Abstract
O6-Alkylguanine-DNA alkyltransferase (AGT) is a widely distributed, unique DNA repair protein that acts as a single agent to directly remove alkyl groups located on the O6-position of guanine from DNA restoring the DNA in one step. The protein acts only once and its alkylated form is degraded rapidly. It is a major factor in counteracting the mutagenic, carcinogenic and cytotoxic effects of agents that form such adducts including N-nitroso-compounds and a number of cancer chemotherapeutics. This review describes the structure, function and mechanism of action of AGTs and of a family of related alkyltransferase-like proteins, which do not alone act to repair O6-alkylguanines in DNA but link repair to other pathways. The paradoxical ability of AGTs to stimulate the DNA-damaging ability of dihaloalkanes and other bis-electrophiles via the formation of AGT-DNA crosslinks is also described. Other important properties of AGTs include the ability to provide resistance to cancer therapeutic alkylating agents and the availability of AGT inhibitors such as O6-benzylguanine that might overcome this resistance is discussed. Finally, the properties of fusion proteins in which AGT sequences are linked to other proteins are outlined. Such proteins occur naturally and synthetic variants engineered to react specifically with derivatives of O6-benzylguanine are the basis of a valuable research technique for tagging proteins with specific reagents.
1. Introduction
This review article outlines the multifaceted roles of AGT (O6-alkylguanine-DNA alkyltransferase). It has been known for 30 years that this protein provides a unique means of DNA repair, but only in the last decade have other properties of interest to toxicology emerged including its ability to actually enhance DNA damage by some bifunctional agents and the existence of a large family of related proteins whose function is still being clarified. AGT is also a target for the design of cancer therapeutic strategies and has been used to develop a useful research tool for specific labeling of proteins There is a vast literature on alkyltransferase-mediated DNA repair and this article contains only a fraction of the important citations of work in this field. Further documentation can be found in earlier review articles (1-10).
2. Brief history
The first AGT to be fully characterized was the Ada gene product from E. coli, which was shown by Lindahl and colleagues to repair O6-methylguanine in DNA by a direct transfer of the methyl group from the DNA to a Cys residue in the protein itself (11, 12) (Figure 1). This inactivates the protein, which can act only once, and thus leads to a saturability of DNA repair capacity until de novo synthesis of new AGT molecules. The biochemical study of the Ada protein was facilitated by the fact that it is highly inducible in response to alkylation damage and could be purified in relatively large amounts after such treatment (13, 14). Subsequent studies showed that there was a second AGT in E. coli, which is the product of the Ogt gene (15, 16). This protein is not inducible and is present in small amounts but is responsible for repair of low levels of alkylation damage. Both proteins are frequently termed MGMT (O6-methylguanine-DNA methyltransferase) since they were discovered by their action on methylated DNA and O6-methylguanine is their preferred substrate. It was also shown that they were able to repair the minor methylation product O4-methylthymine in a similar transfer reaction (15-17). Exposure to endogenous methylating agents such as S-adenosylmethionine and N-nitroso-compounds generated in vivo may provide a background generation of alkylation damage that requires the existence of AGTs to maintain the integrity of DNA (18, 19).
Figure 1.
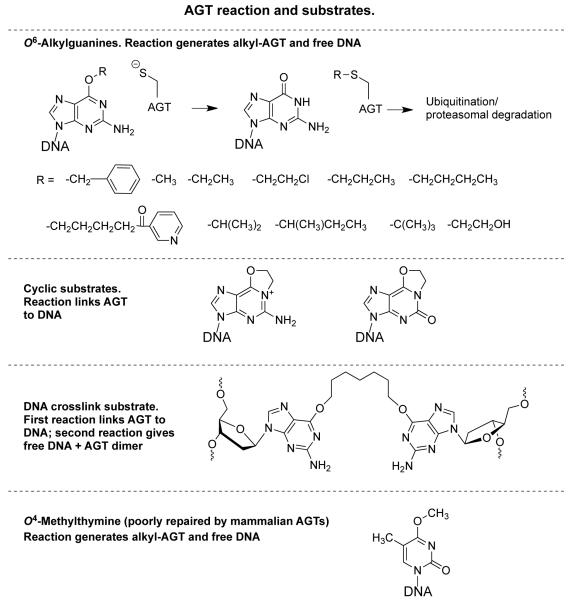
AGT reaction and substrates
At the same time as the Ada protein was being characterized, it was clear that a similar protein existed in mammalian tissues since the disappearance of O6-methylguanine from rat tissue DNA after treatment with dimethylnitrosamine (DMN) or N-methyl-N-nitrosurea (MNU) was saturable at higher doses of the alkylating agent (20, 21), and a protein that brought about O6-methylguanine removal was detected in rat extracts (22, 23). Even in liver, which has the highest content of AGT, this protein was a very minor component and mammalian AGT is not induced by alkylation damage and is only slightly increased by certain stimuli such as partial hepatectomy (1, 2, 24) and glucocorticoids (25). It was therefore difficult to purify sufficient material for a full characterization but subsequent studies with semi-purified material from human and rat liver indicated that a similar alkyl transfer reaction directly restoring the DNA did occur (26, 27). Cloning of the gene and the use of recombinant DNA technology provided adequate protein for a complete characterization (28-30). Despite the limited amounts of purified material available, it could be demonstrated that the mammalian protein was effective at repairing larger adducts such as O6-ethylguanine (31, 32). Later studies have shown repair of a number of bulky adducts, including O6-[4-oxo-4-(3-pyridyl)butyl]guanine formed by the tobacco-derived carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) and O6-benzylguanine (33-36), leading to the more generic name AGT (Figure 1). The best substrate for DNA repair by the human AGT is the O6-benzylguanine adduct (36).
3. DNA repair by AGTs
3.1. Prevention of mutations
Many studies in which AGTs have been expressed in cultured cells and microorganisms have shown that the expression of AGT greatly reduces the incidence of mutations caused by exposure to methylating agents such as MNU or N-methyl-N′-nitro-N-nitrosoguanidine (MNNG). The mutations prevented are virtually all G:C to A:T transitions caused by O6-methylguanine, which is readily copied by DNA polymerases with the frequent incorrect insertion of thymine (10, 37-41). Recent studies have also shown that O6-methylguanine in DNA causes misincorporation of uridine by RNA polymerases and generates altered proteins at the level of transcription (42).
3.2. Prevention of cytoxicity
AGT provides powerful protection against killing by these methylating agents. This is also due to its ability to repair O6-methylguanine. The toxicity of this adduct results from the recognition of O6-methylguanine:thymine pair by the mismatch repair system (43, 44). This recognition leads to homologous recombination, sister chromatid exchanges and cell death, either via direct signaling of the complex to apoptotic pathways or via abortive repeated excision and resynthesis of the thymine-containing daughter DNA strand (43, 45-48). Therefore, the absence of AGT-mediated repair in a mismatch deficient background after exposure to alkylating agents is particularly deleterious since there is increased survival of cells that have an increased chance of developing mutations.
Recently, transgenic mice either lacking AGT expression or overexpressing AGT were used to demonstrate that AGT protects the developing brain from methylazoxymethanol (49). Neuronal cultures derived from mice with the MGMT gene (encoding AGT) deleted were more sensitive to this genotoxic agent. Granule cell development and motor function were more drastically affected in mice lacking AGT than in wild type mice. Conversely, transgenic increase in AGT provided resistance to this damage (49).
3.3. Prevention of tumor development
Alterations in AGT levels caused by genetic manipulations have been used to provide convincing evidence of its critical role after exposure to alkylating agents. Transgenic overexpression of AGT in the thymus prevents the production of thymic lymphomas after exposure to MNU (50) even in a p53 deficient background (51). Subsequent studies using other tissue specific promoters have shown reduced incidence of tumors in the liver after treatment with DMN (52), in the skin exposed to MNU or 1-(4-amino-2-methyl-5-pyrimidinyl)methyl-3-(2-chloroethyl)-3-nitrosourea (ACNU) (53, 54), the colon following azoxymethane (55), and the lung after exposure to NNK (56). Gene disruption leading to a total loss of AGT activity gave rise to an increased tumor incidence in the liver in response to DMN (57), in the colon in response to azoxymethane (58), and increases in thymic lymphomas and lung adenomas in response to NMU (59). These results provide convincing evidence that AGT is highly protective against carcinogenic methylating agents and that O6-methylguanine is a critical DNA lesion initiating tumors produced by such agents.
3.4. Mechanism of DNA repair
Sequences encoding AGTs are readily recognized in genome data bases since virtually all AGTs contain the signature protein sequence -(I/V)PCHR(V/I)(I/V)- surrounding the Cys site that serves as the alkyl acceptor (Figure 2). These proteins, which consist of a small single polypeptide chain, have been found in most bacteria, archea, fungi and animals but not in plants. Notably, AGT is also absent from the fission yeast Schizosaccharomyces pombe. Crystal structures are available for a number for AGTs including the Ada-C domain from E. coli (60), human (61-63) and the archaeon Thermococcus kodakaraensis KOD1 (64). These proteins show a similar overall structure despite limited sequence identity that is confined to the active site pocket and DNA binding domain.
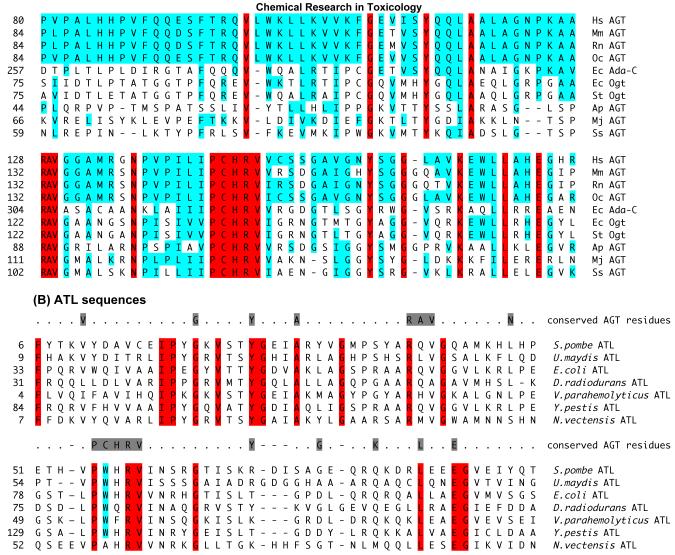
Figure 2.
Key sequences in AGTs and ATLs. (A) shows the amino acid sequence alignment of the C-terminal domain containing the active site of AGT proteins from different species. The AGT sequences are from Homo sapiens (Hs), Mus musculus (Mm), Rattus norvegicus (Rn), Oryctolagus cuniculus (Oc), Escherichia coli (Ec), Salmonella typhimurium (St), Aeropyrum pernix (Ap), Methanococcus jannaschii (Mj) and Sulfolobus solfataricus (Ss). The amino acids that are conserved in the above AGTs are shown in red. The amino acids that similar or identical to human AGT protein are indicated in blue. The numbers on the left side of the sequences indicate the position of the amino acid in the AGT primary sequence. (B) shows amino acid sequence alignment of ATL proteins from different species and a comparison with conserved AGT residues. The ATL sequences are from Schizosaccharomyces pombe, Ustilago maydis, Escherichia coli, Deinococcus radiodurans, Vibrio parahaemolyticus, Yersinia pestis and Nematostella vectensis. The amino acids that are highly conserved in ATLs are indicated in red. The tryptophan residue that replaces the AGT alkyl acceptor site cysteine is indicated in blue. The amino acids that are highly conserved in AGTs from figure 1 are shaded grey and shown at the top of the aligned ATL sequences. The numbers on the left side of the sequences indicate the position of the amino acid in the primary sequence.
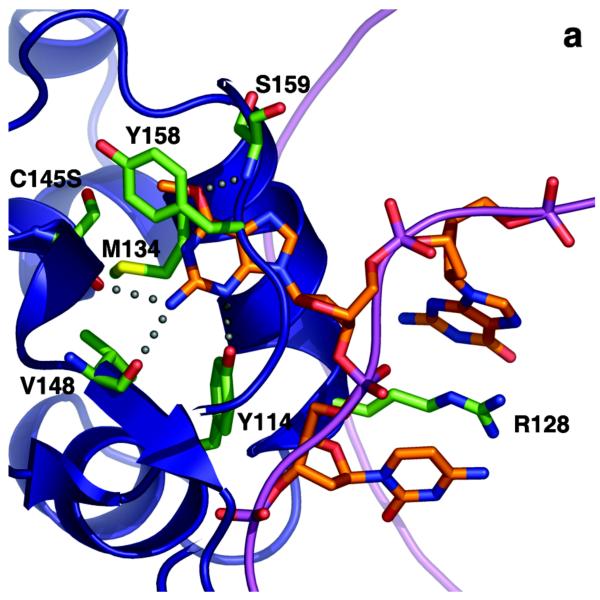
The AGT structure consists of two domains. The C-terminal domain contains the active site pocket and DNA binding region, which are coupled by an asparagine hinge formed by conserved residue Asn137 (amino acid numbers given throughout are for the human AGT). The N-terminal domain, which in human AGT contains a bound zinc atom, plays an important structural function and can stabilize the complex formed when the two domains are expressed separately (65). AGT binds to DNA using a helix-turn-helix motif but unusually attacks the minor groove. The recognition helix is small and contains hydrophobic residues allowing it to pack closely within the DNA minor groove with minimal sequence-specificity (6, 62). The binding of AGT alters the DNA structure, bending the chain by about 15°, widening the minor groove and displacing the target O6-methylguanine base into the active site pocket (Figure 3A). This displacement is caused by a rotation promoted by a tyrosine residue (Tyr114), either by steric (62) or by electrostatic effects (66), and is stabilized by an arginine finger (Arg128) that replaces the base in the DNA helix. These residues are conserved in AGTs (Figure 2) although the Tyr residue is sometimes replaced by Phe, which can act in the same way.
Figure 3.
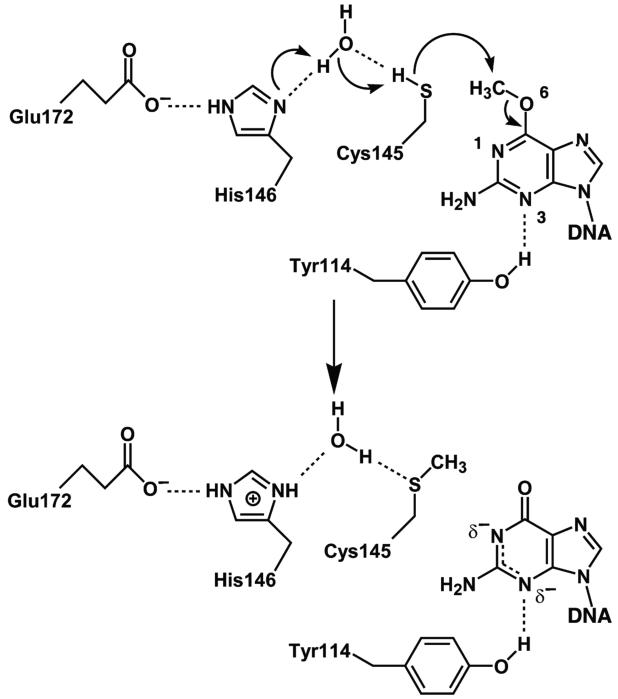
AGT:substrate complex and reaction mechanism. (A) Complex of C145S AGT mutant bound to DNA containing O6-methylguanine showing key residues Y114, R128, C145S and M134 in green. The DNA is shown in orange and the protein ribbon in blue. (B) Reaction mechanism of AGT showing activation of Cys 145 via interactions with Glu172-His146-water-C145. Redrawn with permission from refs (61, 62).
The Cys that forms the acceptor site is rendered highly reactive via its interaction through a hydrogen bond network made up of conserved residues Cys145-water-His146-Glu172 (61). It is thus effectively converted into a thiolate anion (67) (Figure 3B). The O6-methylguanine is positioned by the hydrophobic cleft in the active site pocket for the displacement reaction that brings about restoration of the guanine in DNA. Quantum chemical models support this mechanism where His146 acts as a water-mediated general base to activate Cys145, which then performs a nucleophilic attack to dealkylate the guanine base (68-70). Kinetic studies of the reaction are consistent with a model in which AGT binds and scans DNA, flips the O6-alkylguanine residue, transfers the alkyl group and releases the dealkylated DNA (36). The alkyl transfer reaction was found to be rate limiting for methyl but not benzyl adducts (36, 70).
Although most small DNA adducts are removed from DNA via the action of glycosylases that cleave the N-glycosidic bond to form an abasic site and release the base, there is no known glycosylase for O6-alkylguanine. It may be difficult to provide an active site that has the very high degree of discrimination to distinguish between guanine and O6-alkylguanine. This discrimination is necessary to prevent the removal of a significant number of guanine residues. Such loss would lead to toxicity in an analogous way to that demonstrated with overproduction of known glycosylases and an inbalance in the activities of the enzymes in the base excision pathway (71-75). The alkyl transfer mechanism adopted by AGT avoids this problem since it merely relies on placing the alkyl adduct in the correct position to the reactive Cys for transfer to occur. If a normal base is transiently placed in this position, there is no reaction and DNA scanning can continue with no deleterious effects. This may explain the unusual reaction mechanism employed by AGTs. Although the fact that the protein can be used only once is a disadvantage, this mechanism has the advantage that it does not rely on absolute discrimination of the target base or on other proteins for rapid and efficient repair and maintaining a balance with other repair proteins.
The studies described above provide a convincing explanation for the mechanism of alkyl transfer once a substrate has been recognized and bound at the active site. However, it is not yet well understood how AGT, which requires no cofactors or energy source to carry out repair, carries out its DNA scanning function to allow rapid repair even when there are only a few sites of repairable damage. E. coli Ada AGT slides along DNA at near to the one-dimensional diffusion limit (76). There is evidence that movement of human AGT along the DNA occurs preferentially from the 5′ to 3′ end (62, 77). A local asymmetry in binding may allow such movement (78) and it is known that human AGT binds to DNA in a cooperative fashion (79-81). These complexes contain overlapping protein molecules where there is little contact between the nth protein and proteins n+1 and n+2, but the N-terminal surface of the nth protein is positioned to contact the C-terminal surface of protein n+3. Such binding could facilitate rapid directional scanning and the efficient repair of lesions contained within the chromatin structure.
The ability of AGT to bind into the minor groove of DNA and its compact shape and size may allow easy access to chromatin DNA. This unusual binding can also allow interactions with other proteins that may also assist in repair. Numerous proteins that may interact with AGT were identified using MS techniques (82) but, with the exception of the interactions with papillomavirus oncoprotein E6 protein (83) or BRCA2 (84), the consequences have not been described. Mutations of BRCA2 that delete a 29-amino-acid region in a conserved domain prevent its binding to human AGT and resulted in an increased sensitivity to the presence of O6-methylguanine (84).
Structures of wild type human AGT and double-stranded oligodeoxynucleotides show that the protein binds over approximately a seven base pair region (62) and optimal repair of DNA requires substrates of 8-12 nucleotides (1, 2). Experiments using a footprinting technique indicated a larger region of DNA contact (85) but this can be explained by the cooperative binding of multiple AGT molecules. Lesions contained within double stranded DNA are the best substrates for AGTs but single stranded DNA is also repaired efficiently. O6-Methylguanine present in RNA is a very poor substrate for AGT and is unlikely to be repaired significantly in vivo (86). RNA repair is prevented by a steric clash of the 2′ hydroxyl group with Gly131Cα, which is only 3.5Å from the ribose C2 atom (6, 62). This is advantageous as it prevents the AGT pool from being used up after exposure to alkylating agents by acting on the less important RNA substrate thus allowing all of the AGT to be employed for correction of DNA.
