The data indicate that association of the 8-kDa domain with the ssDNA, in the absence of magnesium, is accompanied by an average net release of ~15 water molecules, which is slightly less than determined for the solution containing MgCl2 (see above) (see Discussion).
Figure 8.
a. Log-log plots of the net release of water molecules from the 8-kDa domain ssDNA complexes in the presence of glycerol or DMSO and in the absence of magnesium. Fluorescence titrations of the ssDNA 10-mer, dεA(pεA)9, with the rat pol β8 -kDa domain (λex = 325 nm,λ em = 410 nm) in buffer C (pH 7.0, 10°C), containing different glycerol concentrations (w/v): 5 % (▲);10 % (■);15% (❑); 17.5% (●); 20%(❍). b. Fluorescence titrations of the ssDNA 16-mer, dεA(pεA)15, with the rat pol β8 -kDa domain in buffer C (pH 7.0, 10°C), containing 100 mM NaCl and different glycerol concentrations (w/v): 10 % (■);15% (❑); 17.5% (●); 20%(❍). The concentration of the ssDNA 10-and the 16 -mer is 1 × 10−6 M. The solid lines are nonlinear least -squares fits of the titration curves, using the single-site binding isotherm defined by eq 8. c. The dependence of the logarithm of the binding constants, K10 (■) and K16 (❑), upon the logarithm of water concentration, in buffer C (pH 7.0, 10°C), containing 100 mM NaCl, in the presence of glycerol. The solid lines are the linear least-squares fits, which provides the slopes, ∂LogK10/∂Log[H2O] = −14.9 ± 2.3 and ∂LogK16/∂Log[H2O] = −15.9 ± 2.3, for the 10-and 16 -mer, respectively (details in text).
Fluorescence titrations the ssDNA 14-mer, which exclusively engage the 8-kDa domain, forming the (pol β)5 binding mode, and the ssDNA 20 -mer, which forms the (pol β)16 binding mode, respectively, with the intact rat pol β, are shown in Figures 9a and 9b. The behavior of the intact enzyme is different from the corresponding data obtained for the 8-kDa domain (Figures 8a and 8b). The macroscopic affinities of the pol β-ssDNA oligomers associations only moderately increase at higher glycerol concentrations, as indicated by the shift of the titration curves toward the lower total protein concentrations. Strikingly different are the values of the ΔFmax in the complex, which are very modestly affected by the presence of glycerol. The solid lines in figures 9a, and 9b are nonlinear least-squares fits of the experimental titration curves to eq. 8 with KN and ΔFmax as fitting parameters.
Figure 9.
a. Glycerol effect on the rat pol β-ssDNA interactions in the presence of magnesium. Fluorescence titrations of the ssDNA 14-mer, dεA(pεA)13, with the intact rat pol β (λex = 325 nm,λem = 410 nm) in buffer C (pH 7.0, 10°C), containing 1 mM MgCl2 and different glycerol concentrations (w/v): 10 % (■);15% (❑); 17.5% (●); 20%(❍). b. Analogous fluorescence titrations of the ssDNA 20-mer, dεA(pεA)19, with the intact pol β. The concentration of the ssDNA 14-and the 20 -mer is 1 × 10−6M. The solid lines are nonlinear least -squares fits of the titration curves, using the single-site binding isotherm defined by eq 8. b. The dependence of the logarithm of the binding constants, K14 (❑) and K20 (■), upon the logarithm of water concentration. The solid lines are the linear least-squares fits, which provide the slopes, ∂LogK13/∂Log[H2O] = −6.7± 1.3 and ∂LogK20/∂Log[H2O] = −9.8 ± 1.9, for the 14-and 20 -mer, respectively (details in text).
The dependences of the logarithm of the macroscopic binding constants, K14 and K 20 upon the logarithm of [H2O] (log-log plot), are shown in Figure 9c. The plots are linear in examined solute concentration ranges. The slopes of the plots are ∂logK14/∂log[H2O] = −6.7 ± 1.3 and ∂logK20/∂log[H2O] = −9.8 ± 1.9, respectively. Corresponding values of the slope in the absence of magnesium, are ∂logK14/∂log[H2O] = −8.4 ± 1.7 and ∂logK20/∂log[H2O] = −5.9 ± 1.2, respectively (Table 2). As observed for the isolated 8-kDa domain, very similar values of the slopes for both ssDNA oligomers, were obtained in the presence of DMSO (data not shown) (Table 2). Thus, the data indicate that association of intact pol β with the ssDNA, in the presence and absence of Mg2+, is accompanied by a significantly smaller net release of water molecules than observed for the isolated 8-kDa domain (Tables 1 and 2) (see Discussion).