3.5. Fate of the alkylated form of AGT
The structural alteration caused by the addition of an alkyl group to Cys145 of human AGT leads to recognition by ubiquitin ligases and degradation by the proteasome (87-89). Such degradation may be needed for continued repair since the alkylated form or inactive mutants of AGT such as C145A interfere with repair by active AGT molecules (90). Alkylation of AGT at Cys145 leads to a sterically driven movement of a helical region that contains Met134 and Arg128 and an opening of the Asn137 hinge. These changes, which presumably reveal a site for ubiquitination, are brought about the close contact of the alkyl group with the carbonyl oxygen of Met134 (see Fig 3A) and the steric collision with Asn137 (61). This destabilizes the protein (91) but does not greatly reduce the binding to DNA (92) so the ubiquitination may be needed for release. It was reported that S. cerevisiae AGT is targeted for degradation by a synergistic action of both the Ubr1/Rad6-dependent N-end rule pathway and the Ufd4/Ubc4-dependent ubiquitin fusion degradation (UFD) pathway (93). Details of the site of ubiquitination and the ligase involved are not known for human AGT. Constructs where a GFP sequence is placed at the N-terminus of the human AGT sequence are active and their alkylation products are degraded (94) suggesting that a site other than the amino terminus can be used. The interactions of human AGT with E6 (83) and with BRCA2 may be related to its turnover (84). BRCA2 degradation is also enhanced by methylation damage and alkylation of AGT (84).
3.6. Repair of O4-methylthymine by AGTs
AGTs also repair O4-methylthymine in DNA (Figure 1). This adduct is formed at lower levels than O6-methylguanine but may be an even more potent inducer of mutations (95-99). However, only some AGTs, such as Ogt, repair O4-methylthymine efficiently. Ada is less effective (100-102) and human and other mammalian AGTs have much less activity (103, 104). Indeed, although some AGT-mediated loss of O4-methylthymine in rat liver was reported, albeit after removal of O6-methylguanine was virtually complete (105), expression of human AGT in E. coli actually retards the removal of O4-methylthymine (104, 106). This is due to its interference with the slow removal of O4-methylthymine by nucleotide excision repair (NER). When a strain in which NER is eliminated was used, a weak activity of the human AGT to prevent T:A to C:G mutations caused by MNNG was revealed (106). This contrasts with the total reduction in these mutations when Ogt was expressed at a similar level.
The key factor in whether AGTs can repair O4-methylthymine appears to be the ability of amino acid residues in the active site pocket to position the target O4-methylthymine sufficiently close to the reactive Cys acceptor site. Mutants of human AGT containing alterations in the active site residues can repair O4-methylthymine (106-108). An effective human AGT mutant was obtained by replacing the sequence -V149CSSGAVGN157- with the corresponding Ogt sequence -I143GRNGTMTG151- (106). This protein resembled Ogt in effectively preventing T:A to C:G mutations due to O4-alkylthymine formed by methylating, ethylating or propylating agents (106). This contrasts with the inability of mammalian AGTs to repair larger O4-alkylthymine residues (106, 109, 110).
3.7. Repair of larger O6-alkylguanine lesions by AGTs
Another important species difference in AGTs relates to the ability to repair different alkyl groups attached to the O6-position of guanine. The mechanism for repair described above requires only that the carbon of the alkyl group liked to the oxygen of guanine be placed in proximity to the reactive Cys. Indeed, human and rodent AGTs can repair a variety of O6-alkylguanine lesions (1, 31, 33-36, 111) (Figure 1). The relative rates of repair are benzyl >> methyl > ethyl> n-propyl, n-butyl. Other adducts such as 4-oxo-4-(3-pyridyl)butyl-, iso-butyl, tert-butyl are also repaired but at slower rates (31, 32). However, repair can only take place if the 2′-deoxyguanosine with the alkyl adduct can be accommodated in the active site pocket. Some AGTs have a steric restriction in this space (33, 60, 61) and are less able to repair bulky adducts (1, 2, 33, 102, 112). For example, the S. cerevisiae AGT and the E. coli Ada AGT are virtually inactive with O6-benzylguanine adducts in DNA as a substrate (17, 31, 33, 34, 100). The active site of these AGTs is restricted by the bulky side chain of a tryptophan residue and the absence of the Pro140 residue that is present in human AGT, whose role is described in section 6.1.
O6-(2-Chloroethyl)guanine is an important AGT substrate. Although the repair cannot be assayed directly due to the instability of this adduct, it is clear that it is repaired very rapidly since expression of human AGT provides effective protection against chloroethylating compounds such as the anti-cancer drugs ACNU, N,N′-bis(2-chloroethyl)-N-nitrosourea (BCNU), 4-methyl-N-(2-chloroethyl)-N′-cyclohexyl-N-nitrosourea (MeCCNU) and cloretazine (113-118). As first described by Ludlum and colleagues (119, 120), the cytotoxicity of these compounds is due to the formation of DNA interstrand crosslinks. These result from a non-enzymatic reaction of an initial O6-(2-chloroethyl)guanine adduct to form 1, O6-ethanoguanine, which then reacts with the cytosine in the complementary strand of DNA to produce a 1-(3-deoxycytidyl)-2-(1-deoxyguanosinyl)ethane interstrand crosslink. This reaction occurs very quickly from the initial monoadduct and its prevention by AGT shows that O6-(2-chloroethyl)guanine is a good AGT substrate.
The 1-(3-deoxycytidyl)-2-(1-deoxyguanosinyl)ethane crosslink does not involve the O6-position and is unaffected by AGT. However, cyclic derivatives that do contain O6-adducts such as 1, O6-ethanoguanine or 1, O6-ethanoxanthine are substrates for the protein (Figure 1). In this case, reaction leads to the covalent attachment of the AGT protein to the DNA (62, 121). Human AGT is actually able to repair an interstrand crosslink DNA damage where the two DNA strands are joined by a 7-carbon alkyl linker via the guanine-O6 in each strand (122, 123). Such a heptane crosslink was repaired with initial formation of an AGT-DNA complex and further reaction of a second AGT molecule yielding a human AGT dimer and free DNA (see Figure 1). Although this crosslink has never been characterized from treated cells, it is likely to arise after exposure to the developmental drug hepsulfam since human AGT expression protected cells from killing by this drug (123). AGT was not effective in repairing a similar four-carbon crosslink that could be produced by the drug busulfan. This is due to the limited conformational flexibility preventing sufficient rotation of the butyl-linked substrate to allow the access of the protein to the adduct. The Ada protein, which has a more restricted active site, could not repair either the seven-or the four-carbon O6-2′-deoxyguanosine-alkyl-O6-2′-deoxyguanosine interstrand DNA cross-links (122).
Surprisingly, in view of the rapid repair of O6-(2-chloroethyl)guanine by AGT, O6-hydroxyethylguanine is a very poor substrate (32, 35). One possible reason for this is that the hydroxyethyl-group may favor the adoption of the anti-conformation of the O6-adduct (35). The crystal structures of O6-methylguanine in DNA bound to human AGT shows that it is located with the methyl group in the syn-position pointing towards Cys145 (62) (Figures 1 and 3A). This conformation, which is needed for reaction, would be favored by the hydrophobic nature of the binding pocket. The hydrophilic group of O6-hydroxyethylguanine may not readily assume this conformation (35).
3.8. Sequence specificity of repair by AGTs
AGT-mediated repair of O6-methylguanine shows only slight sequence specificity as expected from structural data showing binding via the minor groove with few potentially sequence-specific interactions but there are exceptions to this rule [reviewed by (124)]. There was a slightly slower rate when the O6-methylguanine was 3′ to guanine residue (1, 124-126). A somewhat larger effect has been reported for a neighboring 5-methylcytosine but these effects were highly dependent on the overall sequence context (124, 127, 128). Interestingly, a greater sequence dependence was seen in the repair of O6-[4-oxo-4-(3-pyridyl)butyl]guanine adducts by mammalian AGTs (35, 124, 129). This may be due to sequence effects changing the orientation of the O6-[4-oxo-4-(3-pyridyl)butyl]guanine adduct in the active site pocket. This could affect the proximity to the reactive Cys or the propensity to adopt the syn conformation of the alkyl group needed for repair. Sequence specificity of repair of O6-[4-oxo-4-(3-pyridyl)butyl]guanine in the 12th codon of the H-ras gene was greater with rodent AGTs than with human AGT (129). It should be noted that, because of the stoichiometric nature of the AGT reaction and its saturation when all molecules of AGT are used up, it is possible that even a small difference in substrate preference may lead to some lesions having a significantly higher chance of being unrepaired. With agents such as NNK that can generate both methyl and 4-oxo-4-(3-pyridyl)butyl adducts (130), the least preferred adduct/sequence may be rendered highly persistent since de novo synthesis of AGT would be required for its repair.
4. DNA damage mediated by AGT
4.1. Effect on damage caused by dihaloalkanes
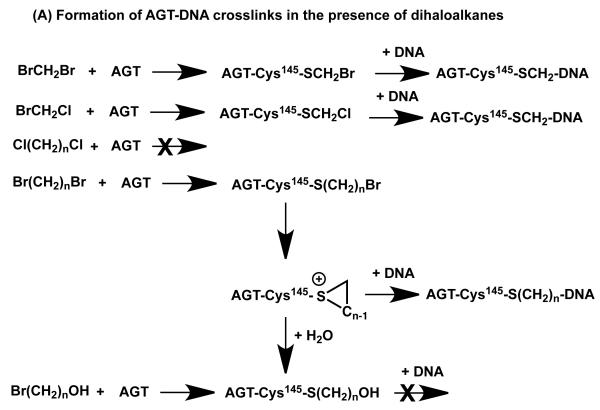
After exposure of E. coli to dibromomethane or 1,2-dibromoethane, there was an increased incidence of cell killing and mutations when human AGT, Ada-C or Ogt were expressed (131-133). More detailed investigation of the mechanism of this paradoxical enhancement of DNA damage by a DNA repair protein showed that 1,2-dibromoethane reacts with the reactive Cys in AGT to generate a AGT-S-(2-bromoethyl) intermediate, which rearranges into a highly reactive half-mustard (134). This is unstable and in vitro in the absence of DNA it rapidly forms an inert -S(CH2)2-OH adduct. In the presence of DNA, the DNA-binding property of AGT brings this intermediate into close contact with DNA. It then reacts, predominantly at guanine residues, to form a covalent adduct, thus crosslinking the AGT to DNA (Figure 4A). DNA-AGT crosslinks formed in this way have been detected in cells treated with 1,2-dibromoethane.
Figure 4.
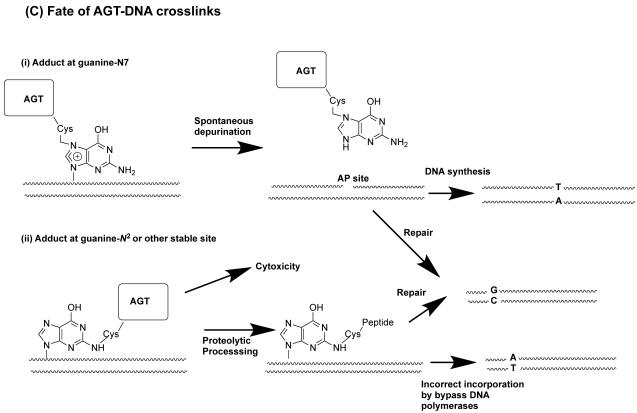
Formation of DNA-AGT crosslinks in the presence of dihaloalkanes and other bis-electrophiles. (A) Reactions of dihaloalkanes to generate crosslinks at Cys145. (B) Reactions of bis-electrophiles to generate crosslinks at Cys145 and Cys150. The chemical can react directly with AGT at either Cys145 or Cys150 as shown in 1A and 1B respectively. Further reaction with DNA then forms the AGT-DNA crosslink. The bis-electrophile may also react with DNA directly to generate a reactive species that can then react with AGT to produce the AGT-DNA crosslink at either Cys residue as shown in 2A and 2B. (C) The AGT-crosslinks can cause mutations or cytoxicity. (i) Attachment at the guanine N7 position generates an unstable linkage that can spontaneously depurinate to form an AP site which on DNA replication can then lead to the observed G:C to T:A transversions. (ii) More stable adducts, possibly at the guanine N2, although this has not been characterized, can be processed and copied by bypass polymerases to lead to the observed G:C to A:T transitions.
An N7-guanine adduct formed in this way was characterized by MS analysis (Figure 4C). The release of the adduct by spontaneous depurination or further processing of the DNA damage to reduce the size of the protein-DNA conjugate allows synthesis by bypass DNA polymerases causing mutations (135, 136). Toxicity may result from an inability to bypass the protein-DNA conjugate efficiently. An essentially similar mechanism occurs with dibromomethane forming a AGT-S-CH2Br intermediate (137) (Figure 4A). In both cases, virtually all of the mutations were at G:C sites and both G to T transversions (which can be derived from AP sites after depurination of the adduct) and G to A transitions were produced. The N7-adducts formed by dihaloalkanes and other bis-electrophiles (see below section 4.2) are the only DNA-AGT derivatives that have been fully characterized, but there are multiple possibilities for other adducts that could lead to the transition mutations, including an adduct at the N2 position (Figure 4C).
A similar mechanism of protein-DNA crosslinking and AGT-enhanced mutagenicity and toxicity was readily demonstrated for other dihaloalkanes containing bromine or iodine, as well as BrCH2Cl and Br(CH2)2Cl (Figure 4A). Among a series of α,ω-disubstituted dihaloalkanes containing Br or I, the initial reaction with AGT increased with methylene chain length. Dichloroalkanes were ineffective at generating such AGT-DNA adducts due to the lack of reaction with AGT (136-138).
Mutants of AGT in which Cys145 was altered to alanine or serine did not promote the genotoxicity and mutagenicity of dihaloalkanes and in vitro experiments showed that there was no formation of an AGT-protein complex with these proteins (134). This is consistent with the mechanism described above in which the ability to react with the reactive Cys is a critical step. Studies with other mutants at key residues needed for the displacement of DNA bases into the active site pocket such as Arg128 and Tyr 114 also blocked the ability of AGT to mediate the genotoxicity of 1,2-dibromoethane. The ability to form the AGT-Cys145S-(CH2)2Br adduct was not impaired with these mutant proteins but the production of a covalent adduct with DNA was greatly diminished (139).
4.2. Effect on damage produced by other bis-electrophiles
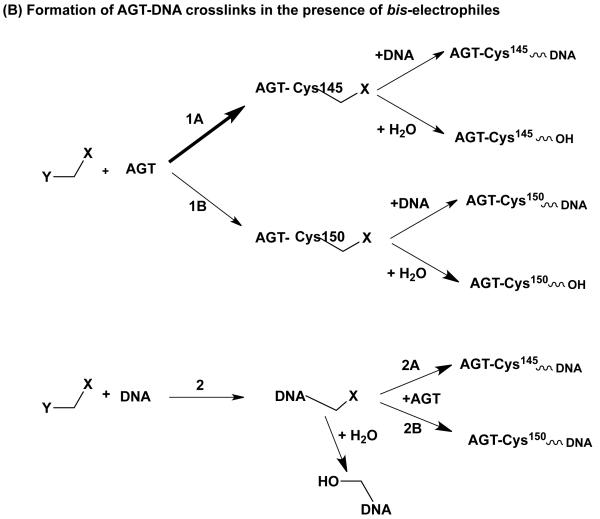
Subsequently, it was shown that a number of other bis-electrophiles can also cause AGT-mediated DNA damage by virtue of their ability to generate DNA-AGT crosslinks. These compounds include 1,2,3,4-diepoxybutane (138, 140, 141), nitrogen mustards (142) and epibromohydrin (143).
Although one pathway to such crosslink formation is similar with these bis-electrophiles, there are two significant differences with the results with dihaloalkanes (Figure 4B). Firstly, these compounds can also react initially with DNA to form a reactive DNA-adduct, which then reacts with AGT (140, 141). Incubation of AGT with N7-(2′-hydroxy-3′,4′-epoxybut-1′-yl)-2′-deoxyguanosine led to a covalent adduct at Cys145 and the same adduct was seen when AGT was linked to DNA in the presence of 1,2,3,4-diepoxybutane (140). Similarly, N7-guanine adducts were characterized as a result of the reaction of AGT, DNA and chlorambucil and mechlorethamine (142). This contrasts with studies of the 1,2-dibromoethane-derived AGT-DNA cross-links where in vitro experiments using [14C]-1,2-dibromoethane showed no reaction when incubated with DNA alone but labeling of AGT and subsequent crosslinking when DNA was added as the last component to the reaction mix (134, 135).
Secondly, the interaction with human AGT can also occur at a second Cys residue in the binding pocket (Cys150) (140, 142). Thus, some AGT-DNA crosslinks were formed with the C145A(S) mutants and only the double mutation C145A/C150S abolished the ability of AGT to increase mutagenesis (141, 143). Although increased transitions and transversions at G:C pairs were observed, there was also a significant increase in transversions at A:T sites when 1,2,3,4-diepoxybutane was used (141). This may be due to the ability of 1,3-1,2,3,4-diepoxybutane to react with DNA at A:T pairs (144).
4.3. AGT-mediated enhancement of toxicity and mutations in mammalian cells
Most of the studies showing that AGT enhances the genotoxicity of bis-electrophiles in vivo have been carried out with bacterial cells as hosts for expression of bacterial or mammalian AGTs since these systems are easier to manipulate, but there is evidence of a similar effect in mammalian cells. Chinese hamster lung fibroblasts, which contain little or no endogenous AGT, were sensitized to the growth inhibitory effects of dihaloalkanes by expression of E. coli AGTs (145). More extensive studies carried out with CHO cells stably transfected with a plasmid that expresses human AGT showed that the cytotoxicity of 1,2-dibromoethane, dibromomethane and epibromohydrin was significantly increased by the presence of AGT (146). Mutations caused by these agents in the hypoxanthine-guanine phosphoribosyltransferase gene were increased by 2-3-fold in the presence of AGT. Both base substitution mutations and the formation of large deletions (> or = 3 exons) were increased. The increases in mutations were statistically significant but much less than those seen in the bacterial systems whereas the effects on increased toxicity were similar. Some of this difference may be due to the reporter systems involved, although it is probably related to the greater ability of the bacterial cells to replicate DNA, albeit with errors, when it contains DNA-protein crosslinks. The strong effect of AGT expression to increase the cytotoxicity of these bifunctional agents in mammalian cells would be consistent with a lack of capacity to replicate DNA in the presence of such damage. At present, the mechanisms by which protein-DNA crosslinks are dealt with are poorly understood but are thought to involve proteolysis, NER and homologous recombination (147, 148).
AGT-DNA crosslinks were detected in nuclear extracts after treatment of AGT-transfected-CHO or Hela cell extracts with mechlorethamine (149) or 1,2,3,4-diepoxybutane (150) respectively. Numerous other proteins were also detected as DNA-conjugates in these analyses. It is possible that with these reactive drugs, the contribution to overall genotoxicity by the AGT-mediated route is masked by the ability to form DNA-DNA crosslinks, DNA-monoadducts and other protein-DNA crosslinks. It should be noted that many proteins contain sites reactive to electrophiles and theoretically can be conjugated to DNA by bis-electrophiles as observed in the experiments with nuclear extracts (149, 150). Such reaction has been noted for glyceraldehyde 3-phosphate dehydrogenase and histone 2b but their expression in E. coli did not increase mutagenesis by dihaloalkanes or 1,2,3,4-diepoxybutane (151, 152). At present, AGT is the only protein that clearly enhances bis-electrophile-induced genotoxicity. This may be due to a combination of the reactivity of its active site residue, the ability of its DNA-binding domain to displace a base into the activity site pocket close to the reactive cysteine derivative and possibly to the cellular processing of the AGT-DNA crosslink, which could differ from responses to other DNA-bound proteins. Therefore, although AGT is only present in small amounts, these may be sufficient to influence the toxic effects of bis-electrophiles. Evidence supporting this concept is provided by studies showing that AGT enhances the neurotoxic effects of mechlorethamine in mice and cultured cells (49).
5. Chemically induced loss of AGT activity
5.1. Reactants causing inactivation of AGT
In addition to forming the DNA-AGT crosslinks, exposure to agents that react readily with the active site Cys causes a direct loss of AGT activity. A significant fraction of the AGT-S-X formed reacts with water rather than DNA but this reaction inactivates AGT (Figure 4). This inactivation can occur with monofunctional agents such as 2-bromoethanol (134) (Figure 4A) and these could render cells more sensitive to alkylation damage. Exposures to aldehydes such as acrolein, formaldehyde and acetaldehyde also inactivate AGT by reaction at the active site Cys (153-155). Direct alkylation also occurs with many alkylating agents that form adducts repairable by AGT (1, 156) but these agents react much more extensively with DNA and there is no evidence that the direct loss of AGT is important.