DISCUSSION
The Rat Pol β8 -kDa Domain Has the Potential to Engage Much Large Fragments of the DNA Than When It Is Embedded in the Intact Enzyme
Studies described in this work provide an insight into the complex energetics of the primary DNA-binding subsite of the rat pol β, located on 8-kDa domain of the enzyme. The site-size of the isolated rat pol β8 -kDa domain DNA-binding site, determined using a series of ssDNA oligomers, is ~9 and ~13 in the presence or absence of Mg2+, respectively. This is a much larger site-size than the interacting area in the (pol β)5 binding mode, where the enzyme exclusively interacts with the DNA using the 8 -kDa domain and engages only ~5 nucleotides, independent of the presence of magnesium cations [11–15]. Recall, the (pol β)5 binding mode is an equilibrium complex. Kinetic data would be necessary to assess the presence of intermediates, differing in the site-sizes in the 8-kDa domain complex with the DNA. Nevertheless, the fact that the 8-kDa domain is able to engage a significantly larger fragment of the nucleic acid than in the (pol β)5 binding mode, strongly suggests that such intermediates are present in the initial recognition process. The much smaller site-size of the (pol β)5 binding mode seems to be a result of internal equilibria in the formed complex between the intact enzyme of the DNA, leading to the optimal engagement and orientation of the intact enzyme in interactions with the nucleic acid (see below).
Crystallographic and NMR studies indicate that the 8-kDa domain contains a helix-hairpin-helix motif (HhH) which is proposed to be a nonspecific, ssDNA-binding motif [9,10]. The motif contains two α-helices connected by a short interhelical loop (Figures 1a and 1b). The loop can make contact with the two nucleic acid phosphate groups via hydrogen bonds. It is clear that the motif alone, although a part of the binding site, cannot account for either the site-size of ~9 or ~13 nucleotides in the complex with the nucleic acid. However, on both sides of the HhH motif, there are patches of 8–7 lysine and arginine residues. Both patches, together with the HhH loop, can form a DNA-binding site of the domain that would account for the determined site-sizes.
As we discussed above, to efficiently engage both positively -charged patches, binding of the nucleic acid to the 8-kDa domain would require a significant bending of the bound ssDNA, particularly, in the absence of magnesium. Such a profound bending of the nucleic acid is clearly seen in FRET measurements (Figure 3b). In the presence of Mg2+, the bending is weak and strongly suggesting that one of the patches partially releases the nucleic acid, leading to the observed lower site-size of the complex, in these solution conditions (see above). Notice, the domain can also engage the lysine residue 87 in interactions with the ssDNA, which is located in the tether connecting the domain with the rest of the polymerase molecule. This is particularly true for the complex with the 16-mer and in the absence of Mg2+. Large differences between enthalpies of binding of the 8-kDa domain to the 10-and to the 16 -mer, in the presence and, particularly, in the absence of magnesium, indicate that the 16-mer involves additional interactions, possibly with the lysine 87 (Table 1).
Previous data also indicated that the magnesium effect on the site-size of the 8-kDa domain -ssDNA complex results from the specific binding of Mg2+ to the domain, which induces conformational changes in the protein [15]. The findings that magnesium significantly affects the apparent enthalpy and numbers of the released water molecules in the association reaction of the domain with the ssDNA corroborate this conclusion (see below) (Table 1). The conformational changes, leading to the decreased site-size of the complex, may originate from a reorientation of the HhH motif, which could partially hinder the access of the nucleic acid to one of the positive charged patches on both sides of the motif and/or to the lysine residue 87 for the longer DNA (Figure 1). In this capacity, magnesium binding to the domain seems to be a part of the more intricate mechanism of optimally orienting the domain, with respect to the rest of the enzyme molecule, from the initial engagement of the nucleic acid, leading to the formation of the final, equilibrium (pol β)5 binding mode (see below).