5.2. Degradation of AGT after reaction with nitric oxide
Exposure to nitric oxide readily nitrosylates the active site Cys residue of AGT. This is theoretically a reversible reaction but the presence of S-nitrosylcysteine at Cys145 causes the structural change leading to polyubiquitination of the protein and its proteasomal degradation (157). Thus, conditions that enhance the generation of nitric oxide such as the formation of reactive oxygen species during inflammatory responses cause loss of AGT activity. The ability of S-nitrosoglutathione reductase to limit S-nitrosylation may be important in preventing such inactivation (158). Mice lacking this reductase showed impaired repair of O6-alkylguanine. It was suggested that the incidence of hepatocellular carcinoma, which is associated with elevated expression of inducible nitric oxide synthase, may be increased by lack of AGT-mediated DNA repair particularly if S-nitrosoglutathione reductase activity is low (158).
6. Resistance to chemotherapy
Since AGT-mediated removal of O6-alkylguanine adducts prevents cell killing by methylating agents such as dacarbazine and temozolomide, and by chloroethylating agents, such as BCNU, ACNU and MeCCNU, the presence of high levels of AGT in tumor cells provides a powerful resistance mechanism to cancer chemotherapy using these agents.
AGT activity has also been linked to resistance to 6-thioguanine (159). A melanoma cell refractory to this drug expressed high levels of AGT and was sensitized by inhibition of AGT. After incorporation into DNA, 6-thioguanine is readily methylated by S-adenosylmethionine to form S6-methylthioguanine, which is then recognized by MMR leading to cell death (160, 161). AGT does bind to S6-methylthioguanine, but this adduct is a very poor substrate for AGT repair (162, 163) so it is possible that binding of AGT prevents the toxicity without direct adduct repair.
6.1. Irreversible inhibitors (pseudosubstrates)
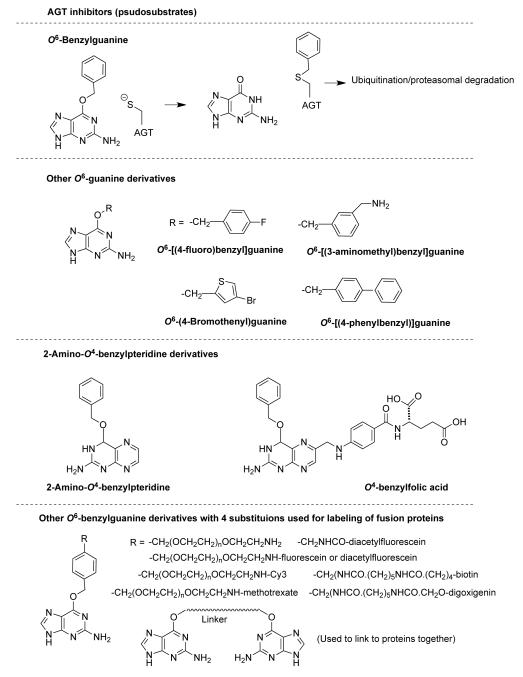
Several procedures aimed at reducing tumor AGT activity have been suggested but the only one that has reached clinical trials is the use of irreversible inhibitors (1, 164-167). This work was initiated through in the laboratory of the late R. C. Moschel. His synthesis of the pseudosubstrate O6-benzylguanine and the finding that it was able to effectively inhibit human AGT and thus sensitize tumor cells to killing by BCNU provided a proof of principle for this concept (115). O6-Benzylguanine was designed as an inactivator of AGT based on the hypothesis that it would be accepted as a substrate and form S-benzylcysteine at the active site releasing guanine (Figure 5). Since benzyl- is better than methyl- as a reactant in such displacement reactions, it was hoped that the faster rate would compensate for the lack of DNA-mediated binding to the active site. Experimental studies have confirmed that this reaction mechanism is correct since inactivation is time dependent and accompanied by the formation of S-benzylcysteine at the active site and stoichiometric release of guanine (61, 168). Another important reason that O6-benzylguanine is effective as an AGT inhibitor is that the benzyl group interacts with the Pro140 residue in human AGT to facilitate its binding in the correct orientation within the active site (61). This Pro is not conserved in AGTs and those AGTs lacking this residue are less readily inactivated by O6-benzylguanine. Some AGTs including those from S. cerevisiae and E. coli Ada not only lack the Pro residue but also have smaller active site pockets, which are unable to accept O6-benzylguanine, rendering them almost totally resistant (33, 168). It was therefore fortunate that the original studies with O6-benzylguanine were carried out with human AGT and human cells rather than bacterial and yeast models, which have been heavily advocated as more convenient surrogates for drug development. It is also noteworthy that mammalian cell membranes are readily penetrated by the lipophilic O6-benzylguanine whereas the E. coli membrane limits the uptake unless its lipopolysaccharide is mutated (169). O6-Benzylguanine is now a commercially available reagent useful for experimental studies examining the role of AGT activity in mammals. Benzylation of AGT via reaction with O6-benzylguanine has the same effect in causing the structural change as that seen after alkylation following DNA repair, and therefore also leads to rapid degradation of the protein (87).
Figure 5.
Inhibitors of AGT activity. References to the inhibitors and many related compounds are found in (112, 116, 165, 167, 170-174, 270). Although not shown here, additions to the N9 position are well tolerated and can be used to design additional inhibitors with improved solubility and other useful properties. The bottom section shows some of the compounds that can be used to label SNAP tags with useful chemical probes as described in section 10.1. Many additional compounds and variants of these probes have also been synthesized (254-262).
Numerous other AGT inhibitors based on the same principles have been described (112, 116, 165, 167, 170) including some such as O6-[3-(aminomethyl)benzyl]guanine and O6-benzylfolate that are significantly more potent (167, 171-173) (Figure 5) but only O6-benzylguanine and O6-(4-bromothenyl)guanine (170, 174) have entered clinical trials. Early trials showed that these inhibitors alone were non-toxic. Phase II trials in combination with therapeutic alkylating agents showed only limited responses. A significant problem is an enhanced hematopoietic toxicity, which limits the amount of the alkylating agent that can be given in these combinations (167, 175-177). These inhibitors are not tumor specific and loss of AGT activity in bone marrow increases the myelosuppressive action of the alkylating agents.
6.2. Tumor specific inhibition
Several approaches to overcome this problem have been proposed. A simple solution is to use regional administration techniques where possible to limit the effect on sensitive normal tissues. This can be accomplished for brain tumors by using implanted polymers that slowly release BCNU (Gliadel wafers) along with systemic treatment with O6-benzylguanine (178). The more potent and water-soluble AGT inhibitors (171, 172) may also be useful for direct application to some localized tumors. Another method would be to use derivatives or prodrugs that are more tumor specific (171, 173, 179, 180).
6.3. O6-Benzylguanine-resistant mutants of AGT for protection and selection
There has been much interest in using the expression of O6-benzylguanine-resistant mutants of AGT to protect bone marrow stem cells. It is relatively easy to generate mutants of the human AGT sequence to provide such resistance (169, 181-183). Even some single amino acid changes, such as a P140K alteration, produces resistance without greatly affecting the ability to repair DNA damage (164, 169). This change not only removes the Pro residue that aids binding, but also restricts the size of the active site pocket and places a charged side chain in this pocket. These alterations increase the IC50 for the inhibitor 0.2 μM to >2 mM. Expression of this resistant mutant using viral vectors can provide good resistance of bone marrow cells to the combination of O6-benzylguanine and alkylating agents (4, 184, 185). This approach to overcoming the toxicity of such combinations in cancer therapy may be limited by the cost, complexity and concerns over safety of the vector delivery systems. However, the use of viruses containing an expressible P140K-AGT and another desired gene may have general value in gene therapy. Treatment with O6-benzylguanine and an alkylating agent provides a powerful selection method allowing for enrichment of cells that express the desired gene (186-190).
7. Consequences of low AGT levels
7.1. Effects on carcinogenesis or cancer prognosis and therapy
There are no clear studies with human populations showing conclusively that a lack of AGT expression increases the probability of developing cancer, and it should be noted that mice with the inactivated MGMT gene do not show an increased spontaneous tumor incidence in the absence of exposure to DNA damaging agents (191, 192). However, in light of the overwhelming experimental data showing that AGT protects against mutations caused by alkylating agents and that humans are exposed to such agents, it seems highly probable that low AGT expression levels would be linked to a rise in tumor development if the appropriate groups were followed.
There is very good data based on both retrospective and prospective studies that the lack of AGT expression increases the likelihood of a positive response to cancer therapy with either chloroethylating or methylating agents (193-196). This is in keeping with the multitude of studies showing that AGT activity protects from cell killing by these agents and provides resistance to them.
7.2. Epigenetic silencing of the MGMT gene
It has been known for many years that some tumor cell lines lack AGT expression (197, 198). These cells are often described as Mer− (Methyl Excision Repair negative). Early studies by Brent and others indicated that many of these cell lines did not express AGT due to epigenetic silencing of the MGMT gene linked to methylation at certain sites in the promoter region (199, 200). Although this process is still not fully understood, particularly in the early stages of silencing, many studies support the concept that specific gene methylation at key sites is an important factor in this inactivation (201-205). Susceptibility to such inactivating methylation is increased by a SNP in the MGMT gene (206, 207). Methylation may also be increased by p53 and reduced by estradiol (117, 208-210).
The frequency of the Mer− status in immortalized cell lines is quite high. The reason for this is unknown. It is possible that lack of AGT expression may aid in unrestricted growth. In contrast, the incidence of a total absence of AGT content in primary tumor samples is quite low although levels in human tumors and normal tissues are highly variable and a greater proportion of tumors may contain a small population of cells that lack AGT. Nevertheless, the rapid development of methods for assaying DNA methylation has provided a means of generating a myriad published studies describing methylation of the MGMT gene promoter and possible correlations with therapeutic response. At least 275 such papers have been published in the last two years. Some, but not all, of these studies suggest that there is a correlation between MGMT methylation status and susceptibility to tumor development, and between MGMT methylation and a favorable response to therapy [see (196, 211-219) and references therein]. However, in only a limited number of cases, has the actual AGT activity or mRNA level been measured and not all of the methylation sites measured have been shown to cause a lack of MGMT expression. Also, correlations with response have been seen even when treatment did not include drugs forming adducts repaired by AGT. Silencing by DNA methylation may involve many genes, and MGMT promoter methylation may only be an indicator of such events rather than being directly responsible for them. Altered methylation may simply reflect a more general increase in aggressive tumor growth and resistance to therapy rather than indicate that the AGT status is affected. Experiments where AGT activity and/or protein have been measured show this is not always associated with the methylation pattern measured or resistance to alkylation. The fact that most tumor samples are also heterogeneous with different levels of AGT expression in various cells is another indication that global measurements of MGMT gene methylation status are unlikely to be useful in guiding therapy.
Recently, it has been observed that some antiepileptic drugs including levetiracetam decrease AGT expression and protein by activating the HDAC1-corepressor complex to enhance p53 binding on the MGMT promoter (210). This may be clinically useful since it increases the response to temozolomide, and also provides a warning that use of such drugs in patients with malignant gliomas must be controlled for in clinical trials.
8. Polymorphisms affecting AGT
A number of genetic variants in the human MGMT gene, which is located on chromosome 10 at 10q26, have been reported. Because of the central role of AGT in response to carcinogenic and therapeutic alkylating agents, there have been many studies that have attempted to correlate the alterations with either cancer incidence or response to therapy of cancer [see (207, 220-224) and references therein]. The results of these studies do not provide a clear or consistent picture. This may be due to the variants having only a small effect and the limited sample size of the affected group in the populations investigated, although recent attempts to deal with this problem by meta analysis have been published (225).
Genetic variations located within the promoter and enhancer regions of AGT can alter expression levels and thus affect sensitivity to toxic and therapeutic alkylating agents (206, 207, 226, 227). It is currently difficult to evaluate the significance of these changes experimentally since many factors may influence AGT levels particularly in the cells such as lymphocytes that are readily available from patients.
There are two relatively common SNPs affecting the AGT protein sequence in 20-25% of the population. These are L84F and I143V/K178R (the changes at codons for 143 and 178 are in almost perfect disequilibrium). Many surveys have been reported in which these alterations have been examined for correlation with alterations to cancer risk or response to treatment. The L84F polymorphism did not change any of the properties of the purified AGT protein in repair of a variety of DNA substrates (129, 220, 228) or the response of cells to the methylating agent temozolomide (94). Despite this, a meta analysis study based on more than 30,000 samples indicated that there was an increased risk of cancer in individuals with the L84F alteration (225) and lymphocytes from these individuals showed an increase in sensitivity to the production of chromosome aberrations by NNK (229) and an increased mutation frequency in smokers (230). It is possible that some other property of AGT is revealed in these studies but the possibility of a linkage of the polymorphism to some other genetic factor is not ruled out.
The I143V/K178R AGT has an alteration close to Cys145 acceptor site but it is unlikely that this change has a significant effect on activity towards methyl adducts since this residue is not conserved and is Val in some AGTs. However, the I143V change did cause a difference from wild type human AGT in being less sensitive to sequence considerations in the repair of O6-[4-oxo-4-(3-pyridyl)butyl]guanine (129). Based on assays of peripheral blood mononuclear cells, it is also possible that the I143V/K178R variant may be present at a higher steady state level (226). This could indicate increased synthesis, decreased inactivation or degradation of the polymorphic form. These results would support the concept that this variant may protect against cancer and a reduced incidence was seen in a study of heavy smokers and lung cancer (231). Despite this, meta analysis data suggested that there actually may be a slight increase in lung cancer incidence and no association with overall cancer risk (225).
The W65C and G160R variants in AGT are very uncommon (c.1% or less) but may have more drastic effects. The W65C is highly unstable and rapidly degraded (228). The G160R variant was much less effective than wild type human AGT in the repair of DNA containing bulky adducts and was resistant to inactivation with the free base O6-benzylguanine (34, 129, 228). This is consistent with relatively large and highly charged side chain of the Arg in the substrate binding pocket that discriminates against larger adducts. These changes are therefore likely to reduce DNA repair capacity, but their rarity suggests that they will not have a significant impact on human disease.
9. Bridge to other DNA repair pathways and transcription
9.1. Alkyltransferase-like proteins (ATLs)
A protein termed ATL with some primary sequence similar to the C-terminal domain of AGT is present in many species including bacteria, fungi, archea and sea anemones (232, 233) (Figure 1 lower panel). ATLs are found in a number of species that lack AGTs including S. pombe, T, thermophilus and Deinococcus radiodurans. ATLs lack the Cys at the -PCHR-acceptor site, which is replaced most commonly by Trp (c 89%) but also by Ala (9%) or occasionally by other amino acids. They do contain the DNA binding domain with the conserved Arg and Tyr residues (Figure 1). ATL proteins have been characterized from E. coli (234, 235), Vibrio parahaemolyticus (236), S. pombe (234, 235, 237),Thermus thermophilus (238, 239) and the starlet sea anemone Nematostella vectensis (S. Kanugula and A. E. Pegg, unpublished).
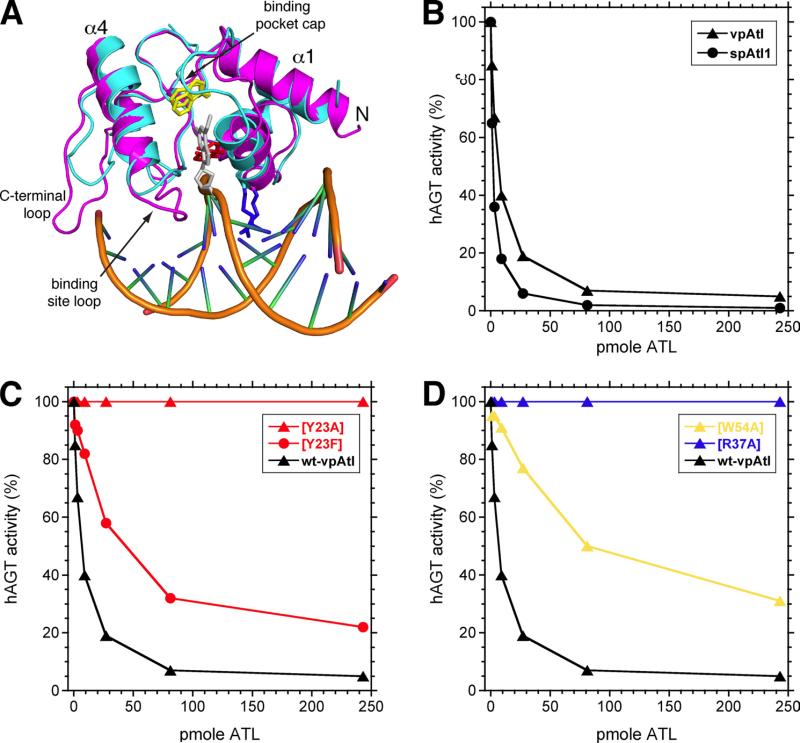
Structures of the ATLs from S. pombe (234) and V. parahaemolyticus (236) have been obtained by X-ray crystallography and NMR respectively (Figure 6A). These structures show that ATLs have a very similar shape to the C-terminal domain of AGTs and bind to DNA in a similar manner to AGT but bend the DNA more extensively by c. 45° (234, 236). In in vitro assays, ATLs bind tightly to DNA with O6-alkylguanine adducts and block repair by AGTs. As expected, binding is reduced and interference with AGT-mediated repair is reduced by mutation of the key Arg and Tyr residues (Figure 6B-D). ATLs have no intrinsic alkyltransferase, glycosylase, or endonuclease activities (234-238).
Figure 6.
Properties of ATLs. Panel A shows overlay of the V. parahaemolyticus ATL (vpARL) structure (cyan) with the crystal structure of S. pombe ATL (spATL) bound to DNA containing O6-methylguanine (grey) The side chains of Tyr23, Arg37, and Trp54 are shown in red, blue, and yellow, respectively. Panels B-D show the inhibition of human AGT activity by ATLs and mutants at key residues. Panel B effect of vpAtl (triangles) and spAtl1 (circles). Panel C shows effect of mutating Tyr23 in vpAtl; wild type (black) triangles, Y23A (red triangles), and Y23F (red circles). D shows the effect of mutating Arg37 and Trp54 in vpAtl; wild type (black), R37A (blue) and W54A (gold). Reproduced from Figure 3 of reference (236).
In vivo expression of ATLs in strains lacking AGTs enhances repair of O6-alkylguanine and reduces mutations caused by methylating, ethylating, propylating and hydroxyethylating agents (234, 239, 240). This protective effect requires an intact NER system and interactions between ATL and proteins in the NER complex have been demonstrated (234, 239, 240). These results collectively support the hypothesis that ATLs enhances repair of O6-alkylguanine adducts by forming a complex that allows recognition by the NER system. Since O6-alkylguanine adducts cause little distortion of the DNA structure, they are poorly recognized by the NER system in the absence of ATLs.
The E. coli ATL is not very effective in promoting repair of O6-methylguanine but is much more active on larger adducts such as hydroxyethyl-, 1-hydroxypropyl-, 2-hydroxypropyl-and n-propyl- (234, 240, 241)(S. Kanugula and A. E. Pegg, unpublished). The two E. coli AGTs are very efficient in the repair of O6-methylguanine but are ineffective with bulkier adducts and the ATL/NER pathway may be needed for efficient repair of such O6-adducts including O6-hydroxyethylguanine. The structures of ATLs bound to DNA with either an O6-methylguanine or an O6-[4-oxo-4-(3-pyridyl)butyl]guanine adduct show that the binding pocket is significantly larger than that of AGTs and can readily accept bulkier adducts (233, 234).