The Intrinsic Affinity of the DNA-Binding Subsite of the Rat Pol β8 -kDa Domain Is Driven By Large Entropy Changes Resulting From the Release of Water Molecules From the Domain
A characteristic feature of the 8-kDa domain interactions with the ssDNA is zero or positive apparent enthalpy changes accompanying the binding (Table 1). In other words, the interactions are mainly driven by the apparent entropy changes. The insight into the nature of the observed entropy change comes from the examination of the solvent effect on the intrinsic affinities (Figures 6a, 6b, 6c, and 6d). In the absence of magnesium, formation of the complexes with the 10-and 16 -mer is accompanied by the release of ~15 – 16 water molecules, while in the presence of Mg+2, binding of the ssDNA oligomers induces the release of ~19 water molecules. Such large numbers of the released water molecules substantially contribute to the observed apparent entropy changes. The fact that oligomers, differing by 60% in length, induce the release of the same number of the water molecules strongly suggests that the water release mostly originates from the binding subsite of the domain and not from the nucleic acid.
The increase of the fluorescence of the ssDNA etheno-derivatives reflects the conformation of the nucleic acid (strong separation and immobilization of the bases) and much less the polarity of the environment [11–15,21,22]. Conformational changes of the DNA occurring in the complex, as indicated by the observed fluorescence changes, cannot contribute to the observed, positive entropy changes. Strong immobilization of the bases, as indicated by the large increase of the etheno-derivative fluorescence, should lead to negative rather than positive entropy changes. The larger number of released water molecules in the presence of magnesium than in the absence of Mg2+, may be surprising, as the site-size of the complex is diminished from ~13 to ~9 nucleotides. First, it additionally indicates that the number of the released water molecules is independent of the length of the interacting nucleic acid, which corroborates the conclusion that the water release originates from the DNA-binding site of the domain. Second, it indicates that the conformation of the binding site is different in the presence of Mg2+, leading to the different thermodynamic response upon association with the DNA (see above).
Different Enthalpy Changes and Numbers of Released Water Molecules in the Formation of the (Pol β)5 Binding Mode, as Compared To the Isolated 8 -kDa Domain - ssDNA Complexes, Indicate a Significant Communication Between the 8-kDa Domain and the Catalytic 31-kDa Domain of the Intact Pol β
Both the 8-kDa domain and the (pol β)5 binding mod e, where the enzyme exclusively engages the 8-kDa domain in interactions with the nucleic acid, have similar intrinsic affinities for the ssDNA, in the range of ~8 × 104M−1- ~2 × 105 M−1 (see above). Thus, the intrinsic affinities alone would not provide any indications as to the existence of a mutual interdependence of the 8-kDa domain and the catalytic 31-kDa domain in the intact enzyme. Moreover, the very fact that the enzyme forms different binding modes, where it engages different domains, would indicate a pronounced autonomy of the domains [11–15]. On the other hand, the apparent enthalpy changes accompanying the formation of the (pol β)5 binding mode and the isolated 8-kDa domain association with the DNA are very significantly different (Table 1). With the exception of the 16-mer binding to the 8-kDa domain in the absence of magnesium, binding of the nucleic acid to the domain, embedded in the intact enzyme structure, is accompanied by much more positive enthalpy changes than determined for the isolated domain. The differences are particularly strong in the presence of magnesium (Table 1).
Furthermore, the number of water molecules released upon complex formation is approximately twice as low for the (pol β)5 binding mode than that determined for the isolated domain (Table 1 and Table 2). These data indicate that transition for the initial complex formed by the enzyme with the ssDNA to the (pol β)5 binding mode (see above), where, in both cases, only the 8-kDa domain enters into interactions with the DNA, is accompanied by the net uptake of water molecules (17,18]. Because the transition involves dramatic change of the protein - DNA site-size, from ~9 or ~13 to ~5 nucleotides, the water uptake most probably results from the release of the part of the potential binding site of the domain from interactions with the DNA.
These results also indicate the existence of a considerable communication between the catalytic 31-kDa domain and the 8-kDa domain of the intact enzyme, which imposes the reorientation of the 8-kDa domain and change of the domain -ssDNA site -size, in spite of the apparent autonomous behavior reflected in the formation of different binding modes with the ssDNA [11–15]. Such a communication must be accomplished through the 14-amino acid tether connecting the domains and controlled by binding of the Mg2+ cations. So far, the tether was thought to be a flexible region without a clear functional role. The obtained data strongly suggest that the tether plays a much more active role in the functioning of the pol β than previously thought, as a conduit of communication signals. The different behavior of the 16-mer, in the absence of magnesium, in terms of the enthalpy changes, may result from the fact that the oligomer is long enough to engage in interactions the lysine residue 87 located in the tether (see above). Second, the large values of the unfavorable positive enthalpy changes and smaller number of water molecules in the formation of the (pol β)5 binding mode by the intact enzyme indicate that, unlike in the case of the isolated 8-kDa domain, the favorable entropy changes originate from the structural changes of the entire enzyme molecule and less from the solute exchange reactions (see below).