There may be some species specificity in the repair of O6-alkylguanine by the ATL/NER mechanism. S. pombe and Thermus thermophilus do not contain any AGTs so these species may rely on the ATL/NER pathway for repair of O6-methylguanine (237, 239) and their ATLs promote repair of this adduct efficiently. Inactivation of the ATL gene in these species causes increased sensitivity to MNNG and other alkylating agents.
Although the hypothesis that ATL exerts its effects via interaction with NER proteins has strong experimental support, it is quite possible that it also interacts with other DNA repair pathways including MMR. Binding of ATL not only blocks AGT-mediated repair but also MMR-initiated DNA degradation (241). Spontaneous mutations in T. thermophilus lacking NER are reduced to background levels when ATL is also eliminated (239).
ATLs have not yet been identified in vertebrates. The AGTs in these organisms are better able to repair bulky O6-alkylguanine adducts so it is possible that the ATL gene has been lost. However, as described above, vertebrate AGTs are very inefficient at the repair of O6-hydroxyethylguanine, which is repaired by the E. coli ATL/NER pathway and there is an ATL in the starlet sea anemone N. vectensis. This protein binds to O6-alkylguanine adducts in DNA in similar way to the E. coli, V. parahaemolyticus and S. pombe ATLs but species specificity in the interactions with NER proteins and the lack of genetic tests available for this organism have prevented studies of its potential in vivo substrates (S. Kanugula and A. E. Pegg, unpublished). It is certainly possible that further investigation will reveal proteins that perform the same role as ATL in vertebrates. There is a clear parallel to the XPE/DDB2 component of the NER pathway that is required only for the recognition of certain types of DNA damage (242, 243).
9.2. AGTs as transcriptional regulators
The Ada protein present in E. coli and some other gram-negative bacteria, which was the first characterized AGT, contains an N-terminal domain (N-Ada) that is responsible for regulating the adaptive response by increasing repair capacity after alkylation damage (14, 244, 245). This domain is joined by a readily cleaved linked region to the C-terminal domain (C-Ada), which is responsible for the AGT activity. The N-Ada region contains a second Cys acceptor site, which attacks the methylphosphotriesters that are formed in DNA by alkylating agents (246). This Cys, located at residue Cys38 in E. coli Ada (245), is highly reactive as a result of its activation by coordination with a Zn atom, which also bound to three other Cys residues (247). In fact, only one of the two methylphosphotriesters diastereomers, the Sp form, is recognized and it seems more likely that transcriptional activation rather than DNA repair is the function of this reaction. The formation of S-methylcysteine by this reaction activates a latent transcriptional-activation of the N-Ada domain, which then binds to DNA regions containing the ada promoter sequence and activates the expression of genes including Ada itself that provide resistance to alkylating agents (245, 248, 249).
Mammalian AGTs lack any domain corresponding to N-Ada and not inducible by alkylation damage. However, it is intriguing that the N-terminal domain of the human AGT contains a bound zinc coordinated with residues Cys5, Cys24, His29, and His85 (61). This domain when expressed separately can react with O6-alkylguanine if zinc is added although much more weakly than the true AGT activity of the C-terminal domain (65). The presence of zinc is not essential for activity of the complete form of human AGT but its presence increased the rate of repair and provided conformational stability (250).
There are reports suggesting that AGT may play a role in mammalian transcription. Immunostaining with specific antibodies to AGT showed that it is concentrated at active transcription sites and interacts with CREB binding protein CBP/300 (251, 252). The alkylated form of AGT blocks the ability of the estrogen-receptor to stimulate transcription (252). It was suggested that alkylation exposed the region containing the sequence -V98LWKLLKVV106- in AGT. allowing it to bind to the estrogen receptor and thus preventing association with the steroid receptor coactivator-1 and the stimulation of cell growth. Mutation of this AGT region blocked the effect and its similarity to the -LXXLL-motif of steroid receptor coactivator-1 supports this hypothesis although further structural and biochemical studies will be needed to establish its significance. Recently, it was shown that AGT expression reduces angiogenesis with a decrease in some tyrosine kinases (253).
10. AGT and ATL-fusions to other proteins or histones
10.1. Experimental use of AGT fusions for protein tagging
The ability of the active site of AGT to react with O6-benzylguanine to form a covalent adduct at Cys145 has been taken advantage of to generate a reagent for the specific labeling of recombinant proteins (254-256). Since the para-position of benzyl group is directed towards the opening of the active site pocket it can be modified with quite large additions without affecting the ability to react with AGT (257). Such additions can be used to devise cell permeable O6-benzylguanine derivatives that will react and covalently label the protein with a variety of useful chemical probes including fluoroscein, biotin, methotrexate, Ca2 -sensing indicators, and reagents for studying protein isolation, protein immobilization and protein:protein crosslinking and interactions (258-262) (Figure 5). Mutations of the AGT sequence to prevent DNA binding and enhance reaction with such derivatives and convey stability were selected using molecular evolution screening techniques to obtain a protein that can be expressed as a fusion with any cloned sequence and used for biochemical investigations (255, 263, 264). This reagent and the derivatives are now commercially available as a SNAP-tag. Recently, AGT mutant proteins that react with O2-benzylcytosine have been selected and can provide a second protein, which can be expressed as a fusion and tagged in a similar way using derivatives of O2-benzylcytosine (265). This allows for simultaneous labeling with different probes (266) and should be a valuable addition to this technique.
10.2. AGT and ATL fusions with Endo V
Many archea that live under extreme conditions contain a protein termed AGTEndoV. This consists of a single polypeptide bifunctional protein having an N-terminal domain encoding an AGT activity and a C-terminal domain encoding an EndoV activity (267). EndoV is an endonuclease, which nicks DNA at the second phosphodiester bond 3′ to a deaminated base substrate, commencing an excision/polymerase repair pathway (268). Both AGT and Endo V functions are active when recombinant AGTendoV is assayed in vitro. AGTEndoV protected from both oxidative damage and MNNG when expressed in E. coli. In fact, it was actually slightly more effective in preventing mutations than human AGT expressed at the same level. This suggests that the combination of activities provides a more efficient repair system (267). One explanation for this, and for the combination of two DNA proteins in the same polypeptide chain, would be that the presence of two distinct DNA binding domains aids in the rapid scanning of DNA and the recognition of DNA lesions.
A number of archea genomes contain ATL EndoV fusions (234). This could also facilitate DNA scanning for lesions and the EndoV domain might have an XPG-like function contributing to NER in these organisms.
A protein termed c-AGT2 is present in C. elegans and related nematodes, which also contain a conventional AGT termed cAGT-1. The cAGT2 protein has AGT activity although it lacks the N-terminal domain of most AGTs. The region equivalent to the C-terminal domain of most AGTs, which contains the DNA binding and active site is located in the N-terminal region of c-AGT2. This is fused to a C-terminal domain that has a sequence resembling histone 1C (269). The histone domain may provide a structural role similar to that of the AGT N-terminal domain (65) but could also accelerate DNA scanning or play an entirely different role relating to gene expression.
11. Conclusions
The mechanism of action and function of AGTs to repair DNA and prevent mutations and cytotoxicity are now generally well understood although some fundamental questions remain. Answering these questions may provide significant general advances in understanding the responses to DNA damage as well as filling in these gaps concerning AGTs. The ability of AGTs to repair some O6-alkylguanine adducts in DNA so rapidly is a particularly interesting property, but the mechanism for such rapid scanning is not understood. The cooperative binding of AGT to DNA may aid in this scanning. Further study of this and of the AGTs or ATLs that occur as fusions with other proteins providing a second DNA binding domain, which are found in organisms that grow under extreme conditions, are needed. Also, the interaction of AGTs with other proteins is an important area in which experimental clarification is required. These proteins could be involved in DNA repair either by enhancing the scanning for lesions or via contributing to the removal and degradation of alkyl-AGT. They may also link AGT to a variety of other cellular functions.
The existence of ATLs and the evidence that O6-alkylguanines can be repaired efficiently by NER after recognition and DNA distortion brought about by the binding of ATLs may explain the absence of AGTs in some microorganisms and the ability of others to repair O6-alkyl adducts in DNA that are too large for the active site of their AGTs. Repair via the ATL/NER pathway may permit transcription-coupled repair of O6-alkylguanines, which does not occur with AGT alone. It is possible that in mammalian cells AGT itself might act as a recognition signal for some adducts such as S6-methylthioguanine and O6-hydroxyethylguanine. Clues provided by the studies of ATLs suggest that AGT might also be able to act to promote repair of adducts, which it binds to but does not repair well, is worth considering. However, it is still unclear why neither AGTs nor ATLs have been detected in plant cells and how plants respond to DNA damaging agents that form O6-alkylguanines.
The ability of bis-electrophiles to crosslink AGT to DNA not only indicates an additional route for their genotoxicity but also provides a useful method to generate protein-DNA crosslinks for experimental purposes in order to study their further processing, repair and mutagenic and cytotoxic properties. Not only does this reaction generate defined lesions each containing a known protein sequence but, by using AGT mutants and fusion to other polypeptides, these adducts can be manipulated experimentally. As long as the reactive Cys and the DNA binding domain of the AGT are maintained, any required protein sequence could be crosslinked in this way. The extent to which normal cellular levels of AGT contribute to the toxicity and mutagenicity of dihaloalkanes and other bis-electrophiles as compared to other routes of metabolism such as glutathione S-transferase conjugates, P450-dependent activation and direct formation of DNA adducts is not yet clear. The availability of mice that lack or overexpress AGT due to inactivation or increased expression of the MGMT gene provides a means to evaluate this. The importance of AGT-DNA crosslinks in the toxicity and mutagenicity of bis-electrophiles can only be demonstrated conclusively by conducting studies in both mutant mouse lines.
At present, despite a large number of publications, none of the AGT polymorphisms or very rare variants have been clearly linked to human disease or cancer therapy and there is no clear parallel between the properties imparted by these MGMT gene variations and the phenotypes attributed to them. This work may be clarified by studies with larger numbers and better-defined human populations that will enable confounding factors to be excluded. It is also possible that linkages between the polymorphisms and some other functions brought about either via AGT interactions or presently unknown gene linkages may contribute to the epidemiological findings.
It should be noted that the MGMT gene is very large (>170kb) and contains 4 long introns, each exceeding 40 kb. It is possible that some of these sequences have effects either on AGT expression or the expression of other genes. Global estimations of the methylation status of the MGMT gene promoter region are not likely to be useful as sole markers of the response to cancer therapy whereas actual measurements of AGT protein and activity, although more methodologically challenging, may be more helpful. The problems of cell heterogeneity in tumor tissues also raise difficulties in these studies. It has been shown that an experimental tumor in mice containing a small percentage of Mer+ tumor cells mixed with Mer− tumor cells becomes almost exclusively Mer+ after therapy with an alkylating agent. Nevertheless, it is possible that more detailed comparisons where both AGT protein levels and specific sites of gene methylation are correlated with therapeutic outcome using methylating or chloroethylating agents may allow refinement of this type of screening to identify patients with a good chance of response. These comparisons might also be useful for identifying patients who should be considered for treatment with AGT inhibitors. Similarly, analysis of selected and better-defined populations might also be helpful in establishing a protective role of AGT against human cancer.
The preliminary results suggesting a role of a mammalian AGT as a transcriptional regulator are intriguing and further study of this possibility is needed, particularly in the light of the clear evidence from bacteria that a zinc-conjugated alkylation site motif can be used as an activation switch for such regulation. The relatively rapid degradation of the alkyl-AGT may limit the extent to which this type of regulation can occur, but the t1/2 of the alkyl form is several hours, which, while unstable compared to the unmodified protein (t1/2 >24 h), is quite sufficient for such regulation. More details of the conformational changes in the alkylated form of AGT and its interaction with other proteins as well as the mechanism of ubiquitination and the sites of ubiquitin attachment would be very helpful.
O6-Benzylguanine is an established research tool to abolish AGT activity and its derivatives are also useful reagents for labeling the modified AGT polypeptide used for protein tagging. It remains to be seen whether O6-benzylguanine or related compounds will prove to be useful drugs to enhance chemotherapy or whether O6-benzylguanine-resistant AGT mutant vectors can be used clinically. However, these studies have provided important experimental evidence supporting the underlying concepts that will be a framework for future studies.
Acknowledgements
I thank Michael G. Fried, Thomas E. Spratt, Sreenivas Kanugula and Natalia A. Loktionova and my other collaborators and colleagues for their contributions to this work. I am one of the listed inventors on the patents describing O6-benzylguanine and related compounds and their ability to inactivate alkyltransferases and improve chemotherapy as well as a patent on O6-benzylguanine-resistant AGTs.
Funding Support. Work on AGT was supported by NIH grants CA-018138, CA-071976 and CA-097209.
References
- (1).Pegg AE, Dolan ME, Moschel RC. Structure, function and inhibition of O6-alkylguanine-DNA alkyltransferase. Progr. Nucleic Acids Res. Mol. Biol. 1995;51:167–223. doi: 10.1016/s0079-6603(08)60879-x. [DOI] [PubMed] [Google Scholar]
- (2).Pegg AE. Repair of O6-alkylguanine by alkyltransferases. Mutat. Res. 2000;462:83–100. doi: 10.1016/s1383-5742(00)00017-x. [DOI] [PubMed] [Google Scholar]
- (3).Margison G, Povey AC, Kaina B, Santibáñez-Koref MF. Variability and regulation of O6-alkylguanine-DNA alkyltransferase. Carcinogenesis. 2003;24:625–635. doi: 10.1093/carcin/bgg005. [DOI] [PubMed] [Google Scholar]
- (4).Gerson SL. MGMT: its role in cancer aetiology and cancer therapeutics. Nat. Rev. Cancer. 2004;4:296–307. doi: 10.1038/nrc1319. [DOI] [PubMed] [Google Scholar]
- (5).Mishina Y, Duguid EM, He C. Direct reversal of DNA alkylation damage. Chem. Rev. 2006;106:215–232. doi: 10.1021/cr0404702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Tubbs JL, Pegg AE, Tainer JA. DNA binding, nucleotide flipping, and the helix-turn-helix motif in base repair by O6-alkylguanine-DNA alkyltransferase and its implications for cancer chemotherapy. DNA Repair, (Amst.) 2007;6:1100–1115. doi: 10.1016/j.dnarep.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Eker AP, Quayle C, Chaves I, van der Horst GT. DNA repair in mammalian cells: Direct DNA damage reversal: elegant solutions for nasty problems. Cell. Mol. Life Sci. 2009;66:968–980. doi: 10.1007/s00018-009-8735-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Dalhus B, Laerdahl JK, Backe PH, Bjoras M. DNA base repair - recognition and initiation of catalysis. FEMS Microbiol. Rev. 2009;33:1044–1078. doi: 10.1111/j.1574-6976.2009.00188.x. [DOI] [PubMed] [Google Scholar]
- (9).Sharma S, Salehi F, Scheithauer BW, Rotondo F, Syro LV, Kovacs K. Role of MGMT in tumor development, progression, diagnosis, treatment and prognosis. Anticancer Res. 2009;29:3759–3768. [PubMed] [Google Scholar]
- (10).Shrivastav N, Li D, Essigmann JM. Chemical biology of mutagenesis and DNA repair: cellular responses to DNA alkylation. Carcinogenesis. 2010;31:59–70. doi: 10.1093/carcin/bgp262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Olsson M, Lindahl T. Repair of alkylated DNA in Escherichia coli. Methyl group transfer from O6-methylguanine to a protein cysteine residue. J. Biol. Chem. 1980;255:10569–10571. [PubMed] [Google Scholar]