The Low Affinity of the DNA-Binding Subsite on the 31-kDa Domain, As Compared To the DNA-Binding Subsite On the 8-kDa Domain, Results from the Diminished Number of the Released Water Molecules From the Subsite
Recall, formation of the (pol β)16 binding mode includes besides the 8-kDa domain, an additional engagement of the DNA-binding subsite located on the catalytic 31-kDa domain of the intact pol β[11–15]. Nevertheless, the affinity of the DNA-binding subsite on the catalytic domain is at least ~2 orders of magnitude lower than the affinity of the DNA -binding subsite on the 8-kDa domain [11–15]. Moreover, the intrinsic binding constants for the (pol β)16 and (pol β)5 binding modes, K16 ≈ 1.8 × 105 M−1 and K5 ≈ 7.5 × 104 M−1, differ only by a factor of ~2.5, in the presence of magnesium (Figure 9c). The DNA -binding subsite on the 31-kDa domain makes only a weak favorable contribution to the intrinsic affinity of the total DNA-binding site. The data obtained in this work indicate that formation of the (pol β)16 binding mode is accompanied by slightly lower positive enthalpy changes, as compared to the formation of the (pol β)5 binding mode, both in the presence and absence of magnesium (Table 2). The simplest explanation of this behavior is that the additional engagement of the subsite on the 31-kDa domain in the (pol β)16 binding mode is accompanied by a modest negative enthalpy change, which partly compensates the large positive enthalpy change of the 8-kDa domain -ssDNA interactions in the same complex (Table 2).
However, the most striking feature is the very modest number of water molecules released upon the formation of the (pol β)16 binding mode. Thus, engagement of the 8 -kDa domain in the (pol β)5 binding mode is accompanied by the release of ~8.4 and ~6.7 water molecules, while the additional engagement of the 31-kDa domain in the (pol β)16 binding mode leads to the release of ~5. 9 and ~9.8 water molecules, in the absence and presence of Mg2+, respectively (Table 2). Within experimental accuracy, these values are the same for both binding modes and indicate that engagement of the subsite on 31-kDa domain is not accompanied by a release of additional water molecules. As a result, the entropy changes remain almost unaffected, as compared to the formation of the (pol β)5 binding mode and do not con tribute to an increase of the free energy of binding of the intact enzyme. In other words, the small favorable contribution of the 31-kDa domain to the affinity of the (pol β)16 binding mode results from the modest negative enthalpy change accompanying the engagement of its subsite in interactions with the DNA. As we discussed above, the entropy changes accompanying the (pol β)5 binding mode formation originate from structural changes of the entire pol β and less from the solute exchange reactions (see below). Small numbers of the released water molecules upon formation of the (pol β)16 binding mode indicate that these structural changes are preserved in this mode (Table 2).
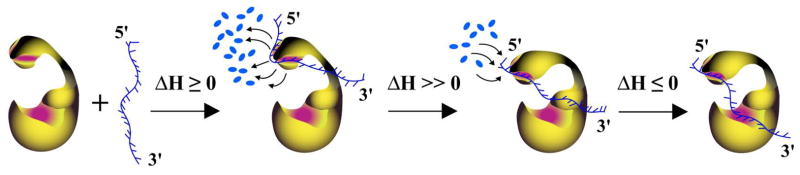
Schematic representation of the observed behavior of the pol β-ssDNA system, base d on the data obtained in this work, is shown in Figure 10. The enzyme engages in the initial complex the 8-kDa domain in a process accompanied by the release of a large number (~16 – 19) water molecules and characterized by the zero or small positive ΔHo. The entire DNA binding subsite of the 8-kDa domain interacts with the nucleic acid. Transition to the (pol β)5 binding mode includes reorientation of the 8-kDa domain with respect to the 31-kDa domain, the release of the part of the 8-kDa domain binding subsite from the interactions with the DNA. The process is accompanied by the net uptake of ~ 10 water molecules by the domain, most probably by the part of its binding subsite freed from the nucleic acid, and a large positive ΔHo. The final engagement of the total DNA-binding site in forming the (pol β)16 binding mode involves association of the DNA-binding subsite on the 31-kDa domain with the nucleic acid, which occurs without additional water release and is accompanied by a small negative ΔHo.
Figure 10.
Schematic model of the pol β-ssDNA complex formation. In the initial complex, the enzyme engages only the 8-kDa domain in a process accompanied by the release of the large number, ~16 – 19, water molecules and characterized by zero or a small positive ΔHo. The entire potential DNA binding subsite of the 8-kDa domain is involved in interactions with the nucleic acid. Transition to the (pol β)5 binding mode includes reorientation of the 8 -kDa domain with respect to the 31-kDa domain. The process is accompanied by the net uptake of ~ 10 water molecules by the 8-kDa domain and a large positive ΔHo. The final engagement of the total DNA-binding site in forming the (pol β)16 binding mode, involves engagement of the DNA -binding subsite on the 31-kDa domain in interactions with the nucleic acid, without additional water release and accompanied by a small negative ΔHo (details in text).
The difference in the sign of the thermodynamic function, not only in its value, between the two DNA-binding subsites of pol β, characterizing the association with the nucleic acid, indicate a profound difference in the nature of interactions of both subsites with the DNA. The high intrinsic affinity of the DNA-binding subsite on the 8-kDa domain is necessary to make the primary initial contact with the DNA. It is a recognition-initiation subsite with flexible adjustment of its site-size, allowing the enzyme to change its orientations on the nucleic acid. The binding is predominantly entropy-driven, resulting mostly from the release of the water molecules. The established contact does not have to dramatically change during the catalysis on small ssDNA gaps. The dominant role of water molecules in affecting its intrinsic affinity indicates that the 8-kDa domain can considerably increase its affinity for the DNA in conditions that mimic the diminished solvent activity, e.g., by molecular crowding in the cell.
On the other hand, the DNA-binding subsite on the 31-kDa domain contains the active site of the polymerase. It changes its engagements with the nucleic acid with every synthesized base pair. The low intrinsic affinity of the subsite on the 31-kDa domain should facilitate these transitions. Moreover, structural changes that increase the entropy of the complex of the intact enzyme with the nucleic acid, initiated in the reorientation of the enzyme molecule in the formation of the (pol β)5 binding mode and preserved in the (pol β)16 binding mode, indicate the presence of a very dynamic and flexible structure of the complex, which should facilitate the catalysis [52,53]. Nevertheless, small negative enthalpy changes of the engagement of the subsite on the 31-kDa domain in interactions with the DNA, strongly suggest that the subsite undergoes conformational changes necessary to adjust to the changing structure of its DNA counterpart.
Acknowledgments
We wish to thank Mrs. Gloria Drennan Bellard for reading the manuscript. This work was supported by NIH Grant GM58565 (to W. B.).
Abbreviations
- ssDNA
single-stranded DNA
- dsDNA
double-stranded DNA
- HhH
helix-hairpin-helix, DMSO, dimethyl sulfoxide
- FRET
fluorescence resonance energy transfer
- CP
7-Diethylamino-3-(4′-maleimidylphenyl)-4-methylcoumarin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kornberg A, Baker TA. DNA Replication. W.H. Freeman; New York: 1992. [Google Scholar]
- 2.Friedberg EC, Walker GC, Siede W. DNA Repair and Mutagenesis. ASM Press; Washington, D.C: 1995. [Google Scholar]
- 3.Fry M, Loeb LA. Animal Cell DNA Polymerases. CRC Press, Inc; Boca Raton, FL: 1986. [Google Scholar]
- 4.Hübscher U, Nasheuer HP, Syväoja JE. Eukaryotic DNA polymerases, a growing family. Trends Biochem Sci. 2000;25:143–147. doi: 10.1016/s0968-0004(99)01523-6. [DOI] [PubMed] [Google Scholar]