- (12).Lindahl T, Demple B, Robins P. Suicide inactivation of the E. coli O6-methylguanine-DNA methyltransferase. EMBO J. 1982;1:1359–1363. doi: 10.1002/j.1460-2075.1982.tb01323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Samson L, Cairns J. A new pathway for DNA repair in Escherichia coli. Nature. 1977;267:281–283. doi: 10.1038/267281a0. [DOI] [PubMed] [Google Scholar]
- (14).Samson L. The suicidal DNA repair methyltransferases of microbes. Mol. Microbiol. 1992;6:825–831. doi: 10.1111/j.1365-2958.1992.tb01533.x. [DOI] [PubMed] [Google Scholar]
- (15).Potter PM, Wilkinson MC, Fitton J, Carr FJ, Brennand J, Cooper DP, Margison GP. Characterization and nucleotide sequence of ogt, the O6-alkylguanine-DNA-alkyltransferase gene of E. coli. Nucleic Acids Res. 1987;15:9177–9193. doi: 10.1093/nar/15.22.9177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Rebeck GW, Smith CM, Goad DL, Samson L. Characterization of the major DNA repair methyltransferase activity in unadapted Escherichia coli and identification of a similar activity in Salmonella typhimurium. J. Bacteriol. 1989;171:4563–4568. doi: 10.1128/jb.171.9.4563-4568.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Wilkinson MC, Potter PM, Cawkwell L, Georgiadis P, Patel D, Swann PF, Margison GP. Purification of the E. coli ogt gene product to homogeneity and its rate of action on O6-methylguanine, O6-ethylguanine and O4-methylthymine in dodecadeoxyribonucleotides. Nucleic Acids Res. 1989;17:8475–8484. doi: 10.1093/nar/17.21.8475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Rydberg B, Lindahl T. Nonenzymatic methylation of DNA by the intracellular methyl group donor S-adenosyl-L-methionine is a potentially mutagenic reaction. EMBO J. 1982;1:211–216. doi: 10.1002/j.1460-2075.1982.tb01149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Farmer PB. Studies using specific biomarkers for human exposure assessment to exogenous and endogenous chemical agents. Mutat. Res. 1999;428:69–81. doi: 10.1016/s1383-5742(99)00033-2. [DOI] [PubMed] [Google Scholar]
- (20).Pegg AE. Alkylation of rat liver DNA by dimethylnitrosamine: effect of dosage on O6-methylguanine levels. J. Natl. Cancer Inst. 1977;58:681–687. doi: 10.1093/jnci/58.3.681. [DOI] [PubMed] [Google Scholar]
- (21).Pegg AE, Hui G. Removal of methylated purines from rat liver DNA after administration of dimethylnitrosamine. Cancer Res. 1978;38:2011–2017. [PubMed] [Google Scholar]
- (22).Pegg AE. Dimethylnitrosamine inhibits enzymatic removal of O6-methylguanine from DNA. Nature, (London) 1978;274:182–184. doi: 10.1038/274182a0. [DOI] [PubMed] [Google Scholar]
- (23).Pegg AE. Enzymatic removal of O6-methylguanine from DNA by mammalian cell extracts. Biochem. Biophys. Res. Commun. 1978;84:166–173. doi: 10.1016/0006-291x(78)90278-4. [DOI] [PubMed] [Google Scholar]
- (24).Pegg AE, Perry W, Bennett RA. Partial hepatectomy increases the ability of rat liver extracts to catalyze removal of O6-methylguanine from alkylated DNA. Biochem. J. 1981;197:195–201. doi: 10.1042/bj1970195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Horiguchi M, Kim J, Matsunaga N, Kaji H, Egawa T, Makino K, Koyanagi S, Ohdo S. Glucocorticoid-dependent expression of O6-methylguanine-DNA methyltransferase gene modulates dacarbazine-induced hepatotoxicity in mice. J. Pharmacol. Exp. Ther. 2010;333:782–787. doi: 10.1124/jpet.110.165597. [DOI] [PubMed] [Google Scholar]
- (26).Pegg AE, Roberfroid M, von Bahr C, Foote RS, Mitra S, Bresil H, Likachev A, Montesano R. Removal of O6-methylguanine from DNA by human liver fractions. Proc. Natl. Acad. Sci. U.S.A. 1982;79:5162–5165. doi: 10.1073/pnas.79.17.5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Pegg AE, Wiest L, Foote RS, Mitra S, Perry W. Purification and properties of O6-methylguanine-DNA transmethylase from rat liver. J. Biol. Chem. 1983;258:2327–2333. [PubMed] [Google Scholar]
- (28).Koike G, Maki H, Takeya H, Hayakawa H, Sekiguchi M. Purification, structure and biochemical properties of human O6-methylguanine-DNA methyltransferase. J. Biol. Chem. 1990;265:14754–14762. [PubMed] [Google Scholar]
- (29).Tano K, Shiota S, Collier J, Foote RS, Mitra S. Isolation and structural characterization of a cDNA clone encoding the human DNA repair protein for O6-alkylguanine. Proc. Natl. Acad. Sci. U.S.A. 1990;87:686–690. doi: 10.1073/pnas.87.2.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Sakumi K, Shiraishi A, Hayakawa H, Sekiguchi M. Cloning and expression of cDNA for rat O6-methylguanine-DNA methyltransferase. Nucleic Acids Res. 1991;19:5597–5601. doi: 10.1093/nar/19.20.5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Morimoto K, Dolan ME, Scicchitano D, Pegg AE. Repair of O6-propylguanine and O6-butylguanine in DNA by O6-alkylguanine-DNA alkyltransferases from rat liver and E. coli. Carcinogenesis. 1985;6:1027–1031. doi: 10.1093/carcin/6.7.1027. [DOI] [PubMed] [Google Scholar]
- (32).Pegg AE, Scicchitano D, Dolan ME. Comparison of the rates of repair of O6-alkylguanines in DNA by rat liver and bacterial O6-alkylguanine-DNA alkyltransferase. Cancer Res. 1984;44:3806–3811. [PubMed] [Google Scholar]
- (33).Goodtzova K, Kanugula S, Edara S, Pauly GT, Moschel RC, Pegg AE. Repair of O6-benzylguanine by the Escherichia coli Ada and Ogt and the human O6-alkylguanine-DNA alkyltransferase. J. Biol. Chem. 1997;272:8332–8339. doi: 10.1074/jbc.272.13.8332. [DOI] [PubMed] [Google Scholar]
- (34).Mijal RS, Thomson NM, Moschel RC, Kanugula S, Fang Q, Pegg AE, Peterson LA. The repair of the tobacco-specific nitrosamine derived adduct, O6-[4-oxo-4-(3-pyridyl)butyl]guanine by O6-alkylguanine-DNA alkyltransferase variants. Chem. Res. Toxicol. 2004;17:424–434. doi: 10.1021/tx0342417. [DOI] [PubMed] [Google Scholar]
- (35).Coulter R, Blandino M, Tomlinson JM, Pauly GT, Krajewska M, Moschel RC, Peterson LA, Pegg AE, Spratt TE. Differences in the rate of repair of O6-alkylguanines in different sequence contexts by O6-alkylguanine-DNA alkyltransferase. Chem. Res. Toxicol. 2007;20:1966–1971. doi: 10.1021/tx700271j. [DOI] [PubMed] [Google Scholar]
- (36).Zang H, Fang Q, Pegg AE, Guengerich FP. Kinetic analysis of steps in the repair of damaged DNA by human O6-alkylguanine DNA-alkyltransferase. J. Biol. Chem. 2005;280:30873–30881. doi: 10.1074/jbc.M505283200. [DOI] [PubMed] [Google Scholar]
- (37).Loechler EL, Green CL, Essigmann JM. In vivo mutagenesis by O6-methylguanine built into a unique site in a viral genome. Proc. Natl. Acad. Sci. U.S.A. 1984;81:6271–6275. doi: 10.1073/pnas.81.20.6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Warren JJ, Forsberg LJ, Beese LS. The structural basis for the mutagenicity of O6-methylguanine lesions. Proc. Natl. Acad. Sci. U S A. 2006;103:19701–19706. doi: 10.1073/pnas.0609580103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Woodside AM, Guengerich FP. Effect of the O6-substituent on misincorporation kinetics catalyzed by DNA polymerases at O6-methylguanine and O6-benzylguanine. Biochemistry. 2002;41:1027–1038. doi: 10.1021/bi011495n. [DOI] [PubMed] [Google Scholar]
- (40).Choi JY, Chowdhury G, Zang H, Angel KC, Vu CC, Peterson LA, Guengerich FP. Translesion synthesis across O6-alkylguanine DNA adducts by recombinant human DNA polymerases. J. Biol. Chem. 2006;281:38244–38256. doi: 10.1074/jbc.M608369200. [DOI] [PubMed] [Google Scholar]
- (41).Pence MG, Egli M, Choi JY, Guengerich FP. Structural basis for proficient incorporation of dTTP opposite O6-methylguanine by human DNA polymerase {iota} J. Biol. Chem. 2010;285:40666–40672. doi: 10.1074/jbc.M110.183665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Burns JA, Dreij K, Cartularo L, Scicchitano DA. O6-Methylguanine induces altered proteins at the level of transcription in human cells. Nucleic Acids Res. 2010;38:8178–8187. doi: 10.1093/nar/gkq706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Klapacz J, Meira LB, Luchetti DG, Calvo JA, Bronson RT, Edelmann W, Samson LD. O6-Methylguanine-induced cell death involves exonuclease 1 as well as DNA mismatch recognition in vivo. Proc. Natl. Acad. Sci. U. S. A. 2009;106:576–581. doi: 10.1073/pnas.0811991106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Schroering AG, Kothandapani A, Patrick SM, Kaliyaperumal S, Sharma VP, Williams KJ. Prolonged cell cycle response of HeLa cells to low-level alkylation exposure. Cancer Res. 2009;69:6307–6314. doi: 10.1158/0008-5472.CAN-09-0899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Karran P. Mechanism of tolerance to DNA damaging therapeutic drugs. Carcinogenesis. 2001;22:1931–1937. doi: 10.1093/carcin/22.12.1931. [DOI] [PubMed] [Google Scholar]
- (46).Jiricny J. The multifaceted mismatch-repair system. Nat. Rev. Mol. Cell Biol. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- (47).Quiros S, Roos WP, Kaina B. Processing of O6-methylguanine into DNA double-strand breaks requires two rounds of replication whereas apoptosis is also induced in subsequent cell cycles. Cell Cycle. 2010;9:168–178. doi: 10.4161/cc.9.1.10363. [DOI] [PubMed] [Google Scholar]
- (48).Rajesh P, Rajesh C, Wyatt MD, Pittman DL. RAD51D protects against MLH1-dependent cytotoxic responses to O6-methylguanine. DNA Repair, (Amst.) 2010;9:458–467. doi: 10.1016/j.dnarep.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Kisby GE, Olivas A, Park T, Churchwell M, Doerge D, Samson LD, Gerson SL, Turker MS. DNA repair modulates the vulnerability of the developing brain to alkylating agents. DNA Repair, (Amst.) 2009;8:400–412. doi: 10.1016/j.dnarep.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Dumenco LL, Allay E, Norton K, Gerson SL. The prevention of thymic lymphomas in transgenic mice by human O6-alkylguanine-DNA alkyltransferases. Science. 1993;259:219–222. doi: 10.1126/science.8421782. [DOI] [PubMed] [Google Scholar]
- (51).Reese JS, Allay E, Gerson SL. Overexpression of human O6-alkylguanine DNA alkyltransferase, (AGT) prevents MNU induced lymphomas in heterozygous p53 deficient mice. Oncogene. 2001;20:5258–5263. doi: 10.1038/sj.onc.1204700. [DOI] [PubMed] [Google Scholar]
- (52).Nakatsuru Y, Matsukuma S, Nemoto N, Sugano H, Sekiguchi M, Ishikawa T. O6-methylguanine-DNA methyltransferase protects against nitrosamine-induced hepatocarcingenesis. Proc. Natl. Acad. Sci. U.S.A. 1993;90:6468–6472. doi: 10.1073/pnas.90.14.6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Becker K, Dosch J, Gregel CM, Martin BA, Kaina B. Targeted expression of human O6-methylguanine-DNA methyltransferase, (MGMT) in transgenic mice protects against tumor initiation in two-stage- skin carcinogenesis. Cancer Res. 1996;56:3244–3249. [PubMed] [Google Scholar]
- (54).Becker K, Gregel CM, Kaina B. The DNA repair protein O6-methylguanine-DNA methyltransferase protects against skin tumor formation induced by antineoplastic chloroethylnitrosourea. Cancer Res. 1997;57:3335–3338. [PubMed] [Google Scholar]
- (55).Zaidi NH, Pretlow TP, O’Riordan MA, Dumenco LL, Allay E, Gerson SL. Transgenic expression of human MGMT protects against azoxymethane-induced aberrant crypt foci and G to A mutations in the K-ras oncogene of mouse colon. Carcinogenesis. 1995;16:451–456. doi: 10.1093/carcin/16.3.451. [DOI] [PubMed] [Google Scholar]
- (56).Liu L, Qin X, Gerson SL. Reduced lung tumorigenesis in human methylguanine DNA-methyltransferase transgenic mice achieved by expression of transgene within the target cell. Carcinogenesis. 1999;20:279–284. doi: 10.1093/carcin/20.2.279. [DOI] [PubMed] [Google Scholar]
- (57).Iwakuma T, Sakumi K, Nakatsuru Y, Kawate H, Igarashi H, Shiraishi A, Tsuzuki T, Ishikawa T, Sekiguchi M. High incidence of nitrosamine-induced tumorigenesis in mice lacking DNA repair methyltransferase. Carcinogenesis. 1997;18:1631–1635. doi: 10.1093/carcin/18.8.1631. [DOI] [PubMed] [Google Scholar]
- (58).Bugni JM, Meira LB, Samson LD. Alkylation-induced colon tumorigenesis in mice deficient in the Mgmt and Msh6 proteins. Oncogene. 2009;28:734–741. doi: 10.1038/onc.2008.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Sakumi K, Shiraishi A, Shimizu S, Tsuzuki T, Ishikawa T, Sekiguchi M. Methylnitrosourea-induced tumorigenesis in MGMT gene knockout mice. Cancer Res. 1997;57:2415–2418. [PubMed] [Google Scholar]
- (60).Wibley JEA, Pegg AE, Moody PCE. Crystal structure of the human O6-alkylguanine-DNA alkyltransferase. Nucleic Acids Res. 2000;28:393–401. doi: 10.1093/nar/28.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Daniels DS, Mol CD, Arvai AS, Kanugula S, Pegg AE, Tainer JA. Active and alkylated human AGT structures: a novel zinc site, inhibitor and extrahelical binding. EMBO J. 2000;19:1719–1730. doi: 10.1093/emboj/19.7.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Daniels DS, Woo TT, Luu KX, Noll DM, Clarke ND, Pegg AE, Tainer JA. Novel modes of DNA binding and nucleotide flipping by the human DNA repair protein AGT. Nat. Struct. Mol. Biol. 2004;11:714–720. doi: 10.1038/nsmb791. [DOI] [PubMed] [Google Scholar]
- (63).Duguid EM, Rice PA, He C. The structure of the human AGT protein bound to DNA and its implications for damage detection. J. Mol. Biol. 2005;350:657–666. doi: 10.1016/j.jmb.2005.05.028. [DOI] [PubMed] [Google Scholar]
- (64).Hashimoto H, Inoue T, Nishioka M, Fujiwara S, Takagi M, Imanaka T, Kai Y. Hyperthermostable protein structure maintained by intra- and inter-helix ion pairs in archael O6-methylguanine-DNA methyltransferase. J. Mol. Biol. 1999;292:707–716. doi: 10.1006/jmbi.1999.3100. [DOI] [PubMed] [Google Scholar]
- (65).Fang Q, Kanugula S, Pegg AE. Function of domains of human O6-alkylguanine-DNA alkyltransferase. Biochemistry. 2005;44:15396–15405. doi: 10.1021/bi051460d. [DOI] [PubMed] [Google Scholar]
- (66).Hu J, Ma A, Dinner AR. A two-step nucleotide-flipping mechanism enables kinetic discrimination of DNA lesions by AGT. Proc. Natl. Acad. Sci. U. S. A. 2008;105:4615–4620. doi: 10.1073/pnas.0708058105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Guengerich FP, Fang Q, Liu L, Hachey DL, Pegg AE. O6-Alkylguanine-DNA alkyltransferase: Low pKa and reactivity of cysteine 145. Biochemistry. 2003;42:10965–10970. doi: 10.1021/bi034937z. [DOI] [PubMed] [Google Scholar]
- (68).Georgieva P, Himo F. Density functional theory study of the reaction mechanism of the DNA repairing enzyme alkylguanine alkyltransferase. Chem. Phys. Lett. 2008;463:214–218. [Google Scholar]
- (69).Shukla PK, Mishra PC. Repair of O6-methylguanine to guanine by cysteine in the absence and presence of histidine and by cysteine thiolate anion: a quantum chemical study. Phys Chem Chem Phys. 2009;11:8191–8202. doi: 10.1039/b908295f. [DOI] [PubMed] [Google Scholar]
- (70).Hou Q, Du L, Gao J, Liu Y, Liu C. QM/MM study on the reaction mechanism of O6-alkylguanine-DNA alkyltransferase. J. Phys. Chem. B. 2010;114:15296–15300. doi: 10.1021/jp106714m. [DOI] [PubMed] [Google Scholar]
- (71).Glassner BJ, Rasmussen LJ, Najarian MT, Posnick LM, Samson LD. Generation of a strong mutator phenotype in yeast by imbalanced base excision repair. Proc. Natl. Acad. Sci. U.S.A. 1998;95:9997–10002. doi: 10.1073/pnas.95.17.9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Posnick LM, Samson LD. Imbalanced base excision repair increased spontaneous mutation and alkylation sensitivity in Escherichia coli. J. Bacteriol. 1999;181:6763–6771. doi: 10.1128/jb.181.21.6763-6771.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Berdal KG, Johansen RF, Seeberg E. Release of normal bases from intact DNA by a native DNA repair enzyme. EMBO J. 1998;17:363–367. doi: 10.1093/emboj/17.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Nilsen H, Krokan HE. Base excision repair in a network of defence and tolerance. Carcinogenesis. 2001;22:987–998. doi: 10.1093/carcin/22.7.987. [DOI] [PubMed] [Google Scholar]
- (75).Klapacz J, Lingaraju GM, Guo HH, Shah D, Moar-Shoshani A, Loeb LA, Samson LD. Frameshift mutagenesis and microsatellite instability induced by human alkyladenine DNA glycosylase. Mol. Cell. 2010;37:843–853. doi: 10.1016/j.molcel.2010.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Lin Y, Zhao T, Jian X, Farooqui Z, Qu X, He C, Dinner AR, Scherer NF. Using the bias from flow to elucidate single DNA repair protein sliding and interactions with DNA. Biophys. J. 2009;96:1911–1917. doi: 10.1016/j.bpj.2008.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Luu KX, Kanugula S, Pegg AE, Pauly GT, Moschel RC. Repair of oligodeoxyribonucleotides by O6-alkylguanine-DNA alkyltransferase. Biochemistry. 2002;41:8689–8697. doi: 10.1021/bi025857i. [DOI] [PubMed] [Google Scholar]
- (78).Zhao T, Dinner AR. Apparent directional scanning for DNA repair. Biophys. J. 2008;94:47–52. doi: 10.1529/biophysj.107.110619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Rasimas JJ, Kar SR, Pegg AE, Fried MG. Interactions of human O6-alkylguanine-DNA alkyltransferase, (AGT) with short single-stranded DNAs. J. Biol. Chem. 2007;282:3357–3366. doi: 10.1074/jbc.M608876200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Melikishvili M, Rasimas JJ, Pegg AE, Fried MG. Interactions of human O6-alkylguanine-DNA alkyltransferase, (AGT) with short double-stranded DNAs. Biochemistry. 2008;47:13754–13763. doi: 10.1021/bi801666c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Adams CA, Melikishvili M, Rodgers DW, Rasimas JJ, Pegg AE, Fried MG. Topologies of complexes containing O6-alkylguanine-DNA alkyltransferase and DNA. J. Mol. Biol. 2009;389:248–263. doi: 10.1016/j.jmb.2009.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Niture SK, Doneanu CE, Velu CS, Bailey NI, Srivenugopal KS. Proteomic analysis of human O6-methylguanine-DNA methyltransferase by affinity chromatography and tandem mass spectrometry. Biochem. Biophys. Res. Commun. 2005;337:1176–1184. doi: 10.1016/j.bbrc.2005.09.177. [DOI] [PubMed] [Google Scholar]