- 5.Matsumoto Y, Kim K. Excision of deoxyribose phosphate residues by DNA polymerase beta during DNA repair. Science. 1995;269:699–702. doi: 10.1126/science.7624801. [DOI] [PubMed] [Google Scholar]
- 6.Matsumoto Y, Kim K, Katz DS, Feng J-A. Catalytic center of DNA polymerase beta for excision of deoxyribose phosphate groups. Biochemistry. 1998;37:6456–6464. doi: 10.1021/bi9727545. [DOI] [PubMed] [Google Scholar]
- 7.Wiebauer K, Jiricny J. Mismatch-specific thymine DNA glycosylase and DNA polymerase beta mediate the correction of G.T mispairs in nuclear extracts from human cells. Proc Natl Acad Sci USA. 1990;87:5842–5845. doi: 10.1073/pnas.87.15.5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Budd ME, Campbell JL. DNA polymerases required for repair of UV-induced damage in Saccharomyces cerevisiae. Mutation Research. 1997;384:157–167. doi: 10.1128/mcb.15.4.2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu HY, Horton JK, Gryk MR, Prasad R, Naron JM, Sun DA, Hecht SM, Wilson SH, Mullen GP. Identification of small molecule synthetic inhibitors of DNA polymerase beta by NMR chemical shift mapping. J Biol Chem. 2004;279:39736–44. doi: 10.1074/jbc.M402842200. [DOI] [PubMed] [Google Scholar]
- 10.Sawaya MR, Pelletier H, Kumar A, Wilson SH, Kraut JA. Crystal Structure of the rat DNA polymerase beta: evidence a common polymerase mechanism. Science. 1994;264:1930–1935. doi: 10.1126/science.7516581. [DOI] [PubMed] [Google Scholar]
- 11.Rajendran S, Jezewska MJ, Bujalowski W. Human DNA polymerase β recognizes single-stranded DNA using two different binding modes. J Biol Chem. 1998;273:31021–31031. doi: 10.1074/jbc.273.47.31021. [DOI] [PubMed] [Google Scholar]
- 12.Jezewska MJ, Rajendran S, Bujalowski W. Transition between different binding modes in rat DNA polymerase β-ssDNA complexes. J Mol Biol. 1998;284(28):1113–1131. doi: 10.1006/jmbi.1998.2252. [DOI] [PubMed] [Google Scholar]
- 13.Rajendran S, Jezewska MJ, Bujalowski W. Recognition of template primer and gapped DNA substrates by human DNA polymerase β. J Mol Biol. 2001;308:477–500. doi: 10.1006/jmbi.2001.4571. [DOI] [PubMed] [Google Scholar]
- 14.Jezewska MJ, Rajendran S, Bujalowski W. Energetics and specificity of rat DNA polymerase β interactions with template -primer and gapped DNA substrates. J Biol Chem. 2001;276:16123–16136. doi: 10.1074/jbc.M010434200. [DOI] [PubMed] [Google Scholar]
- 15.Jezewska MJ, Rajendran S, Bujalowski W. Interactions of the 8-kDa domain of rat DNA polymerase β with ssDNA. Biochemistry. 2001;40:3295–3307. doi: 10.1021/bi002749s. [DOI] [PubMed] [Google Scholar]
- 16.Jezewska MJ, Galletto R, Bujalowski W. Tertiary conformation of the template-primer and gapped DNA substrates in complexes with rat polymerase β. Fluorescence energy transfer studies using the multiple donor-acceptor approach. Biochemistry. 2003;42:11864–11878. doi: 10.1021/bi030111l. [DOI] [PubMed] [Google Scholar]
- 17.Jezewska MJ, Galletto R, Bujalowski W. Rat polymerase β gapped DNA Interactions: antagonistic effects of the 5′ terminal PO4 group and magnesium on the enzyme binding to the gapped DNAs with different ssDNA gaps. Cell Biochem Biophys. 2003;38:125–160. doi: 10.1385/CBB:38:2:125. [DOI] [PubMed] [Google Scholar]
- 18.Bujalowski W, Jezewska MJ, Galletto R. Dynamics of Gapped DNA Recognition by Human Polymerase β. J Biol Chem. 2002;277:20316–20327. doi: 10.1074/jbc.M200918200. [DOI] [PubMed] [Google Scholar]
- 19.Edelhoch H. Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry. 1967;6:1948–1954. doi: 10.1021/bi00859a010. [DOI] [PubMed] [Google Scholar]
- 20.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 21.Baker BM, Vanderkooi J, Kallenbach NR. Base Stacking in a Fluorescent Dinucleoside Monophosphate: εApεA. Biopolymers. 1978;17:1361–1372. [Google Scholar]