- (83).Srivenugopal KS, Ali-Osman F. The DNA repair protein, O6-methylguanine-DNA methyltransferase is a proteolytic target for the E6 human papillomavirus oncoprotein. Oncogene. 2002;21:5940–5945. doi: 10.1038/sj.onc.1205762. [DOI] [PubMed] [Google Scholar]
- (84).Philip S, Swaminathan S, Kuznetsov SG, Kanugula S, Biswas K, Chang S, Loktionova NA, Haines DC, Kaldis P, Pegg AE, Sharan SK. Degradation of BRCA2 in alkyltransferase-mediated DNA repair and its clinical implications. Cancer Res. 2008;68:9973–9981. doi: 10.1158/0008-5472.CAN-08-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Hazra TK, Roy R, Biswas T, Grabowski DT, Pegg AE, Mitra S. Specific recognition of O6-methylguanine in DNA by active site mutants of human O6-methylguanine-DNA methyltransferase. Biochemistry. 1997;36:5769–5776. doi: 10.1021/bi963085i. [DOI] [PubMed] [Google Scholar]
- (86).Pegg AE, Morimoto K, Dolan ME. Investigation of the specificity of O6-alkylguanine-DNA alkyltransferase. Chem. Biol. Interact. 1988;65:275–281. doi: 10.1016/0009-2797(88)90112-3. [DOI] [PubMed] [Google Scholar]
- (87).Pegg AE, Wiest L, Mummert C, Stine L, Moschel RC, Dolan ME. Use of antibodies to human O6-alkylguanine-DNA alkyltransferase to study the content of this protein in cells treated with O6-benzylguanine or N-methyl-N′-nitro-N-nitrosoguanidine. Carcinogenesis. 1991;12:1679–1683. doi: 10.1093/carcin/12.9.1679. [DOI] [PubMed] [Google Scholar]
- (88).Srivenugopal KS, Yuan XH, Friedman HS, Ali-Osman F. Ubiquitination-dependent proteolysis of O6-methylguanine-DNA alkyltransferase in human and murine tumor cells following inactivation with O6-benzylguanine or 1,3 bis(2-chloroethyl)-1-nitrosourea. Biochemistry. 1996;35:1328–1334. doi: 10.1021/bi9518205. [DOI] [PubMed] [Google Scholar]
- (89).Xu-Welliver M, Pegg AE. Degradation of the alkylated form of the DNA repair protein O6-alkylguanine-DNA alkyltransferase. Carcinogenesis. 2002;23:823–830. doi: 10.1093/carcin/23.5.823. [DOI] [PubMed] [Google Scholar]
- (90).Edara S, Kanugula S, Pegg AE. Expression of the inactive C145A mutant human O6-alkylguanine-DNA alkyltransferase in E. coli increases cell killing and mutations by N-methyl-N′-nitro-N-nitrosoguanidine. Carcinogenesis. 1999;20:103–108. doi: 10.1093/carcin/20.1.103. [DOI] [PubMed] [Google Scholar]
- (91).Rasimas JJ, Dalessio PA, Ropson IJ, Pegg AE, Fried MG. Active-site alkylation destabilizes human O(6)-alkylguanine DNA alkyltransferase. Protein Sci. 2004;13:301–305. doi: 10.1110/ps.03319404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (92).Rasimas JJ, Pegg AE, Fried MG. DNA binding mechanism of O6-alkylguanine-DNA alkyltransferase: Effects of protein- and DNA-alkylation on complex stability. J. Biol. Chem. 2003;278:7973–7980. doi: 10.1074/jbc.M211854200. [DOI] [PubMed] [Google Scholar]
- (93).Hwang CS, Shemorry A, Varshavsky A. Two proteolytic pathways regulate DNA repair by cotargeting the Mgt1 alkylguanine transferase. Proc. Natl. Acad. Sci. U. S. A. 2009;106:2142–2147. doi: 10.1073/pnas.0812316106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (94).Remington M, Chtchetinin J, Ancheta K, Nghiemphu PL, Cloughesy T, Lai A. The L84F polymorphic variant of human O6-methylguanine-DNA methyltransferase alters stability in U87MG glioma cells but not temozolomide sensitivity. Neuro. Oncol. 2009;11:22–32. doi: 10.1215/15228517-2008-080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (95).Dosanjh MK, Essigmann JM, Goodman MF, Singer B. Comparative efficiency of forming m4T•G versus m4T•A base pairs at a unique site by use of Escherichia coli DNA polymerase I, (Klenow fragment) and Drosophila melanogaster polymerase α-primase complex. Biochemistry. 1990;29:4698–4702. doi: 10.1021/bi00471a026. [DOI] [PubMed] [Google Scholar]
- (96).Singer B, Dosanjh MK. Site-directed mutagenesis for quantitations of base-base interactions at defined sites. Mutat. Res. 1990;233:45–51. doi: 10.1016/0027-5107(90)90150-3. [DOI] [PubMed] [Google Scholar]
- (97).Preston BD, Singer B, Loeb LA. Comparison of the relative mutagenicities of O-alkylthymines site-specifically incorporated into fX174 DNA. J. Biol. Chem. 1987;262:13821–13827. [PubMed] [Google Scholar]
- (98).Pauly GT, Hughes SH, Moschel RC. Comparison of mutagenesis by O6-methyl- and O6-ethylguanine and O4-methylthymine in Escherichia coli using double-stranded and gapped plasmids. Carcinogenesis. 1998;19:457–461. doi: 10.1093/carcin/19.3.457. [DOI] [PubMed] [Google Scholar]
- (99).Pauly GT, Moschel RC. Mutagenesis by O6-methyl-, O6-ethyl-, and O6-benzylguanine and O4-methylthymine in human cells: effects of O6-alkylguanine-DNA alkyltransferase and mismatch repair. Chem. Res. Toxicol. 2001;14:894–900. doi: 10.1021/tx010032f. [DOI] [PubMed] [Google Scholar]
- (100).Harris LC, Margison GP. Expression in mammalian cells of the Escherichia coli O6-alkylguanine-DNA-alkyltransferase gene Ogt reduces the toxicity of alkylnitrosoureas. Br. J. Cancer. 1993;67:1196–1202. doi: 10.1038/bjc.1993.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (101).Sassanfar M, Dosanjh MK, Essigmann JM, Samson L. Relative efficiencies of the bacterial, yeast, and human DNA methyltransferases for the repair of O6-methylguanine and O4-methylthymine. J. Biol. Chem. 1991;266:2767–2771. [PubMed] [Google Scholar]
- (102).Graves RJ, Li BFL, Swann P. Repair of O6-methylguanine, O6-ethylguanine, O6-isopropylguanine and O4-methylthymine in synthetic oligodeoxynucleotides by Escherichia coli ada gene O6-alkylguanine-DNA-alkyltransferase. Carcinogenesis. 1989;10:661–666. doi: 10.1093/carcin/10.4.661. [DOI] [PubMed] [Google Scholar]
- (103).Paalman SR, Noll DM, Clarke ND. Formation of a covalent complex between methylguanine methyltransferase and DNA via disulfide bond formation between the active site cysteine and thiol-containing analog of guanine. Nucleic Acids Res. 1997;25:1795–1801. doi: 10.1093/nar/25.9.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (104).Samson L, Han S, Marquis JC, Rasmussen LJ. Mammalian DNA repair methyltransferases shield O4-methylthymine from nucleotide excision repair. Carcinogenesis. 1997;18:919–924. doi: 10.1093/carcin/18.5.919. [DOI] [PubMed] [Google Scholar]
- (105).O’Toole SM, Pegg AE, Swenberg JA. Repair of O6-methylguanine and O4-methylthymidine in F344 rat liver following treatment with 1,2-dimethylhydrazine and O6-benzylguanine. Cancer Res. 1993;53:3895–3898. [PubMed] [Google Scholar]
- (106).Fang Q, Kanugula S, Tubbs JL, Tainer JA, Pegg AE. Repair of O4-alkylthymine by O6-alkylguanine-DNA alkyltransferases. J.Biol. Chem. 2010;285:8185–8195. doi: 10.1074/jbc.M109.045518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (107).Encell LP, Loeb LA. Redesigning the substrate specificity of human O6-alkylguanine-DNA alkyltransferase. Mutants with enhanced repair of O4-methylthymine. Biochemistry. 1999;38:12097–12103. doi: 10.1021/bi9913606. [DOI] [PubMed] [Google Scholar]
- (108).Encell LP, Loeb l. A. Enhanced in vivo repair of O4-methylthymine by a mutant human DNA alkyltransferase. Carcinogenesis. 2000;21:1397–1402. [PubMed] [Google Scholar]
- (109).Bronstein MS, Cochrane JE, Craft TR, Swenberg JA, Skopek TR. Toxicity, mutagenicity and mutational spectra of N-ethyl-N-nitrosourea in human cell lines with different DNA repair phenotypes. Cancer Res. 1991;51:5188–5197. [PubMed] [Google Scholar]
- (110).Bronstein MS, Skopek TR, Swenberg JA. Efficient repair of O6-ethylguanine, but not O4-ethylthymine or O2-ethylthymine, is dependent upon O6-alkylguanine-DNA alkyltransferase and nucleotide excision repair activities in human cells. Cancer Res. 1992;52:2008–2011. [PubMed] [Google Scholar]
- (111).Dolan ME, Scicchitano D, Singer B, Pegg AE. Comparison of repair of methylated pyrimidines in poly(dT) by extracts from rat liver and Escherichia coli. Biochem. Biophys. Res. Commun. 1984;123:324–330. doi: 10.1016/0006-291x(84)90416-9. [DOI] [PubMed] [Google Scholar]
- (112).Margison GP, Santibáñez-Koref MF. O6-Alkylguanine-DNA alkyltransferase: role in carcinogenesis and chemotherapy. BioEssays. 2002;24:255–266. doi: 10.1002/bies.10063. [DOI] [PubMed] [Google Scholar]
- (113).Erickson LC, Laurent G, Sharkey NA, Kohn KW. DNA cross-linking and monoadduct repair in nitrosourea-treated human tumor cells. Nature. 1980;288:727–729. doi: 10.1038/288727a0. [DOI] [PubMed] [Google Scholar]
- (114).Brent TP. Suppression of cross-link formation in chloroethylnitrosourea-treated DNA by an activity in extracts of human leukemic lymphoblasts. Cancer Res. 1984;44:1887–1892. [PubMed] [Google Scholar]
- (115).Dolan ME, Moschel RC, Pegg AE. Depletion of mammalian O6-alkylguanine-DNA alkyltransferase activity by O6-benzylguanine provides a means to evaluate the role of this protein in protection against carcinogenic and therapeutic alkylating agents. Proc. Natl. Acad. Sci. U. S. A. 1990;87:5368–5372. doi: 10.1073/pnas.87.14.5368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (116).Dolan ME, Pegg AE. O6-Benzylguanine and its role in chemotherapy. Clin. Cancer Res. 1997;3:837–847. [PubMed] [Google Scholar]
- (117).Ishiguro K, Shyam K, Penketh PG, Sartorelli AC. Role of O6-alkylguanine-DNA alkyltransferase in the cytotoxic activity of cloretazine. Mol. Cancer Ther. 2005;4:1755–1763. doi: 10.1158/1535-7163.MCT-05-0169. [DOI] [PubMed] [Google Scholar]
- (118).Ishiguro K, Zhu YL, Shyam K, Penketh PG, Baumann RP, Sartorelli AC. Quantitative relationship between guanine O6-alkyl lesions produced by Onrigin and tumor resistance by O6-alkylguanine-DNA alkyltransferase. Biochem. Pharmacol. 2010;80:1317–1325. doi: 10.1016/j.bcp.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (119).Tong WP, Kirk MC, Ludlum DB. Formation of the cross-link 1-[N3-deoxycytidyl],2-N1-deoxyguanosinyl]-ethane in DNA treated with N,N1-bis(2-chloroethyl)-N-nitrosourea. Cancer Res. 1982;42:3102–3105. [PubMed] [Google Scholar]
- (120).Ludlum DB. The chloroethylnitrosoureas: Sensitivity and resistance to cancer chemotherapy at the molecular level. Cancer Invest. 1997;15:588–598. doi: 10.3109/07357909709047601. [DOI] [PubMed] [Google Scholar]
- (121).Gonzaga PE, Potter PM, Niu T, Yu D, Ludlum DB, Rafferty JA, Margison GP, Brent TP. Identification of the cross-link between human O6-methylguanine-DNA methyltransferase and chloroethylnitrosourea-treated DNA. Cancer Res. 1992;52:6052–6058. [PubMed] [Google Scholar]
- (122).Fang Q, Noronha AM, Murphy SP, Wilds CJ, Tubbs JL, Tainer JA, Chowdhury G, Guengerich FP, Pegg AE. Repair of O6-G-alkyl-O6-G interstrand cross-links by human O6-alkylguanine-DNA alkyltransferase. Biochemistry. 2008;47:10892–10903. doi: 10.1021/bi8008664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (123).McManus FP, Fang Q, Booth JD, Noronha AM, Pegg AE, Wilds CJ. Synthesis and characterization of an O6-2′-deoxyguanosine-alkyl-O6-2′-deoxyguanosine interstrand cross-link in a 5′-GNC motif and repair by human O6-alkylguanine-DNA alkyltransferase. Org. Biomol. Chem. 2010;8:4414–4426. doi: 10.1039/c0ob00093k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (124).Guza R, Pegg AE, Tretyakova N. Effects of sequence context on O6-alkylguanine-DNA alkyltransferase repair of O6-alkyldeoxyguanosine adducts. In: Stone MP, editor. Structural biology of DNA damage and repair. Vol 1041. ACS; Washington DC: 2010. pp. 73–101. (ACS Symposium Series). [Google Scholar]
- (125).Dolan ME, Oplinger M, Pegg AE. Sequence specificity of guanine alkylation and repair. Carcinogenesis. 1988;9:2139–2143. doi: 10.1093/carcin/9.11.2139. [DOI] [PubMed] [Google Scholar]
- (126).Guza R, Rajesh M, Fang Q, Pegg AE, Tretyakova N. Kinetics of O6-methyl-2′-deoxyguanosine repair by O6-alkylguanine DNA alkyltransferase within K-ras gene-derived DNA sequences. Chem. Res. Toxicol. 2006;19:531–538. doi: 10.1021/tx050348d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (127).Bentivegna SS, Bresnick E. Inhibition of human O6-methylguanine-DNA methyltransferase by 5-methylcytosine. Cancer Res. 1994;54:327–329. [PubMed] [Google Scholar]
- (128).Guza R, Ma L, Fang Q, Pegg AE, Tretyakova N. Cytosine methylation effects on the repair of O6-methylguanines within CG dinucleotides. J. Biol. Chem. 2009;284:22601–22610. doi: 10.1074/jbc.M109.000919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (129).Mijal RS, Kanugula S, Vu CC, Fang Q, Pegg AE, Peterson LA. DNA sequence context affects repair of the tobacco-specific adduct O6-[4-oxo-4-(3-pyridyl)butyl]guanine by human O6-alkylguanine-DNA alkyltransferases. Cancer Res. 2006;66:4968–4974. doi: 10.1158/0008-5472.CAN-05-3803. [DOI] [PubMed] [Google Scholar]
- (130).Hecht SS. Progress and challenges in selected areas of tobacco carcinogenesis. Chem. Res. Toxicol. 2008;21:160–171. doi: 10.1021/tx7002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (131).Abril N, Luque-Romero FL, Prito-Alamo MJ, Margison GP, Pueyo C. Ogt Alkyltransferase enhances dibromoalkane mutagenicity in excision repair-deficient Escherichia coli K-12. Mol. Carcinog. 1995;12:110–117. doi: 10.1002/mc.2940120208. [DOI] [PubMed] [Google Scholar]
- (132).Abril N, Luque-Romero L, Prieto-Alamo M-J, Rafferty JA, Margison GP, Pueyo C. Bacterial and mammalian DNA alkyltransferases sensitize Escherichia coli to the lethal and mutagenic effects of dibromoalkanes. Carcinogenesis. 1997;18:1883–1888. doi: 10.1093/carcin/18.10.1883. [DOI] [PubMed] [Google Scholar]
- (133).Liu H, Xu-Welliver M, Pegg AE. The role of human O6-alkylguanine-DNA alkyltransferase in promoting 1,2-dibromoethane-induced genotoxicity in Escherichia coli. Mutat. Res. 2000;452:1–10. doi: 10.1016/s0027-5107(00)00062-2. [DOI] [PubMed] [Google Scholar]
- (134).Liu L, Pegg AE, Williams KM, Guengerich FP. Paradoxical enhancement of the toxicity of 1,2-dibromoethane by O6-alkylguanine-DNA alkyltransferase. J. Biol. Chem. 2002;277:37920–37928. doi: 10.1074/jbc.M205548200. [DOI] [PubMed] [Google Scholar]
- (135).Liu L, Hachey DL, Valadez G, Williams KM, Guengerich FP, Loktionova NA, Kanugula S, Pegg AE. Characterization of a mutagenic DNA adduct formed from 1,2-dibromoethane by O6-alkylguanine-DNA alkyltransferase. J. Biol. Chem. 2004;279:4250–4259. doi: 10.1074/jbc.M311105200. [DOI] [PubMed] [Google Scholar]
- (136).Guengerich FP. Principles of covalent binding of reactive metabolites and examples of activation of bis-electrophiles by conjugation. Arch. Biochem. Biophys. 2005;433:369–378. doi: 10.1016/j.abb.2004.07.035. [DOI] [PubMed] [Google Scholar]
- (137).Liu L, Williams KM, Guengerich FP, Pegg AE. O6-Alkylguanine-DNA alkyltransferase has opposing effects in modulating the genotoxicity of dibromomethane and bromomethyl acetate. Chem. Res. Toxicol. 2004;17:742–752. doi: 10.1021/tx049958o. [DOI] [PubMed] [Google Scholar]
- (138).Valadez JG, Liu L, Loktionova NA, Guengerich FP, Pegg AE. Activation of bis-electrophiles to mutagenic conjugates by human O6-alkylguanine-DNA alkyltransferase. Chem. Res. Toxicol. 2004;17:972–982. doi: 10.1021/tx049897u. [DOI] [PubMed] [Google Scholar]
- (139).Liu L, Watanabe K, Fang Q, Williams KM, Guengerich FP, Pegg AE. Effect of alterations of key active site residues in O6-alkylguanine-DNA alkyltransferase on Its ability to modulate the genotoxicity of 1,2-dibromoethane. Chem. Res. Toxicol. 2007;20:155–163. doi: 10.1021/tx600257g. [DOI] [PubMed] [Google Scholar]
- (140).Loeber R, Rajesh M, Fang Q, Pegg AE, Tretyakova N. Cross-Linking of the human DNA repair protein O6-alkylguanine DNA alkyltransferase to DNA in the presence of 1,2,3,4-diepoxybutane. Chem. Res. Toxicol. 2006;19:645–654. doi: 10.1021/tx0600088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (141).Kalapila AG, Loktionova NA, Pegg AE. Alkyltransferase-mediated toxicity of 1,3-butadiene diepoxide. Chem. Res. Toxicol. 2008;21:1851–1861. doi: 10.1021/tx800178t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (142).Loeber R, Michaelson E, Fang Q, Campbell C, Pegg AE, Tretyakova N. Cross-linking of the DNA repair protein O6-alkylguanine DNA alkyltransferase to DNA in the presence of antitumor nitrogen mustards. Chem. Res. Toxicol. 2008;21:787–795. doi: 10.1021/tx7004508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (143).Kalapila AG, Loktionova NA, Pegg AE. Effect of O6-alkylguanine-DNA alkyltransferase on genotoxicity of epihalohydrins. Environ. Mol. Mutagen. 2009;50:502–514. doi: 10.1002/em.20491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (144).Seneviratne U, Antsypovich S, Goggin M, Dorr DQ, Guza R, Moser A, Thompson C, York DM, Tretyakova N. Exocyclic deoxyadenosine adducts of 1,2,3,4-diepoxybutane: synthesis, structural elucidation, and mechanistic studies. Chem. Res. Toxicol. 2010;23:118–133. doi: 10.1021/tx900312e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (145).Abril N, Margison GP. Mammalian cells expressing Escherichia coli O6-alkylguanine-DNA alkyltransferases are hypersenstivie to dibromoalkanes. Chem. Res. Toxicol. 1999;12:544–551. doi: 10.1021/tx980250h. [DOI] [PubMed] [Google Scholar]
- (146).Kalapila AG, Pegg AE. Alkyltransferase-mediated toxicity of bis-electrophiles in mammalian cells. Mutat. Res. 2010;684:35–42. doi: 10.1016/j.mrfmmm.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (147).Barker S, Weinfeld M, Murray D. DNA-protein crosslinks: their induction, repair, and biological consequences. Mutat. Res. 2005;589:111–135. doi: 10.1016/j.mrrev.2004.11.003. [DOI] [PubMed] [Google Scholar]
- (148).Wood RD. Mammalian nucleotide excision repair proteins and interstrand crosslink repair. Environ. Mol. Mutagen. 2010;51:520–526. doi: 10.1002/em.20569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (149).Loeber RL, Michaelson-Richie ED, Codreanu SG, Liebler DC, Campbell CR, Tretyakova NY. Proteomic analysis of DNA-protein cross-linking by antitumor nitrogen mustards. Chem. Res. Toxicol. 2009;22:1151–1162. doi: 10.1021/tx900078y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (150).Michaelson-Richie ED, Loeber RL, Codreanu SG, Ming X, Liebler DC, Campbell C, Tretyakova NY. DNA-protein cross-linking by 1,2,3,4-diepoxybutane. J. Proteome Res. 2010;9:4356–4367. doi: 10.1021/pr1000835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (151).Loecken EM, Guengerich FP. Reactions of glyceraldehyde 3-phosphate dehydrogenase sulfhydryl groups with bis-electrophiles produce DNA-protein cross-links but not mutations. Chem. Res. Toxicol. 2008;21:453–458. doi: 10.1021/tx7003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (152).Loecken EM, Dasari S, Hill S, Tabb DL, Guengerich FP. The bis-electrophile diepoxybutane cross-links DNA to human histones but does not result in enhanced mutagenesis in recombinant systems. Chem. Res. Toxicol. 2009;22:1069–1076. doi: 10.1021/tx900037u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (153).Krokan H, Grafstrom RC, Sundqvist K, Esterbauer H, Harris CC. Cytotoxicity, thiol depletion and inhibition of O6-methylguanine-DNA methyltransferase by various aldehydes in cultured human bronchial fibroblasts. Carcinogenesis. 1985;6:1755–1759. doi: 10.1093/carcin/6.12.1755. [DOI] [PubMed] [Google Scholar]
- (154).Grafstrom RC, Pegg AE, Harris CC, Sundqvist K, Krokan H. O6-Methylguanine-DNA methyltransferase activity and aldehyde induced inhibition of O6-methylguanine repair in human lung cells. In: Myrnes B, Krokan H, editors. Repair of DNA lesions introduced by N-Nitroso Compounds. Norwegian University Press; Oslo, Norway: 1986. pp. 154–175. [Google Scholar]
- (155).Hayes RB, Klein S, Suruda A, Schulte P, Boeniger M, Stewart P, Livingston GK. O6-Alkylguanine DNA alkyltransferase activity in student embalmers. Am. J. Ind. Med. 1997;31:361–365. doi: 10.1002/(sici)1097-0274(199703)31:3<361::aid-ajim13>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- (156).Brent TP. Inactivation of purified human O6-alkylguanine-DNA alkyltransferase by alkylating agents or alkylated DNA. Cancer Res. 1986;46:2320–2323. [PubMed] [Google Scholar]
- (157).Liu L, Xu-Welliver M, Kanugula S, Pegg AE. Inactivation and degradation of O6-alkylguanine-DNA alkyltransferase after reaction with nitric oxide. Cancer Res. 2002;62:3037–3043. [PubMed] [Google Scholar]
- (158).Wei W, Li B, Hanes MA, Kakar S, Chen X, Liu L. S-nitrosylation from GSNOR deficiency impairs DNA repair and promotes hepatocarcinogenesis. Sci. Transl. Med. 2010;2:19ra13. doi: 10.1126/scitranslmed.3000328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (159).Gefen N, Brkic G, Galron D, Priel E, Ozer J, Benharroch D, Gopas J. Acquired resistance to 6-thioguanine in melanoma cells involves the repair enzyme O6-methylguanine-DNA methyltransferase, (MGMT) Cancer Biol. Ther. 2010;9:49–55. doi: 10.4161/cbt.9.1.10285. [DOI] [PubMed] [Google Scholar]
- (160).Swann PF, Waters TR, Moulton DC, Xu YZ, Zheng Q, Edwards M, Mace E. Role of postreplicative DNA mismatch repair in the cytotoxic action of thioguanine. Science. 1996;273:1109–1111. doi: 10.1126/science.273.5278.1109. [DOI] [PubMed] [Google Scholar]
- (161).Waters TR, Swann PF. Cytotoxic mechanism of 6-thioguanine: hMutSa, the human mismatch binding heterodimer, binds to DNA containing S6-methylguanine. Biochemistry. 1997;36:2501–2506. doi: 10.1021/bi9621573. [DOI] [PubMed] [Google Scholar]
- (162).Spratt TE, de los Santos H. Reaction of O6-alkylguanine-DNA alkyltransferase with O6-methylguanine analogues: evidence that the oxygen of O6-methylguanine is protonated by the protein to effect methyl transfer. Biochemistry. 1992;31:3688–3694. doi: 10.1021/bi00129a018. [DOI] [PubMed] [Google Scholar]
- (163).Spratt TE, Campbell CR. Synthesis of oligonucleotides containing analogs of O6-methylguanine and reaction with O6-alkylguanine-DNA alkyltransferase. Biochemistry. 1994;33:11364–11371. doi: 10.1021/bi00203a035. [DOI] [PubMed] [Google Scholar]
- (164).Pegg AE, Xu-Welliver M, Loktionova NA. The DNA repair protein O6-alkylguanine-DNA alkyltransferase as a target for cancer chemotherapy. In: Ehrlich M, editor. DNA alterations in cancer: genetic and epigenetic changes. Eaton Publishing; Natick, MA: 2000. pp. 471–488. E. [Google Scholar]
- (165).Pegg AE, Dolan ME. Overcoming resistance to alkylating agents by inhibitors of O6-alkylguanine-DNA alkyltransferase. In: Panasci LC, Alaoui-Jamali M, editors. DNA Repair in Cancer Therapy. Humana Press; NJ: 2004. pp. 143–177. [Google Scholar]
- (166).Liu L, Gerson SL. Targeted modulation of MGMT: clinical implications. Clin. Cancer Res. 2006;12:328–331. doi: 10.1158/1078-0432.CCR-05-2543. [DOI] [PubMed] [Google Scholar]
- (167).Kaina B, Margison GP, Christmann M. Targeting O6-methylguanine-DNA methyltransferase with specific inhibitors as a strategy in cancer therapy. Cell. Mol. Life Sci. 2010;67:3663–3681. doi: 10.1007/s00018-010-0491-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (168).Pegg AE, Boosalis M, Samson L, Moschel RC, Byers TL, Swenn K, Dolan ME. Mechanism of inactivation of human O6-alkylguanine-DNA alkyltransferase by O6-benzylguanine. Biochemistry. 1993;32:11998–12006. doi: 10.1021/bi00096a009. [DOI] [PubMed] [Google Scholar]
- (169).Xu-Welliver M, Kanugula S, Pegg AE. Isolation of human O6-alkylguanine-DNA alkyltransferase mutants highly resistant to inactivation by O6-benzylguanine. Cancer Res. 1998;58:1936–1945. [PubMed] [Google Scholar]
- (170).McElhinney RS, Donnelly DJ, McCormick JE, Kelly J, Watson AJ, Rafferty JA, Elder RH, Middleton MR, Willington MA, McMurry TBH, Margison GP. Inactivation of O6-alkylguanine-DNA alkyltransferase 1. Novel O6-(hetarylmethyl)guanines having basic rings in the side chain. J. Med. Chem. 1998;41:5265–5271. doi: 10.1021/jm9708644. [DOI] [PubMed] [Google Scholar]
- (171).Nelson ME, Loktionova NA, Pegg AE, Moschel RC. Amino-O4-benzylpteridine derivatives: potent inactivators of O6-alkylguanine-DNA alkyltransferase. J. Med. Chem. 2004;47:3887–3891. doi: 10.1021/jm049758+. [DOI] [PubMed] [Google Scholar]
- (172).Pauly GT, Loktionova NA, Fang Q, Vankayala SL, Guida WC, Pegg AE. Substitution of aminomethyl at the meta-position enhances the Inactivation of O6-alkylguanine-DNA alkyltransferase by O6-benzylguanine. J. Med. Chem. 2008;51:7144–7153. doi: 10.1021/jm800675p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (173).Javanmard S, Loktionova NA, Fang Q, Pauly GT, Pegg AE, Moschel RC. Inactivation of O6-alkylguanine-DNA alkyltransferase by folate esters of O6-benzyl-2′-deoxyguanosine and of O6-[4-(hydroxymethyl)benzyl]guanine. J. Med. Chem. 2007;50:5193–5201. doi: 10.1021/jm0705859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (174).Middleton MR, Kelly J, Thatcher N, Donnelly DJ, McElhinney RS, McMurry TBH, McCormick JE, Margison GP. O6-(4-Bromothenyl)guanine improves the therapeutic index of temozolomide against A375M melanoma xenografts. Int. J. Cancer. 2000;85:248–252. doi: 10.1002/(sici)1097-0215(20000115)85:2<248::aid-ijc16>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- (175).Quinn JA, Jiang SX, Carter J, Reardon DA, Desjardins A, Vredenburgh JJ, Rich JN, Gururangan S, Friedman AH, Bigner DD, Sampson JH, McLendon RE, Herndon JE, 2nd, Threatt S, Friedman HS. Phase II trial of Gliadel plus O6-benzylguanine in adults with recurrent glioblastoma multiforme. Clin. Cancer Res. 2009;15:1064–1068. doi: 10.1158/1078-0432.CCR-08-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (176).Meany HJ, Warren KE, Fox E, Cole DE, Aikin AA, Balis FM. Pharmacokinetics of temozolomide administered in combination with O6-benzylguanine in children and adolescents with refractory solid tumors. Cancer Chemother. Pharmacol. 2009;65:137–142. doi: 10.1007/s00280-009-1015-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (177).Watson AJ, Middleton MR, McGown G, Thorncroft M, Ranson M, Hersey P, McArthur G, Davis ID, Thomson D, Beith J, Haydon A, Kefford R, Lorigan P, Mortimer P, Sabharwal A, Hayward O, Margison GP. O6-Methylguanine-DNA methyltransferase depletion and DNA damage in patients with melanoma treated with temozolomide alone or with lomeguatrib. Br. J. Cancer. 2009;100:1250–1256. doi: 10.1038/sj.bjc.6605015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (178).Guerin C, Olivi A, Weingart JD, Lawson HC, Brem H. Recent advances in brain tumor therapy: local intracerebral drug delivery by polymers. Invest. New Drugs. 2004;22:27–37. doi: 10.1023/b:drug.0000006172.65135.3e. [DOI] [PubMed] [Google Scholar]
- (179).Wei G, Loktionova NA, Pegg AE, Moschel RC. Beta-glucuronidase-cleavable prodrugs of O6-benzylguanine and O6-benzyl-2′-deoxyguanosine. J. Med. Chem. 2005;48:256–261. doi: 10.1021/jm0493865. [DOI] [PubMed] [Google Scholar]
- (180).Chuk MK, Cole DE, McCully C, Loktionova NA, Pegg AE, Parker RJ, Pauly G, Widemann BC, Balis FM, Fox E. Plasma and CNS pharmacokinetics of O4-benzylfolic acid and metabolite in a non-human primate model. Cancer Chemother. Pharmacol. 2011 doi: 10.1007/s00280-010-1407-9. in press PMID:20725726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (181).Christians FC, Loeb LA. Novel human DNA alkyltransferases obtained by random substitution and genetic selection in bacteria. Proc. Natl. Acad. Sci. U.S.A. 1996;93:6124–6128. doi: 10.1073/pnas.93.12.6124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (182).Loktionova NA, Xu-Welliver M, Crone T, Kanugula S, Pegg AE. Mutant forms of O6-alkylguanine-DNA alkyltransferase protect CHO cells from killing by BCNU plus O6-benzylguanine or O6-8-oxo-benzylguanine. Biochem. Pharmacol. 1999;58:237–244. doi: 10.1016/s0006-2952(99)00095-7. [DOI] [PubMed] [Google Scholar]
- (183).Xu-Welliver M, Pegg AE. Point mutations at multiple sites including highly conserved amino acids maintain activity but render O6-alkylguanine-DNA alkyltransferase insensitive to O6-benzylguanine. Biochem J. 2000;347:519–526. doi: 10.1042/0264-6021:3470519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (184).Reese JS, Roth JC, Gerson SL. Bone marrow-derived cells exhibiting lung epithelial cell characteristics are enriched in vivo using methylguanine DNA methyltransferase-mediated drug resistance. Stem Cells. 2008;26:675–681. doi: 10.1634/stemcells.2007-0803. [DOI] [PubMed] [Google Scholar]
- (185).Milsom MD, Jerabek-Willemsen M, Harris CE, Schambach A, Broun E, Bailey J, Jansen M, Schleimer D, Nattamai K, Wilhelm J, Watson A, Geiger H, Margison GP, Moritz T, Baum C, Thomale J, Williams DA. Reciprocal relationship between O6-methylguanine-DNA methyltransferase P140K expression level and chemoprotection of hematopoietic stem cells. Cancer Res. 2008;68:6171–6180. doi: 10.1158/0008-5472.CAN-08-0320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (186).Wang D, Worsham DN, Pan D. Co-expression of MGMT(P140K) and alpha-L-iduronidase in primary hepatocytes from mucopolysaccharidosis type I mice enables efficient selection with metabolic correction. J. Gene Med. 2008;10:249–259. doi: 10.1002/jgm.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (187).Kiem HP, Wu RA, Sun G, von Laer D, Rossi JJ, Trobridge GD. Foamy combinatorial anti-HIV vectors with MGMTP140K potently inhibit HIV-1 and SHIV replication and mediate selection in vivo. Gene Ther. 2010;17:37–49. doi: 10.1038/gt.2009.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (188).Beard BC, Trobridge GD, Ironside C, McCune JS, Adair JE, Kiem HP. Efficient and stable MGMT-mediated selection of long-term repopulating stem cells in nonhuman primates. J. Clin. Invest. 2010;120:2345–2354. doi: 10.1172/JCI40767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (189).Giordano FA, Sorg UR, Appelt JU, Lachmann N, Bleier S, Roeder I, Kleff V, Flasshove M, Zeller WJ, Allgayer H, von Kalle C, Fruehauf S, Moritz T, Laufs S. Clonal inventory screens uncover monoclonality following serial transplantation of MGMTP140K transduced stem cells and dose-intense chemotherapy. Hum. Gene Ther. 2011 doi: 10.1089/hum.2010.088. in press. PMID:21319998. [DOI] [PubMed] [Google Scholar]
- (190).Lin Y, Cheung P, Roth JC, Wilson DL, Gerson SL. Imaging Stem Cell-derived Persistent Foci After In Vivo Selection of Lentiviral MGMT-P140K Transduced Murine Bone Marrow Cells. Mol. Ther. 2011 doi: 10.1038/mt.2010.315. In press. PMID:21304493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (191).Nagasubramanian R, Hansen RJ, Delaney SM, Cherian MM, Samson LD, Kogan SC, Dolan ME. Survival and tumorigenesis in O6-methylguanine DNA methyltransferase-deficient mice following cyclophosphamide exposure. Mutagenesis. 2008;23:341–346. doi: 10.1093/mutage/gen018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (192).Wirtz S, Nagel G, Eshkind L, Neurath MF, Samson LD, Kaina B. Both base excision repair and O6-methylguanine-DNA methyltransferase protect against methylation-induced colon carcinogenesis. Carcinogenesis. 2010;31:2111–2117. doi: 10.1093/carcin/bgq174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (193).Chen ZP, Yarosh D, Garcia Y, Tampieri D, Mohr G, Malapetsa A, Langleben A, Panasci LC. Relationship between O6-methylguanine-DNA methyltransferase levels and clinical response induced by chloroethylnitrosourea therapy in glioma patients. Can. J. Neurol. Sci. 1999;26:104–109. [PubMed] [Google Scholar]
- (194).Jaeckle KA, Eyre HJ, Townsend JJ, Schulman S, Knudson HM, Belanich M, Yarosh DB, Bearman SI, Giroux DJ, Schold SC. Correlation of tumor O6-methylguanine-DNA methyltransferase levels with survival of malignant astrocytoma patients treated with bis-chloroethylnitrosourea: A Southwest Oncology Group study. J. Clin. Oncol. 1998;16:3310–3315. doi: 10.1200/JCO.1998.16.10.3310. [DOI] [PubMed] [Google Scholar]
- (195).Chinot OL, Barrie M, Fuentes S, Eudes N, Lancelot S, Metellus P, Muracciole X, Braguer D, Ouafik L, Martin PM, Dufour H, Figarella-Branger D. Correlation between O6-methylguanine-DNA methyltransferase and survival in inoperable newly diagnosed glioblastoma patients treated with neoadjuvant temozolomide. J. Clin. Oncol. 2007;25:1470–1475. doi: 10.1200/JCO.2006.07.4807. [DOI] [PubMed] [Google Scholar]
- (196).Busch C, Geisler J, Lillehaug JR, Lonning PE. MGMT expression levels predict disease stabilisation, progression-free and overall survival in patients with advanced melanomas treated with DTIC. Eur. J. Cancer. 2010;46:2127–2133. doi: 10.1016/j.ejca.2010.04.023. [DOI] [PubMed] [Google Scholar]
- (197).Day RS, III, Ziolkowski CHJ, Scudiero DA, Meyer SA, Lubiniecki AS, Girardi AJ, Galloway SM, Bynum GD. Defective repair of alkylated DNA by human tumor and SV-40-transformed human cell strains. Nature, (London) 1980;288:724–727. doi: 10.1038/288724a0. [DOI] [PubMed] [Google Scholar]
- (198).Sklar R, Strauss B. Removal of O6-methylguanine from DNA of normal and xeroderma pigmentosum-derived lymphoblastoid lines. Nature, (London) 1981;289:417–420. doi: 10.1038/289417a0. [DOI] [PubMed] [Google Scholar]
- (199).von Wronski M, Harris LC, Tano K, Mitra S, Bigner DD, Brent TP. Cytosine methylation and suppression of O6-methylguanine-DNA methyltransferase promoter activity in human rhabdomyosarcoma cell lines and xenografts. Oncol. Res. 1992;4:167–174. [PubMed] [Google Scholar]
- (200).von Wronski MA, Brent TP. Effect of 5-azacytidine on expression of the human DNA repair enzyme O6-methylguanine-DNA methyltransferase. Carcinogenesis. 1994;15:577–582. doi: 10.1093/carcin/15.4.577. [DOI] [PubMed] [Google Scholar]
- (201).Danam RP, Howell SR, Brent TP, Harris LC. Epigenetic regulation of O6-methylguanine-DNA methyltransferase gene expression by histone acetylation and methyl-CpG binding proteins. Mol. Cancer Ther. 2005;4:61–69. [PubMed] [Google Scholar]
- (202).Danam RP, Howell SR, Remack JS, Brent TP. Heterogeneous methylation of the O6-methylguanine-DNA methyltransferase promoter in immortallized IMR90 cell lines. Int. J. Oncol. 2001;18:1187–1193. doi: 10.3892/ijo.18.6.1187. [DOI] [PubMed] [Google Scholar]
- (203).Danam RP, Qian XC, Howell SR, Brent TP. Methylation of selected CpGs in the human O6-methylguanine-DNA methyltransferase promoter region as a marker of gene silencing. Mol. Carcinog. 1999;24:85–89. [PubMed] [Google Scholar]
- (204).Everhard S, Tost J, El Abdalaoui H, Criniere E, Busato F, Marie Y, Gut IG, Sanson M, Mokhtari K, Laigle-Donadey F, Hoang-Xuan K, Delattre JY, Thillet J. Identification of regions correlating MGMT promoter methylation and gene expression in glioblastomas. Neuro Oncol. 2009;11:348–356. doi: 10.1215/15228517-2009-001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (205).Watts GS, Pieper RO, Costello JF, Peng Y-M, Dalton WS, Futscher BW. Methylation of discrete regions of the O6-methylguanine-DNA methyltransferase, (MGMT) CpG island is associated with heterochromatinization of the MGMT transcription start site and silencing of the gene. Mol. Cell. Biol. 1997;17:5612–5619. doi: 10.1128/mcb.17.9.5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (206).Candiloro IL, Dobrovic A. Detection of MGMT promoter methylation in normal individuals is strongly associated with the T allele of the rs16906252 MGMT promoter single nucleotide polymorphism. Cancer Prev. Res. 2009;2:862–867. doi: 10.1158/1940-6207.CAPR-09-0056. [DOI] [PubMed] [Google Scholar]
- (207).Leng S, Bernauer A, Hong C, Do K, Yingling CM, Flores KG, Tessema M, Tellez CS, Willink R, Burki EA, Picchi MA, Stidley CA, Prados MD, Costello J, Gilliland FD, Crowell RE, Belinsky SA. The A/G Allele of Rs16906252 Predicts for MGMT Methylation and Is Selectively Silenced in Premalignant Lesions from Smokers and in Lung Adenocarcinomas. Clin. Cancer Res. 2011 doi: 10.1158/1078-0432.CCR-10-3026. in press. PMID:21355081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (208).Lai JC, Cheng YW, Goan YG, Chang JT, Wu TC, Chen CY, Lee H. Promoter methylation of O6-methylguanine-DNA-methyltransferase in lung cancer is regulated by p53. DNA Repair, (Amst.) 2008;7:1352–1363. doi: 10.1016/j.dnarep.2008.04.016. [DOI] [PubMed] [Google Scholar]