- 22.Tolman GL, Barrio JR, Leonard NJ. Chloroacetaldehyde-modified di-nucleoside phosphates. Dynamic fluorescence quenching and quenching due to Intramolecular complexation. Biochemistry. 1974;13:4869–4878. doi: 10.1021/bi00721a001. [DOI] [PubMed] [Google Scholar]
- 23.Lohman TM, Bujalowski W. Thermodynamic methods for model-independent determination of equilibrium binding isotherms for protein-DNA interactions: spectroscopic approaches to monitor binding. Meth Enzym. 1991;208:258–290. doi: 10.1016/0076-6879(91)08017-c. [DOI] [PubMed] [Google Scholar]
- 24.Bujalowski W, Jezewska MJ. Spectrophotometry & Spectrofluorimetry. In: Gore MG, editor. A Practical Approach. Oxford University Press; Oxford: 2000. [Google Scholar]
- 25.Bujalowski W. Thermodynamic and kinetic methods of analyses of protein nucleic acid interactions. From simpler to more complex systems. Chem Rev. 2006;(106):556–606. doi: 10.1021/cr040462l. [DOI] [PubMed] [Google Scholar]
- 26.Yang M, Millar DP. Fluorescence resonance energy transfer as a probe of DNA structure and function. Meth Enzym. 1997;278:417–444. doi: 10.1016/s0076-6879(97)78022-4. [DOI] [PubMed] [Google Scholar]
- 27.Vamosi G, Clegg RM. The helix-coil transition of DNA duplexes and hairpins observed by multiple fluorescence parameters. Biochemistry. 1992;37:14300–14316. doi: 10.1021/bi9727601. [DOI] [PubMed] [Google Scholar]
- 28.Trakselis MA, Alley SC, Able-Santos E, Benkovic SJ. Creating a dynamic picture of the sliding clamp during T4 DNA polymerase holoenzyme assembly by using fluorescence resonance energy transfer. Proc Natl Acad Sci USA. 2001;98:8368–8375. doi: 10.1073/pnas.111006698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jezewska MJ, Rajendran S, Bujalowska D, Bujalowski W. Does ssDNA pass through the inner channel of the protein hexamer in the complex with the E. coli DnaB helicase? fluorescence energy transfer studies. J Biol Chem. 1998;273:10515–10529. doi: 10.1074/jbc.273.17.10515. [DOI] [PubMed] [Google Scholar]
- 30.Lakowicz JR. Principles of Fluorescence Spectroscopy. Plenum Press; New York: 1999. [Google Scholar]
- 31.Berman HA, Yguerabide J, Taylor P. Fluorescence energy transfer on acethylcholinesterase: special relationship between peripheral site and active Center. Biochemistry. 1980;19:2226–2235. doi: 10.1021/bi00551a036. [DOI] [PubMed] [Google Scholar]
- 32.Jezewska MJ, Bujalowski PJ, Bujalowski W. Interactions of the DNA polymerase X of African swine fever virus with double-stranded DNA. Functional structure of the complex. J Mol Biol. 2007;373:75–95. doi: 10.1016/j.jmb.2007.06.054. [DOI] [PubMed] [Google Scholar]
- 33.Bujalowski W, Jezewska MJ. Thermodynamic analysis of the structure-function relationship in the total DNA-binding site of enzyme - DNA complexes. Meth Enzym. 2009;466:294–324. doi: 10.1016/S0076-6879(09)66013-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jezewska MJ, Bujalowski W. A General method of analysis of ligand binding to competing macromolecules using the spectroscopic signal originating from a reference macromolecule. Application to Escherichia coli replicative helicase DnaB protein -nucleic Acid interactions. Biochemistry. 1996;35:2117–2128. doi: 10.1021/bi952344l. [DOI] [PubMed] [Google Scholar]
- 35.Jezewska MJ, Rajendran S, Bujalowski W. Interactions of Escherichia coli replicative helicase PriA protein with single-stranded DNA. Biochemistry. 2000;39:10454–10467. doi: 10.1021/bi001113y. [DOI] [PubMed] [Google Scholar]
- 37.Jezewska MJ, Rajendran S, Bujalowski W. Complex of Escherichiacoli primary replicative helicase DnaB protein with a replication fork. Recognition and structure. Biochemistry. 1998;37:3116–3136. doi: 10.1021/bi972564u. [DOI] [PubMed] [Google Scholar]