- (209).Lai JC, Wu JY, Cheng YW, Yeh KT, Wu TC, Chen CY, Lee H. O6-Methylguanine-DNA methyltransferase hypermethylation modulated by 17beta-estradiol in lung cancer cells. Anticancer Res. 2009;29:2535–2540. [PubMed] [Google Scholar]
- (210).Bobustuc GC, Baker CH, Limaye A, Jenkins WD, Pearl G, Avgeropoulos NG, Konduri SD. Levetiracetam enhances p53-mediated MGMT inhibition and sensitizes glioblastoma cells to temozolomide. Neuro. Oncol. 2010;12:917–927. doi: 10.1093/neuonc/noq044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (211).Glas M, Happold C, Rieger J, Wiewrodt D, Bahr O, Steinbach JP, Wick W, Kortmann RD, Reifenberger G, Weller M, Herrlinger U. Long-term survival of patients with glioblastoma treated with radiotherapy and lomustine plus temozolomide. J. Clin. Oncol. 2009;27:1257–1261. doi: 10.1200/JCO.2008.19.2195. [DOI] [PubMed] [Google Scholar]
- (212).Preusser M. MGMT analysis at DNA, RNA and protein levels in glioblastoma tissue. Histol. Histopathol. 2009;24:511–518. doi: 10.14670/HH-24.511. [DOI] [PubMed] [Google Scholar]
- (213).Cao VT, Jung TY, Jung S, Jin SG, Moon KS, Kim IY, Kang SS, Park CS, Lee KH, Chae HJ. The correlation and prognostic significance of MGMT promoter methylation and MGMT protein in glioblastomas. Neurosurgery. 2009;65:866–875. doi: 10.1227/01.NEU.0000357325.90347.A1. [DOI] [PubMed] [Google Scholar]
- (214).Weller M, Felsberg J, Hartmann C, Berger H, Steinbach JP, Schramm J, Westphal M, Schackert G, Simon M, Tonn JC, Heese O, Krex D, Nikkhah G, Pietsch T, Wiestler O, Reifenberger G, von Deimling A, Loeffler M. Molecular predictors of progression-free and overall survival in patients with newly diagnosed glioblastoma: A prospective translational study of the German glioma network. J. Clin. Oncol. 2009;27:5743–5750. doi: 10.1200/JCO.2009.23.0805. [DOI] [PubMed] [Google Scholar]
- (215).Kurzwelly D, Glas M, Roth P, Weimann E, Lohner H, Waha A, Schabet M, Reifenberger G, Weller M, Herrlinger U. Primary CNS lymphoma in the elderly: temozolomide therapy and MGMT status. J.Neurooncol. 2010;97:389–392. doi: 10.1007/s11060-009-0032-0. [DOI] [PubMed] [Google Scholar]
- (216).Weller M, Stupp R, Reifenberger G, Brandes AA, van den Bent MJ, Wick W, Hegi ME. MGMT promoter methylation in malignant gliomas: ready for personalized medicine? Nat.Rev. Neurol. 2010;6:39–51. doi: 10.1038/nrneurol.2009.197. [DOI] [PubMed] [Google Scholar]
- (217).Svrcek M, Buhard O, Colas C, Coulet F, Dumont S, Massaoudi I, Lamri A, Hamelin R, Cosnes J, Oliveira C, Seruca R, Gaub MP, Legrain M, Collura A, Lascols O, Tiret E, Flejou JF, Duval A. Methylation tolerance due to an O6-methylguanine DNA methyltransferase, (MGMT) field defect in the colonic mucosa: an initiating step in the development of mismatch repair-deficient colorectal cancers. Gut. 2010;59:1516–1526. doi: 10.1136/gut.2009.194787. [DOI] [PubMed] [Google Scholar]
- (218).Sonoda Y, Yokosawa M, Saito R, Kanamori M, Yamashita Y, Kumabe T, Watanabe M, Tominaga T. O6-Methylguanine DNA methyltransferase determined by promoter hypermethylation and immunohistochemical expression is correlated with progression-free survival in patients with glioblastoma. Int. J. Clin. Oncol. 2010;15:352–358. doi: 10.1007/s10147-010-0065-6. [DOI] [PubMed] [Google Scholar]
- (219).Kreth S, Thon N, Eigenbrod S, Lutz J, Ledderose C, Egensperger R, Tonn JC, Kretzschmar HA, Hinske LC, Kreth FW. O-Methylguanine-DNA Methyltransferase, (MGMT) mRNA Expression Predicts Outcome in Malignant Glioma Independent of MGMT Promoter Methylation. PLoS ONE. 2011;6:e17156. doi: 10.1371/journal.pone.0017156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (220).Pegg AE, Fang Q, Loktionova NA. Human variants of O6-alkylguanine-DNA alkyltransferase. DNA Repair, (Amst.) 2007;6:1071–1078. doi: 10.1016/j.dnarep.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (221).Liu Y, Scheurer ME, El-Zein R, Cao Y, Do KA, Gilbert M, Aldape KD, Wei Q, Etzel C, Bondy ML. Association and interactions between DNA repair gene polymorphisms and adult glioma. Cancer Epidemiol. Biomarkers Prev. 2009;18:204–214. doi: 10.1158/1055-9965.EPI-08-0632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (222).Hazra A, Chanock S, Giovannucci E, Cox DG, Niu T, Fuchs C, Willett WC, Hunter DJ. Large-scale evaluation of genetic variants in candidate genes for colorectal cancer risk in the Nurses’ Health Study and the Health Professionals’ Follow-up Study. Cancer Epidemiol. Biomarkers Prev. 2008;17:311–319. doi: 10.1158/1055-9965.EPI-07-0195. [DOI] [PubMed] [Google Scholar]
- (223).Doecke J, Zhao ZZ, Pandeya N, Sadeghi S, Stark M, Green AC, Hayward NK, Webb PM, Whiteman DC. Polymorphisms in MGMT and DNA repair genes and the risk of esophageal adenocarcinoma. Int. J. Cancer. 2008;123:174–180. doi: 10.1002/ijc.23410. [DOI] [PubMed] [Google Scholar]
- (224).Huang SH, Chang PY, Liu CJ, Lin MW, Hsia KT. O6-Methylguanine-DNA methyltransferase gene coding region polymorphisms and oral cancer risk. J. Oral Pathol. Med. 2010;39:645–650. doi: 10.1111/j.1600-0714.2009.00880.x. [DOI] [PubMed] [Google Scholar]
- (225).Zhong Y, Huang Y, Zhang T, Ma C, Zhang S, Fan W, Chen H, Qian J, Lu D. Effects of O6-methylguanine-DNA methyltransferase, (MGMT) polymorphisms on cancer: a meta-analysis. Mutagenesis. 2010;25:83–95. doi: 10.1093/mutage/gep050. [DOI] [PubMed] [Google Scholar]
- (226).Margison GP, Heighway J, Pearson S, McGown G, Thorncroft MR, Watson AJ, Harrison KL, Lewis SJ, Rohde K, Barber PV, O’Donnell P, Povey AC, Santibanez-Koref MF. Quantitative trait locus analysis reveals two intragenic sites that influence O6-alkylguanine-DNA alkyltransferase activity in peripheral blood mononuclear cells. Carcinogenesis. 2005;26:1473–1480. doi: 10.1093/carcin/bgi087. [DOI] [PubMed] [Google Scholar]
- (227).Krzesniak M, Butkiewicz D, Samojedny A, Chorazy M, Rusin M. Polymorphisms in TDG and MGMT genes - epidemiological and functional study in lung cancer patients from Poland. Ann. Hum. Genet. 2004;68:300–312. doi: 10.1046/j.1529-8817.2004.00079.x. [DOI] [PubMed] [Google Scholar]
- (228).Fang Q, Loktionova NA, Moschel RC, Javanmard S, Pauly GT, Pegg AE. Differential inactivation of polymorphic variants of human O6-alkylguanine-DNA alkyltransferase. Biochem. Pharmacol. 2008;75:618–626. doi: 10.1016/j.bcp.2007.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (229).Hill CE, Wickliffe JK, Wolfe KJ, Kinslow CJ, Lopez MS, Abdel-Rahman SZ. The L84F and the I143V polymorphisms in the O6-methylguanine-DNA-methyltransferase, (MGMT) gene increase human sensitivity to the genotoxic effects of the tobacco-specific nitrosamine carcinogen NNK. Pharmacogenet. Genomics. 2005;15:571–578. doi: 10.1097/01.fpc.0000167332.38528.a5. [DOI] [PubMed] [Google Scholar]
- (230).Hill CE, Wickliffe JK, Guerin AT, Kinslow CJ, Wolfe KJ, Ammenheuser MM, Abdel-Rahman SZ. The L84F polymorphism in the O6-methylguanine-DNA-methyltransferase, (MGMT) gene is associated with increased hypoxanthine phosphoribosyltransferase, (HPRT) mutant frequency in lymphocytes of tobacco smokers. Pharmacogenet. Genomics. 2007;17:743–753. doi: 10.1097/FPC.0b013e3281111eb1. [DOI] [PubMed] [Google Scholar]
- (231).Crosbie PA, McGown G, Thorncroft MR, O’Donnell PN, Barber PV, Lewis SJ, Harrison KL, Agius RM, Santibanez-Koref MF, Margison GP, Povey AC. Association between lung cancer risk and single nucleotide polymorphisms in the first intron and codon 178 of the DNA repair gene, O6-alkylguanine-DNA alkyltransferase. Int. J. Cancer. 2008;122:791–795. doi: 10.1002/ijc.23059. [DOI] [PubMed] [Google Scholar]
- (232).Margison GP, Butt A, Pearson SJ, Wharton S, Watson AJ, Marriott A, Caetano CM, Hollins JJ, Rukazenkova N, Begum G, Santibanez-Koref MF. Alkyltransferase-like proteins. DNA Repair, (Amst.) 2007;6:1222–1228. doi: 10.1016/j.dnarep.2007.03.014. [DOI] [PubMed] [Google Scholar]
- (233).Tubbs JL, Tainer JA. Alkyltransferase-like proteins: molecular switches between DNA repair pathways. Cell. Mol. Life Sci. 2010;67:3749–3762. doi: 10.1007/s00018-010-0405-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (234).Tubbs JL, Latypov V, Kanugula S, Butt A, Melikishvili M, Kraehenbuehl R, Fleck O, Marriott A, Watson AJ, Verbeek B, McGown G, Thorncroft M, Santibanez-Koref MF, Millington C, Arvai AS, Kroeger MD, Peterson LA, Williams DM, Fried MG, Margison GP, Pegg AE, Tainer JA. Flipping of alkylated DNA damage bridges base and nucleotide excision repair. Nature. 2009;459:808–813. doi: 10.1038/nature08076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (235).Pearson SJ, Ferguson J, Santibanez-Koref M, Margison GP. Inhibition of O6-methylguanine-DNA methyltransferase by an alkyltransferase-like protein from Escherichia coli. Nucleic Acids Res. 2005;33:3837–3844. doi: 10.1093/nar/gki696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (236).Aramini JM, Tubbs JL, Kanugula S, Rossi P, Ertekin A, Maglaqui M, Hamilton K, Ciccosanti CT, Jiang M, Xiao R, Soong TT, Rost B, Acton TB, Everett JK, Pegg AE, Tainer JA, Montelione GT. Structural basis of O6-alkylguanine recognition by a bacterial alkyltransferase-like DNA repair protein. J Biol. Chem. 2010;285:13736–13741. doi: 10.1074/jbc.M109.093591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (237).Pearson SJ, Wharton S, Watson AJ, Begum G, Butt A, Glynn N, Williams DM, Shibata T, Santibanez-Koref MF, Margison GP. A novel DNA damage recognition protein in Schizosaccharomyces pombe. Nucleic Acids Res. 2006;34:2347–2354. doi: 10.1093/nar/gkl270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (238).Morita R, Nakagawa N, Kuramitsu S, Masui R. An O6-methylguanine-DNA methyltransferase-like protein from Thermus thermophilus Interacts with a nucleotide excision repair protein. J. Biochem. 2008;144:267–277. doi: 10.1093/jb/mvn065. [DOI] [PubMed] [Google Scholar]
- (239).Onodera T, Morino K, Tokishita SI, Morita R, Masui R, Kuramitsu S, Ohta T. Role of alkyltransferase-like, (ATL) protein in repair of methylated DNA lesions in Thermus thermophilus. Mutagenesis. 2011;26:303–308. doi: 10.1093/mutage/geq093. [DOI] [PubMed] [Google Scholar]
- (240).Mazon G, Philippin G, Cadet J, Gasparutto D, Fuchs RP. The alkyltransferase-like ybaZ gene product enhances nucleotide excision repair of O6-alkylguanine adducts in E. coli. DNA Repair, (Amst.) 2009;8:697–703. doi: 10.1016/j.dnarep.2009.01.022. [DOI] [PubMed] [Google Scholar]
- (241).Mazon G, Philippin G, Cadet J, Gasparutto D, Modesti M, Fuchs RP. Alkyltransferase-like protein, (eATL) prevents mismatch repair-mediated toxicity induced by O6-alkylguanine adducts in Escherichia coli. Proc. Natl. Acad. Sci. USA. 2010;107:18050–18055. doi: 10.1073/pnas.1008635107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (242).Jones KL, Zhang L, Seldeen KL, Gong F. Detection of bulky DNA lesions: DDB2 at the interface of chromatin and DNA repair in eukaryotes. IUBMB Life. 2010;62:803–811. doi: 10.1002/iub.391. [DOI] [PubMed] [Google Scholar]
- (243).Hanawalt PC, Ford JM, Lloyd DR. Functional characterization of global genomic DNA repair and its implications for cancer. Mutat. Res. 2003;544:107–114. doi: 10.1016/j.mrrev.2003.06.002. [DOI] [PubMed] [Google Scholar]
- (244).Sedgwick B, Lindahl T. Recent progress on the Ada response for inducible repair of DNA alkylation damage. Oncogene. 2002;21:8886–8894. doi: 10.1038/sj.onc.1205998. [DOI] [PubMed] [Google Scholar]
- (245).He C, Hus JC, Sun LJ, Zhou P, Norman DP, Dotsch V, Wei H, Gross JD, Lane WS, Wagner G, Verdine GL. A methylation-dependent electrostatic switch controls DNA repair and transcriptional activation by E. coli ada. Mol. Cell. 2005;20:117–129. doi: 10.1016/j.molcel.2005.08.013. [DOI] [PubMed] [Google Scholar]
- (246).Jones GD, Le Pla RC, Farmer PB. Phosphotriester adducts, (PTEs): DNA’s overlooked lesion. Mutagenesis. 2010;25:3–16. doi: 10.1093/mutage/gep038. [DOI] [PubMed] [Google Scholar]
- (247).Myers LC, Terranova MP, Ferentz AE, Wagner G, Verdine GL. Repair of DNA methylphosphotriesters through a metalloactivated cysteine nucleophile. Science. 1993;261:1164–1167. doi: 10.1126/science.8395079. [DOI] [PubMed] [Google Scholar]
- (248).Myers LC, Verdine GL, Wagner G. Solution structure of the DNA methyl phosphotriester repair domain of Escherichia coli Ada. Biochemistry. 1993;32:14089–14094. doi: 10.1021/bi00214a003. [DOI] [PubMed] [Google Scholar]
- (249).Myers LC, Jackow F, Verdine GL. Metal dependence of transcriptional switching in Escherichia coli ada. J. Biol. Chem. 1995;270:6664–6670. doi: 10.1074/jbc.270.12.6664. [DOI] [PubMed] [Google Scholar]
- (250).Rasimas JJ, Kanugula S, Dalessio PM, Ropson IJ, Pegg AE. Effects of zinc occupancy on human O6-alkylguanine-DNA alkyltransferase. Biochemistry. 2003;42:980–990. doi: 10.1021/bi026970b. [DOI] [PubMed] [Google Scholar]
- (251).Ali RB, Teo AKC, Oh H-K, Chuang LSH, Ayi T-C, Li BFL. Implication of localization of human DNA repair enzyme O6-methylguanine-DNA methyltransferase at active transcription sites in transcription-repair coupling of the mutagenic O6-methylguanine lesion. Mol. Cell. Biol. 1998;18:1660–1669. doi: 10.1128/mcb.18.3.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (252).Teo AKC, Oh HK, Ali RB, Li BFL. The modified human DNA repair enzyme O6-methylguanine-DNA methyltransferase is a negative regulator of estrogen receptor-mediated transcription upon alkylation DNA damage. Mol. Cell. Biol. 2001;21:7105–7114. doi: 10.1128/MCB.21.20.7105-7114.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (253).Chahal M, Xu Y, Lesniak D, Graham K, Famulski K, Christensen JG, Aghi M, Jacques A, Murray D, Sabri S, Abdulkarim B. MGMT modulates glioblastoma angiogenesis and response to the tyrosine kinase inhibitor sunitinib. Neuro. Oncol. 2010;12:822–833. doi: 10.1093/neuonc/noq017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (254).Damoiseaux R, Keppler A, Johnsson K. Synthesis and applications of chemical probes for human O6-alkylguanine-DNA alkyltransferase. Chembiochem. 2001;4:285–287. doi: 10.1002/1439-7633(20010401)2:4<285::AID-CBIC285>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- (255).Gronemeyer T, Chidley C, Juillerat A, Heinis C, Johnsson K. Directed evolution of O6-alkylguanine-DNA alkyltransferase for applications in protein labeling. Protein Eng. Des. Sel. 2006;19:309–316. doi: 10.1093/protein/gzl014. [DOI] [PubMed] [Google Scholar]
- (256).Hinner MJ, Johnsson K. How to obtain labeled proteins and what to do with them. Curr. Opin. Biotechnol. 2010;21:766–776. doi: 10.1016/j.copbio.2010.09.011. [DOI] [PubMed] [Google Scholar]
- (257).Moschel RC, McDougall MG, Dolan ME, Stine L, Pegg AE. Structural features of substituted purine derivatives compatible with depletion of human O6-alkylguanine-DNA alkyltransferase. J. Med. Chem. 1992;35:4486–4491. doi: 10.1021/jm00101a028. [DOI] [PubMed] [Google Scholar]
- (258).Keppler A, Gendreizig S, Gronemeyer T, Pick H, Vogel H, Johnsson K. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat. Biotechnol. 2003;21:86–89. doi: 10.1038/nbt765. [DOI] [PubMed] [Google Scholar]
- (259).Kindermann M, George N, Johnsson N, Johnsson K. Covalent and selective immobilization of fusion proteins. J. Am. Chem. Soc. 2003;125:7810–7811. doi: 10.1021/ja034145s. [DOI] [PubMed] [Google Scholar]
- (260).Keppler A, Kindermann M, Gendreizig S, Pick H, Vogel H, Johnsson K. Labeling of fusion proteins of O6-alkylguanine-DNA alkyltransferase with small molecules in vivo and in vitro. Methods. 2004;32:437–444. doi: 10.1016/j.ymeth.2003.10.007. [DOI] [PubMed] [Google Scholar]
- (261).Kamiya M, Johnsson K. Localizable and highly sensitive calcium indicator based on a BODIPY fluorophore. Anal. Chem. 2010;82:6472–6479. doi: 10.1021/ac100741t. [DOI] [PubMed] [Google Scholar]
- (262).Bannwarth M, Correa IR, Sztretye M, Pouvreau S, Fellay C, Aebischer A, Royer L, Rois E, Johnsson K. Indo-1 derivatives for local calcium sensing. ACS Chem. Biol. 2009;4:179–190. doi: 10.1021/cb800258g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (263).Juillerat A, Gronemeyer T, Keppler A, Gendreizig S, Pick H, Vogel H, Johnsson K. Directed evolution of O6-alkylguanine-DNA akyltransferase for efficient Labeling of fusion proteins with small molecules In vivo. Chem. Biol. 2003;10:313–317. doi: 10.1016/s1074-5521(03)00068-1. [DOI] [PubMed] [Google Scholar]
- (264).Juillerat A, Heinis C, Sielaff I, Barnikow J, Jaccard H, Kunz B, Terskikh A, Johnsson K. Engineering substrate specificity of O6-alkylguanine-DNA alkyltransferase for specific protein labeling in living cells. Chembiochem. 2005;6:1263–1269. doi: 10.1002/cbic.200400431. [DOI] [PubMed] [Google Scholar]
- (265).Gautier A, Juillerat A, Heinis C, Correa IR, Jr., Kindermann M, Beaufils F, Johnsson K. An engineered protein tag for multiprotein labeling in living cells. Chem. Biol. 2008;15:128–136. doi: 10.1016/j.chembiol.2008.01.007. [DOI] [PubMed] [Google Scholar]
- (266).Provost CR, Sun L. Fluorescent labeling of COS-7 expressing SNAP-tag fusion proteins for live cell imaging. J. Vis.Exp. 2011 doi: 10.3791/1876. in press PMID.20485262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (267).Kanugula S, Pauly GT, Moschel RC, Pegg AE. A bifunctional DNA repair protein from Ferroplasma acidarmanus exhibits O6-alkylguanine-DNA alkyltransferase and endonuclease V activities. Proc. Natl. Acad. Sci. U.S,A. 2005;102:3617–3622. doi: 10.1073/pnas.0408719102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (268).Kow YW. Repair of deaminated bases in DNA. Free Radic. Biol. Med. 2002;33:886–893. doi: 10.1016/s0891-5849(02)00902-4. [DOI] [PubMed] [Google Scholar]
- (269).Kanugula S, Pegg AE. A novel DNA repair alkyltransferase from Caenorhabditis elegans. Environ. Mol. Mutagenesis. 2001;38:235–243. doi: 10.1002/em.1077. [DOI] [PubMed] [Google Scholar]
- (270).Chae M-Y, McDougall MG, Dolan ME, Swenn K, Pegg AE, Moschel RC. Substituted O6-benzylguanine derivatives and their inactivation of human O6-alkylguanine-DNA alkyltransferase. J. Med. Chem. 1994;37:342–347. doi: 10.1021/jm00029a005. [DOI] [PubMed] [Google Scholar]