- 38.Jezewska MJ, Galletto R, Bujalowski W. Interactions of the RepA helicase hexamer of plasmid RSF1010 with the ssDNA. Quantitative analysis of stoichiometries, Intrinsic affinities, cooperativities, and heterogeneity of the total ssDNA-binding site. J Mol Biol. 2004;343:115–136. doi: 10.1016/j.jmb.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 39.Szymanski MR, Jezewska MJ, Bujalowski W. Interactions of the Escherichia coli Primosomal PriB Protein with the Single-Stranded DNA. Stoichiometries, Intrinsic Affinities, Cooperativities, and Base Specificities. J Mol Biol. 398;2010:8–25. doi: 10.1016/j.jmb.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McGhee JD, von Hippel PH. Theoretical aspects of DNA -protein intera ctions: cooperative and noncooperative binding of large ligands to a one-dimensional homogeneous lattice. J Mol Biol. 1974;86:469–489. doi: 10.1016/0022-2836(74)90031-x. [DOI] [PubMed] [Google Scholar]
- 41.Epstein IR. Cooperative and non-cooperative binding of large ligands to a finite one-dimensional lattice. A model for ligand - oligonucleotide interactions. Biophys Chem. 1978;8:327–339. doi: 10.1016/0301-4622(78)80015-5. [DOI] [PubMed] [Google Scholar]
- 42.Bujalowski W, Lohman TM, Anderson CF. On the cooperative binding of large ligands to a one-dimensional homogeneous lattice: the generalized three-state lattice model. Biopolymers. 1989;28:1637–1643. doi: 10.1002/bip.360280912. [DOI] [PubMed] [Google Scholar]
- 43.Saenger W. Principles of Nucleic Acid Structure. Springer-Verlag; New York: 1984. [Google Scholar]
- 44.Connors KA. The Study of Reaction Rates in Solution. VCH Publishers; New York: 1990. Chemical Kinetics. [Google Scholar]
- 45.Tanford C. Extension of the theory of linked functions to incorporate the effects of protein hydration. J Mol Biol. 1969;39:539–544. doi: 10.1016/0022-2836(69)90143-0. [DOI] [PubMed] [Google Scholar]
- 46.Timasheff SN. Protein - Solvent Interactions, protein hydration, and modulation of biochemical reactions by solvent components. Proc Natl Acad Sc USA. 2002;99:9721–9726. doi: 10.1073/pnas.122225399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parsegian VA, Rand RP, Rau DC. Osmotic stress, crowding, preferential hydration, and binding: A comparison of perspectives. Proc Natl Acad Sc USA. 2000;97:3987–3992. doi: 10.1073/pnas.97.8.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Record MT, Jr, Lohman TM, deHaseth PL. Ion effects on ligand - nucleic acid interactions. J Mol Biol. 1976;107:145–158. doi: 10.1016/s0022-2836(76)80023-x. [DOI] [PubMed] [Google Scholar]
- 49.Record MT, Jr, Anderson CF, Lohman TM. Thermodynamic analysis of ion effects on the binding and conformational equilibria of proteins and nucleic acids: the roles of ion association or release, screening, and ion effects on water activity. Quart Rev Biophys. 1978;11:103–178. doi: 10.1017/s003358350000202x. [DOI] [PubMed] [Google Scholar]
- 50.Capp MW, Pegram LM, Saecker RM, Kratz M, Riccardi D, Wendorff T, Cannon JG, Record MT., Jr Interactions of the osmolyte glycine betaine with molecular surfaces in water: thermodynamics, structural interpretation, and prediction of m-values. Biochemistry. 2009;43:4810372–9. doi: 10.1021/bi901273r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pegram LM, Record MT., Jr Quantifying the roles of water and solutes (denaturants, osmolytes and Hofmeister salts) in protein and model processes using the solute partitioning model. Methods Mol Biol. 2009;490:179–193. doi: 10.1007/978-1-59745-367-7_8. [DOI] [PubMed] [Google Scholar]
- 52.Morales JC, Kool ET. functional hydrogen-bonding map of the minor groove binding tracks of six DNA polymerases. Biochemistry. 2000;39:12979–12988. doi: 10.1021/bi001578o. [DOI] [PubMed] [Google Scholar]
- 53.Washington MT, Wolfle WT, Spratt TS, Prakash L, Prakash S. Yeast DNA polymerase η makes functional contacts with the DNA minor groove only at the incoming nucleotide triphosphate. Proc Natl Acad Sci USA. 2003;100:5113–5118. doi: 10.1073/pnas.0837578100. [DOI] [PMC free article] [PubMed] [Google Scholar]